Abstract
Background:
Chemotherapy resistance is a major determinant of poor overall survival rates in high-grade serous ovarian cancer (HGSC). We have previously shown that gene expression alterations affecting the NF-κB pathway characterise chemotherapy resistance in HGSC, suggesting that the regulation of an immune response may be associated with this phenotype.
Methods:
Given that intrinsic drug resistance pre-exists and is governed by both tumour and host factors, the current study was performed to examine the cross-talk between tumour inflammatory microenvironment and cancer cells, and their roles in mediating differential chemotherapy response in HGSC patients. Expression profiling of a panel of 184 inflammation-related genes was performed in 15 chemoresistant and 19 chemosensitive HGSC tumours using the NanoString nCounter platform.
Results:
A total of 11 significantly differentially expressed genes were found to distinguish the two groups. As STAT1 was the most significantly differentially expressed gene (P=0.003), we validated the expression of STAT1 protein by immunohistochemistry using an independent cohort of 183 (52 resistant and 131 sensitive) HGSC cases on a primary tumour tissue microarray. Relative expression levels were subjected to Kaplan–Meier survival analysis and Cox proportional hazard regression models.
Conclusions:
This study confirms that higher STAT1 expression is significantly associated with increased progression-free survival and that this protein together with other mediators of tumour–host microenvironment can be applied as a novel response predictive biomarker in HGSC. Furthermore, an overall underactive immune microenvironment suggests that the pre-existing state of the tumour immune microenvironment could determine response to chemotherapy in HGSC.
Keywords: ovarian cancer, chemoresistance, predictive biomarker, tumour microenvironment
Epithelial ovarian cancer is a leading cause of morbidity and mortality from gynaecologic malignancies, with ∼22 000 new cases each year in North America. High-grade serous ovarian cancer (HGSC) is the most prevalent histological type characterised by late detection, metastasis and resistance to chemotherapy (Marcus et al, 2014). Approximately 50% of patients exhibit resistance to chemotherapy within 6 months of treatment that eventually results in death because of tumour relapse (Weberpals et al, 2011; Marcus et al, 2014). A number of reports (Sakamoto et al, 2001; Selvanayagam et al, 2004; Bernardini et al, 2005), aimed at identifying mechanisms leading to primary chemoresistance, have revealed differential gene expression signatures in the drug-resistant patients. Recently, we showed that gene expression alterations in chemotherapy-resistant HGSC affected, among others, the NF-κB pathway, drawing attention for a need to understand immunoregulation in the context of differential drug response in this tumour (Koti et al, 2013).
Chemotherapy resistance can be defined as being either primary (innate) or secondary (acquired). Primary chemotherapy resistance is conferred by pre-existing gene expression that is inherent within the tumour, in chemotherapy-naive tumour cells or in the immediate microenvironment, that provides a state of resistance to a range of drugs. Secondary chemotherapy resistance is conferred to the tumour as a direct result of alterations that are acquired or induced by selection following exposure to chemotherapeutic agents. Chemotherapy resistance remains a major determinant of poor survival rates in HGSC, but there has been little progress in developing novel biomarkers of therapeutic response. This failure may partially be attributed to most studies being directed towards the tumour cell-intrinsic events and ignoring the contributory effects of variation in the local immune response. The role of the tumour microenvironment in the survival of tumour cells, and the dual roles of cancer immunoediting via tumour-promoting inflammation and suppression, is becoming well recognised (Zhang et al, 2003; Galon et al, 2006; Schreiber et al, 2011; Baxevanis et al, 2013). There is also increasing awareness that pre-existing adaptive immune status affect response to subsequent therapy across various cancers. A diverse tumour immunome was recently shown to be associated with varying clinical outcomes in colorectal cancer (Bindea et al, 2013). The abundance of tumour-infiltrating lymphocytes (TILs) correlates with survival across multiple cancers. In HGSC, ratios of CD8+ T cells to T regulatory cells have been associated with disease outcome (Preston et al, 2013). Higher survival rates have been reported in tumours containing both CD8+ T and CD20+ B cells (Milne et al, 2009; Nelson, 2010). In addition to the cellular infiltration, recent studies also suggest a significant role of CXC chemokines that recruit TILs in ovarian cancer (Kryczek et al, 2009; Rainczuk et al, 2012). Although a number of studies have addressed the immune cell infiltration in the HGSC tumour microenvironment, there is insufficient information on the role of cytokine/chemokine mediators that putatively lead to a variable adaptive immune response to HGSC.
Based on our previous findings implicating NF-κB to be one of the key pathways involved in chemotherapy resistance, we hypothesised that a pre-existing differential tumour immune microenvironment might lead to a variable response to chemotherapy in HGSC. We therefore analyzed differences in the tumour inflammatory environment using digital multiplexed gene expression profiling in a cohort of sensitive and resistant HGSC tumours, with an overall objective of identifying predictive markers of response to primary chemotherapy resistance. These data suggested involvement of T helper type 1 (Th1)-related factors such as signal transducer and activator of transcription 1 (STAT1), and other factors associated with an adaptive immune response that could be associated with the drug response phenotype as this protein is a key mediator of Th1 response following interferon stimulation. The rationale for determining STAT 1 expression in tumour tissues was because variation in cytokine activation and expression could arise from both tumour and a variety of the microenvironment cellular populations. Solid tumours contain a diversity of cell types recruited into close proximity (reviewed in Junttila and de Sauvage, 2013), so that overall immunohistochemical quantification of STAT1 was performed. The immunohistochemistry results were obtained from an independent cohort of chemo-naive HGSC and they confirmed that STAT1 protein levels were significantly associated with progression-free survival, and implicate a role of the inflammatory tumour microenvironment in mediating chemotherapy response in this tumour.
