Abstract
Background:
MicroRNA-210 (miR-210) is an oncogenic miRNA previously associated with prognosis in human gliomas, an incurable tumour type of the central nervous system. Here miR-210 was investigated as a potential serum biomarker in the diagnosis and prognosis of glioma.
Methods:
Serum was immediately prepared from blood samples collected from patients with glioma grades I–IV at primary diagnosis (n=136) and healthy controls (n=50) from February 2007 to March 2014 in the Department of Neurosurgery of the First Affiliated Hospital of Wannan Medical College (Wuhu, China). Total RNA was isolated from serum. cDNA was synthesised with primers specific for miR-210 and miR-16-1 (internal control), and quantitative real-time RT-PCR was performed. Results were statistically analysed to determine the role of miR-210 in the diagnosis and prognosis of human glioma patients.
Results:
An approximately seven-fold increase in miR-210 expression was detected in serum samples from glioblastoma patients relative to healthy controls. A threshold expression value (2.259) was chosen from receiver operator characteristic curves (ROC), and the low and high miR-210 expression groups were analysed by multivariate Cox proportional hazard regression and Kaplan–Meier analyses. Results revealed an association of high serum miR-210 expression with tumour grade and poor patient outcome (P-values <0.001).
Conclusions:
Serum miR-210 is a promising diagnostic and prognostic biomarker that can be detected in the peripheral blood of patients with glioma.
Keywords: glioma, microRNA-210, diagnosis, prognosis, biomarker
Gliomas are the most common malignant primary intracranial tumours of the central nervous system. These tumours are highly aggressive with a high relapse rate and mortality. Unfortunately, there are currently no effective therapies to treat them (Omuro and DeAngelis, 2013; Ahluwalia and Chang, 2014). Although early detection with circulating biomarkers is an established method in the diagnosis and treatment of many cancer types, diagnostic tests for gliomas are currently limited to MRI and CT (Shankar et al, 2013; Xiao et al, 2014), which are generally performed when gliomas have already progressed to a late or advanced stage. Therefore, to have a set of markers for early detection is still a primary goal to improve diagnosis and treatment of human gliomas.
MicroRNAs (miRNAs) are small (17–24 nucleotides) noncoding single-stranded RNAs that regulate the expression of multiple target genes either by degrading a specific mRNA(s) or inhibiting translation (Nikaki et al, 2012). Many studies have demonstrated that miRNA expression is frequently dysregulated in a cancer-specific manner. This finding presents the opportunity to develop miRNAs as potential biomarkers for the diagnosis and prognosis of human cancers (Ho et al, 2010; Redova et al, 2013; Joosse et al, 2014; Lai et al, 2014; Ulivi and Zoli, 2014). MiRNAs have become particularly appealing as biomarkers as some have also been detected in blood and thus easily exploited for noninvasive detection of cancer (Toiyama et al, 2013; Zhang et al, 2014; Wang et al, 2014b). For example, serum expression of miR-210 has been investigated as a biomarker for the early detection and prognosis of colorectal cancer (Toiyama et al, 2013). The prevalence of circulating miRNAs has also been observed in ovarian cancer and breast cancer and, furthermore, assessed for their potential utility as biomarkers for diagnosis and prognosis of these diseases (Zhang et al, 2014; Wang et al, 2014b).
MiR-210 is an oncogenic miRNA that is a target of hypoxia inducible factors 1 and 2 (Gee et al, 2014). This miRNA has been shown to be upregulated in human tumours derived from most tissue types, including non-small cell lung cancer (NSCLC) (Eilertsen et al, 2014), paragangliomas (Merlo et al, 2013), pancreatic cancer (Takikawa et al, 2013), oral tumours (Scapoli et al, 2010), hepatocellular cancer (Zhan et al, 2014), adrenocortical carcinoma (Szabo et al, 2014), colon cancer (CRC) (Pizzini et al, 2013), ovarian cancer (Li et al, 2014), breast cancer (Wang et al, 2014a), and renal cell cancer (Iwamoto et al, 2014). Recently, high miR-210 expression was also found in biopsy material from human gliomas and, furthermore, associated with poor survival of patients (Qiu et al, 2013; Lai et al, 2014). However, screens for circulating miR-210 levels in glioma patients have not yet been performed.
Here miR-210 was investigated as a candidate circulating biomarker for the early detection and prognosis of gliomas. The goals of the study were first to determine the feasibility of detecting and quantifying the level of expression of miR-210 in serum from glioma patients and, second, to determine the relevance of miR-210 expression to prognosis. In order to evaluate serum miR-210 expression as a potential biomarker for the diagnosis and prognosis of glioma patients, analysis was performed on serum samples from a set of GBM patients and healthy controls for proof of the methodology followed by examination of a larger cohort of samples from patients with gliomas of different histological grades.
Materials and methods
Ethics statement
Patient samples were collected and stored in the Wannan Medical College tumour bank (Wuhu, China). Written informed consent was obtained from all of the patients for procedures approved by the Research Ethics Committee of Wannan Medical College. Patient data and samples were treated according to the ethical and legal standards adopted by the Declaration of Helsinki 2013.
Study design
Blood samples (5 ml; n=186) were collected from glioma patients (n=136) and healthy donors (n=50) from February 2007 to March 2014 in the Department of Neurosurgery of the First Affiliated Hospital of Wannan Medical College. None of the patients had received chemotherapy or radiotherapy prior to surgery. To prepare serum, blood samples were left to clot at room temperature for 30 min immediately following collection and subsequently centrifuged at 3000 g and 4 °C for 10 min. Serum was transferred to 1.5-ml tubes and stored at −80 °C for further processing.
