Abstract
During aerobic respiration, Bacillus subtilis utilizes three terminal oxidases, cytochromes aa3, caa3, and bd. Cytochrome bd is encoded by the cydABCD operon. We report here the first identification of a regulator for the cydABCD operon, YdiH. While working with ΔresDE mutant strains, we identified colonies which contained suppressor mutations (cmp) which bypassed the requirement for ResD for all phenotypes not associated with cytochrome aa3 or caa3. Mapping identified a class of Tn10 insertions which were close to the cmp locus (Tn10-2) and a second class (Tn10-1) which was inserted in cydD, a gene which appears to be essential to the cmp phenotype. Sequencing of the cmp loci from four independent ΔresDE cmp isolates yielded four loss-of-function alleles of ydiH, a gene encoding a protein with homology to AT-rich DNA-binding proteins. Additionally, we determined that cytochrome bd was aberrantly expressed in the ΔresDE cmp background. Together these data led to the hypothesis that YdiH serves as a negative regulator of cydABCD expression, a hypothesis supported by both gel-shift and DNase I footprinting analyses. YdiH protected the cydA promoter region at three 22-bp repeats located in the long 5′ untranslated region (193 bp). Induction of the cydABCD operon in a ΔresDE background showed that expression of the terminal oxidase bd was responsible for the bypass phenotype observed in a ΔresDE cmp strain, indicating that cytochrome bd expression complemented the loss of cytochromes aa3 and caa3 in the ΔresDE strain.
Bacillus subtilis utilizes a branched electron transport chain under aerobic conditions. To date, three terminal oxidases have been identified in B. subtilis. Both cytochromes aa3 (26) and caa3 (5) have been identified as heme-copper oxidases. The third oxidase has been shown to be a member of the cytochrome bd family (35). The cydABCD operon of B. subtilis encodes cytochrome bd and a putative ABC transporter required for the production of functional cytochrome bd (35). This oxidase is produced under conditions of low oxygen tension and in cells grown in the presence of glucose (35). A single cydA transcriptional start site with a putative −10 and −35 consensus for a σA promoter has been found in cells grown to stationary phase in nutrient sporulation medium with phosphate buffer and glucose (NSMPG) (35). A perfect 16-bp palindromic sequence, upstream of the translation start site for cydA, was proposed as a potential operator binding site for a regulatory protein (35). To date, no regulators have been reported for the cydABCD operon. It was originally reported that the quinol oxidases (either cytochrome aa3 or bd), are required for aerobic growth in B. subtilis (34). However, further evidence has shown that a strain deficient in the production of both cytochrome aa3 and cytochrome bd, a derivative of the B. subtilis 168 strain (24), can be constructed and grown aerobically (37). A putative fourth terminal oxidase, YthAB, has been found in B. subtilis and is a member of the cytochrome bd family (34).
YdiH was identified during the course of sequencing the B. subtilis genome. Based on homology, YdiH was proposed to be a member of a family of AT-rich DNA-binding proteins (16) that includes p25, a recently characterized DNA-binding protein from Thermus aquaticus YT-1 (6).
The ResD/ResE two-component signal transduction system plays a role in the regulation of both aerobic respiration and anaerobic respiration. ResD regulates the expression of fnr (21), hmp (21), nasDEF (21), hemN (12), hemZ (12), and the sbo-alb operon (20) under anaerobic conditions and has a role in the regulation of ctaA (23, 38), ctaBCDEF (17), resABCDE (31), and petCBD (31) under aerobic conditions. Because ResDE is essential for expression of both ctaA and ctaB, which is required for heme A biosynthesis, ΔresDE strains lack cytochromes aa3 and caa3.
In the course of our work with ΔresDE strains, we have found that these strains develop secondary mutations which have been given the designation cmp mutations. The ΔresDE strains bearing the cmp mutation are complemented for a number of phenotypes typically associated with a ΔresDE strain. In the course of identifying and characterizing these cmp mutations, we have implicated YdiH, a previously uncharacterized putative DNA-binding protein, in the regulation of the cydABCD operon. In this paper, we report the characterization of YdiH as a negative regulator for cydABCD transcription. Additionally, we show that the absence of terminal oxidases in the ΔresDE strain is responsible for a number of phenotypes previously reported for that strain.
MATERIALS AND METHODS
Strains and plasmids.
