Abstract
The ATM protein kinase, is a serine/threonine protein kinase that is recruited and activated by DNA double-strand breaks, mediates responses to ionizing radiation in mammalian cells. Here we show that ATM is held inactive in unirradiated cells as a dimer and phosphorylates the opposite strand of the dimer in response to DNA damage. Cellular irradiation induces rapid intermolecular autophosphorylation of serine 1981 that causes dimer dissociation and initiates cellular ATM kinase activity. ATM cannot phosphorylate the substrates when it could not undergo dimer monomer transition. After DNA repair, the active monomer will undergo dephosphorylation to form dimer again and dephosphorylation is critical for dimer reformation. Our work reveals novel function of ATM dimer monomer transition and explains why ATM dimer monomer transition plays such important role for ATM cellular activity during DNA repair.
Keywords: ATM activation, Intermolecular phosphorylation, Dephosphorylation, DNA repair
1. Introduction
Ataxia telangiectasia (A-T) is an inherited disease characterized by immune deficiencies, neurodegeneration, susceptibility to cancer, and sensitivity to ionizing radiation [1,2]. The A-T gene product, the ATM protein, is activated in response to DNA doublestrand breaks (DSB) [3–6]and ATM become phosphorylated on Ser 1981 [7]. ATM autophosphorylation initiates the conversion of the inactive ATM dimer to an active monomeric ATM [7]. ATM then phosphorylates multiple DNA damage response proteins, including Nbs1, P53, Chk2, and SMC1 [3,5,8–13]. The phosphorylation of these proteins by ATM is essential for correct activation of cell cycle check points and for the initiation of DNA repair [5]. Consequently, cells lacking functional ATM protein exhibit defects in DNA repair and loss of cell cycle checkpoints [8,14], which results in increased sensitivity to ionizing radiation [15–18].
Although the downstream signaling pathway activated by ATM is well characterized, the mechanism of ATM activation in response to DSB remains to be elucidated. Previous work showed that the phosphorylation of ATM does not directly regulate the activity of the kinase, but instead disrupts ATM dimer, and the dimer monomer transition plays important role during ATM activation [7]. However, a key question has not been answered in almost a decade since this dimer monomer model was identified: why ATM activation undergoes dimer monomer transition and why dimer dissociation or monomer formation is so important? We identified here that ATM phosphorylated the opposite strand of ATM during intermolecular autophosphorylation and only monomer of ATM can phosphorylate the substrates of ATM including P53 and Chk2 [19–21]. ATM monomer could form dimer again after dephosphorylation.
2. Materials and methods
2.1. Cells and antibodies
GM5849 A-T cells (Coriell Institute, NJ) were cultured according to the suppliers’ recommendations. Cells were transfected using Lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen, CA). Clonogenic cell survival assays were done as previously described [16–18]. Antibodies used were ATM antibodies 5C2 and 2C1 (Genetex, San Antonio, TX), phospho-Ser 1981 (Rockland, Gilbertsville, PA), P53 (Calbiochem), anti-phospho-Ser 15 P53 (EMD Biosciences), H2AX (Oncogene Science), anti-cH2AX (Cell Signaling), anti-phospho-Thr 68 Chk2 (Cell Signaling Technology), anti-Tip60 (Santa Cruz), anti-HA (Abcam), anti-Myc (Cell Signaling).
2.2. Mutagenesis
Point mutations were inserted by site-directed mutagenesis to create restriction sites for SpeI (nucleotide 9279: A9281TG9282A) and EcoR1 (nucleotide 9373: A8378C) in the ATM cDNA. The C terminus of ATM was removed by SpeI-EcoR1 digestion, and oligonucleotides with overhanging SpeI-EcoR1 sites encoding the indicated mutations were inserted.
2.3. Immunoprecipitation and Western blot analysis
Cells (1 × 107) were lysed in ATM lysis buffer (20 mM Hepes at pH 7.4, 150 mM NaCl, 0.2% Tween 20, 1.5 mM MgCl2, 1 mM EGTA, 2 mM DTT, 50 mM NaF, 500 lM NaVO4, 1 mM PMSF, 1 μg/ml aprotinin and 1 μg/ml leupeptin) and cleared by centrifugation. Antibodies against ATM (PC116; EMD Biosciences) or Tip60 (HA or Tip60; Abcam and Upstate Biotechnology) were used for immunoprecipitation, and immune complexes collected on protein-A agarose beads. Immunoprecipitates were washed three times in ATM lysis buffer, and once each in high salt buffer (100 mM Tris at pH 7.4, 600 mM NaCl, 1 mM DTT and 1 mM PMSF), and base buffer (10 mM Hepes at pH 7.4, 10 mM MgCl 2, 50 mM NaCl, 1 mM DTT and 1 mM PMSF).