A training phase was performed first on a small set of preoperative serum samples collected from WHO grade IV astrocytoma (glioblastoma; GBM; n=10) patients and normal healthy donors (n=10) as the controls. In the validation phase, serum samples (n=126) were analysed from patients with astrocytic tumours of all grades (WHO grades I–IV) in order to establish the utility of the assay for differential diagnosis. Pathological diagnoses included low-grade gliomas (n=48), pilocytic astrocytomas (WHO grade I, PA; n=13) and diffuse astrocytomas (WHO grade II, DA; n=35), and high-grade gliomas (n=78), anaplastic astrocytomas (WHO grade III, AA; n=46) and GBM (n=32). The cohort consisted of 74 males and 52 females with a median age of 44 years (age range: 15–73 years). The clinicopathological features of all the patients included in the validation phase are summarised in Table 1. Follow-up information of all eligible patients was updated every 3 months by telephone visit and questionnaire letters. Overall survival (OS) was calculated from the date of the initial surgery to the date of death. Patients who died of diseases not directly related to glioma were excluded from the study.
Table 1. Association between miR-210 expression and different clinicopathological features of gliomas.
| Clinicopathological parameters | No. of cases | miR-210 expression | P-value |
|---|---|---|---|
| Sex; male | 70 | 9.34±7.02 | 0.42 |
| Sex; female | 56 | 8.40±6.02 | |
| Age; <44 years | 74 | 9.20±6.64 | 0.44 |
| Age; ⩾44 years | 52 | 8.29±6.26 | |
| WHO grade; I; PA | 13 | 2.87±1.41 | <0.001 |
| WHO grade; II; DA | 35 | 4.99±2.79 | |
| WHO grade; III; AA | 46 | 10.88±6.16 | |
| WHO grade; IV; GBM | 32 | 12.48±7.29 | |
| Tumour size; ⩾5 cm | 59 | 8.30±5.97 | 0.40 |
| Tumour size; <5 cm | 67 | 9.28±6.90 | |
| KPS; <90 | 53 | 9.65±6.62 | 0.09 |
| KPS; ⩾90 | 73 | 7.69±6.15 |
Abbreviations: AA=anaplastic astrocytomas; DA=diffuse astrocytomas; GBM=glioblastoma multiforme; KPS=Karnofsky Performance Scale; PA=pilocytic astrocytomas; WHO=World Health Organisation. The median age was 44 years.
RNA isolation
MiRNAs were extracted from 400 μl of serum using the miRcute miRNA Isolation Kit (Tiangen Biotech Co., LTD.; Beijing, China) according to the manufacturer's protocols. Briefly, lysis solution (400 μl) was added to an equal volume of serum (400 μl). The aqueous phase containing total RNA was extracted with chloroform and transferred to an elution column where protein was removed from the bound RNA. MiRNAs were finally eluted in RNase-free water (30 μl), quantified with the NanoDrop 2000 spectrophotometer (NanoDrop Technologies; Houston, TX, USA), and stored at −80 °C. Only samples with an A260/A280 ratio between 1.8 and 2.0 were utilised for further analysis.
Reverse transcription (RT)
cDNA specific for miR-210 and miR-16-1 (internal control) was synthesised from 3 to 4 μg of total RNA in a reaction mixture containing miR-210 (5′-GTCTGTATGGTTGTTCTGCTCTCTGTCTCATCCCTATCTACAACCATACAGACTCAGCCGCTG-3′)/miR-16-1 (5′-GTCGTATCCAGAGCAGGGTCCGAGGTACACGTTCGCTCTGGATACGACCGCCAATATT-3′) RT primers (Genepharma; Shanghai, China) and M-MLV reverse transcriptase on the DNA Engine Opticon 2 Real-Time Cycler (MJ Research, Inc.; Waltham, MA, USA) with the following cycle profile: 16 °C for 30 min, 42 °C for 30 min, 85 °C for 10 min. Synthesised cDNA was stored at −80 °C for further analysis.
Real-time quantitative reverse transcription-PCR (qRT-PCR)
Each qRT-PCR included 0.8 μl miR-210/miR-16-1 primer set (5 μM) (miR-210, forward: 5′-TTGACCTGTGCGTGTGACA-3′, reverse: 5′-TATGGTTGTTCTGCTCTCTGTCTC-3′ miR-16-1, forward: 5′-CGCCTGTAGCAGCACGTAA-3′, reverse: 5′-CAGAGCAGGGTCCGAGGTA-3′ Genepharma), 20 μl RT-PCR master mix (Genepharma), 0.2 μl Taq DNA polymerase (5 U μl−1; Genepharma), 4 μl of cDNA, and water to a final volume of 40 μl. PCR was performed on the DNA Engine Opticon 2 Real-Time Cycler (MJ Research, Inc., Waltham, MA, USA) with the following cycle profile: 95 °C for 3 min, followed by 45 cycles of 95 °C for 12 s, and 62 °C for 40 s. Cycle threshold (Ct) values were determined, and the expression levels were calculated in triplicate from the equation 2−ΔCt where the raw data of the target miRNA were normalised to the Ct of miR-16-1, which served as the internal control (Li et al, 2013b; Zanutto et al, 2014). PCR products were run on agarose gels to determine size, and dissociation curves were subsequently utilised to examine specificity of the qPCR assay.
Statistical analyses
All data were analysed using MedCalc version 13.0.0 (Broekstraat 52, 9030; Mariakerke, Belgium). Data were expressed as means±s.d. Differences between two groups were assessed by the Mann–Whitney U test, and multiple comparisons between more than two groups were conducted with the Kruskal–Wallis test.