Table 1 lists the strains and plasmids used in this study. Escherichia coli DH5α was the host for all plasmid constructions. E. coli BL21(DE3)/pLysS (Novagen) served as the host for overexpression of the YdiH protein. B. subtilis JH642 served as the host for all strain constructions.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | Lab stock | |
| BL21(DE3)pLysS | Novagen | |
| Bacillus | ||
| JH642 | pheA1 trpC2 | J. A. Hoch |
| 1A601 | purB83::Tn917 Mlsr | 33 |
| ΔtatCY-ΔtatCD | ΔtatCY::Spcr ΔtatCD::Knr | 14 |
| MH1562 | phoB::Tn917 Mlsr | 15 |
| MH5202 | pheA1 trpC2 ΔresDE::Tetr | 31 |
| MH5857 | pheA1 trpC2 ΔresDE::Tetrcmp amyE::phoD-lacZ Cmr | This study |
| MH5859 | pheA1 trpC2 ΔresDE::Tetrcmp amyE::phoD-lacZ Cmr | This study |
| MH5863 | pheA1 trpC2 ydhQΩpMS7 Ermr | This study |
| MH5867 | pheA1 trpC2 rrnE-23s::Tn10-2 SpcrydhQΩpMS7 Ermr | This study |
| MH5874 | pheA1 trpC2 ΔtatCY::SpcrrrnE-23s::Tn10-2 Knr | This study |
| MH5878 | pheA1 trpC2 amyE::pMS35cydA-lacZ Cmr | This study |
| MH5879 | pheA1 trpC2 ΔresDE::Tetrcmp amyE::cydA-lacZ Cmr | This study |
| MH5880 | pheA1 trpC2 ΔresDE::TetramyE::cydA-lacZ Cmr | This study |
| MH5881 | pheA1 trpC2 ydhQΩpMS7 ErmrtatCY::Spcr | This study |
| MH5882 | pheA1 trpC2 cydD::Tn10-1 Spcr ΔresDE::Tetrcmp | This study |
| MH5883 | pheA1 trpC2 rrnE-23s::Tn10-2 Spcr ΔresDE::Tetr | This study |
| MH5884 | pheA1 trpC2 JH642ΩpMS38 | This study |
| MH5885 | pheA1 trpC2 MH5884 ΔresDE::Tetr | This study |
| MH5887 | pheA1 trpC2 ΔresDE::Tetrcmp | Guofu Sun |
| MH5888 | pheA1 trpC2 ΔresDE::Tetrcmp | Ruth Chestnut |
| MH5889 | pheA1 trpC2 ΔtatCY::Spcr | This study |
| MH5890 | pheA1 trpC2 rrnE-23s::Tn10-2 Knr | This study |
| MH5891 | pheA1 trpC2 ydiHΩpMS45 SpcramyE::cydA-lacZ Cmr | This study |
| MH5893 | pheA1 trpC2 ΔresDE::TetrydiHΩpMS45 SpcramyE::cydA-lacZ Cmr | This study |
| Plasmids | ||
| pCR2.1 | Vector for cloning PCR products | Invitrogen |
| pIC333 | Vector for Tn10 mutagenesis, Spcr Ermr | 29 |
| pDH32 | Vector for construction of promoter-lacZ fusions; Ampr Cmr | 28 |
| pAT110 | Vector for mutant generation; Ermr | 32 |
| pDG1727 | Vector for mutant generation; Spcr | 9 |
| pVK73 | Antibiotic switching vector from Spcr to Knr | 3 |
| pET16b | Vector for protein overexpression and His-tagging Ampr | Novagen |
| pMS4 | Internal fragment of ydhQ in pCR2.1 | This study |
| pMS7 | Fragment of ydhQ in pAT110; Ermr Ampr | This study |
| pMS34 | Fragment of cydA promoter in pCR 2.1; Ampr | This study |
| pMS37 | Fragment of cydA in pCR2.1; Ampr | This study |
| pMS38 | Pspac-cydABCD in pDH88; Ampr Cmr | This study |
| pMS35 | cydA-lacZ fusion in pDH32; Ampr Cmr | This study |
| pMS40 | Internal fragment of ydiH in pCR2.1; Ampr | This study |
| pMS45 | Internal fragment of ydiH in pDG1727; Ampr Spcr | This study |
| pMS41 | Coding sequence of ydiH in pCR2.1; Ampr | This study |
| pMS43 | Coding sequence of ydiH in pET16b; Ampr | This study |