2.4. Kinase assays
Extracts were immunoprecipitated as above. Immunoprecipitates were washed once in kinase buffer (10 mM Hepes, pH 7.4/ 10 mM MgCl2/50 mM NaCl/10 mM MnCl2) and incubated in 50 μl of kinase buffer containing 50 μM ATP, P53 peptide (2 μg of EPPLS-EPPLSQEAFADLWKK), and 10 μCi of [γ-32P] ATP (1 Ci = 37 GBq) for 30 min at 30 °C. Reactions were terminated with 30% acetic acid (20 μl), spotted onto P81 paper, washed in 15% acetic acid, airdried, and counted.
2.5. HAT assays
Extracts were immunoprecipitated as above, except that the high salt wash was omitted. Immunoprecipitates were washed twice in HAT assay buffer (50 mM Tris, pH 8/10% glycerol/ 0.1 mM EDTA/1 mM DTT), and incubated in 60 ll of HAT assay buffer containing acetyl-CoA (100 μM) and biotinylated histone H4 peptide (0.5 μg) for 30 min at 30 °C. An aliquot of the reaction was immobilized onto streptavidin plates and acetylation detected by using a HAT ELISA according to the manufacturer’s instructions (Upstate Biotechnology). HAT activity is expressed as the change in absorbance relative to the reference wavelength (450–540 nm).
2.6. Immunofluorescence
Cells (on cover slides) were fixed in phosphate-buffered saline (PBS) containing 2% paraformaldehyde. Cells were permeabilized in 0.2% Triton X-100 in PBS for 5 min, and then blocked in fetal bovine serum for 20 min. Primary antibodies were prepared in 10% fetal bovine serum supplemented with 0.2% saponin. After a 1 h incubation with primary antibody, cells were washed three times with 0.2% Tween-20 and incubated for 1 h in secondary antibody (conjugated to either Texas Red or FITC; Santa Cruz Biotechnology, CA). Slides were mounted with Fluoromount-G (Southern Biotech, AL). Images were collected with an AxioImager Z1 microscope (Carl Zeiss, Inc.) equipped with a color digital camera (Axiocam MRc Rev.3; Carl Zeiss, Inc.) and Plan Apochromat oil M27 lens (636, NA 1.4) Acquisition software.
3. Results
3.1. ATM phosphorylates the opposite strand in the intermolecular dimer in response to DSB
Although it has been demonstrated that ATM undergo dimer monomer transition during DNA repair and it is critical for ATM to be functional, it is very difficult to address if ATM phosphorylate the same strand or the opposite strand. Every single molecular of ATM has both kinase domain and phosphorylation site, which make it impossible to rule out one possibility from another. We use both Myc tagged kinase dead ATM and HA tagged Ser 1981 mutation in our work. Previous work showed that the 350amino-acid kinase domain located at the carboxy terminus of ATM and the kinase dead mutation the ATMKD does not possess kinase activity and does not autophosphorylate [17,22]. Following DNA exposure to DNA damage, ATMKD does not undergo dimer monomer transition and remain dimeric form [17]. Previous work also showed that ATM1981 can not undergo dimer monomer transition. Although mutation of serine 1981 did not abolish ATM kinase activity in vitro [7], expression of S1981A-ATM fails to complement AT cells, and effectively inhibit the cellular activity of endogenous ATM.
