Abstract
The events that occur after the binding of the enzymatic E colicins to Escherichia coli BtuB receptors that lead to translocation of the cytotoxic domain into the periplasmic space and, ultimately, cell killing are poorly understood. It has been suggested that unfolding of the coiled-coil BtuB receptor binding domain of the E colicins may be an essential step that leads to the loss of immunity protein from the colicin and immunity protein complex and then triggers the events of translocation. We introduced pairs of cysteine mutations into the receptor binding domain of colicin E9 (ColE9) that resulted in the formation of a disulfide bond located near the middle or the top of the R domain. After dithiothreitol reduction, the ColE9 protein with the mutations L359C and F412C (ColE9 L359C-F412C) and the ColE9 protein with the mutations Y324C and L447C (ColE9 Y324C-L447C) were slightly less active than equivalent concentrations of ColE9. On oxidation with diamide, no significant biological activity was seen with the ColE9 L359C-F412C and the ColE9 Y324C-L447C mutant proteins; however diamide had no effect on the activity of ColE9. The presence of a disulfide bond was confirmed in both of the oxidized, mutant proteins by matrix-assisted laser desorption ionization-time of flight mass spectrometry. The loss of biological activity of the disulfide-containing mutant proteins was not due to an indirect effect on the properties of the translocation or DNase domains of the mutant colicins. The data are consistent with a requirement for the flexibility of the coiled-coil R domain after binding to BtuB.
Colicins are plasmid-encoded antibacterial proteins that are secreted as part of the stress response system of Escherichia coli to kill other bacteria. They are classified into types on the basis of the cell surface receptor on the target cells to which they bind. All E colicins bind to the product of the chromosomal btuB gene, an outer membrane protein which is an essential component of the high-affinity transport system for vitamin B12 in E. coli, and require the outer membrane protein OmpF as a coreceptor (19). Based on immunity tests (9, 38) the E colicins have been subdivided into nine subtypes, colicin E1 (ColE1) to ColE9. These fall into one of three cytotoxic classes: membrane-depolarizing or pore-forming agents such as ColE1 (10); DNases such as ColE2, ColE7, ColE8, and ColE9 (9); and RNases such as ColE3, ColE4, ColE5, and ColE6 (25, 28). In common with most colicins, the enzymatic E-type colicins consist of three functional domains. The killing activity is contained in the C-terminal domain, which can be isolated as a stable and active protein (15, 21, 29, 37). The central section contains the receptor-binding (R) domain, while the N-terminal T domain is responsible for translocation of the cytotoxic domain into the cytoplasm of the target cell (1, 14). After binding to their outer membrane receptors, group A colicins, such as ColE9, are translocated across the membrane in a process which is mediated by the tol system (23). Translocation requires a specific pentapeptide sequence in the T domain, known initially as the TolA box (31), but which is now known to interact with TolB (4, 7). ColE9 contains a TolB box from residues 35 to 39, DGSGW, which has been shown by mutagenesis to be important for its killing action (14) and for the interaction of the T domain with the translocation protein TolB (7). The mechanism by which TolB recognizes and specifically binds to the TolB box sequence is unknown; however, recent nuclear magnetic resonance (NMR) experiments have shown that the T domain of ColE9 contains a large structurally disordered region that possesses a high degree of flexibility (8). The recent X-ray structure of the RNase ColE3 (35) did not reveal any resolved electron density for residues 1 to 83, a region of the T domain whose sequence is highly conserved in the enzymatic E colicins and, thus, might be expected to be similarly flexible.
The mechanism by which the cytotoxic C-terminal domains of enzymatic E colicins are translocated to the cytoplasm of E. coli cells, across the outer membrane, the periplasmic space, and the cytoplasmic membrane is an impressive feat and is probably unique in bacteria. The events that take place after receptor binding are speculative but presumably require the entry of at least part of the T domain of a tol-dependent colicin into the periplasm of the target E. coli cell, where it can interact with Tol proteins such as TolB (6), and in some way open a pathway in the outer membrane that allows entry of the cytotoxic domain. Information on the mechanism by which the enzymatic domain reaches the cytoplasm is very limited; however, it was recently reported that the DNase domains of ColE9 and ColE2 exhibit channel-forming activity in planar lipid bilayers that is linked to toxin translocation across the cytoplasmic membrane of E. coli cells (27).
Here we report that the introduction of cysteine residues in the R domain of ColE9, in positions where a disulfide bond can be formed, inhibits colicin activity without significantly affecting BtuB binding, or binding to TolB. The addition of dithiothreitol (DTT) to the oxidized proteins containing disulfide bonds restored colicin activity. These observations are consistent with a requirement for a conformational change in the R domain that is essential for colicin activity.
MATERIALS AND METHODS
Plasmids, bacterial strains, and media.
E. coli JM83 (ara [Δlac-proAB] rpsL φ80lacZΔM15) was used as the host strain for cloning and mutagenesis. E. coli BL21(DE3) or ER2566 (Novagen) was used as the host strain for the expression vector pET21a (Novagen), which has a strong, IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible T7 polymerase promoter and a C-terminal polyhistidine tag (His-tag) to facilitate the purification of overexpressed proteins as ColE9/Im9 complexes. E. coli DH5α (Invitrogen) was used as a colicin-sensitive strain to determine growth inhibition by ColE9 and mutant proteins. E. coli 113/3 is a metE mutant of the W strain of E. coli (ATCC 9637) (11). All cultures were routinely grown in Luria-Bertani (LB) broth, or on plates of LB agar, supplemented where required with ampicillin (100 μg ml−1). Plasmid pCS4, that encodes the ceaI gene, together with the ceiI immunity gene with a C-terminal His-tag, under the control of an inducible T7 promoter has been previously described (15) and was used as the template DNA to introduce cysteine mutations into the ceaI gene.
Site-directed mutagenesis.
Site-directed mutants of the R domain of ColE9 were constructed by using the megaprimer PCR method (34). A mutagenic primer was used in conjunction with a suitable forward or reverse primer to obtain a PCR product, which was then used as a megaprimer in a second-stage PCR. The final PCR product containing the desired mutation in the ceai gene was isolated by using the QIAEX-II gel extraction kit (QIAGEN) and cloned into the required plasmid vector.