cydA-lacZ promoter fusions were constructed in pDH32, which has unique EcoRI and BamHI sites upstream of a promoterless lacZ gene to correctly orient the promoter DNA fragment. Primers FMH741 (5′-CCGAATTC−305TAGCAGCCGACATAAATAAG−285-3′; EcoRI site underlined) and FMH742 (5′-CCGGATCC7CACTCATGCTTTCTCCTCC ATTTCC−18-3′; BamHI site underlined) were used to amplify a 312-bp fragment of the cydA promoter region spanning from −305 to +7 relative to the start of translation of CydA, using JH642 chromosomal DNA as template. The resulting fragment was ligated into pCR2.1 (Invitrogen) to create pMS34. The cydA promoter fragment was released from pMS34 by digestion with EcoRI and BamHI and cloned into the EcoRI and BamHI sites of pDH32 to create pMS35. DNA sequencing confirmed the sequence of the cydA promoter. MH5878 (cydA-lacZ) was created by transformation of JH642 with pMS35 linearized by digestion with PstI such that the promoter fusion was integrated into the amyE locus by double-crossover homologous recombination. Transformants were selected on chloramphenicol, and the required insertion was confirmed by the amyE mutant phenotype. Transformation of chromosomal DNA from MH5878 (cydA-lacZ) to MH5887 (ΔresDE cmp) produced MH5879 (cydA-lacZ ΔresDE cmp). Transformation of chromosomal DNA from MH5202 (ΔresDE) into MH5878 (cydA-lacZ) produced MH5880 (cydA-lacZ ΔresDE).
To create a mutation in ydiH, we cloned an internal fragment of the gene to pDG1727, which allows for the creation of an insertion-duplication mutation. Primers FHM788 (5′-42ACGGCTGCCGCTTTACTATC62-3′) and FHM789 (5′-557TGTTCCGG CACATTCAAACG537-3′) were used to amplify a 516-bp internal fragment of ydiH, using JH642 chromosomal DNA as a template. The resulting PCR product was cloned to pCR2.1 to create pMS40. The ydiH fragment was released from pMS40 by digestion with EcoRV and BamHI and cloned into the EcoRV and BamHI sites of pDG1727 to create pMS45. pMS45 was transformed to MH5878 (cydA-lacZ) to create MH5891 (ΩydiH cydA-lacZ). MH5893 (ΩydiH ΔresDE cydA-lacZ) was created by transforming chromosomal DNA from MH5202 (ΔresDE) into MH5891 (ΩydiH cydA-lacZ).
Inducible expression of the cydABCD operon was achieved by cloning in pDH88, which contains an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Pspac promoter. Primers FMH764 (5′-CCAAGCTT−23TAACCGGAAATGGAGGAG−5-3′; HindIII site underlined) and FMH765 (5′-CCGCATCG307ACGCCAATAATTGCTTCAATC286-3′; SphI site underlined) were used to amplify a 346-bp fragment containing positions −23 to +307 relative to the initiation of translation of CydA. This fragment contains the ribosome binding site for the cydABCD operon. The resulting PCR product was cloned to pCR2.1 to create pMS37. The cydA fragment was released from pMS37 by digestion with HindIII and SphI and cloned into complementary sites in pDH88 to yield pMS38. pMS38 was transformed to JH642 to yield MH5884 (Pspac-cydABCD). Transformation of chromosomal DNA from MH5202 (ΔresDE) to MH5884 (Pspac-cydABCD) yielded MH5885 (ΔresDE Pspac-cydABCD).
Due to the high rate of appearance of spontaneous suppressor mutations in a ΔresDE background, special precautions were taken in the constructions of all strains bearing a resDE mutation. Chromosomal DNA from MH5202 (ΔresDE) was used to transform the appropriate background whenever a strain with a resDE mutation was required. This was done to avoid the isolation of strains with cmp mutations during the production of competent cells of MH5202 (ΔresDE). All transformants were screened to confirm that the correct phenotype was present for each construction.