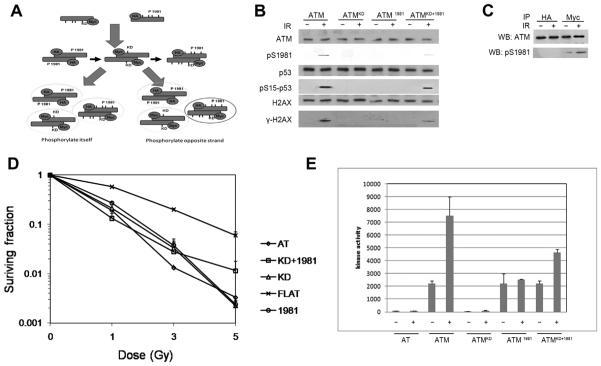
If ATM phosphorylates the same strand within dimer in response of DSB, we should not detect any phosphorylation of ATM after cotransfection of myc tagged ATMKD and HA tagged ATM1981 into AT cells (Fig. 1A). Interestingly, we detected phosphorylation of ATM S1981 in ATMKD and ATM1981 cotransfected cells, whereas neither ATMKD or ATM1981 can be autophosphorylated (Fig. 1B). We further showed that after immunoprecipitation with HA and myc antibody, only myc tagged ATM, ATMKD, can be phosphorylated (Fig. 1C). These data strongly suggest that ATM molecular phosphorylate the opposite strand but not the same strand (Fig. 1A). Further work showed that ATMKD+1981 can not only undergo autophosphorylation but can phosphorylate P53 and H2AX (Fig. 1B). Clonogenic assay showed that they can partially complement AT cells (Fig. 1D). The phosphorylated ATMKD+1981 showed partial stimulated kinase activity in response to DNA damage (Fig. 1E).
Fig. 1.
ATM dimer phosphorylates the opposite strand instead of the same strand within the dimer. (A) Schematic model of experiment design. The left arrow shows the possibility of self-autophosphorylation of the dimer; the right arrow shows the possibility of interactive autophosphorylation. GM5849 ATM negative cells were stably cotransfected with HA and Myc-tagged ATM constructs. Cells were treated with IR. Cells expressing ATMKD and ATM1981 was immunoprecipitated either HA or Myc antibody. Only Myc tagged ATMKD can be phosphorylated and since ATMKD lack kinase activity, thus it can only be phosphorylated by the counterpart strand of the dimer which can only be ATM1981. (B) GM5849 ATM negative cells were stably cotransfected with HA and Myc-tagged ATM constructs. Cells were treated with IR (+). Cells expressing ATMKD and ATM1981 was immunoprecipitated and analyzed by Western blot analysis for ATM, phospho-ATM (pS1981), P53 and phospho-P53 (pS15-P53), and H2AX and phospho- H2AX (cH2AX). (C) GM5849 ATM negative cells were stably cotransfected with HA and Myc-tagged ATM constructs. Cells were treated with IR (+). Cells expressing ATMKD and ATM1981 was immunoprecipitated either HA or Myc antibody. ATM and ps1981 ATM level were measured. Only Myc tagged ATMKD can be phosphorylated. (D) Cells were irradiated at the indicated dose, and the number of surviving colonies measured 12 days later. Data represent the mean ± s.d. from three independent assays. (E) GM5849 ATM negative cells were stably cotransfected with HA and Myc-tagged ATM constructs. Cells were treated with IR (+) as indicated time course. ATM was immunoprecipitated. The intrinsic kinase activity of ATM was then measured using P53 peptide (containing serine 15) as the substrate.
Comparing to the autophosphorylation in full length ATM transfected cells, the autophosphorylation is very weak in the ATMKD and ATM1981 cotransfected cells (Fig. 1B). We also detected much weak P53 and H2AX phosphorylation in the cotransfected cells (Fig. 1B), as well as kinase activity (Fig. 1E). These results are consistent with the model that ATM phosphorylates the opposite strand (Fig. 1A). After cotransfection into AT cells, myc-ATMKD and HA-ATM1981 will form three types of dimers: myc-ATMKD plus myc-ATM1981, myc-ATMKD plus HA-ATM1981, HA-ATM1981 plus HA-ATM1981 (Fig. 1A). If ATM molecular phosphorylates the same strand, none of these dimers will be phosphorylated. However, if we can detected any phosphorylation, that could only be myc-ATMKD plus HA-ATM1981 dimer, and it can only be the opposite strand phosphorylation model.