Materials and methods
Ethics statement
This study was approved by Queen's University Institutional Ethics Review Board, the CHUM institutional ethics committee (Comité d'éthique de la recherche du Centre hospitalier de l'Université de Montréal) and Ottawa Health Research Institute (OHRI) Research Ethics Board. Informed consent from all patients was obtained before sample collection.
Patient samples for discovery study
Fresh frozen tumour tissue samples from a cohort of 34 locally advanced (stage IIa–IV) HGSC tumours were accrued from the Ontario Tumour Bank (Ontario Institute for Cancer Research) and the OHRI. All samples were collected from patients who were naive to chemotherapy and radiotherapy at the time of primary cytoreductive surgery. Samples were stored at −80 °C immediately following collection until further processing. All samples had ≥70% tumour content and were histologically classified using WHO criteria, and disease staging was performed as per the International Federation of Gynaecology and Obstetrics (FIGO) guidelines. Our discovery cohort consisted of 15 patients who exhibited progressive disease within 6 months of initiation or within 8 months of completion of chemotherapy classified as intrinsically resistant/partially resistant (R/PR) and 19 patients showing no relapse until 18 months considered intrinsically sensitive (S) to chemotherapy (Koti et al, 2013; Park et al, 2013).
NanoString-based gene expression profiling
Total RNA was isolated from fresh frozen tumour tissues using Qiagen RNeasy kit (Qiagen Inc., Toronto, ON, Canada) as per the manufacturer's instructions. The RNA concentration and purity was estimated spectrophometrically on NanoDrop ND-100 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
All the RNA samples included in the study passed the quality control requirements (as assessed by the RNA integrity number or the OD 260/280 ratio) of the platform. Digital multiplexed NanoString nCounter analysis system (NanoString Technologies, Seattle, WA, USA)-based gene expression profiling was performed on 100 ng total RNA from each sample as input material according to the manufacturer's instructions at the Ontario Genomics Institute, Toronto. Tumour RNA samples were subjected to analysis by nCounter Human inflammation panel consisting of 184 human inflammation genes with 6 six housekeeping controls (Supplementary Table 1) in the pre-built panel (Nanostring Technologies). In this assay, single-target transcripts in the reaction are represented in the form of colour-coded barcodes. The resulting material was incorporated into an overnight hybridisation reaction, carried out by combining 5 μl of the total RNA sample with 20 μl of nCounter Reporter probes in hybridisation buffer and 5 μl of nCounter Capture probes for a total reaction volume of 30 μl. The hybridisations were incubated at 65 °C for ∼16–20 h. During the overnight hybridisation reaction, probe pairs are present in large excess to ensure that each target finds a probe pair. Abundances of specific target molecules can then be quantified using the nCounter Digital Analyzer to count the individual fluorescent barcodes and assess target molecules present in each sample. For each assay, a high-density scan (encompassing 600 fields of view) was performed. Following hybridisation, the cartridges were analysed in the Digital Analyzer that counts (that represent the number of molecules counted) the barcodes and further tabulates them.
NanoString data analysis
Normalisation of raw data was performed using the nSolver software (NanoString Technologies) as we previously reported (Martin et al, 2014). The raw NanoString counts were initially subjected to normalisation for all target RNAs in all samples based on built-in positive controls. This step accounts for intersample, experimental variation such as hybridisation efficiency and post-hybridisation processing. The geometric mean of each of the controls is calculated that indicates the overall assay efficiency. The housekeeping genes are then used for mRNA content normalisation. To facilitate downstream statistical analysis, values of <0 were blanketed and considered equal to 1. Following the initial normalisation steps, data were imported to Graphpad Prism software (GraphPad Software, Inc., La Jolla, CA, USA) for statistical analysis. Multiple t-tests were performed with correction for multiple comparisons using Sidak–Bonferroni method. An expression difference with a P-value of <0.05 was considered statistically significant. Pearson's correlation analysis was performed using Graphpad Prism software to derive correlation coefficient between the expression of STAT1 and CXCL10 genes.
Technical validation of NanoString data by real-time quantitative PCR (qRT–PCR) in discovery cohort
Total RNA was subjected to cDNA synthesis using RT2 first-strand cDNA synthesis kit (Qiagen Inc.) as per the manufacturer's instructions. Custom multiplexed PCR array (SABiosciences) consisting of CFL1, CREB1, CXCL10, STAT1, MAP3K7, MKNK1, RIPK1 and MYD88 gene targets was used to perform qRT–PCR on samples from the discovery cohort. The GAPDH and GUSB genes were included as housekeeping controls. The PCR array design also included pre-built reaction control and positive PCR control. All qRT–PCR experiments were performed in duplicates on LC-450 light cycler (Roche Diagnostics, Mississauga, ON, Canada). Data analysis was performed using relative quantification by the ΔΔCt method (Schmittgen and Livak, 2008; Koti et al, 2013).
Independent validation of STAT1 expression on a HGSC tissue microarray (TMA) by immunohistochemistry (IHC)
A TMA corresponding to an independent cohort of 213 primary chemo-naive HGSC tumours from patients who underwent surgery at the CHUM, was subjected to IHC to determine the expression of STAT1. Clinicopathological details of the cohort are summarised in Table 1. Areas of tumour were selected by a gynaecologic pathologist on haematoxylin and eosin-stained slides. Formalin-fixed, paraffin-embedded (FFPE) tumour blocks were cored using a 0.6 mm diameter needle and arrayed 1 mm apart in a grid pattern within a recipient paraffin block. The TMAs were sectioned at 4 μm and the slides were stained using the BenchMark XT automated stainer (Ventana Medical System Inc., Tucson, AZ, USA). The STAT1 antibody specificity was verified using western blot analysis (Supplementary Figure 2). The optimal concentration of rabbit anti-human polyclonal STAT1 primary antibody (Abcam, Cambridge, UK; no. ab2415) was determined by serial dilutions. Antigen retrieval was carried out with Cell Conditioning 1 (Ventana Medical System Inc.; no. 950-124) for 60 min. The slides were incubated with primary STAT1 antibody (1 : 2000) at 37 °C for 60 min. Reactions were carried out using the ultraView DAB detection kit (Ventana Medical System Inc.; no. 760-500). Slides were counterstained with haematoxylin and bluing reagent (Ventana Medical System Inc.; no. 760-2021; no. 760–2037) for 4 min.