Receiver operating characteristic (ROC) curve analysis and the area under the curve (AUC) statistic were used as measures of the accuracy of miR-210 for correctly identifying glioma patients. A multivariable logistic regression model was used to calculate odds ratios (ORs) for age- and sex-adjusted cases associated with gliomas according to serum miR-210 levels.
The Kaplan–Meier method and log-rank test were used to estimate survival of the different groups. Multivariate Cox proportional hazard regression analysis was used to assess hazard ratios of survival outcome according to grade of glioma and serum miR-210 levels, unadjusted and adjusted, for potential confounding factors for death, including age, sex, pathological grade, KPS, and tumour size. Assumptions of proportionality were confirmed for the Cox proportional hazards analyses by generating Kaplan–Meier survival curves (high vs low miR-210 expression; high- vs low-grade pathological diagnosis) and by ensuring that the two curves did not intersect each other. Differences were considered statistically significant when P<0.05.
Results
Training phase evaluation of miR-210 expression in serum from GBM patients
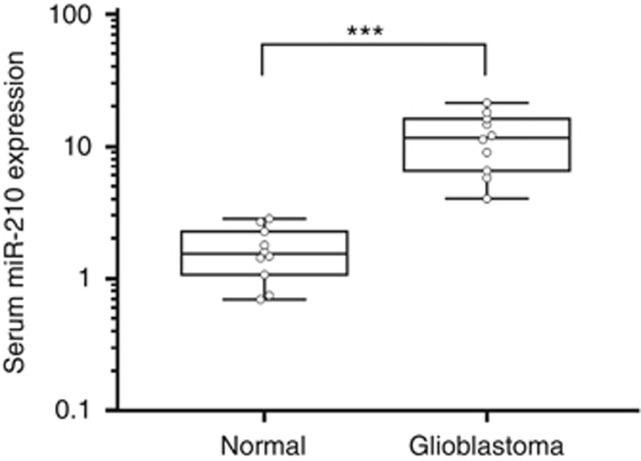
To determine the feasibility of miR-210 detection in serum, quantitative analysis of miR-210 and miR-16-1 were first performed on a subset of serum samples (n=10) from patients with GBM, the most aggressive form of glioma, and serum samples (n=10) prepared from the blood of healthy control subjects. Not only was miR-210 detectable by qRT-PCR in cDNA synthesised from a small volume of serum (400 μl), but an approximately seven-fold difference in expression levels was also evident. The increased expression level of miR-210 in GBM samples relative to control samples was statistically significant (P<0.001; Figure 1).
Figure 1.
MiR-210 is increased in serum from glioblastoma patients relative to healthy controls. Initial screening for miR-210 expression in the training phase, using a small subset of serum specimens from glioma patients. Box plots are shown for miR-210 levels in serum from normal control subjects (n=10) and glioblastoma patients (n=10). Boxes represent interquartile range, and the horizontal line across each box represents the median value. The y axis (log10 scale) represents relative expression of miR-210 where the data were normalised to miR-16-1 expression in sera. Statistical analysis was performed using Mann–Whitney U tests. ***P<0.001.
Serum miR-210 expression levels as a potential diagnostic biomarker for glioma
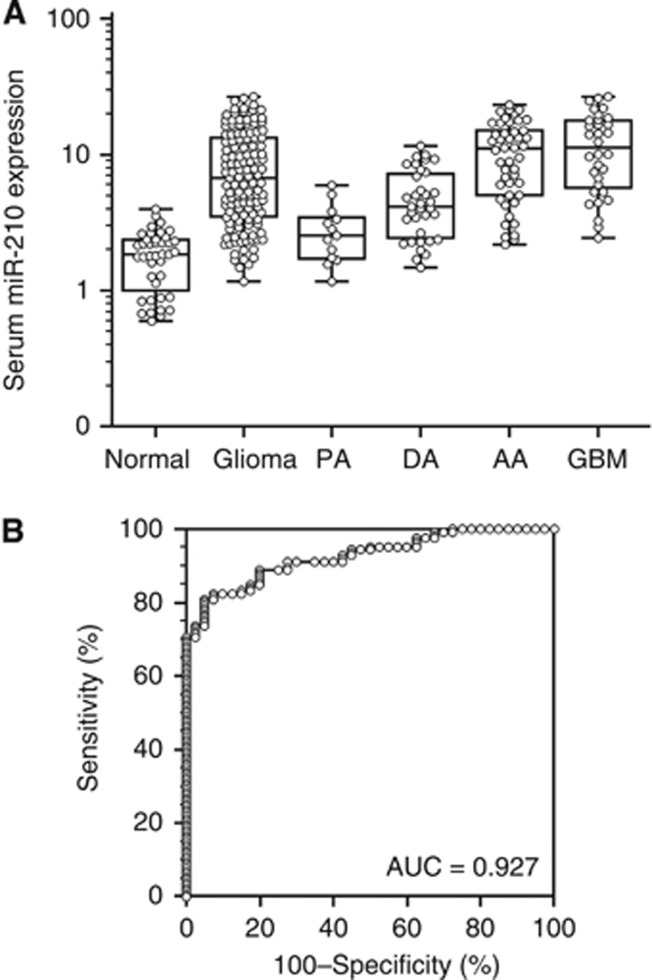
The training phase demonstrated that the miR-210 was detectable in serum samples from glioma patients by the methodology employed. Analysis was subsequently validated on serum samples from a cohort of patients with gliomas of all grades. To evaluate the diagnostic potential of miR-210, expression was determined in a total of 166 serum samples, including those from patients with glioma (n=126) and normal control subjects (n=40). In the cohort overall, the expression levels of serum miR-210 were four-fold increased in patients with glioma relative to controls (P<0.001; Figure 2A). To determine whether this difference was due to gliomas in general or a specific grade of glioma, the expression data were subsequently segregated based on pathological diagnosis. Increased expression of serum miR-210 in high-grade glioma patients (III–IV) remained statistically significant relative to controls in this analysis (P<0.001; Figure 2A). Only a trend for increasing serum miR-210 expression was apparent in low-grade glioma (grade I–II) patients relative to controls. When patient samples were compared based on histological grade, a statistically significant increase in serum miR-210 expression was evident in high-grade relative to low-grade samples (P<0.001; Figure 2A).