3.2. Dimer-monomer transition plays a key role for ATM in vivo activation and cellular activities
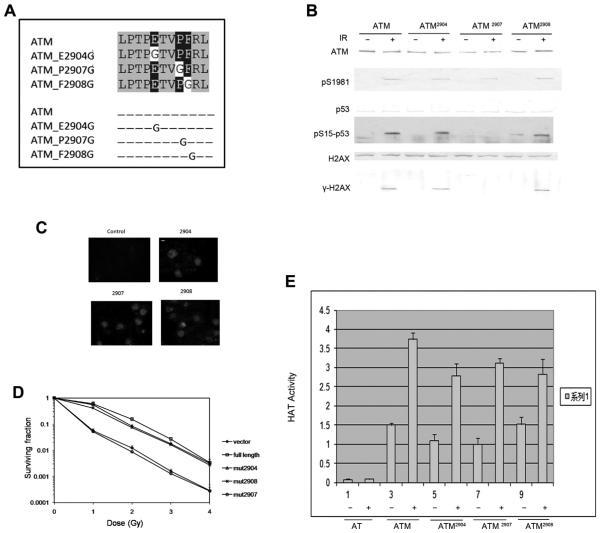
The PETPVFRLT box located at the C terminus of ATM and it is conserved between ATM, ATR and DNA-PK. ATM, ATR and DNA-PK are all large proteins that belong to the phosphoinositde 3-kinase (PI(3)K) superfamily. The function of the PETPVFRLT box remains unclear. We generated 3 point mutations in the PETPERLT box, which including ATM2904, ATM2907 and ATM2908 (Fig. 2A). ATM 2907 mutation has been found in AT patient but the mechanism is not clear how ATM2907 cause the loss of ATM activity. After trans-fection of AT cells with ATM2904, ATM2907 and ATM2908, we detected normal ATM phosphorylation, P53 phosphorylation and H2AX phosphorylation (Fig. 2B) in all the mutants. But interestingly, the ATM2907 mutant showed IR radiation sensitivity and behaved like AT cells which lack ATM protein (Fig. 2D). To rule out the possibility of ATM expression and nuclear location change, we detected ATM level and expression by both Western blot and immunofluorescence, no detectable difference was detected between ATM full length transfected cells and these mutants (Fig. 2C and D). Our previous work showed that Tip60 acetyltransferase bind to ATM and make contribution to ATM activation [16,17], and C terminus of ATM could be the binding site of Tip60. So we detected ATM associated Tip60 HAT activity in these cells. We observed normal HAT activity in response to DNA damage in all the mutants and wide type cells, which indicate that the PET-PVFRLT box does not affect the association of Tip60 and ATM (Fig. 2E). These observations showed PETPVFRLT box play an important role for ATM activation but not autophosphorylation and it is not due to the binding to Tip60.
Fig. 2.
Mutagenesis study of the function of PETPVFRLT box of ATM C terminal reveals its role in dimer monomer transition. (A) PETPVFRLT box of ATM C terminal. ATM2904 mutation, ATM2907 mutation and ATM2908 mutation comprising the last 33 amino acids at the C terminus of each of the proteins are shown. Blue box: identical; Red box: similar. (B) GM5849 ATM negative cells were stably cotransfected with mutations of ATM2904, ATM2907, ATM2908 constructs. Cells were treated with IR (+). Cells were immunoprecipitated and analyzed by Western blot analysis for ATM, phospho-ATM (pS1981), P53 and phospho-P53 (pS15-P53), and H2AX and phospho-H2AX (cH2AX). (C) GM5849 ATM negative cells were stably cotransfected with mutations of ATM2904, ATM2907, ATM2908 constructs. Immunofluorescent staining to detect ATM was then carried out. It shows efficient expression of ATM in the nuclear of GM 5849 cells. Scale bar equals 10 μM. Scale bar, 10 μM. (D) Cells were irradiated at the indicated dose, and the number of surviving colonies measured 12 days later. Data represent the mean ± s.d. from three independent assays. (E) GM5849 cells expressing vector, ATM2904, or ATM2907, ATM2908 were exposed to bleomycin and ATM immunoprecipitated with ATM antibody. ATM-associated HAT activity was measured as above. Results are shown as mean ± s.d. (n = 6). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
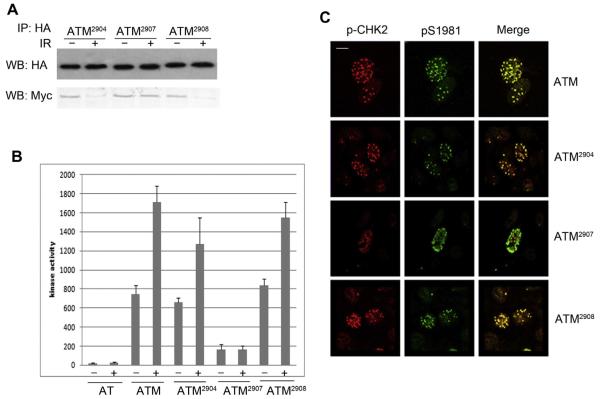
To further address the mechanism that how the PETPVFRLT box affect ATM function, we detected the dimer monomer transition and we found that ATM2907 remain dimeric after IR treatment (Fig. 3A). We then performed kinase assay using P53 peptide as the substrate. Interestingly, we could not detect any kinase activity in the ATM2907 mutant, which indicated that only monomeric form of ATM can phosphorylate the substrate (Fig. 3B). This observation was further confirmed by the weak Chk2 phosphorylation in ATM2907 mutant (Fig. 3C). We further use immunofluorescence to detect ATM phosphorylation and Chk2 phosphorylation and observed ATM foci after IR treatment in ATM2907 mutant (Fig. 3C), which indicated that although ATM2907 mutant cannot undergo dimer monomer transition, the autophosphorylated dimer can still be recruited to damaged site. Thus monomeric form is not required for ATM recruitment.