Table 1. Clinicopathological details of high-grade serous ovarian cancer patients in validation cohort.
| Low STAT1, n (%) | High STAT1, n (%) | Overall, n (%) | |
|---|---|---|---|
|
Age | |||
| N | 88 | 95 | 183 |
| Mean | 60 | 62 | 61 |
| Median | 60 | 63 | 62 |
| s.d. | 11.0 | 9.6 | 10.4 |
| Min | 34 | 38 | 34 |
| Max | 82 | 81 | 82 |
|
FIGO | |||
| I/II | 12 (13.6%) | 21 (22.1%) | 33 (18%) |
| III/IV | 76 (86.4%) | 74 (77.9%) | 150 (82%) |
|
Residual disease | |||
| Unknown | 15 (17%) | 9 (9.5%) | 24 (13.1%) |
| ≤1 cm | 37 (42%) | 47 (49.5%) | 83 (45.4%) |
| >1 cm | 36 (40.9%) | 39 (41.1%) | 76 (41.5%) |
|
Treatment | |||
| Platinum/taxane | 70 (79.5%) | 85 (89.5%) | 155 (84.5%) |
| Platinum/taxane+additional agenta | 4 (4.5%) | 7 (7.4%) | 11 (6%) |
| Platinum | 2 (2.3%) | 1 (1%) | 3 (1.6%) |
| Taxane | 1 (1.1%) | 1 (1%) | 2 (1.1%) |
| Cisplatin/topotecan | 5 (5.7%) | 1 (1%) | 6 (3.3%) |
| Cyclophosphamide/carboplatin | 6 (6.8%) | 0 | 6 (3.3%) |
Abbreviations: FIGO=International Federation of Gynaecology and Obstetrics; STAT1=signal transducer and activator of transcription 1.
Clinicopathological details of the high-grade serous ovarian cancer (HGSC) cohort of 183 patients used for independent validation of STAT1 expression by immunohistochemistry.
Additional agent: placebo or ganitumab (AMG479) or nintedanib (BIBF1120) or bevacizumab.
TMA scoring and IHC data analysis
The TMA was scanned, digitally conserved, and manually visualised. Overall staining (including epithelial and stromal compartments) was scored according to the staining intensity in each core (value of 0 for absent, 1 for weak, 2 for moderate and 3 for strong). Each array was independently analysed in a blinded study by two independent observers. Inter-rating correlation coefficient was 83%. The average score from the two independent observers, for each respective core, was used for analysis. A total of 183 cases, for which clinical data were available, were subjected to further statistical analysis. Pearson's correlation coefficient was used to estimate the correlation between clinical data and the marker. Survival curves were plotted using the Kaplan–Meier curve analysis and the log-rank test was used to test for significant differences. Receiver operative characteristic (ROC) curves (Supplementary Figure 3) were used to determine the threshold value for the marker corresponding to the best sensitivity and specificity for patient progression-free survival. All statistical analyses were done using Statistical Package for the Social Science software version 16.0 (SPSS, Inc., Chicago, IL, USA). A P-value of <0.05 was considered significant.
Results
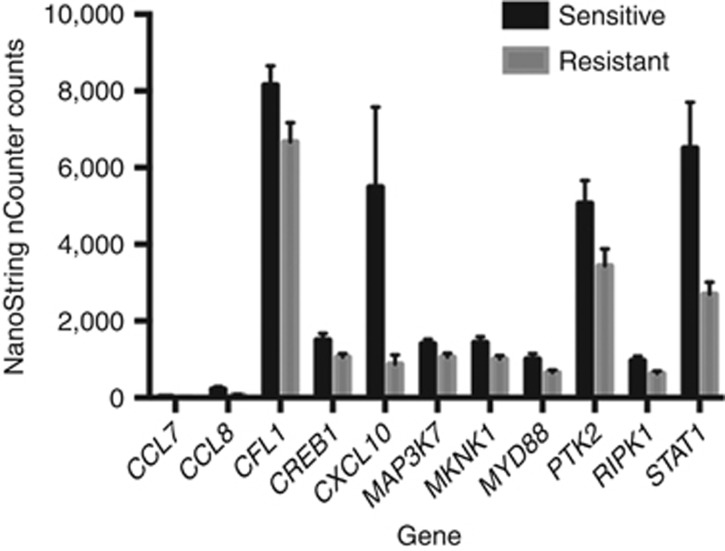
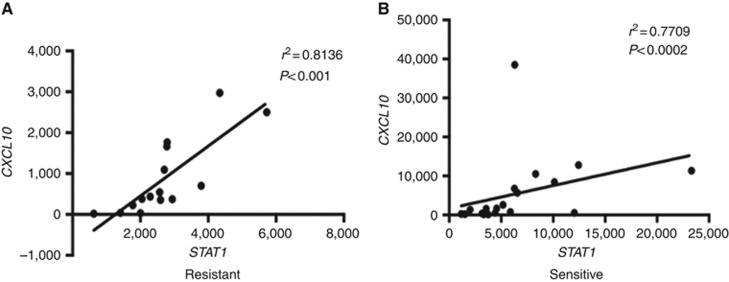
In this study, inflammatory gene expression profiling, by NanoString technology, of HGSCs that were clinically classified as chemotherapy resistant and sensitive displayed significant (P<0.05) different expression levels in eleven genes. STAT1, CXCL10, CREB1, MKNK1, MAP3K7, CFL1, PTK2, RIPK1, MYD88, CCL8 and CCL7 were overexpressed in the sensitive cohort compared with the resistant (Figure 1), consistent with the rationale that these chemokines and immune factors could be active in the host microenvironment and promote a more favourable drug response. To determine the reproducibility of these findings, technical validation of 8 of the 11 genes was performed using qRT–PCR. Technical validation of the differentially expressed genes showed full concordance with the NanoString-based gene expression findings (Supplementary Figure 1). Of these eight genes more highly expressed in the sensitive cohort, the interferon (IFN)-inducible protein (IP-10)/CXCL10 is one of the major targets of STAT1 activation by IFNγ. We therefore performed Pearson's correlation analysis comparing the expressions of these two genes in the resistant (Figure 2A) and sensitive (Figure 2B) tumours to determine their respective correlation coefficients. This analysis revealed a strong positive correlation between the expression of STAT1 and CXCL10 genes within the resistant and sensitive groups with r2=0.81 (P<0.001) and r2 =0.77 (P<0.0002), respectively. Moreover, the greatly increased expression levels of STAT1 in the sensitive tumours is consistent with this protein having a functional role in mediating an improved response to chemotherapy, underscoring the importance of confirming these findings at the protein level.