Figure 2.
Increased expression of miR-210 in serum samples from gliomas. (A) Box plots represent serum miR-210 levels in healthy control subjects (Normal; n=40), all patients with astrocytic gliomas (Glioma; n=126), and patients separated based on histological WHO grades I–V (PA, n=13; DA, n=35; AA, n=46; GBM, n=32). The y axis (log10 scale) represents miR-210 expression normalised to miR-16-1. Boxes represent the interquartile range, and the horizontal line across each box represents the median value. Statistically significant differences were determined using the Mann–Whitney U test and the Kruskal–Wallis test. (B) ROC curve analysis based on serum miR-210 levels for distinguishing glioma patients from normal controls. Serum miR-210 yielded an area under the curve (AUC) value of 0.927 (95% confidence interval=0.889–0.964), with 91.27% sensitivity and 72.50% specificity in distinguishing glioma patients from normal controls.
The diagnostic potential and discriminatory accuracy of serum miR-210 was evaluated by ROC curve analysis and the corresponding AUC values. ROC analyses revealed that serum miR-210 levels were robust in discriminating patients with glioma from healthy controls, with an AUC value of 0.927 (95% CI=0.889–0.964; Figure 2B). The highest accuracy was at a cutoff expression value of 2.259, where the negative predictive value, positive predictive value, sensitivity, and specificity to identify a patient with glioma were 72.5, 91.3, 91.27, and 72.50%, respectively. In addition, multivariate logistic regression analysis revealed that miR-210 expression in serum was a potential diagnostic biomarker for the identification of patients with gliomas after adjustment for patient's age and sex (P<0.001; Table 2). The OR of 28.19 indicated that increased serum miR-210 levels were strongly linked to a diagnosis of glioma in patients.
Table 2. Multivariate logistic analyses for serum miR-210 levels and various clinical parameters in patients with glioma.
| Parameter Glioma patients vs control subjects | OR (95%CI) | P |
|---|---|---|
| Sex, female vs male | 0.61 (0.45–1.58) | 0.31 |
| Age, <44 vs ⩾44 years | 1.15 (0.45–2.93) | 0.77 |
| MiR-210 in serum, ⩽2.259 vs >2.259 | 28.19 (10.97–72.41) | <0.001 |
Abbreviations: CI=confidence interval; OR=odds ratio.
The cutoff value of serum miR-210 in glioma patients vs control subjects was derived from receiver operating characteristic curves with Youden's index.
Association between serum miR-210 expression and prognosis of glioma patients
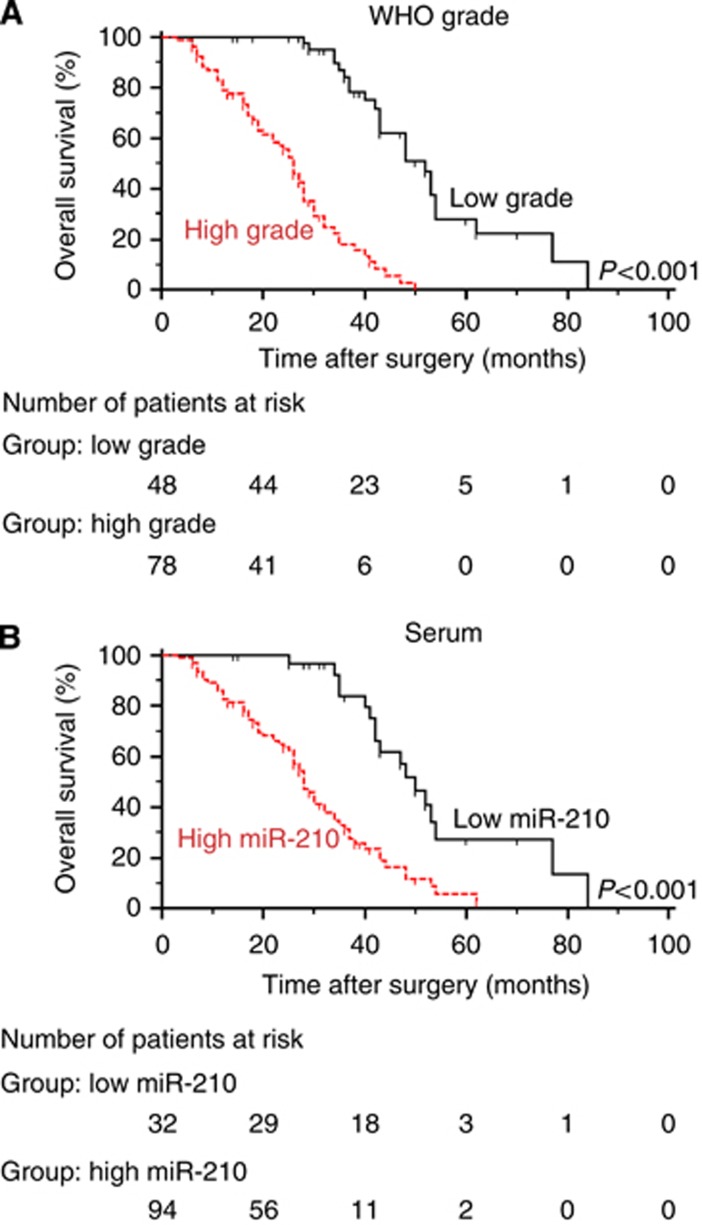
The previous analyses demonstrated that miR-210 expression was associated with glioma grade. Such results indicated that miR-210 expression might potentially also be linked to prognosis as higher-grade gliomas generally portend a worse survival in patients than lower-grade cases. In this cohort, Kaplan–Meier analysis indeed demonstrated that patients with a diagnosis of a high-grade glioma had significantly shorter OS than those with low-grade gliomas (P<0.001; Figure 3A; Table 3). The threshold for high or low miR-210 expression was determined based on the ROC curves for gliomas vs controls with Youden's index. When Kaplan–Meier analysis was performed with these values for high and low miR-210 expression (low<2.259<high), high expression of serum miR-210 was found to be associated with worse OS (P<0.001; log-rank test; Figure 3B). However, no significant associations were found for OS and sex, age at diagnosis, or tumour size.