Colicin activity assays.
Colicin titers were determined by spotting 2-μl samples of purified ColE9/Im9 protein complex (serially diluted in 50 mM K2HPO4 · KH2PO4 buffer [pH 7.0]; 0.1 mg of bovine serum albumin ml−1) onto a large, square LB agar plate containing ampicillin (100 μg ml−1) that was overlaid with a soft agar lawn of the sensitive indicator strain E. coli DH5α (pUC18) (36). The colicin titer is the lowest concentration of colicin complex that produces a clear zone of inhibition in the indicator lawn.
Protein purification.
pET vectors (Novagen) encoding ColE9 mutant proteins along with a polyhistidine-tagged Im9 were transformed into E. coli ER2566 or BL21(DE3) cells. ColE9/Im9 complexes were purified as previously described by metal chelate chromatography (14) with an elution buffer containing 1 M imidazole, 0.5 M NaCl, and 20 mM Tris-HCl (pH 7.9). Protein concentrations were determined by absorbance at 280 nm.
Diamide oxidation and DTT reduction.
ColE9 mutant protein complexes were dialysed overnight against reducing buffer (50 mM K2HPO4 · KH2PO4 buffer [pH 7.0], 1 mM DTT), with up to four 5-ml fractions in 5 liters of buffer. For diamide oxidation, the protein samples were dialysed overnight against 50 mM K2HPO4 · KH2PO4 buffer, pH 7.0, to remove the DTT. Protein samples were then adjusted to 1 mM with diamide and incubated for 30 min in the dark at room temperature, before overnight dialysis against nonreducing buffer (50 mM K2HPO4 · KH2PO4 buffer, pH 7.0).
MALDI-TOF mass spectrometry.
Tryptic digests were carried out in 25 mM ammonium bicarbonate buffer, pH 8.5. A total of 2 μg of each ColE9/Im9 complex was incubated with 80 ng of trypsin at room temperature for 5 h before the addition of 60 ng of trypsin inhibitor to stop the reaction. Trypsin-digested ColE9/Im9 protein complexes were divided into 15-μl aliquots, one of which was reduced by the addition of DTT to a final concentration of 1 mM; the other was left in its oxidized form. A total of 2 μl of each sample was spotted onto a 96-well matrix-assisted laser desorption ionization (MALDI) plate, each spot being overlaid with 1 μl of alpha-cyano 4-hydroxy cinnamic acid (10 mg ml−1) in 50% acetonitrile, 0.1% trifluoroacetic acid. MALDI-time of flight (TOF) experiments were carried out by using an Applied Biosystems 4700 proteomics analyzer. Differences between the spectra of the oxidized and the reduced digested proteins were analyzed to detect peaks in the oxidized spectra corresponding to two cysteine-containing peptides linked through a disulfide bond.
BtuB binding assays.
We used two different assays of BtuB receptor binding that have both been previously described (30). The biological protection assay determines the ability of ColE9 mutant protein complexes to protect E. coli cells from killing by ColE9/Im9 and was performed as previously described (30) but by using E. coli DH5α cells. In the vitamin B12 competition assay, E. coli 113/3 was grown overnight in L broth at 37°C with aeration. The cells were diluted 1:100 in 50 ml of M9 minimal medium supplemented with 0.2% (wt/vol) glucose, 10 mM MgSO4, 20 mM CaCl2, 100 μg of thiamine ml−1, 1 nM vitamin B12, and various concentrations of ColE9 mutant protein complexes. The cells were grown for approximately 6 h, and measurements of growth were taken at 30-min intervals by determining the optical density at 600 nm.
Nuclease activity.
Duplicate Kunitz assays were performed as described previously (33). Briefly, assays were carried out in volumes of 1 ml containing 50 mM Tris-HCl (pH 7.5), 20 mM MgS04, and 50 μg of calf thymus DNA at a temperature of 25°C. For these assays, only ColE9 and ColE9 mutant proteins were purified in the absence of Im9 under denaturing conditions by using 6 M guanidine hydrochloride as previously described (14). The reaction was initiated by the addition of 50 μg of ColE9, and DNA hydrolysis was monitored at 2-min intervals by the change in hyperchromicity at 260 nm by using a Philips PU 8730 spectrophotometer.
Surface plasmon resonance.
The interaction of the complex of the oxidized ColE9 protein with mutations L359C and F412C (L359C-F412C)/Im9 and of the complex of ColE9 with mutations Y324C and L447C (Y324C-L447C)/Im9 with TolB was compared to the interaction with ColE9/Im9 by using surface plasmon resonance (SPR) (20). TolB (50 μg/ml) was coupled to a CM5 sensor chip by using an amine-coupling kit (Biacore AB, Uppsala, Sweden) with a 6-min contact time. After deactivation, the relative response between charged and uncharged flow cells was 5,000. ColE9/Im9 complex (3 μM) diluted in HBS buffer (0.01 M HEPES [pH 7.4], 0.15 M NaCl, 3 mM EDTA, 0.005% [vol/vol] surfactant P20), pH 7.4, was injected over a TolB chip for 2 min, followed by a 10 mM glycine regeneration. The same experiment was then repeated with oxidized ColE9 L359C-F412C/Im9 and ColE9 Y324C-L447C/Im9 mutant proteins. The experiment was repeated three times with very similar results on each occasion.
CD.
Circular dichroism (CD) spectra were measured on a JASCO J-810 spectropolarimeter, thermostated at 22°C. All protein samples were prepared at a concentration of 3.5 μM in 10 mM potassium phosphate buffer, pH 7.0. Data were collected between 190 and 250 nm in a 1-mm path length. All spectra were corrected for buffer contributions.
RESULTS
Introducing disulfide locks in the R domain.