Fig. 3.
ATM2907 mutation cannot undergo dimer monomer transition and lack functional substrate phosphorylation. (A) ATM negative GM5849 cells were stably cotransfected with HA and Myc-tagged ATM mutation constructs of ATM2904, ATM2907, ATM2908, such that both HA-ATM and Myc-ATM were expressed in the same cells. Cells were treated (+) or nontreated with IR (-) as indicated. Then immunoprecipitation was used with HA antibody. Interaction between HA-ATM and Myc-ATM was then assessed by Western blot (WB) analysis to detect HA-ATM and coprecipitating Myc-ATM. (B) GM5849 ATM negative cells were stably transfected with ATM2904, ATM2907 and ATM2908. Cells were treated with IR (+) as indicated time course. ATM was immunoprecipitated. The intrinsic kinase activity of ATM was then measured using P53 peptide (containing serine 15) as the substrate. (C) GM5849 ATM negative cells were stably transfected with ATM2904, ATM2907 and ATM2908. Cells were irradiated (5 Gy +) and allowed to recover for 20 min. Cells were fixed and immunostained with P-Chk2 antibody (red) and phospho-Ser 1981 antibody to detect active ATM (green). Scale bar, 10 μM. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Dephosphorylation of ATM is essential for ATM dimer re-formation after DNA repair
It has been shown that ATM will undergo autophosphorylation and dimer monomer transition in response to DNA damage. After the DNA damage was repaired, ATM will undergo dephosphorylation and form dimer again [23]. However, it is not clear if there’s correlation between dephosphorylation of ATM and dimer re-formation. Here we showed that the dephosphorylation of ATM is important for ATM dimer re-formation. If ATM dephosphorylation is inhibited, ATM will not form dimer correctly after DNA repair.
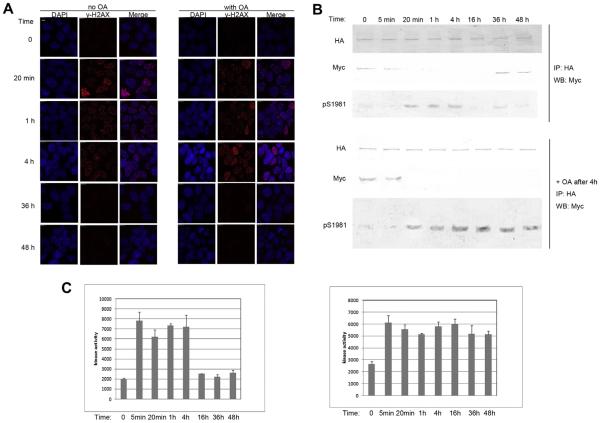
We took the advantage of previous work demonstrating that ATM and protein phosphatase 2A exist as a complex in the cell. Inhibition of protein phosphatase 2A by okadaic acid (OA) leads to rapid accumulation of ATM autophosphorylation on Ser 1981. The immunofluorescence work showed that H2AX phosphorylation and dephosphorylation did not change within examined time course after OA treatment during DNA repair (Fig. 4A). However, we observed that the dephosphorylation of ATM was inhibited (Fig. 4B). Interestingly, we also observed that when dephosphorylation of ATM was inhibited, ATM could not form dimer again and remain monomeric form (Fig. 4B). Further investigation showed that without OA treatment, ATM cannot phosphorylate P53 peptide in kinase assay after dimer re-formation, whereas the monomeric ATM form with accumulated consistent phosphorylation could still phosphorylated P53 peptide in kinase assay (Fig. 4C). These results showed that monomer is the active form of ATM and dimer is the inactive form of ATM. More importantly, not only dimer monomer transition is important for ATM activation, but the dimer re-formation play important role for ATM proper function in the cell.