Figure 3.
Kaplan–Meier curves for overall survival based on pathological diagnosis of the primary tumour and miR-210 expression in matched serum samples. Kaplan–Meier curves displaying overall survival of (A) patients with high-grade (n=78) vs low-grade tumours (n=48; P<0.001), and (B) patients with high-serum (n=94) vs low-serum miR-210 expression (n=32; P<0.001).
Table 3. Univariate and multivariate analyses for overall survival in glioma patients by Cox regression analysis.
| Parameter | Univariate analysis, HR (95%CI) | P | Multivariate analysis, HR (95%CI) | P |
|---|---|---|---|---|
| Age | 1.08 (0.70–1.66) | 0.71 | 0.80 (0.49–1.30) | 0.37 |
| Sex | 1.19 (0.77–1.85 | 0.41 | 1.03 (0.647–1.66) | 0.89 |
| WHO grade | 4.63 (2.95–7.28) | <0.001 | 8.38 (4.43–15.88) | <0.001 |
| Tumour size | 1.07 (0.69–1.64) | 0.77 | 0.80 (0.50–1.28) | 0.36 |
| KPS | 1.56 (1.01–2.39) | 0.04 | 1.16 (0.73–1.84) | 0.52 |
| Serum miR-210 | 3.08 (2.0–4.75) | <0.001 | 3.84 (2.09–7.08) | <0.001 |
Abbreviations: CI=confidence interval; HR=hazard ratio; KPS=Karnofsky Performance Scale, WHO=World Health Organisation.
Univariate and multivariate analyses were used to determine whether the miR-210 expression levels in serum samples from glioma patients and various clinical parameters were independent prognostic factors for patient outcome. Multivariate analysis using Cox regression for all variables demonstrated that serum miR-210 expression (OS: P<0.001, 95% CI 2.09–7.08) and histological grade (OS: P<0.001, 95% CI 4.43–15.88) were independent prognostic factors for patients with glioma. No significant associations were found with sex, age at diagnosis, tumour size, or KPS score (Table 3).
Discussion
MiRNAs are gaining increasing attention as potential biomarkers in cancer as some are detected in extracellular spaces and body fluids creating a novel opportunity for noninvasive assay development (Kawaguchi et al, 2013; Menendez et al, 2013; Srivastava et al, 2014). In this study, miR-210 expression was investigated as one such circulating biomarker for the diagnosis and prognosis of human gliomas. The results demonstrated three major points. First, the overall strategy is feasible as miR-210 was detectable by qRT-PCR of cDNA synthesised from a small volume of serum from patients. Second, serum miR-210 levels could be used to identify glioma patients with high sensitivity and specificity. Finally, high levels of expression were associated with poor prognosis in glioma patients.
Our results are consistent with previous studies where increased expression of miR-210 in glioma biopsies was also associated with worse prognosis (Qiu et al, 2013; Lai et al, 2014). Our approach, however, took these observations an important step forward towards the identification of a set of noninvasive biomarkers for the diagnosis of this disease. MiR-210 thus joins a current trend to include circulating miRNAs as one of these markers. Other circulating miRNAs, miR-128 and miR-342-3p, have also been found to be dysregulated in human gliomas (Roth et al, 2011). An additional seven serum miRNAs were examined in a cohort of malignant astrocytomas and demonstrated potential as noninvasive biomarkers (Yang et al, 2013). The overall strategy supports further investigation of miR-210 as an important circulating biomarker for many other cancer types as well. For example, miR-210 has been identified as a candidate diagnostic and prognostic marker for NSCLC (Li et al, 2013b), and clear cell renal cell carcinoma patients could be identified on the basis of circulating miR-210 expression levels, with 81.0% sensitivity and 79.5% specificity (Zhao et al, 2013).