The minimum R domain of ColE9, as shown by deletion subcloning and assay of the ability of the resulting purified proteins to bind to BtuB, consists of residues 343 to 418 (30). A recent paper reporting the three-dimensional structure of ColB (18) questioned the location of the R domain of ColE9 (30). This suggestion ignores the overwhelming evidence that supports the identification of the R domain: (i) the three-dimensional structure of the complex of BtuB and the 135 residues of the coiled-coil R domain (R135) shows the molecular details of the interaction (22); (ii) the demonstration that a 34-residue peptide, located within the R domain, binds to BtuB with nanomolar affinity (26); and (iii) the demonstration that the 76-residue R domain peptide competes for binding with vitamin B12 (30).
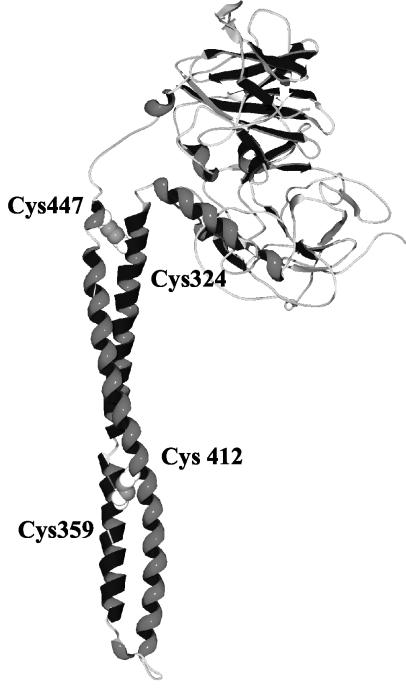
Because of the 100% sequence conservation between the T+R domains of ColE9 and ColE3, we can assume that the structure of these two domains in ColE9 will be identical to that recently reported for ColE3 (35). This is supported by solution NMR data for the 76-residue R domain, which indicate a helical structure in a rod-like, elongated shape (2). The R domain of ColE3 is a 100-Å long antiparallel alacoil of the ROP type with a seven-residue hairpin linking the two arms of the coiled-coil (35). The 76-residue R domain constitutes approximately the bottom half of the alacoil including the hairpin. In order to investigate the role of conformational changes of the R domain in colicin killing, we introduced cysteine mutations in the helices of the R domain of ColE9 in order to create a disulfide bond that would “lock” the conformation of the R domain. Engineered disulfide bonds have been shown to be a useful tool to probe the structural events that accompany the translocation of toxins across membranes (13, 27). We identified two pairs of candidate residues to mutate to cysteines, L359C-F412C and Y324C-L447C (Fig. 1). The L359C-F412C cysteines are estimated to be >1.6 Å apart and were chosen as they are located near the middle of the coiled-coil R domain, while the Y324C-L447C cysteines are estimated to be >1.83 Å apart and are located near the top of the coiled-coil. We reasoned that there might be differences in properties between mutant colicins containing a disulfide bond located near the top of the R domain, where the effect of unfolding might be more significant than that occurring near the middle of the coiled-coil.
FIG. 1.
Structure of ColE3 (35) with the location of the disulfide bond formed between the C324-C447 and C359-C412 mutations in the R domain of ColE9 indicated in spacefill.
We made single cysteine mutations at the four positions and confirmed that there was no effect on the titers of the purified ColE9 mutant protein/Im9 complexes compared to titers of the ColE9/Im9 complex. The double mutants were constructed, and the resulting plasmid constructs were sequenced to confirm the presence of both cysteine mutations in the R domain. Plasmid pBHZ2 encodes ColE9 containing the L359C-F412C mutations, while plasmid pBH29 encodes ColE9 containing the Y324C-L447C mutations. The mutant ColE9 proteins encoded by pBHZ2 and pBH29 (which were called ColE9 L359C-F412C and ColE9 Y324C-L447C, respectively) were then overexpressed as a complex with Im9 and purified by making use of the polyhistidine tag at the C terminus of the Im9 immunity gene as previously described (14). As a control, we constructed plasmid pBH16 which encodes ColE9 containing the mutations Y324C and V405C (Y324C-V405C), positions in which the cysteines should be too far apart to form a disulfide bond. The ColE9 Y324C-V405C/Im9 mutant protein was then purified as a polyhistidine-tagged complex from cells containing plasmid pBH16.
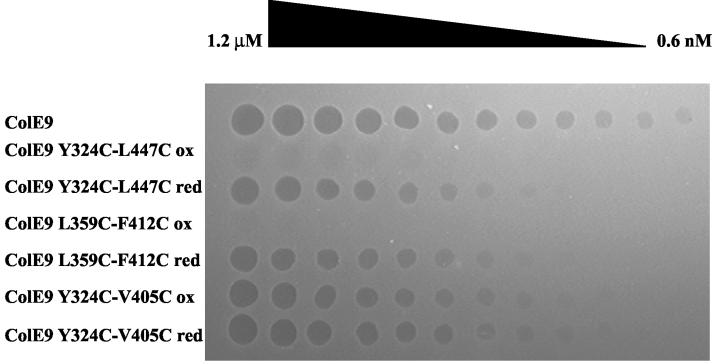
Each of the three purified protein preparations was split into two samples, one of which was oxidized with diamide, and the other was reduced with DTT. The activity of the two samples of each mutant protein was then compared with that of the ColE9/Im9 complex in the large plate assay of biological activity (Fig. 2). After DTT reduction, all three of the mutant proteins were 8- to 16-fold less active than equivalent concentrations of ColE9/Im9. A similar result has been observed with the introduction of two cysteine residues into the DNase domain of ColE9 (27). After oxidation with diamide, no zones of inhibition were visible with ColE9 L359C-F412C/Im9, while oxidized ColE9 Y324C-L447C/Im9 produced very faint zones of inhibition. The activity of ColE9 Y324C-V405C/Im9 was identical in the oxidized or reduced samples.
FIG. 2.
Growth inhibitory activity of disulfide lock mutant proteins. The growth inhibitory activity of the ColE9 L359C-F412C, ColE9 Y324C-L447C, and ColE9 Y324C-V405C mutant proteins is shown compared to ColE9 in a large-plate assay. Aliquots of doubling dilutions of each of the proteins, containing concentrations of between 0.6 nM and 1.2 μM, were spotted onto a large agar plate spread with the indicator E. coli DH5α. A clear zone in the lawn of cells indicates growth inhibition by the colicin protein at that dilution.