In order to create a mutation in ydhQ, we cloned an internal fragment of ydhQ to pAT110 in order to allow the creation of an insertion-duplication mutant. Primers FMH476 (5′-107TTAACTCAAGCCGATGACG136-3′) and FMH477 (5′-580CCAGATG CTGCTGGTCAATA560-3′) were used to amplify a 476-bp internal fragment of ydhQ. The resulting PCR product was cloned to pCR2.1 to create pMS4. The ydhQ fragment was released from pMS4 by digestion with NotI and HindIII and cloned into to NotI and HindIII sites of pAT110 to create pMS7. pMS7 was transformed to JH642 to yield MH5863 (ΩydhQ). MH5889 (ΔtatCY) was created by transforming chromosomal DNA from ΔtatCY-ΔtatCD to JH642 and selecting on spectinomycin. Transformants were screened on plates containing kanamycin to confirm the single mutation. MH5881 (ΔtatCY ΩydhQ) was generated by transforming chromosomal DNA from MH5863 (ΩydhQ) into MH5889 (ΔtatCY).
MH5890 (rrnE-23S::Tn10-2 Knr) was created by transforming the antibiotic-switching vector pVK73 into MH5883 (rrnE-23S::Tn10-2 Spcr) and selecting on kanamycin. Transformants were screened for spectinomycin sensitivity to confirm that the antibiotic switch was successful. Transformation of chromosomal DNA from MH5889 (ΔtatCY) to MH5890 (rrnE-23S::Tn10-2) yielded MH5874 (ΔtatCY rrnE-23S::Tn10-2).
To construct a plasmid for overexpressing YdiH, we amplified the entire coding sequence of ydiH by using the primers FMH790 (5′-TACATATGAATAAGGATCAATCAAAAATTCCGCAGGCGA-3′; NdeI site underlined) and FMH791 (5′-TAGGATCCCTATTCGATTTCCTCTAAAACTGAATAATGC-3′; BamHI site underlined). JH642 was used as the template for PCR. The resulting PCR product was cloned to pCR2.1 to create pMS41. After DNA sequence confirmation, the coding sequence of ydiH was released from pMS41 by digestion with NdeI and BamHI and cloned to the NdeI and BamHI sites of pET16b to create pMS43. pMS43 was transformed to E. coli BL21(DE3)/pLysS, and representative transformants were used for overexpression of the YdiH protein.
Genetic manipulations.
Transformation of B. subtilis was by the two-step transformation method of Cutting and Vander Horn (4). Transformants were selected on tryptose blood agar base medium (TBAB) supplemented with 0.5% glucose (TBABG) and the appropriate antibiotic. Antibiotics were added to the medium for selection of B. subtilis transformants at the following concentrations: chloramphenicol, 5 μg/ml; erythromycin, 1 μg/ml; spectinomycin, 100 μg/ml; tetracycline, 10 μg/ml; kanamycin, 10 μg/ml; and erythromycin, 1 μg/ml, and lincomycin, 25 μg/ml (mls). Preparation of PBS1 transducing lysates and PBS1 transduction were performed by the method of Cutting and Vander Horn (4). Transductants were selected on TBABG plates with the appropriate antibiotic at the concentrations listed above. Transformation of E. coli was done according to the method of Hanahan (10). Transformants were selected on Luria-Bertani (LB) plates containing ampicillin (100 μg/ml).
The plasmid pIC333 was used as the source for Tn10 mutagenesis. Transposition was performed in the wild-type (JH642) background following the method of Steinmetz and Richter (29). Chromosomal DNA from the Tn10 library was used to transform MH5857 (ΔresDE cmp). These transformants were picked to TBAB, TBABG, TBABG-spectinomycin, TBABG-chloramphenicol, and TBABG-tetracycline plates. Any Tn10 insertion that cotransformed with a wild-type copy of the cmp locus was identified based on the reversion from the ΔresDE cmp phenotype to the ΔresDE phenotype on TBAB and TBABG plates. Further confirmation of a Tn10 insertion near the cmp locus was obtained by backcross transformation of Tn10 to a strain bearing the cmp mutation in order to determine the frequency at which each Tn10 was linked to the wild-type copy of the cmp locus. This frequency was based on the number of Tn10-bearing transformants that reverted from the ΔresDE cmp phenotype to the ΔresDE phenotype on plates.
Growth conditions and enzyme assays.
Growth for measurement of cydA-lacZ expression was performed in LB medium supplemented with 0.5% glucose. β-Galactosidase activity was detected by the method of Ferrari et al. (8). One activity unit was defined as 0.33 nmol of o-nitrophenol produced min−1, and the specific activity was calculated as activity per milligram of protein. When appropriate, IPTG was added at a final concentration of 1 mM to the medium. Alkaline phosphate (alkaline phosphatase) activity was measured in cells that had been grown in low-phosphate defined medium (LPDM) as described previously (13).