Figure 1.
Comparison of inflammatory gene expression levels in chemotherapy-resistant vs -sensitive high-grade serous ovarian tumours. The profile of inflammatory markers in the two study cohorts was determined by NanoString nCounter platform. Data normalisation was performed using nSolver software followed by application of Bonferroni test for multiple comparisons using Graphpad Prism software to determine significantly differentially expressed genes. The 11 most significantly differentially expressed genes (P<0.05) are shown.
Figure 2.
The correlation between STAT1 and CXCL10 expression. Pearson's correlation analysis (using Graphpad Prism software) showing significant positive correlation between STAT1 and CXCL10 expression within the resistant (A) and sensitive (B) groups with r2=0.81 (P<0.001) and r2=0.77 (P<0.0002), respectively.
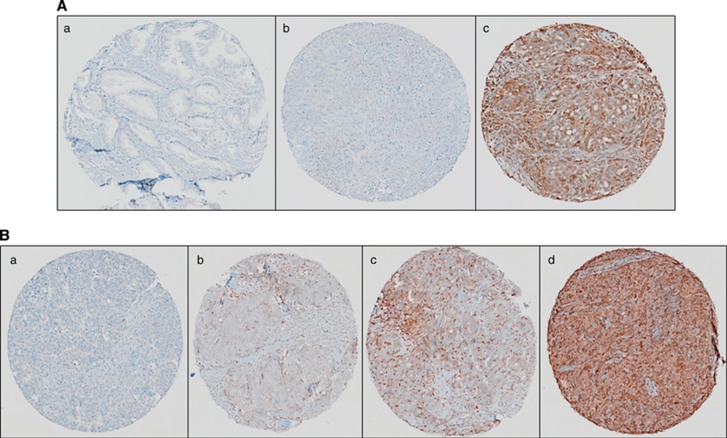
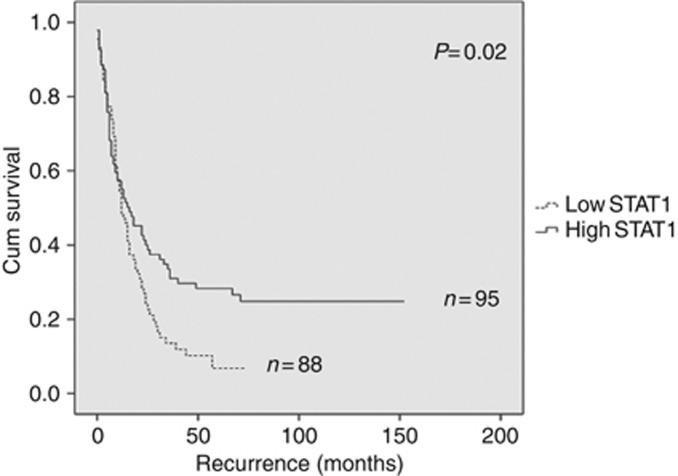
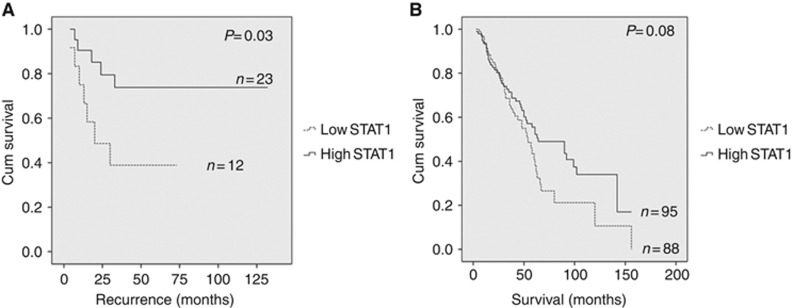
A total of 183 tumours were available for STAT1 IHC staining analysis based on the same classification criteria used previously (Liu and Matulonis, 2006; Wang et al, 2012). Following antibody optimisation (Figure 3A) a semiquantitative score from 0 to 3 (Figure 3B) was utilised to determine the overall staining intensity for the STAT1 protein. Log-rank test analysis revealed a significant (P=0.02) association between low STAT1 expression and decreased progression-free survival (Figure 4). These data provide additional evidence that elevated STAT1 expression together with other genes involved in the Th1 immune response may be investigated more extensively as biomarkers able to predict chemotherapy response in HGSC. Although a significant association between levels of STAT1 expression was noted upon comparison of FIGO stages I and II (Figure 5A), the difference in expression was not significant when stages III and IV were compared. Distinct STAT1 expression at the earlier stages I and II suggests that genotypic heterogeneity during tumour progression and/or the presence of a temporal element underlying the mechanism of cancer immunoediting may abrogate the need for differential STAT1 expression at later stages of the disease. Furthermore, Kaplan–Meier analysis also showed a trend, approaching significance, between decreased STAT1 expression and shorter overall survival compared with patients with longer overall survival (Figure 5B). Additional validation studies in larger cohorts are needed to confirm these findings.