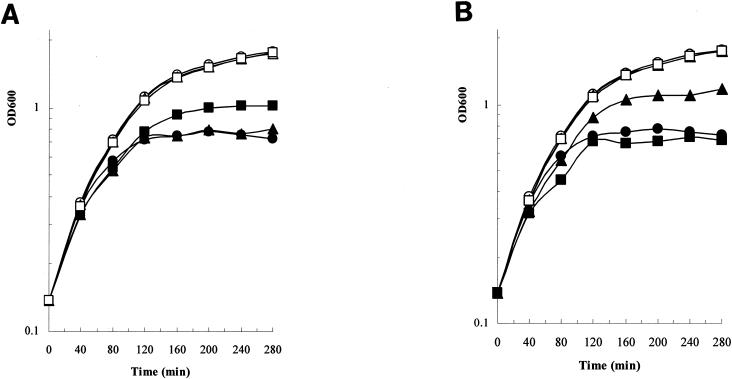
In a liquid growth inhibition experiment, we added ColE9 L359C-F412C/Im9 or ColE9 Y324C-L447C/Im9 mutant proteins that had either been oxidized or reduced to growing E. coli DH5α cells (Fig. 3A). The chosen 30 nM concentration of the mutant proteins was extrapolated from the concentration values required to kill E. coli DH5α cells shown in Fig. 2. The results confirmed the plate assay data that oxidized ColE9 L359C-F412C/Im9 or ColE9 Y324C-L447C/Im9 mutant proteins, unlike the DTT-reduced mutant proteins, had no inhibitory activity. There was a reproducible difference in the relative killing activity in this liquid growth assay, with the ColE9 Y324C-L447C/Im9 mutant protein showing less killing activity than the ColE9 L359C-F412C/Im9 mutant protein. In contrast, the ColE9 Y324C-V405C/Im9 mutant protein, whether oxidized or reduced, exhibited similar growth inhibition to 3 nM ColE9/Im9 (data not shown). The simultaneous addition of DTT and oxidized ColE9 L359C-F412C/Im9 or ColE9 Y324C-L447C/Im9 mutant proteins to E. coli DH5α cells resulted in the timing of growth inhibition (Fig. 3B) that was very similar to that seen in the results shown in Fig. 3A. This indicates that DTT reduction of the oxidized mutant proteins in the growth medium and/or bound to E. coli cells must occur quickly. The ColE9 L359C-F412C/Im9 mutant protein was less active in the presence of DTT than the ColE9 Y324C-L447C/Im9 mutant protein.
FIG. 3.
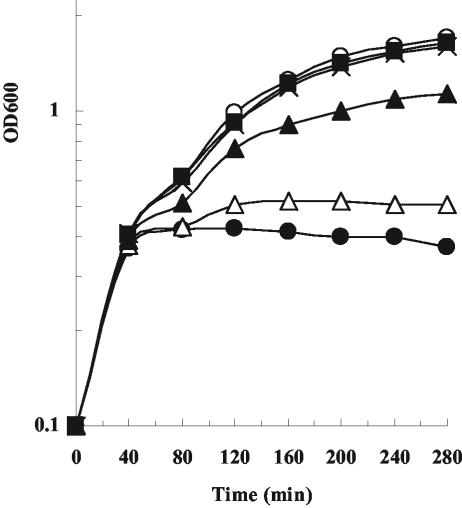
Growth inhibitory activity in liquid medium. (A) The effect on E. coli DH5α cells grown in LB medium, with no additions (○), with 3 nM ColE9 (•), with 30 nM oxidized ColE9 Y324C-L447C mutant protein (□), with 30 nM reduced ColE9 Y324C-L447C mutant protein (▪), with 30 nM oxidized ColE9 L359C-F412C mutant protein (▵), or with 30 nM reduced ColE9 L359C-F412C mutant protein (▴). (B) The effect on E. coli DH5α cells grown in LB medium, with no additions (○), with 1 mM DTT (⋄), with 3 nM ColE9 (•), with 30 nM oxidized ColE9 Y324C-L447C mutant protein plus 1 mM DTT (▪), or with 30 nM oxidized ColE9 L359C-F412C mutant protein plus 1 mM DTT (▴). OD600, optical density at 600 nm.
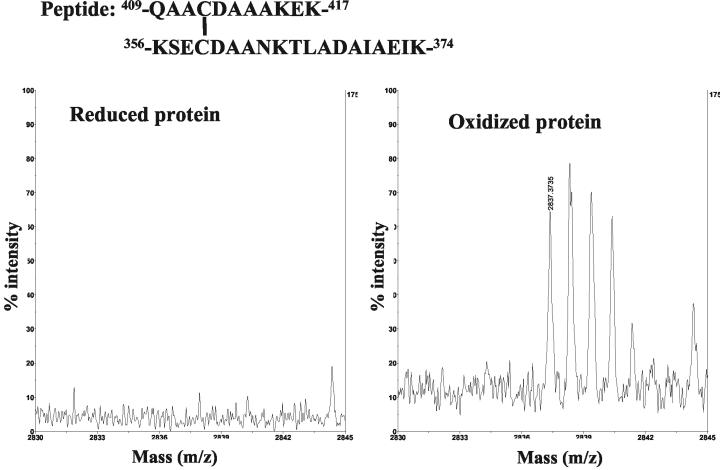
The properties of the ColE9 L359C-F412C/Im9 and ColE9 Y324C-L447C/Im9 mutant proteins were consistent with a disulfide being formed between the two introduced cysteine residues in the R domain. We directly confirmed the presence of a disulfide bond in tryptic digests of the oxidized but not the reduced ColE9 L359C-F412C/Im9 (Fig. 4) and ColE9 Y324C-L447C/Im9 (data not shown) mutant proteins by MALDI-TOF mass spectrometry. The expected mass of the disulfide-linked peptide whose sequence is shown in Fig. 4 is 2836.4, and the observed mass of the central peak arising from the oxidized protein is 2837.4. The series of peaks in the oxidized spectra, differing in size by 1 Da, arise due to variation in the incorporation of natural isotopes. With the reduced protein the expected 1,991-Da peak was observed for the peptide at residues 356 to 374 shown in Fig. 4, and as expected this peak was significantly reduced in the oxidized protein. The 848-Da peak corresponding to the peptide at residues 409 to 417 was not resolved in either spectra.