Phenotypic characterization of mutant strains.
The percentage of heat-resistant spores was determined by the method of Nicholson and Setlow (22). Cytochrome oxidase activity was assayed with TMPD as a substrate as previously described by Muller and Taber (18). Organic acid production was measured on purification agar medium plates by the method of Carls and Hanson (2). Anaerobic growth on plates was carried out in a BBL GasPak jar with BBL GasPak anaerobic system chemicals added according to the manufacturer's instructions. TBABG plates supplemented with 0.2% KNO3 were used as the medium for anaerobic growth.
Preparation and spectrophotometry of solubilized membrane vesicles.
Membrane vesicles were prepared as described by Bisschop and Konings (1), with the following modifications. Cells were collected from stationary-phase cultures grown in LB medium with 0.5% glucose. DNase and RNase were omitted from the lysis procedure. Solubilization of cytochromes and analysis of difference absorption spectra were performed as described by Mueller and Taber (18). Difference absorption spectra (dithionite reduced minus ferricyanide oxidized) were recorded at room temperature at a scan speed of 5 nm/s with a Hitachi U-2000 spectrophotometer. Reduction and oxidation were performed as previously described (18).
Inverse PCR.
In order to determine the site of each Tn10 insertion, inverse PCR was performed. Chromosomal DNA from MH5882 (Tn10-1) or MH5883 (Tn10-2) was subjected to digestion with PstI. Following digestion, each sample was ethanol precipitated and diluted to 10 μl in T4 DNA ligase buffer and treated with T4 DNA ligase overnight to allow for self-ligation. PstI cuts once within the Tn10 insertion and then will cut the chromosomal DNA adjacent to the Tn10 insertion at the next PstI site. The resulting product was used as the template in a two-step inverse PCR. Based on the sequence of pIC333, two sets of primers were developed complementary to each end of the Tn10 remaining. FMH442 (5′-GCTATATCCAGTAAAGTTACAT-3′) and FMH443 (5′-GTGGGAAGGACTATATTCA-3′) were used in the first step of the PCR process. The product generated by these primers was diluted and used as the template for a second PCR. FMH446 (5′-ACATGCTCTTTAGGTA-3′) and FMH447 (5′-CCTCTTGTGAAATTAG-3′) were used to generate the second PCR product. The resulting PCR product was purified following the manufacture's instructions for the Microcon PCR kit (Millipore). DNA sequencing of the purified PCR products was performed by the University of Chicago Cancer Research Sequencing Center. In order to confirm the insertion of Tn10-1 in cydD, we performed PCR with FMH438 (5′-AGCGAGTCAGTGAGCGAGGA-3′) complementary to Tn10 and FMH782 (5′-CGGAGCTGGTGAACAACCTT-3′) complementary to positions −372 to −352 of cydD, using MH5882 as the template for the PCR. The resulting PCR product was purified and sequenced, confirming the insertion of Tn10-1 in cydD. In order to confirm the insertion of Tn10-2 in rrnE-23S, we again used FMH438, which was paired with FMH783 (5′-CCGTGTGCCTACCTACAAGTAGTC-3′) complementary to positions 563 to 583 of rrnE-23S, using MH5883 as the template for PCR. Again the resulting PCR product was purified and sequenced, confirming the insertion of Tn10-2 in rrnE-23S.
PCR identification of the cmp locus.
In order to identify the locus of the cmp mutation, we designed 15 pairs of PCR primers spanning the region from 88 bp upstream of the start of translation of thiL to 98 bp into the coding sequence of tatCY. The sequence and position of all primers used to identify the cmp locus are found in Table 2. These primers were used to amplify 15 overlapping PCR fragments, using MH5857 as the template DNA. All PCRs were performed in triplicate, and the resulting products were purified with the Microcon PCR purification kit. The resulting PCR products were sequenced, and the sequence was compared to the published genome sequence for each region. The corresponding protein sequence for all mutations found was determined with Clone Manager (SciEd Central) and compared to the wild-type protein sequence for the given locus obtained from the B. subtilis genome sequence database, SubtiList (http://genolist.pasteur.fr/SubtiList/).
TABLE 2.