The major unanswered question is how circulating miR-210 relates to the biology of human gliomas. MiR-210 has been found to be upregulated in a variety of other solid tumour tissue types and potentially influences cellular function through diverse pathways as a number of targets of miR-210 have been reported (Gee et al, 2014; Ivan and Huang, 2014), including cytochrome oxidase assembly protein, iron-sulfur cluster scaffold homolog (Chen et al, 2010; Wang et al, 2014c), succinate dehydrogenase complex subunit D (Puissegur et al, 2011), Ephrin-A3 (Wang et al, 2014c), protein tyrosine phosphatase, non-receptor type 1 (PTPN1; (Li et al, 2014), vascular endothelial growth factor (Zeng et al, 2014), B-cell lymphoma 2 (Chio et al, 2013), caspase-8-associated protein-2 (Kim et al, 2012), and E2F transcription factor 3 (Cui et al, 2014). MiR-210 has also been correlated with hypoxia, a biological phenomenon associated with more aggressive tumours. Under hypoxic conditions, miR-210 is frequently increased in hepatocellular carcinoma (HCC) and is involved in hypoxia-induced HCC metastasis. Vacuole membrane protein 1 (VMP1), a putative suppressor of metastasis in HCC, has been identified as a direct target of miR-210. Downregulation of VMP1 by miR-210 has been shown to mediate hypoxia-induced HCC migration and invasion (Ying et al, 2011). MiR-210 has also been shown to target PTPN1, which promoted cell survival and inhibited apoptosis in ovarian cancer in vitro under hypoxia (Li et al, 2014). These findings taken together with its association with gliomas, however, might indicate that serum miR-210 is associated with later events in gliomagenesis and, therefore, may preclude its utility in the early detection of gliomas. The functional implications of miR-210, however, warrant further investigation as a potential molecular target in the development of novel therapeutics against hypoxic tumour cells (Liu et al, 2014).
Although serum miR-210 appears to be promising biomarker, our current study is limited for several reasons. First, it was a retrospective study, thus restricting measurements to the available serum samples, which had been mostly collected at the primary diagnosis. Circulating miRNAs have been exploited in other studies initially for the detection of a cancer but subsequently to monitor patients after surgery and/or during therapeutic treatment. For example, miR-210 was found to be significantly increased in serum from HCC patients before transarterial chemoembolisation but decreased following transarterial chemoembolisation (Zhan et al, 2014). Second, all patients were Chinese, hence the results may not be more generally applicable to populations of other ethnic origins. Finally, clinical parameters, such as pathological diagnosis, are variable between institutions and/or individual clinicians, hence the results with this small cohort may reflect biases inherent in the acquisition of such clinical data. Clearly, these results require validation in prospective studies performed on larger cohorts from multicentre clinical trials.
In summary, our results illustrate that serum miR-210 expression has potential clinical utility as a diagnostic and prognostic biomarker for human glioma patients. Routine use in the clinic could help to select candidate patients for more aggressive adjuvant treatment or the use of novel therapeutic options that may interrupt abnormal expression patterns of specific miRNAs (Auffinger et al, 2013; Li et al, 2013a). Finally, although the results from this study are promising, miR-210 as a routine glioma biomarker in clinical practice requires further validation in prospective studies on larger cohorts at multiple institutions.
Acknowledgments
The authors thank all participants and cooperating clinicians, the Shanghai GenePharma Co., Ltd, and the Translational Medical Centre of First Affiliated Hospital of Wannan Medical College for the support of basic facilities and Hao Ding and Qing-sheng Dong for assistance with the sample collection. This work was supported by grants from Natural Science Foundation of Wuxi (YGZXL1309).
The authors declare no conflict of interest.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
References
- Ahluwalia MS, Chang SM. Medical therapy of gliomas. J Neurooncol. 2014;119:503–512. doi: 10.1007/s11060-014-1495-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffinger B, Thaci B, Ahmed A, Ulasov I, Lesniak MS. MicroRNA targeting as a therapeutic strategy against glioma. Curr Mol Med. 2013;13:535–542. doi: 10.2174/1566524011313040006. [DOI] [PubMed] [Google Scholar]
- Chen Z, Li Y, Zhang H, Huang P, Luthra R. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene. 2010;29:4362–4368. doi: 10.1038/onc.2010.193. [DOI] [PubMed] [Google Scholar]
- Chio CC, Lin JW, Cheng HA, Chiu WT, Wang YH, Wang JJ, Hsing CH, Chen RM. MicroRNA-210 targets antiapoptotic bcl-2 expression and mediates hypoxia-induced apoptosis of neuroblastoma cells. Arch Toxicol. 2013;87:459–468. doi: 10.1007/s00204-012-0965-5. [DOI] [PubMed] [Google Scholar]
- Cui H, Seubert B, Stahl E, Dietz H, Reuning U, Moreno-Leon L, Ilie M, Hofman P, Nagase H, Mari B, Kruger A. Tissue inhibitor of metalloproteinases-1 induces a pro-tumourigenic increase of miR-210 in lung adenocarcinoma cells and their exosomes. Oncogene. 2014;29:300. doi: 10.1038/onc.2014.300. [DOI] [PubMed] [Google Scholar]