FIG. 4.
Presence of a disulfide confirmed by MALDI-TOF. The spectra of the oxidized (right) and reduced (left) ColE9 L359C-F412C mutant protein is shown. The sequence and residue numbers of a disulfide-containing peptide observed in the oxidized ColE9 L359C-F412C protein spectra are indicated at the top of the figure.
Receptor-binding, DNase, and TolB binding activity of the mutant proteins.
The simplest explanation for our findings is that the presence of a disulfide in the R domain prevents a conformational change that is essential for biological activity. There are, however, several alternative explanations for the loss of colicin activity of the oxidized ColE9 L359C-F412C/Im9 and ColE9 Y324C-L447C/Im9 mutant proteins. The presence of the disulfide lock in the R domain could affect the binding of the mutant protein to the BtuB receptor in the outer membrane of E. coli cells, the DNase activity of the mutant protein, or the interaction of the T domain of the mutant protein, especially the TolB box, with TolB, or result in a significant structural change of the colicin molecule. We investigated these possibilities in turn by using a range of experimental techniques.
The relative concentration of the oxidized ColE9 Y324C-L447C/Im9 mutant protein required to protect E. coli DH5α cells from killing by wild-type ColE9 (Fig. 5) was similar to that previously reported for the active site ColE9 H575A/Im9 mutant protein (14). The oxidized L359C-F412C/Im9 protein also protected E. coli DH5α cells from killing by wild-type ColE9/Im9 (data not shown). The oxidized ColE9 L359C-F412C/Im9 or ColE9 Y324C-L447C/Im9 mutant protein inhibited the growth of E. coli 113/3, a metE mutant that is unable to make methionine unless supplied with vitamin B12, in a similar way to that previously reported for the ColE9 H575A/Im9 mutant protein (data not shown).
FIG. 5.
Assay of BtuB receptor binding. In vivo competition assay using E. coli DH5α cells incubated with no additions (○), with 3 nM ColE9 (•), with 300 nM ColE9 L359C-F412C mutant protein (▪), and with 3 nM ColE9 and the ColE9 L359C-F412C mutant protein at a ratio of 1:1 (▵), 1:10 (▴), or 1:100 (X). OD600, optical density at 600 nm.
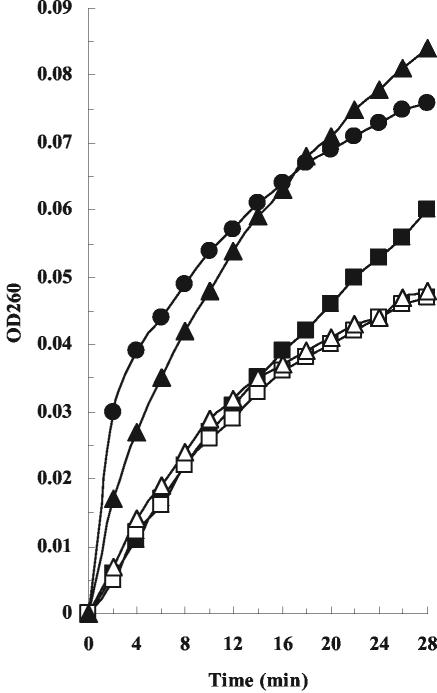
We investigated whether the presence of the disulfide bond affects the DNase activity of the oxidized ColE9 L359C-F412C or ColE9 Y324C-L447C mutant protein by using a Kunitz DNase assay after removal of the bound Im9 protein from the complexes (Fig. 6). As has been reported previously, the increase in absorbance at 260 nm is nonlinear with ColE9 in the presence of Mg2+, although this is the preferred metal ion for double-stranded DNA cleavage (32, 33). Surprisingly, the DNase activity both of the oxidized or reduced ColE9 Y324C-L447C mutant protein and of the oxidized ColE9 L359C-F412C mutant protein was diminished compared with the ColE9 control protein. The activity of the reduced ColE9 L359C-F412C mutant protein was very similar to that of the ColE9 control protein. In the same assay, we also confirmed that the DNase activity of the oxidized ColE9 L359C-F412C or ColE9 Y324C-L447C mutant protein, like that of ColE9, is inhibited by binding Im9 (data not shown).
FIG. 6.
DNase activity assays. The DNase activities of the oxidized (▵) and reduced (▴) ColE9 L359C-F412C and the oxidized (□) and reduced (▪) ColE9 Y324C-L447C mutant proteins were compared to that of ColE9 (•) by using the Kunitz assay. The Im9 protein was removed from the ColE9 proteins to allow the DNase assay to be performed. OD260, optical density at 260 nm.
An interaction between the T domain of ColE3 and TolB was demonstrated by cross-linking experiments (5). An interaction between the T domain of ColE9 and TolB was demonstrated by using the yeast two-hybrid system (7). This interaction was shown by alanine scanning mutagenesis to be dependent upon any one of the three italicized residues in the pentapeptide TolB box sequence DGSGW (14). SPR has been used previously to show an interaction between TolA and the T domains of ColA (12) or ColN (16). We compared the interaction between TolB and the ColE9 L359C-F412C/Im9 and ColE9 Y324C-L447C/Im9 mutant protein complexes with that of ColE9/Im9 by using SPR. The presence of disulfide locks in the two mutant proteins had no inhibitory effect on the interaction between their TolB boxes and TolB (data not shown).
CD provides secondary structure information, allowing the detection of secondary structure perturbations in a protein caused by site-directed mutagenesis. The CD spectra of the oxidized ColE9 L359C-F412C/Im9 and ColE9 Y324C-L447C/Im9 mutant proteins superimpose with the spectra of the native ColE9/Im9 protein (data not shown); therefore, neither the introduction of the pairs of cysteine residues nor the formation of the disulfide bond had any detectable effect on the global secondary structure of ColE9.