Primers used of the identification of the cmp locus
| Primer | Sequence (5′ → 3′) | Starting location (bp) |
|---|---|---|
| FMH617 | CGGGAACATCTTGGATAAT | −88 of thiL |
| FMH618 | TCCGTCACTAACATCATTT | 637 of thiL |
| FMH619 | GCCTGAGCCAAGAGTAAGC | 581 of thiL |
| FMH620 | GTCGGACTGTTTACAATACG | 165 of ydiB |
| FMH621 | GGAGACGTCCTGACATTAGA | 191 of ydiB |
| FMH622 | GCGGTGCCATATCACAATCAT | 142 of ydiC |
| FMH623 | GCGCTGCTTCGAGAAGACAC | 45 of ydiC |
| FMH624 | GCCAGCCGCAAGTAATCCG | 634 of ydiC |
| FMH625 | GCGCCTTCAGAATTGGCTT | 608 of ydiC |
| FMH626 | GCAGTCTCATCACAGCTTGTT | 38 of gcp |
| FMH627 | CGCTTGAAGTGAGGTTTCG | 316 of ydiD |
| FMH628 | CGGTCCACCCGGATATGGC | 539 of gcp |
| FMH629 | GCGGCAGGAGAAGCCTAC | 495 of gcp |
| FMH630 | CCTCAGCCCTTTATCCATCAA | +104 gcp |
| FMH649 | CGCTCCTTAAACAGATCATA | −118 of ydiF |
| FMH650 | GGCTTTGAACCTGTGTAGA | 469 of ydiF |
| FMH651 | GGCGGTTATCAATATGAAGC | 399 of ydiF |
| FMH652 | GAGCATGAATGACACTTCTG | 1040 of ydiF |
| FMH653 | CAGAGCGGAAATGAAGTCCT | 961 of ydiF |
| FMH654 | GGGTCACATAGCAATTCATC | 1811 of ydiF |
| FMH655 | CCAACGACTCCGTCGGATT | 1722 of ydiF |
| FMH656 | CCCATTCGGACAAAAGAGATT | 1903 of ydiF |
| FMH657 | CAGCTGATTAGCTTGTAAGAT | 7 of ydiF |
| FMH658 | GCGCTTCCATTTCCACAC | 335 of ydiG |
| FMH659 | GTGCCATCCGCTTTCATTG | 225 of ydiG |
| FMH660 | CGGATGATACACGCTGTT | 109 of ydiH |
| FMH661 | CGGCTGCCGCTTTACTATCG | 43 of ydiH |
| FMH662 | CGGCATATTTGGGCTCCTC | 6 of tatAY |
| FMH663 | GGTCGGCATTAGGAATCAAG | 494 of ydiH |
| FMH664 | GCGCTACAATCAGCAACC | 79 of tatCY |
Overexpression and purification of YdiH.
E. coli BL21(DE3)pLysS harboring pMS43 was incubated overnight at 37°C in LB medium containing ampicillin (100 μg/ml) and was used to inoculate 1 liter of the same medium at a ratio of 1 to 100. The cells were grown at 30°C until the optical density at 600 nm (OD600) of the culture reached about 0.4. IPTG was then added to the culture at a final concentration of 1 mM, and growth continued for another 3 h. The cells were harvested by centrifugation (5,000 × g) at 4°C and washed once with buffer A (1 M NaCl, 5 mM MgCl2, 10 mM dithiothreitol [DTT], 50 mM Tris-HCl [pH 7.8]). The cell pellets were then suspended in 50 ml of prechilled buffer A on ice, containing 1 mM phenylmethlysulfonyl fluoride) and were immediately subjected to sonication. The disruption of cells was confirmed by phase-contrast microscopy. After centrifugation at 40,000 × g for 1 h at 4°C, the supernatant fraction was filtered through a 0.45-μm-pore-size membrane (Amicon). After adding a 1/50 volume of 0.5 M imidazole in buffer A, the clear cell lysate was mixed with a 2-ml Ni-nitrilotriacetic acid (NTA) (QIAGEN) affinity resin, preequilibrated with buffer A. After gentle shaking at 4°C for 30 min, the mixture was loaded onto an Econo column (inside diameter, 2.5 cm; height, 10 cm; Bio-Rad). The column was washed with buffer A until the elute contained nondetectable protein concentrations according to a Bio-Rad protein assay. The protein bound to the Ni-NTA resin was eluted by using 300 mM imidazole in buffer A. The protein fractions were dialyzed stepwise at 4°C against buffer A containing 20% glycerol with decreasing concentrations of NaCl from 1 M to 0.8, 0.6, 0.4, 0.2, and finally 0.1 M. The purity of YdiH proteins was over 95% as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Preparation of cydA promoter probe.