- Eilertsen M, Andersen S, Al-Saad S, Richardsen E, Stenvold H, Hald SM, Al-Shibli K, Donnem T, Busund LT, Bremnes RM. Positive prognostic impact of miR-210 in non-small cell lung cancer. Lung Cancer. 2014;83:272–278. doi: 10.1016/j.lungcan.2013.11.005. [DOI] [PubMed] [Google Scholar]
- Gee HE, Ivan C, Calin GA, Ivan M. Hypoxamirs and cancer: from biology to targeted therapy. Antioxid Redox Signal. 2014;21:1220–1238. doi: 10.1089/ars.2013.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho AS, Huang X, Cao H, Christman-Skieller C, Bennewith K, Le Q-T, Koong AC. Circulating miR-210 as a novel hypoxia marker in pancreatic cancer. Transl Oncol. 2010;3:109–113. doi: 10.1593/tlo.09256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M, Huang X. MiR-210: fine-tuning the hypoxic response. Adv Exp Med Biol. 2014;772:205–227. doi: 10.1007/978-1-4614-5915-6_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto H, Kanda Y, Sejima T, Osaki M, Okada F, Takenaka A. Serum miR-210 as a potential biomarker of early clear cell renal cell carcinoma. Int J Oncol. 2014;44:53–58. doi: 10.3892/ijo.2013.2169. [DOI] [PubMed] [Google Scholar]
- Joosse SA, Muller V, Steinbach B, Pantel K, Schwarzenbach H. Circulating cell-free cancer-testis mage-a RNA, boris RNA, let-7b and miR-202 in the blood of patients with breast cancer and benign breast diseases. Br J Cancer. 2014;111:909–917. doi: 10.1038/bjc.2014.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi T, Komatsu S, Ichikawa D, Morimura R, Tsujiura M, Konishi H, Takeshita H, Nagata H, Arita T, Hirajima S, Shiozaki A, Ikoma H, Okamoto K, Ochiai T, Taniguchi H, Otsuji E. Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer. Br J Cancer. 2013;108:361–369. doi: 10.1038/bjc.2012.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HW, Mallick F, Durrani S, Ashraf M, Jiang S, Haider KH. Concomitant activation of miR-107/pdcd10 and hypoxamir-210/casp8ap2 and their role in cytoprotection during ischemic preconditioning of stem cells. Antioxid Redox Signal. 2012;17:1053–1065. doi: 10.1089/ars.2012.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai NS, Dong QS, Ding H, Miao ZL, Lin YC. MicroRNA-210 overexpression predicts poorer prognosis in glioma patients. J Clin Neurosci. 2014;21:755–760. doi: 10.1016/j.jocn.2013.06.024. [DOI] [PubMed] [Google Scholar]
- Li L, Huang K, You Y, Fu X, Hu L, Song L, Meng Y. Hypoxia-induced miR-210 in epithelial ovarian cancer enhances cancer cell viability via promoting proliferation and inhibiting apoptosis. Int J Oncol. 2014;44:2111–2120. doi: 10.3892/ijo.2014.2368. [DOI] [PubMed] [Google Scholar]
- Li M, Li J, Liu L, Li W, Yang Y, Yuan J. MicroRNA in human glioma. Cancers. 2013;5:1306–1331. doi: 10.3390/cancers5041306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZH, Zhang H, Yang ZG, Wen GQ, Cui YB, Shao GG. Prognostic significance of serum microRNA-210 levels in nonsmall-cell lung cancer. J Int Med Res. 2013;41:1437–1444. doi: 10.1177/0300060513497560. [DOI] [PubMed] [Google Scholar]
- Liu T, Zhao L, Chen W, Li Z, Hou H, Ding L, Li X. Inactivation of von hippel-lindau increases ovarian cancer cell aggressiveness through the HIF1alpha/miR-210/vmp1 signaling pathway. Int J Mol Med. 2014;33:1236–1242. doi: 10.3892/ijmm.2014.1661. [DOI] [PubMed] [Google Scholar]
- Menendez P, Padilla D, Villarejo P, Palomino T, Nieto P, Menendez JM, Rodriguez-Montes JA. Prognostic implications of serum microRNA-21 in colorectal cancer. J Surg Oncol. 2013;108:369–373. doi: 10.1002/jso.23415. [DOI] [PubMed] [Google Scholar]
- Merlo A, de Quiros SB, de Santa-Maria IS, Pitiot AS, Balbin M, Astudillo A, Scola B, Aristegui M, Quer M, Suarez C, Chiara MD. Identification of somatic vhl gene mutations in sporadic head and neck paragangliomas in association with activation of the HIF-1alpha/miR-210 signaling pathway. J Clin Endocrinol Metab. 2013;98:2013–1636. doi: 10.1210/jc.2013-1636. [DOI] [PubMed] [Google Scholar]
- Nikaki A, Piperi C, Papavassiliou AG. Role of microRNAs in gliomagenesis: targeting miRNAs in glioblastoma multiforme therapy. Expert Opin Investig Drugs. 2012;21:1475–1488. doi: 10.1517/13543784.2012.710199. [DOI] [PubMed] [Google Scholar]
- Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310:1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- Pizzini S, Bisognin A, Mandruzzato S, Biasiolo M, Facciolli A, Perilli L, Rossi E, Esposito G, Rugge M, Pilati P, Mocellin S, Nitti D, Bortoluzzi S, Zanovello P. Impact of microRNAs on regulatory networks and pathways in human colorectal carcinogenesis and development of metastasis. BMC Genomics. 2013;14:1471–2164. doi: 10.1186/1471-2164-14-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puissegur MP, Mazure NM, Bertero T, Pradelli L, Grosso S, Robbe-Sermesant K, Maurin T, Lebrigand K, Cardinaud B, Hofman V, Fourre S, Magnone V, Ricci JE, Pouyssegur J, Gounon P, Hofman P, Barbry P, Mari B. MiR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2011;18:465–478. doi: 10.1038/cdd.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu S, Lin S, Hu D, Feng Y, Tan Y, Peng Y. Interactions of miR-323/miR-326/miR-329 and miR-130a/miR-155/miR-210 as prognostic indicators for clinical outcome of glioblastoma patients. J Transl Med. 2013;11:1479–5876. doi: 10.1186/1479-5876-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redova M, Sana J, Slaby O. Circulating miRNAs as new blood-based biomarkers for solid cancers. Future Oncol. 2013;9:387–402. doi: 10.2217/fon.12.192. [DOI] [PubMed] [Google Scholar]