DISCUSSION
Recognition of the cell surface receptor is the first stage in colicin killing of E. coli cells. The fact that enzymatic colicins bind to the BtuB receptor has been known for decades, and the role of the Tol proteins that are required for translocation of the colicin through the periplasm has received much attention over the last 20 years (3, 24). But the sequence of events that occur after receptor binding and the overall dynamics of the system remain to be elucidated. In this paper we provide evidence that the flexibility of the R domain is essential for the activity of enzymatic colicins and that the structural rigidity of the coiled-coil prevents subsequent events from occurring during cell entry.
The introduction of two cysteine mutations in the R domain of ColE9 results in the formation of a disulfide bond in the diamide-oxidized ColE9 L359C/F412C/Im9 and ColE9 Y324C-L447C/Im9 mutant proteins that abolishes colicin activity. Reduction of the mutant protein by dialysis into DTT-containing buffer restored colicin activity; this result was also observed in a liquid growth inhibition experiment by the addition of DTT to E. coli DH5α cells incubated with the oxidized ColE9 Y324C-L447C/Im9 or ColE9 L359C-F412C/Im9 mutant proteins (Fig. 3B). The differences in growth inhibition of the two mutant proteins seen in each assay could be due to a difference in accessibility of the disulfides to DTT reduction and/or spontaneous oxidation of the disulfide. The presence of the expected disulfide bond in tryptic digests of the diamide-oxidized ColE9 L359C-F412C or ColE9 Y324C-L447C mutant protein, but not in the DTT-reduced proteins, was confirmed by MALDI-TOF mass spectrometry. In contrast to these results, the control ColE9 Y352C-V405C/Im9 mutant protein exhibited almost identical colicin activity after diamide oxidation or DTT reduction. The observation of a disulfide bond in the ColE9 L359C-F412C and ColE9 Y324C-L447C mutant proteins is consistent with the predicted distance between the pairs of cysteine residues when modeled on the three-dimensional structure of ColE3 (35) (Fig. 1).
The loss of biological activity of the oxidized ColE9 L359C-F412C/Im9 or ColE9 Y324C-L447C/Im9 mutant protein complex was not due to a significant effect of the disulfide bond present in the protein on BtuB binding, as the diamide-treated proteins both protected E. coli cells from killing by ColE9/Im9 in similar ratios to those seen previously for the inactive ColE9 H575A/Im9 mutant colicin in a protection experiment (Fig. 5) and also competed for BtuB binding with vitamin B12. The presence of a disulfide bond in the ColE9 L359C-F412C/Im9 or ColE9 Y324C-L447C/Im9 mutant protein had no inhibitory effect on the TolB binding activity of the mutant proteins or on the CD spectrum of the oxidized proteins, but it did have some effect on their DNase activity (Fig. 6). The DNase assay experiments require denaturation of the ColE9 mutant protein in order to remove the bound Im9 protein from the complex, followed by refolding. It is possible that the presence of a disulfide might interfere differently with the refolding process in the two mutant proteins.
Our data clearly show that locking the conformation of the R domain by the presence of a disulfide, located either near the middle or at the top of the coiled-coil, abolishes the biological activity of the mutant colicin. This provides support for a model of ColE9-induced cell killing in which the flexibility of the coiled-coil R domain is essential for the events that occur after BtuB receptor binding. Several authors have previously suggested that the unfolding of the R domain is essential for colicin activity (17, 35), but there is little direct experimental evidence to support the hypothesis. An analysis of ColE1 by differential scanning calorimetry and CD suggested that the R domain has a dominant role in determining the conformation of the other two domains (17). Although the crystal structure of ColE1 does not include the R and T domains, it was proposed that the unfolding of the R domain coiled-coil after binding to BtuB results in unfolding of the T domain, which then interacts with TolC and initiates the translocation process.
Recently the three-dimensional structure of BtuB bound to R135 of ColE3 was determined (22). The perhaps surprising observation from this structure is that the coiled-coil R domain is not buried in a binding pocket in BtuB. The three-dimensional structure shows that 27 residues of R135 (from I369 to T402), located around and including the hairpin, interact with 29 residues of BtuB. Most of the BtuB residues that interact with R135 are located in the exposed loops, but four residues of the N-terminal plug domain of BtuB interact with M383 located in the hairpin of R135. The buried surface area of R135 bound to BtuB is 1,533 Å2, which constitutes 24% of the coiled-coil domain of ColE3. The location of both of the disulfide locks in the coiled-coil domain is outside the region that interacts with BtuB, which explains why the receptor binding of our constructs was not affected.
By using far-UV CD to measure the helical content of R135, it was demonstrated that the helical content decreased by 12% ± 5% on BtuB binding (22). By comparing the Cα displacement of the residues in the bound R135 domain with that of ColE3, disorder of the coiled-coil was observed in the crystal structure of the R domain BtuB complex (22). The Cα displacement increased with the distance from the helical hairpin, with values of 3.5 Å and 6 Å for Y323 and K438, respectively. It was suggested that the function of these observed changes in the conformation of the coiled-coil is to drive the unfolding of the R domain which, if transmitted to the T domain, could lead to the dissociation of the T domain from its interface with Im3, and also the unfolding of the T domain to allow its penetration into the periplasmic space and interaction with Tol proteins (22). There remains uncertainty, however, about the magnitude of the conformational change since there is no evidence that the three-dimensional structure of the truncated R135 polypeptide in solution is identical to that of the complete R domain observed in the X-ray structure of ColE3 (35). A 34-residue peptide, corresponding to residues 366 to 399 of ColE3 containing two cysteines that were introduced to enable formation of a disulfide cross-link for minimization of conformational entropy, was shown to bind to BtuB with nanomolar affinity (26). This BtuB binding peptide, however, also possessed less secondary structure than the same sequence within the complete ColE3 protein. Solution NMR studies of the 76-residue minimum R domain have shown that its helical hairpin structure has multiple, slowly interchanging conformers and a flexible hairpin loop (2). A plausible interpretation of all these data is that, in solution, the coiled-coil R domain and truncated versions thereof can adopt a variety of structures differing in the spatial relationship of the two helices.