Figure 3.
Immunohistochemical analysis of STAT1 in HGSC. (A) The STAT1 antibody (Abcam no. ab2415, polyclonal rabbit anti-human STAT1) optimisation by IHC was performed in (a) normal adjacent to prostate tumour tissue (showing negative staining), (b) HGS ovarian tissue (showing negative staining) and (c) HGS ovarian tissue (showing positive staining). (B) Independent validation of STAT1 expression was performed on an HGSC tissue microarray. Representative IHC images of overall STAT1 expression in HGSC. (a) Tissue punch scored as 0 negative/absent, (b) tissue punch scored as 1 with weak expression, (c) tissue punch scored as 2 for moderate expression and (d) tissue punch scored as 3 for strong expression.
Figure 4.
Kaplan–Meier survival analysis according to STAT1 expression by immunohistochemistry. Survival data were derived from an independent validation cohort of 183 HGSC patients with follow-up information. Kaplan–Meier estimates showing significant (P=0.02) association between increased overall STAT1 expression and longer progression-free survival.
Figure 5.
Kaplan–Meier survival analysis based on STAT1 expression and correlation with FIGO stage and overall survival. (A) Survival analysis based on comparison of FIGO stage I (n=11) and II (n=24), illustrating significant (P=0.03) correlation between levels of STAT1 expression and disease stage in all patients of FIGO stage I and II. N=number of patients. (B) Survival analysis showing association of STAT1 with overall survival showing a trend, approaching significance (P=0.08) towards shorter survival and decreased STAT1 expression. N= number of patients.
Discussion
Activation of both the adaptive and innate immune response to cancers are regulated by IFN (Schreiber et al, 2011). Indeed, the antiproliferative roles of IFN in cancer are well established and have led to novel therapies across various cancers (George et al, 2012). The STAT1 signaling is essential to all three types of IFN pathways and therefore plays an important role in immunosurveillance (Tymoszuk et al, 2014). Specifically, IFN-induced STAT1 activation leads to its binding either to the interferon response elements or Gamma-activated sites, further activating the interferon-induced genes (Leitner et al, 2014). Studies in mice with defective STAT1 activation have shown increased tumour incidence attributed to its tumour-suppressive role in breast tumourigenesis (Levy and Gilliland, 2000; Koromilas and Sexl, 2013). Moreover, in breast cancer tissue biopsies there was selective downregulation of STAT1 protein in tumour cells relative to the surrounding stroma when analysed by immunohistochemistry (Koromilas and Sexl, 2013). These data in breast cancer tissues, and the results reported herein in ovarian cancer, highlight the key role of the microenvironment for tumour development, and the need to perform single-cell resolution analysis in order to determine protein expression in distinct cellular compartments (Koti et al, unpublished). The STAT1 imparts its effects by various mechanisms, such as shaping the immune cell infiltration by changes in expression within the immune compartment (Saha et al, 2010) or by a proliferation arrest because of changes in its expression within the tumour epithelium (Klover et al, 2010; Raven et al, 2011). The IFN-mediated activation of STAT1 also leads to expression of the angiostatic chemokine CXCL10 by multiple cell types including antigen-presenting dendritic cells, macrophages, T cells, fibroblasts and epithelial cells (Groom and Luster, 2011). It is well established that the CXCR3/CXCL10 chemotactic axis is key to trafficking and differentiation of effector Th1 CD4+ cells, NK and CD8+ cells within inflamed tissues (Groom and Luster, 2011; Groom et al, 2012). Furthermore, increased levels of CXCL10 and CCL5/RANTES are associated with enhanced CD8+ T cell infiltration in melanoma, colorectal and gastric cancers (Ohtani et al, 2009; Kunz et al, 1999;, Muthuswamy et al, 2012). Our findings are consistent with recent studies of the immune microenvironment in breast cancer, where a similar trend of decreased CXCL10 and STAT1 expression was associated with relapse (Ascierto et al, 2012). Similar findings correlating increased STAT1 expression in pancreatic (Sun et al, 2014) and colorectal (Simpson et al, 2010) cancers with improved outcomes have been reported. Interestingly, a similar pattern of overexpression of these genes has been observed in other conditions exhibiting immune-mediated tissue destruction such as allograft rejection (Spivey et al, 2011), hepatitis C infection (Zeremski et al, 2008) and autoimmune diseases (Yoshida et al, 2012). However, it is likely that the magnitude of change in expression required for a tumour–host immunological response in cancer in a chemo-naive environment might be quite different to the response needed in other immune-mediated disease conditions. It is noteworthy that activation of cytokine pathways in stromal and immune cells within some models of the chemotherapy-induced tumour micronenvironment may be associated with resistance, rather than sensitivity to chemotherapy (Junttila and de Sauvage, 2013). However, it is likely that the mechanisms underlying natural immunosurveillance may differ to those that are responsible for chemotherapy-induced response in which cell-autonomous cytokine pathways appear to be activated (Sistigu et al, 2014). As recently reviewed (Koromilas and Sexl, 2013), the observed divergence in STAT1 expression in tumour cells in vitro and in vivo reflects the artificial nature of the experimental conditions where the impact of both the stromal and immune microenvironments are lacking. Natural immunosurveillance could be attributed to the tumour cell-intrinsic characteristics such as immunogenic mutational epitopes as well as other immune escape mechanisms that might modulate antitumour immune responses, as described recently in ovarian cancer (Brown et al, 2014). It is also possible that the off-target effects of chemotherapy on the innate and adaptive tumour immune responses (as reviewed recently (Simpson et al, 2010)) alter the pre-existing state of inflammatory mediators in addition to their effect on tumour-infiltrating immune cell distribution. Such effects could further lead to variation in the type of immunological responses elicited by adjuvant chemotherapy treatment.