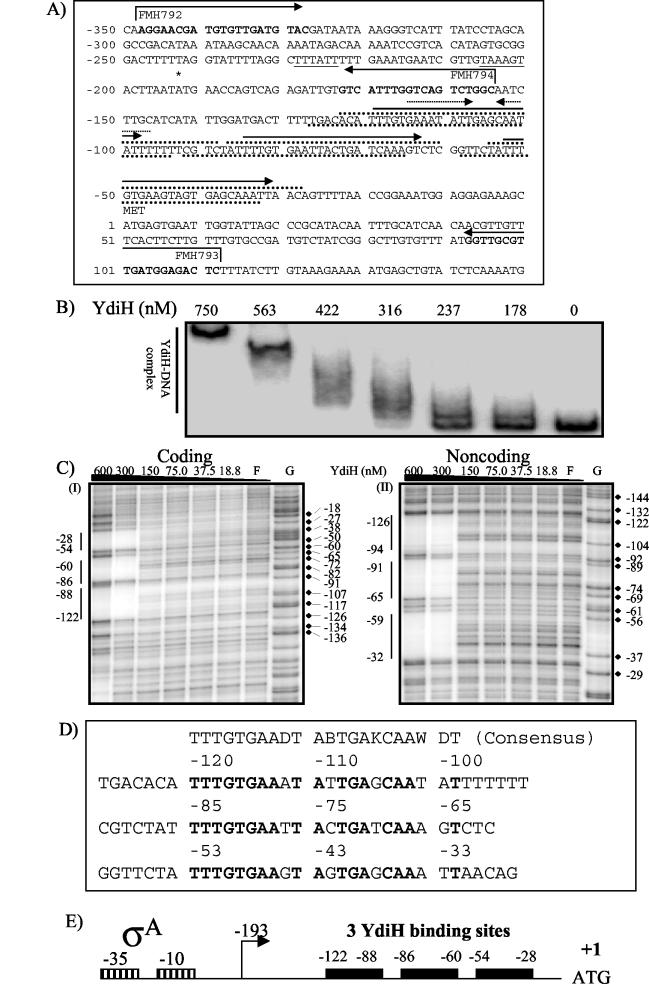
Primers FMH792 and FMH793 were end labeled with T4 polynucleotide kinase (Fermentas) in the presence of [γ-32P]ATP, separated by SDS-PAGE, extracted from the SDS gel slices, and then ethanol precipitated. PCR was conducted with primer pair FMH792 and FMH793 and with JH642 chromosomal DNA as template. For the probes used in the gel shift assay, FMH792 and FMH793 were both radiolabeled. For radiolabeling the coding or noncoding strand, radiolabeled FMH792 and FMH793 or FMH792 and radiolabeled FMH793 were used, respectively. The PCR products were extracted by PAGE and purified by an Elutip-D minicolumn (Schleicher & Schuell, Keene, N.H.) as described in the instruction manual. Similar techniques were used to prepare a probe lacking the YdiH binding site, using primers FMH792 and FMH794. All primer sequences and positions are listed in Fig. 4A.
FIG. 4.
YdiH binds directly to the cydA promoter. (A) Structure of the cydA regulatory region. The major transcriptional start site is shown with an asterisk. The inverted repeats and tandem repeats are indicated as dotted line arrows or solid arrows, respectively. The YdiH protected regions revealed by the DNase I footprinting assay are shown by dotted lines above (coding strand) and below (noncoding strand) the sequence. The consensus sequences for the −35 and −10 σA recognition sequences are underlined. The primer sequences used to generate PCR products are in boldface, and broken arrows indicate the 5′-to-3′ sequence. The numbering is relative to the translation start (ATG) as +1 (also for panels C and D). (B) Gel shift assay of the cydA promoter with YdiH. The probe was the PCR product using radiolabeled primers FMH792 and FMH793 and JH642 chromosomal DNA as template. The primer was labeled as described in Materials and Methods. The concentrations of YdiH used are indicated at the top of each lane. (C) DNase I footprinting of the cydA promoter by using YdiH. Labeled DNA fragments were generated as in panel B. For the coding or noncoding footprint, FMH792 or FMH793 was end labeled, respectively. The YdiH concentrations are shown at the top of each lane. F, free of YdiH; G, Maxam-Gilbert G-sequencing reaction lane as a marker. The vertical lines show the YdiH protected regions. (D) The cydA promoter alignment for directed repeats. The consensus sequence is shown above, and the IUPAC (the International Union of Pure and Applied Chemistry) ambiguity codes for DNA were used: D = A, T, or G; B = T, C, or G; K = G or T; and W = A or T. The identical base pairs among the three repeats are in bold. (E) Graphic representation of YdiH binding on the cydA promoter. Solid black boxes represent the three YdiH binding site on the cydA promoter region. The boxes with vertical lines represent the consensus −10 and −35 for σA identified previously (35). The numbering is relative to the translational start (ATG) as +1. For simplicity, only coding strand protected regions are numbered. The transcription start site is identified with a broken arrow.