Our data support the hypothesis that the unfolding and flexibility of the R domain are necessary for subsequent translocation events. There are a number of unanswered questions resulting from an unfolding model, such as the magnitude of the conformational change in the R domain described above and whether this results in similar changes in the T domain and/or triggers the loss of immunity protein from the colicin/immunity protein complex. We are currently developing experimental tools to explore the early events of the translocation process.
Acknowledgments
We thank all members of our laboratories for their hard work and enthusiastic support of the colicin research project and, along with Phil Bardelang, for constructive comments on the research.
This work was supported by a Programme grant from the Wellcome Trust and by the University of Nottingham.
REFERENCES
- 1.Bénédetti, H., M. Frenette, D. Baty, R. Lloubès, V. Géli, and C. Lazdunski. 1989. Comparison of the uptake systems for the entry of various BtuB group colicins into Escherichia coli. J. Gen. Microbiol. 135:3413-3420. [DOI] [PubMed] [Google Scholar]
- 2.Boetzel, R., E. S. Collins, N. J. Clayden, C. Kleanthous, R. James, and G. R. Moore. 2002. Structural dynamics of the receptor-binding domain of colicin E9. Faraday Discuss. 122:145-162. [DOI] [PubMed] [Google Scholar]
- 3.Bouveret, E., L. Journet, A. Walburger, E. Cascales, H. Bénédetti, and R. Lloubès. 2002. Analysis of the Escherichia coli Tol-Pal and TonB systems by periplasmic production of Tol, TonB, colicin, or phage capsid soluble domains. Biochimie 84:413-421. [DOI] [PubMed] [Google Scholar]
- 4.Bouveret, E., A. Rigal, C. Lazdunski, and H. Bénédetti. 1998. Distinct regions of the colicin A translocation domain are involved in the interaction with TolA and TolB proteins upon import into Escherichia coli. Mol. Microbiol. 27:143-157. [DOI] [PubMed] [Google Scholar]
- 5.Bouveret, E., A. Rigal, C. Lazdunski, and H. Bénédetti. 1997. The N-terminal domain of colicin E3 interacts with TolB which is involved in the colicin translocation step. Mol. Microbiol. 23:909-920. [DOI] [PubMed] [Google Scholar]
- 6.Cao, Z., and P. E. Klebba. 2002. Mechanisms of colicin binding and transport through outer membrane porins. Biochimie 84:399-412. [DOI] [PubMed] [Google Scholar]
- 7.Carr, S., C. N. Penfold, V. Bamford, R. James, and A. M. Hemmings. 2000. The structure of TolB, an essential component of the tol-dependent translocation system, and its protein-protein interaction with the translocation domain of colicin E9. Structure Fold. Des. 8:57-66. [DOI] [PubMed] [Google Scholar]
- 8.Collins, E. S., S. B. Whittaker, K. Tozawa, C. MacDonald, R. Boetzel, C. N. Penfold, A. Reilly, N. J. Clayden, M. J. Osborne, A. M. Hemmings, C. Kleanthous, R. James, and G. R. Moore. 2002. Structural dynamics of the membrane translocation domain of colicin E9 and its interaction with TolB. J. Mol. Biol. 318:787-804. [DOI] [PubMed] [Google Scholar]
- 9.Cooper, P. C., and R. James. 1984. Two new E colicins, E8 and E9, produced by a strain of Escherichia coli. J. Gen. Microbiol. 130:209-215. [DOI] [PubMed] [Google Scholar]
- 10.Cramer, W. A., F. S. Cohen, A. R. Merrill, and H. Y. Song. 1990. Structure and dynamics of the colicin E1 channel. Mol. Microbiol. 4:519-526. [DOI] [PubMed] [Google Scholar]
- 11.Davis, B. D., and E. S. Mingioli. 1950. Mutants of Escherichia coli requiring methionine or vitamin B12. J. Bacteriol. 60:17-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derouiche, R., G. Zeder-Lutz, H. Bénédetti, M. Gavioli, A. Rigal, C. Lazdunski, and R. Lloubès. 1997. Binding of colicins A and E1 to purified TolA domains. Microbiology 143:3185-3192. [DOI] [PubMed] [Google Scholar]
- 13.Duché, D., M. W. Parker, J. M. Gonzalez-Manas, F. Pattus, and D. Baty. 1994. Uncoupled steps of the colicin A pore formation demonstrated by disulfide bond engineering. J. Biol. Chem. 269:6332-6339. [PubMed] [Google Scholar]
- 14.Garinot-Schneider, C., C. N. Penfold, G. R. Moore, C. Kleanthous, and R. James. 1997. Identification of residues in the putative TolA box which are essential for the toxicity of the endonuclease toxin colicin E9. Microbiology 143:2931-2938. [DOI] [PubMed] [Google Scholar]
- 15.Garinot-Schneider, C., A. J. Pommer, G. R. Moore, C. Kleanthous, and R. James. 1996. Identification of putative active-site residues in the DNase domain of colicin E9 by random mutagenesis. J. Mol. Biol. 260:731-742. [DOI] [PubMed] [Google Scholar]
- 16.Gokce, I., E. M. Raggett, Q. Hong, R. Virden, A. Cooper, and J. H. Lakey. 2000. The TolA-recognition site of colicin N. ITC, SPR and stopped-flow fluorescence define a crucial 27-residue segment. J. Mol. Biol. 304:621-632. [DOI] [PubMed] [Google Scholar]
- 17.Griko, Y. V., S. D. Zakharov, and W. A. Cramer. 2000. Structural stability and domain organization of colicin E1. J. Mol. Biol. 302:941-953. [DOI] [PubMed] [Google Scholar]