Collectively, these findings based on elevated STAT1 and CXCL10 gene expression in those tumours that responded to chemotherapy implicate the activity of a pre-existing enhanced Th1-type immune response in the tumour microenvironment. Similarly, in the resistant cohort. the existence of an under reactive immune microenvironment before exposure to chemotherapy may lead to drug resistance because of lack of a synergistic antitumour effect mediated by the immune cells. An under reactive immune microenvironment is incapable of exerting its effect in tumour cell progression because of inadequate immune surveillance, thus leading to immune escape by the tumour cells. It should be noted that the tumour cells' intrinsic ability to modulate the secretion of cytokines and chemokines in the immediate vicinity of proliferating tumour could also provide cellular cross-talk that could contribute towards further modification of the local cellular immune/inflammatory microenvironment. Such a synergistic interaction between these compartments might be expected to lead to cellular variability in the expression levels of STAT1 in the epithelial and stromal elements. Although future longitudinal studies in this area need to be performed in order to correlate the decreased immune activation and its consequence following adjuvant chemotherapy, these initial associations provide insights into the design of novel immunomodulatory therapies that can be applied as adjuvant to current chemotherapy regimens in HGSC. The interferon pathway contributes significantly in modulating the tumour inflammatory microenvironment. Independent validation of STAT1 expression by IHC confirms its predictive role in HGSC and therefore it can be potentially be incorporated in future clinical trials following further validation in larger cohorts. Indeed, in vitro and in vivo studies are warranted to interrogate the mechanistic basis of its expression and its role in therapeutic response.
Overall, our findings provide the basis of mechanisms leading to tumour immune suppression in HGSC and warrant further investigation. In conclusion, the inflammatory gene signature suggestive of a pre-existing under reactive tumour immune microenvironment in chemotherapy-resistant HGSC patients must be explored further and taken into consideration in the design of novel adjuvant immunomodulatory therapies in HGSC.
Acknowledgments
We recognise the generosity of patients who participated in this study. Biological materials were provided by the Ontario Tumor Bank that is funded by the Ontario Institute for Cancer Research and Ottawa Health Research Institute. We are grateful to the Gynecology-Oncology and Pathology services of the CHUMHôpital Notre-Dame for tumour procurement. Tumour banking was supported by the Banque de tissus et données of the Réseau de recherche sur le cancer of the Fond de recherche du Québec Santé (FRQS), associated with the Canadian Tumour Repository Network (CTRNet). A-MMM is a researcher of the Centre derecherche du Centre hospitalier de l'Université de Montréal (CRCHUM) that receives support from the FRQS. The study was supported by Ontario Institute for Cancer Research through funding provided by Government of Ontario to Jeremy Squire, Senate Advisory Research Committee, Queen's University and Cancer Research Society, grants to Madhuri Koti.
Author contributions
The study was conceived and designed by Madhuri Koti, Jeremy Squire and Anne-Marie Mes Masson. Sample preparation and NanoString data validation by qRT–PCR was performed by Madhuri Koti, Alex Siu and Mallikarjun Bidarimath. NanoString data analysis was performed by Kirsteen Mclean from NanoString Technologies and Andrew Edwards. Isabelle Clément and Gulisa Turashvilli performed IHC staining scoring and computational analysis. Madhuri Koti, Isabelle Clément and Jeremy Squire wrote the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Material
References
- Ascierto ML, Kmieciak M, Idowu MO, Manjili R, Zhao Y, Grimes M, Dumur C, Wang E, Ramakrishnan V, Wang XY, Bear HD, Marincola FM, Manjili MH. A signature of immune function genes associated with recurrence-free survival in breast cancer patients. Breast Cancer Res Treat. 2012;131:871–880. doi: 10.1007/s10549-011-1470-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxevanis CN, Papamichail M, Perez SA. Immune classification of colorectal cancer patients: impressive but how complete. Expert Opin Biol Ther. 2013;13:517–526. doi: 10.1517/14712598.2013.751971. [DOI] [PubMed] [Google Scholar]