Fig. 4.
The dephosphorylation of ATM is important for its re-dimerization. (A) Cells cotransfected with HA-ATM and Myc-ATM were treated with solvent or IR (5 Gy +), and 0.5 μM Okadaic acid (OA) was added. Cells were fixed as indicated time course and then underwent immunofluorescent staining of cH2AX antibody. Scale bar, 10 μM. (B) ATM negative GM5849 cells were stably cotransfected with HA and Myc-tagged ATM constructs, such that both HA-ATM and Myc-ATM were expressed in the same cells. Cells were treated (+) or nontreated with IR (-) as indicated. Then immunoprecipitation was used with HA antibody. Interaction between HA-ATM and Myc-ATM was then assessed by Western blot (WB) analysis to detect HA-ATM and coprecipitating Myc-ATM. Levels of ATM and ps1981ATM was measured. Upper panel: No OA treated. Lower panel: OA was added as indicated. (C) GM5849 ATM negative cells were stably cotransfected with HA and Myc-tagged ATM constructs. Cells were treated with IR (+) as indicated time course. ATM was immunoprecipitated. The intrinsic kinase activity of ATM was then measured using P53 peptide (containing serine 15) as the substrate. Left: no OA was added; Right: cells were preincubated with OA after IR.
4. Discussion
Generation of ATM dimer monomer transition monitor Co-IP with two different tags led to the mechanistic insights into the phosphorylation event within the dimer after IR treatment. Our results point to a new mechanism that after DNA damage, the kinase domain of one ATM molecule phosphorylates serine 1981 of an interacting ATM molecule, and the phosphorylated ATM is then dissociated from the complex and is freed to phosphorylate other substrates in the cells. The PETPVFRLT box mutant ATM2907 retains endogenous ATM in a complex. Although it can be phosphorylated but it cannot undergo dimer monomer transition, thus it cannot phosphorylate the substrates and has no cellular activity. After DNA repair, the dephosphorylation of ATM is essential for the active monomer to form inactive dimer again. This mechanism provides an explanation that why ATM pre existed in the cells as dimer since one molecule of ATM cannot phosphorylate itself, the dimer of ATM need one molecule to phosphorylate another. Our work also show that both dimer monomer transition in response to DNA damage and monomer dimer transition after DNA repair are very important for ATM to play proper cellular function.
Since dimer monomer model is proposed in 2003, for almost a decade, most work focused on finding new factors which help to initiate the kinase activity of kinase domain and P1981 phosphorylation [18,24–29]. Here we showed that both kinase activity and P1981 are essential but not enough for ATM cellular activity and function. The dimer monomer transition and the structure change itself help to release the active monomeric form, otherwise the dimer could not phosphorylate the sub-strates and the cells will lack ATM function. Our work also reveals the important function of PETPVFRLT box for dimer dissociation, which indicates that there might be other factors that could contribute to dimer monomer transition except for kinase domain and P1981 phosphorylation site. For example, the phosphorylation site other than P1981 could play important role for this dynamic process [8].
Although a lot work has shown that dephosphorylation is very important for ATM proper function, it is not easy to explain the correlation of ATM dephosphorylation and ATM function defect. Data presented here is consistent with the previous work that dephosphorylation is as important as phosphorylation. Moreover, we proposed based on the data that ATM can only form dimer again after dephosphorylation. This mechanism also provide dynamic model for the regulation of ATM activation in the cells, which might be a little different from the in vitro activation of ATM. In a cell free system, once ATM is activated, it cannot come back to the inactive dimer form, whereas the ATM dimer monomer transition in the cells are reversible, which provide perfect functional base for the dynamic DNA repair process.