- 18.Hilsenbeck, J. L., H. Park, G. Chen, B. Youn, K. Postle, and C. Kang. 2004. Crystal structure of the cytotoxic bacterial protein colicin B at 2.5 A resolution. Mol. Microbiol. 51:711-720. [DOI] [PubMed] [Google Scholar]
- 19.James, R., C. Kleanthous, and G. R. Moore. 1996. The biology of E colicins: paradigms and paradoxes. Microbiology 142:1569-1580. [DOI] [PubMed] [Google Scholar]
- 20.Jonsson, U., L. Fagerstam, B. Ivarsson, B. Johnsson, R. Karlsson, K. Lundh, S. Lofas, B. Persson, H. Roos, I. Ronnberg, and et al. 1991. Real-time biospecific interaction analysis using surface plasmon resonance and a sensor chip technology. BioTechniques 11:620-627. [PubMed] [Google Scholar]
- 21.Kleanthous, C., U. C. Kûhlmann, A. J. Pommer, N. Ferguson, S. E. Radford, G. R. Moore, R. James, and A. M. Hemmings. 1999. Structural and mechanistic basis of immunity towards endonuclease colicins. Nat. Struct. Biol. 6:243-252. [DOI] [PubMed] [Google Scholar]
- 22.Kurisu, G., S. D. Zakharov, M. V. Zhalnina, S. Bano, V. Y. Eroukova, T. I. Rokitskaya, Y. N. Antonenko, M. C. Wiener, and W. A. Cramer. 2003. The structure of BtuB with bound colicin E3 R-domain implies a translocon. Nat. Struct. Biol. 10:948-954. [DOI] [PubMed] [Google Scholar]
- 23.Lazdunski, C. J., E. Bouveret, A. Rigal, L. Journet, R. Lloubès, and H. Bénédetti. 1998. Colicin import into Escherichia coli cells. J. Bacteriol. 180:4993-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lazzaroni, J. C., J. F. Dubuisson, and A. Vianney. 2002. The Tol proteins of Escherichia coli and their involvement in the translocation of group A colicins. Biochimie 84:391-397. [DOI] [PubMed] [Google Scholar]
- 25.Masaki, H., and T. Ohta. 1985. Colicin E3 and its immunity genes. J. Mol. Biol. 182:217-227. [DOI] [PubMed] [Google Scholar]
- 26.Mohanty, A. K., C. M. Bishop, T. C. Bishop, W. C. Wimley, and M. C. Wiener. 2003. Enzymatic E-colicins bind to their target receptor BtuB by presentation of a small binding epitope on a coiled-coil scaffold. J. Biol. Chem. 278:40953-40958. [DOI] [PubMed] [Google Scholar]
- 27.Mosbahi, K., C. Lemaitre, A. H. Keeble, H. Mobasheria, B. Morel, R. James, G. R. Moore, E. J. A. Lea, and C. Kleanthous. 2002. The cytotoxic domain of colicin E9 is a channel-forming endonuclease. Nat. Struct. Biol. 9:476-484. [DOI] [PubMed] [Google Scholar]
- 28.Ogawa, T., K. Tomita, T. Ueda, K. Watanabe, T. Uozumi, and H. Masaki. 1999. A cytotoxic ribonuclease targeting specific transfer RNA anticodons. Science 283:2097-2100. [DOI] [PubMed] [Google Scholar]
- 29.Ohno-Iwashita, Y., and K. Imahori. 1980. Assignment of the functional loci in colicin E2 and E3 molecules by the characterization of their proteolytic fragments. Biochemistry 19:652-659. [DOI] [PubMed] [Google Scholar]
- 30.Penfold, C. N., C. Garinot-Schneider, A. M. Hemmings, G. R. Moore, C. Kleanthous, and R. James. 2000. A 76-residue polypeptide of colicin E9 confers receptor specificity and inhibits the growth of vitamin B12-dependent Escherichia coli 113/3 cells. Mol. Microbiol. 38:639-649. [DOI] [PubMed] [Google Scholar]
- 31.Pilsl, H., and V. Braun. 1995. Novel colicin 10: assignment of four domains to TonB- and TolC-dependent uptake via the Tsx receptor and to pore formation. Mol. Microbiol. 16:57-67. [DOI] [PubMed] [Google Scholar]
- 32.Pommer, A. J., S. Cal, A. H. Keeble, D. Walker, S. J. Evans, U. C. Kühlmann, A. Cooper, B. A. Connolly, A. M. Hemmings, G. R. Moore, R. James, and C. Kleanthous. 2001. Mechanism and cleavage specificity of the H-N-H endonuclease colicin E9. J. Mol. Biol. 314:735-749. [DOI] [PubMed] [Google Scholar]
- 33.Pommer, A. J., R. Wallis, G. R. Moore, R. James, and C. Kleanthous. 1998. Enzymological characterization of the nuclease domain from the bacterial toxin colicin E9 from Escherichia coli. Biochem. J. 334:387-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarkar, G., and S. S. Sommer. 1990. The megaprimer method of site-directed mutagenesis. BioTechniques 8:404-407. [PubMed] [Google Scholar]
- 35.Soelaiman, S., K. Jakes, N. Wu, C. Li, and M. Shoham. 2001. Crystal structure of colicin E3: implications for cell entry and ribosome inactivation. Mol. Cell 8:1053-1062. [DOI] [PubMed] [Google Scholar]
- 36.Wallis, R., G. R. Moore, R. James, and C. Kleanthous. 1995. Protein-protein interactions in colicin E9 DNase-immunity protein complexes. 1. Diffusion-controlled association and femtomolar binding for the cognate complex. Biochemistry 34:13743-13750. [DOI] [PubMed] [Google Scholar]
- 37.Wallis, R., A. Reilly, K. Barnes, C. Abell, D. G. Campbell, G. R. Moore, R. James, and C. Kleanthous. 1994. Tandem overproduction and characterisation of the nuclease domain of colicin E9 and its cognate inhibitor protein Im9. Eur. J. Biochem. 220:447-454. [DOI] [PubMed] [Google Scholar]
- 38.Watson, R., W. Rowsome, J. Tsao, and L. P. Visentin. 1981. Identification and characterization of Col plasmids from classical colicin E-producing strains. J. Bacteriol. 147:569-577. [DOI] [PMC free article] [PubMed] [Google Scholar]