- Bernardini M, Lee CH, Beheshti B, Prasad M, Albert M, Marrano P, Begley H, Shaw P, Covens A, Murphy J, Rosen B, Minkin S, Squire JA, Macgregor PF. High-resolution mapping of genomic imbalance and identification of gene expression profiles associated with differential chemotherapy response in serous epithelial ovarian cancer. Neoplasia. 2005;7:603–613. doi: 10.1593/neo.04760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, Bruneval P, Fridman WH, Becker C, Pages F, Speicher MR, Trajanoski Z, Galon J. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Brown SD, Warren RL, Gibb EA, Martin SD, Spinelli JJ, Nelson BH, Holt RA. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res. 2014;24:743–750. doi: 10.1101/gr.165985.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P, Zinzindohoue F, Bruneval P, Cugnenc PH, Trajanoski Z, Fridman WH, Pages F. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- George PM, Badiger R, Alazawi W, Foster GR, Mitchell JA. Pharmacology and therapeutic potential of interferons. Pharmacol Ther. 2012;135:44–53. doi: 10.1016/j.pharmthera.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Groom JR, Luster AD. CXCR3 in T cell function. Exp Cell Res. 2011;317:620–631. doi: 10.1016/j.yexcr.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groom JR, Richmond J, Murooka TT, Sorensen EW, Sung JH, Bankert K, von Andrian UH, Moon JJ, Mempel TR, Luster AD. CXCR3 chemokine receptor-ligand interactions in the lymph node optimize CD4+ T helper 1 cell differentiation. Immunity. 2012;37:1091–1103. doi: 10.1016/j.immuni.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–354. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- Klover PJ, Muller WJ, Robinson GW, Pfeiffer RM, Yamaji D, Hennighausen L. Loss of STAT1 from mouse mammary epithelium results in an increased Neu-induced tumor burden. Neoplasia. 2010;12:899–905. doi: 10.1593/neo.10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koromilas AE, Sexl V. The tumor suppressor function of STAT1 in breast cancer. JAKSTAT. 2013;2:e23353. doi: 10.4161/jkst.23353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koti M, Gooding RJ, Nuin P, Haslehurst A, Crane C, Weberpals J, Childs T, Bryson P, Dharsee M, Evans K, Feilotter HE, Park PC, Squire JA. Identification of the IGF1/PI3K/NFkB/ERK gene signalling networks associated with chemotherapy resistance and treatment response in high-grade serous epithelial ovarian cancer. BMC Cancer. 2013;13:549. doi: 10.1186/1471-2407-13-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryczek I, Banerjee M, Cheng P, Vatan L, Szeliga W, Wei S, Huang E, Finlayson E, Simeone D, Welling TH, Chang A, Coukos G, Liu R, Zou W. Phenotype, distribution, generation, and functional and clinical relevance of Th17 cells in the human tumor environments. Blood. 2009;114:1141–1149. doi: 10.1182/blood-2009-03-208249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz M, Toksoy A, Goebeler M, Engelhardt E, Brocker E, Gillitzer R. Strong expression of the lymphoattractant C-X-C chemokine Mig is associated with heavy infiltration of T cells in human malignant melanoma. J Pathol. 1999;189:552–558. doi: 10.1002/(SICI)1096-9896(199912)189:4<552::AID-PATH469>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Leitner NR, Lassnig C, Rom R, Heider S, Bago-Horvath Z, Eferl R, Muller S, Kolbe T, Kenner L, Rulicke T, Strobl B, Muller M. Inducible, dose-adjustable and time-restricted reconstitution of STAT1 deficiency in vivo. PLoS One. 2014;9:e86608. doi: 10.1371/journal.pone.0086608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DE, Gilliland DG. Divergent roles of STAT1 and STAT5 in malignancy as revealed by gene disruptions in mice. Oncogene. 2000;19:2505–2510. doi: 10.1038/sj.onc.1203480. [DOI] [PubMed] [Google Scholar]
- Liu J, Matulonis U. Rational use of cytotoxic chemotherapy for recurrent ovarian cancer. J Natl Compr Canc Netw. 2006;4:947–953. doi: 10.6004/jnccn.2006.0078. [DOI] [PubMed] [Google Scholar]
- Marcus CS, Maxwell GL, Darcy KM, Hamilton CA, McGuire WP. Current approaches and challenges in managing and monitoring treatment response in ovarian cancer. J Cancer. 2014;5:25–30. doi: 10.7150/jca.7810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JW, Chilton-MacNeill S, Koti M, van Wijnen AJ, Squire JA, Zielenska M. Digital expression profiling identifies RUNX2, CDC5L, MDM2, RECQL4, and CDK4 as potential predictive biomarkers for neo-adjuvant chemotherapy response in paediatric osteosarcoma. PLoS One. 2014;9:e95843. doi: 10.1371/journal.pone.0095843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne K, Kobel M, Kalloger SE, Barnes RO, Gao D, Gilks CB, Watson PH, Nelson BH. Systematic analysis of immune infiltrates in high-grade serous ovarian cancer reveals CD20, FoxP3 and TIA-1 as positive prognostic factors. PLoS One. 2009;4:e6412. doi: 10.1371/journal.pone.0006412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuswamy R, Berk E, Junecko BF, Zeh HJ, Zureikat AH, Normolle D, Luong TM, Reinhart TA, Bartlett DL, Kalinski P. NF-kappaB hyperactivation in tumor tissues allows tumor-selective reprogramming of the chemokine microenvironment to enhance the recruitment of cytolytic T effector cells. Cancer Res. 2012;72:3735–3743. doi: 10.1158/0008-5472.CAN-11-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BH. CD20+ B cells: the other tumor-infiltrating lymphocytes. J Immunol. 2010;185:4977–4982. doi: 10.4049/jimmunol.1001323. [DOI] [PubMed] [Google Scholar]
- Ohtani H, Jin Z, Takegawa S, Nakayama T, Yoshie O. Abundant expression of CXCL9 (MIG) by stromal cells that include dendritic cells and accumulation of CXCR3+ T cells in lymphocyte-rich gastric carcinoma. J Pathol. 2009;217:21–31. doi: 10.1002/path.2448. [DOI] [PubMed] [Google Scholar]