- Roth P, Wischhusen J, Happold C, Chandran PA, Hofer S, Eisele G, Weller M, Keller A. A specific miRNA signature in the peripheral blood of glioblastoma patients. J Neurochem. 2011;118:449–457. doi: 10.1111/j.1471-4159.2011.07307.x. [DOI] [PubMed] [Google Scholar]
- Scapoli L, Palmieri A, Lo Muzio L, Pezzetti F, Rubini C, Girardi A, Farinella F, Mazzotta M, Carinci F. MicroRNA expression profiling of oral carcinoma identifies new markers of tumor progression. Int J Immunopathol Pharmacol. 2010;23:1229–1234. doi: 10.1177/039463201002300427. [DOI] [PubMed] [Google Scholar]
- Shankar JJ, Woulfe J, Silva VD, Nguyen TB. Evaluation of perfusion CT in grading and prognostication of high-grade gliomas at diagnosis: a pilot study. AJR Am J Roentgenol. 2013;200:W504–W509. doi: 10.2214/AJR.12.8967. [DOI] [PubMed] [Google Scholar]
- Srivastava A, Goldberger H, Dimtchev A, Marian C, Soldin O, Li X, Collins SP, Suy S, Kumar D. Circulatory miR-628-5p is downregulated in prostate cancer patients. Tumour Biol. 2014;35:4867–4873. doi: 10.1007/s13277-014-1638-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo DR, Luconi M, Szabo PM, Toth M, Szucs N, Horanyi J, Nagy Z, Mannelli M, Patocs A, Racz K, Igaz P. Analysis of circulating microRNAs in adrenocortical tumors. Lab Invest. 2014;94:331–339. doi: 10.1038/labinvest.2013.148. [DOI] [PubMed] [Google Scholar]
- Takikawa T, Masamune A, Hamada S, Nakano E, Yoshida N, Shimosegawa T. MiR-210 regulates the interaction between pancreatic cancer cells and stellate cells. Biochem Biophys Res Commun. 2013;437:433–439. doi: 10.1016/j.bbrc.2013.06.097. [DOI] [PubMed] [Google Scholar]
- Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105:849–859. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulivi P, Zoli W. MiRNAs as non-invasive biomarkers for lung cancer diagnosis. Molecules. 2014;19:8220–8237. doi: 10.3390/molecules19068220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhao J, Shi M, Ding Y, Sun H, Yuan F, Zou Z. Elevated expression of miR-210 predicts poor survival of cancer patients: a systematic review and meta-analysis. PLoS One. 2014;9:e89223. doi: 10.1371/journal.pone.0089223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kim S, Kim IM. Regulation of metastasis by microRNAs in ovarian cancer. Front Oncol. 2014;4:143. doi: 10.3389/fonc.2014.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Yin B, Wang B, Ma Z, Liu W, Lv G. MicroRNA-210 promotes proliferation and invasion of peripheral nerve sheath tumor cells targeting EFNA3. Oncol Res. 2014;21:145–154. doi: 10.3727/096504013X13841340689573. [DOI] [PubMed] [Google Scholar]
- Xiao N, Gu W, Wang H, Deng Y, Shi X, Ye L. T1-T2 dual-modal MRI of brain gliomas using pegylated gd-doped iron oxide nanoparticles. J Colloid Interface Sci. 2014;417:159–165. doi: 10.1016/j.jcis.2013.11.020. [DOI] [PubMed] [Google Scholar]
- Yang C, Wang C, Chen X, Chen S, Zhang Y, Zhi F, Wang J, Li L, Zhou X, Li N, Pan H, Zhang J, Zen K, Zhang CY, Zhang C. Identification of seven serum microRNAs from a genome-wide serum microRNA expression profile as potential noninvasive biomarkers for malignant astrocytomas. Int J Cancer. 2013;132:116–127. doi: 10.1002/ijc.27657. [DOI] [PubMed] [Google Scholar]
- Ying Q, Liang L, Guo W, Zha R, Tian Q, Huang S, Yao J, Ding J, Bao M, Ge C, Yao M, Li J, He X. Hypoxia-inducible microRNA-210 augments the metastatic potential of tumor cells by targeting vacuole membrane protein 1 in hepatocellular carcinoma. Hepatology. 2011;54:2064–2075. doi: 10.1002/hep.24614. [DOI] [PubMed] [Google Scholar]
- Zanutto S, Pizzamiglio S, Ghilotti M, Bertan C, Ravagnani F, Perrone F, Leo E, Pilotti S, Verderio P, Gariboldi M, Pierotti MA. Circulating miR-378 in plasma: a reliable, haemolysis-independent biomarker for colorectal cancer. Br J Cancer. 2014;110:1001–1007. doi: 10.1038/bjc.2013.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, He X, Wang Y, Tang Y, Zheng C, Cai H, Liu J, Fu Y, Yang GY. MicroRNA-210 overexpression induces angiogenesis and neurogenesis in the normal adult mouse brain. Gene Therapy. 2014;21:37–43. doi: 10.1038/gt.2013.55. [DOI] [PubMed] [Google Scholar]
- Zhan M, Li Y, Hu B, He X, Huang J, Zhao Y, Fu S, Lu L. Serum microRNA-210 as a predictive biomarker for treatment response and prognosis in patients with hepatocellular carcinoma undergoing transarterial chemoembolization. J Vasc Interv Radiol. 2014;13:00448–00445. doi: 10.1016/j.jvir.2014.04.013. [DOI] [PubMed] [Google Scholar]
- Zhang K, Zhang Y, Liu C, Xiong Y, Zhang J. MicroRNAs in the diagnosis and prognosis of breast cancer and their therapeutic potential (review) Int J Oncol. 2014;45:950–958. doi: 10.3892/ijo.2014.2487. [DOI] [PubMed] [Google Scholar]
- Zhao A, Li G, Peoc'h M, Genin C, Gigante M. Serum miR-210 as a novel biomarker for molecular diagnosis of clear cell renal cell carcinoma. Exp Mol Pathol. 2013;94:115–120. doi: 10.1016/j.yexmp.2012.10.005. [DOI] [PubMed] [Google Scholar]