Acknowledgments
This work was supported by National Basic Research Program of China (973 Program, Grant Nos. 2013CB911001; 2012CB518302), National Natural Science Foundation of China (Grant Nos. 91019024; 31100558) and Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDA01040407).
References
- [1].Meyn MS. Ataxia-telangiectasia, cancer and the pathobiology of the ATM gene. Clin. Genet. 1999;55:289–304. doi: 10.1034/j.1399-0004.1999.550501.x. [DOI] [PubMed] [Google Scholar]
- [2].Rotman G, Shiloh Y. ATM: from gene to function. Hum. Mol. Genet. 1998;7:1555–1563. doi: 10.1093/hmg/7.10.1555. [DOI] [PubMed] [Google Scholar]
- [3].Lavin MF, Birrell G, Chen P, Kozlov S, Scott S, Gueven N. ATM signaling and genomic stability in response to DNA damage. Mutat. Res. 2005;569:123–132. doi: 10.1016/j.mrfmmm.2004.04.020. [DOI] [PubMed] [Google Scholar]
- [4].Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- [5].Bakkenist CJ, Kastan MB. Initiating cellular stress responses. Cell. 2004;118:9–17. doi: 10.1016/j.cell.2004.06.023. [DOI] [PubMed] [Google Scholar]
- [6].Zakian VA. ATM-related genes: what do they tell us about functions of the human gene? Cell. 1995;82:685–687. doi: 10.1016/0092-8674(95)90463-8. [DOI] [PubMed] [Google Scholar]
- [7].Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- [8].Kim ST, Xu B, Kastan MB. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 2002;16:560–570. doi: 10.1101/gad.970602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Matsuoka S, Ballif BA, Smogorzewska A, McDonald 3rd ER, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- [10].Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- [11].Lim DS, Kim ST, Xu B, Maser RS, Lin J, Petrini JH, Kastan MB. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature. 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- [12].Cortez D, Wang Y, Qin J, Elledge SJ. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- [13].Taniguchi T, Garcia-Higuera I, Xu B, Andreassen PR, Gregory RC, Kim ST, Lane WS, Kastan MB, D’Andrea AD. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;109:459–472. doi: 10.1016/s0092-8674(02)00747-x. [DOI] [PubMed] [Google Scholar]
- [14].Fernandes N, Sun Y, Chen S, Paul P, Shaw RJ, Cantley LC, Price BD. DNA damage-induced association of ATM with its target proteins requires a protein interaction domain in the N terminus of ATM. J. Biol. Chem. 2005;280:15158–15164. doi: 10.1074/jbc.M412065200. [DOI] [PubMed] [Google Scholar]
- [15].Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- [16].Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sun Y, Xu Y, Roy K, Price BD. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol. Cell. Biol. 2007;27:8502–8509. doi: 10.1128/MCB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, Price BD. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat. Cell Biol. 2009;11:1376–1382. doi: 10.1038/ncb1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- [20].Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge SJ. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc. Natl. Acad. Sci. U.S.A. 2000;97:10389–10394. doi: 10.1073/pnas.190030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Blasina A, Price BD, Turenne GA, McGowan CH. Caffeine inhibits the checkpoint kinase ATM. Curr. Biol. 1999;9:1135–1138. doi: 10.1016/s0960-9822(99)80486-2. [DOI] [PubMed] [Google Scholar]
- [22].Turenne GA, Paul P, Lafiair L, Price BD. Activation of p53 transcriptional activity requires ATM’s kinase domain and multiple N-terminal serine residues of p53. Oncogene. 2001;20:5100–5110. doi: 10.1038/sj.onc.1204665. [DOI] [PubMed] [Google Scholar]
- [23].Goodarzi AA, Jonnalagadda JC, Douglas P, Young D, Ye R, Moorhead GBG, Lees-Miller SP, Khanna KK. Autophosphorylation of ataxia-telangiectasia mutated is regulated by protein phosphatase 2A. EMBO J. 2004;23:4451–4461. doi: 10.1038/sj.emboj.7600455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes Dev. 2004;18:1423–1438. doi: 10.1101/gad.1200304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/ Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- [27].Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- [28].You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell. Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mochan TA, Venere M, DiTullio RA, Jr., Halazonetis TD. 53BP1, an activator of ATM in response to DNA damage. DNA Repair. 2004;3:945–952. doi: 10.1016/j.dnarep.2004.03.017. [DOI] [PubMed] [Google Scholar]