- Park YT, Jeong JY, Lee MJ, Kim KI, Kim TH, Kwon YD, Lee C, Kim OJ, An HJ. MicroRNAs overexpressed in ovarian ALDH1-positive cells are associated with chemoresistance. J Ovarian Res. 2013;6:18–2215-6-18. doi: 10.1186/1757-2215-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston CC, Maurer MJ, Oberg AL, Visscher DW, Kalli KR, Hartmann LC, Goode EL, Knutson KL. The ratios of CD8(+) T cells to CD4(+)CD25(+) FOXP3(+) and FOXP3(-) T cells correlate with poor clinical outcome in human serous ovarian cancer. PLoS One. 2013;8:e80063. doi: 10.1371/journal.pone.0080063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainczuk A, Rao J, Gathercole J, Stephens AN. The emerging role of CXC chemokines in epithelial ovarian cancer. Reproduction. 2012;144:303–317. doi: 10.1530/REP-12-0153. [DOI] [PubMed] [Google Scholar]
- Raven JF, Williams V, Wang S, Tremblay ML, Muller WJ, Durbin JE, Koromilas AE. Stat1 is a suppressor of ErbB2/Neu-mediated cellular transformation and mouse mammary gland tumor formation. Cell Cycle. 2011;10:794–804. doi: 10.4161/cc.10.5.14956. [DOI] [PubMed] [Google Scholar]
- Saha B, Jyothi Prasanna S, Chandrasekar B, Nandi D. Gene modulation and immunoregulatory roles of interferon gamma. Cytokine. 2010;50:1–14. doi: 10.1016/j.cyto.2009.11.021. [DOI] [PubMed] [Google Scholar]
- Sakamoto M, Kondo A, Kawasaki K, Goto T, Sakamoto H, Miyake K, Koyamatsu Y, Akiya T, Iwabuchi H, Muroya T, Ochiai K, Tanaka T, Kikuchi Y, Tenjin Y. Analysis of gene expression profiles associated with cisplatin resistance in human ovarian cancer cell lines and tissues using cDNA microarray. Hum Cell. 2001;14:305–315. [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- Selvanayagam ZE, Cheung TH, Wei N, Vittal R, Lo KW, Yeo W, Kita T, Ravatn R, Chung TK, Wong YF, Chin KV. Prediction of chemotherapeutic response in ovarian cancer with DNA microarray expression profiling. Cancer Genet Cytogenet. 2004;154:63–66. doi: 10.1016/j.cancergencyto.2004.01.024. [DOI] [PubMed] [Google Scholar]
- Simpson JA, Al-Attar A, Watson NF, Scholefield JH, Ilyas M, Durrant LG. Intratumoral T cell infiltration, MHC class I and STAT1 as biomarkers of good prognosis in colorectal cancer. Gut. 2010;59:926–933. doi: 10.1136/gut.2009.194472. [DOI] [PubMed] [Google Scholar]
- Sistigu A, Yamazaki T, Vacchelli E, Chaba K, Enot DP, Adam J, Vitale I, Goubar A, Baracco EE, Remedios C, Fend L, Hannani D, Aymeric L, Ma Y, Niso-Santano M, Kepp O, Schultze JL, Tuting T, Belardelli F, Bracci L, La Sorsa V, Ziccheddu G, Sestili P, Urbani F, Delorenzi M, Lacroix-Triki M, Quidville V, Conforti R, Spano JP, Pusztai L, Poirier-Colame V, Delaloge S, Penault-Llorca F, Ladoire S, Arnould L, Cyrta J, Dessoliers MC, Eggermont A, Bianchi ME, Pittet M, Engblom C, Pfirschke C, Preville X, Uze G, Schreiber RD, Chow MT, Smyth MJ, Proietti E, Andre F, Kroemer G, Zitvogel L. Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat Med. 2014;20:1301–1309. doi: 10.1038/nm.3708. [DOI] [PubMed] [Google Scholar]
- Spivey TL, Uccellini L, Ascierto ML, Zoppoli G, De Giorgi V, Delogu LG, Engle AM, Thomas JM, Wang E, Marincola FM, Bedognetti D. Gene expression profiling in acute allograft rejection: challenging the immunologic constant of rejection hypothesis. J Transl Med. 2011;9:174–5876-9-174. doi: 10.1186/1479-5876-9-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Yang S, Sun N, Chen J. Differential expression of STAT1 and p21 proteins predicts pancreatic cancer progression and prognosis. Pancreas. 2014;43:619–623. doi: 10.1097/MPA.0000000000000074. [DOI] [PubMed] [Google Scholar]
- Tymoszuk P, Charoentong P, Hackl H, Spilka R, Muller-Holzner E, Trajanoski Z, Obrist P, Revillion F, Peyrat JP, Fiegl H, Doppler W. High STAT1 mRNA levels but not its tyrosine phosphorylation are associated with macrophage infiltration and bad prognosis in breast cancer. BMC Cancer. 2014;14:257–2407-14-257. doi: 10.1186/1471-2407-14-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZC, Birkbak NJ, Culhane AC, Drapkin R, Fatima A, Tian R, Schwede M, Alsop K, Daniels KE, Piao H, Liu J, Etemadmoghadam D, Miron A, Salvesen HB, Mitchell G, DeFazio A, Quackenbush J, Berkowitz RS, Iglehart JD, Bowtell DD, Australian Ovarian Cancer Study Group. Matulonis UA. Profiles of genomic instability in high-grade serous ovarian cancer predict treatment outcome. Clin Cancer Res. 2012;18:5806–5815. doi: 10.1158/1078-0432.CCR-12-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weberpals JI, Koti M, Squire JA. Targeting genetic and epigenetic alterations in the treatment of serous ovarian cancer. Cancer Genet. 2011;204:525–535. doi: 10.1016/j.cancergen.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Arakawa F, Higuchi F, Ishibashi Y, Goto M, Sugita Y, Nomura Y, Niino D, Shimizu K, Aoki R, Hashikawa K, Kimura Y, Yasuda K, Tashiro K, Kuhara S, Nagata K, Ohshima K. Gene expression analysis of rheumatoid arthritis synovial lining regions by cDNA microarray combined with laser microdissection: up-regulation of inflammation-associated STAT1, IRF1, CXCL9, CXCL10, and CCL5. Scand J Rheumatol. 2012;41:170–179. doi: 10.3109/03009742.2011.623137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeremski M, Petrovic LM, Chiriboga L, Brown QB, Yee HT, Kinkhabwala M, Jacobson IM, Dimova R, Markatou M, Talal AH. Intrahepatic levels of CXCR3-associated chemokines correlate with liver inflammation and fibrosis in chronic hepatitis C. Hepatology. 2008;48:1440–1450. doi: 10.1002/hep.22500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, Makrigiannakis A, Gray H, Schlienger K, Liebman MN, Rubin SC, Coukos G. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348:203–213. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.