Gel shift assays.
The cydA probes were prepared as described above. In each reaction, the desired concentration of protein and the probe (20,000 cpm) were incubated in buffer {25 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 6.1, 25 mM NaCl, 0.5 mM DTT, 2 mM EDTA, 2 mM MgCl2, 5% glycerol} for 30 min at room temperature. The samples were loaded onto a 4% polyacrylamide gel made in 1× Tris-borate-EDTA. The gel was run at 4°C for 1.5 h, vacuum dried, and detected by PhosphorImager analysis.
DNase I footprinting of the cydA promoter.
In each reaction, required protein and the probes (20,000 cpm) were incubated at room temperature for 30 min in buffer (10 mM HEPES, pH 6.1, 50 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 1 mM DTT, 10% glycerol). DNase I (3 μl of 0.04 U/μl in 5 mM MgCl2, 5 mM CaCl2) was added to each reaction mixture, and digestion was conducted for 60 s for protein-containing samples and 30 s for protein-free samples. The reaction was stopped, and the DNA fragments were purified by phenol extraction followed by ethanol precipitation. The DNA fragments were run on a 4% polyacrylamide gel containing 7 M urea and detected by PhosphorImager analysis and/or X-ray film (Fuji) radiography.
RESULTS
Identification and characterization of cmp mutants.
B. subtilis strains containing ΔresDE mutations grew poorly on solid TBAB medium and, with time, developed opaque segregates within the lysing ΔresDE colonies. These segregates had an improved growth phenotype. Pure cultures were isolated from the opaque papillae or sectors and named cmp mutants because they compensate for the ΔresDE poor growth phenotype. We asked what other ΔresDE phenotypes were suppressed by a cmp mutation (Table 3). ΔresDE cmp mutant strains did not produce the pink colony color on TBABG or accumulate acid, characteristics of a ΔresDE strain (31). They also oxidize the terminal oxidase substrate, TMPD, and sporulate similar to a wild-type strain. Previous reports (30) have shown that ResD is required for 80% of the wild-type level of alkaline phosphatase induction. We have found that strains bearing the cmp mutation bypass the requirement for ResD for full alkaline phosphatase induction (Table 3). However, the cmp mutation does not compensate for the loss of resDE for anaerobic growth (19-21) or ctaA expression (23, 38). Both ctaA and genes required for anaerobic growth require ResD as a transcription activator, suggesting that the nature of the cmp mutation is not to simply fill the role of ResD as a transcriptional regulator for Res regulon members.
TABLE 3.
Phenotypic characterization of mutants used in this study
| Strain (relevant genotype) | Growtha | Red pigmentationb | Sporulation (%)c | Oxidization of TMPDd | Accumulation of organic acidse | Anaerobic growthf | ctaA-lacZ expressiong | Alkaline phosphatase induction (%)h |
|---|---|---|---|---|---|---|---|---|
| JH642 (wild type) | + | − | 50 | + | − | + | + | 100 |
| MH5202 (ΔresDE) | +/− | + | 0.9 | − | + | − | − | 20 |
| MH5857 (cmp ΔresDE) | + | − | 47 | + | − | − | − | 100 |
| MH5893 (ΩydiH ΔresDE) | + | − | 33 | + | − | − | NDi | 100 |
| MH5891 (ΩydiH) | + | − | 44 | + | − | + | ND | 100 |
| MH5882 (cydD::Tn10-1 ΔresDE cmp) | +/− | + | 0.3 | − | + | − | ND | 20 |
| MH5885 (ΔresDE Pspac-cydABCD with IPTG) | + | − | 20 | + | − | − | ND | ND |
| MH5885 (ΔresDE Pspac-cydABCD without IPTG) | +/− | + | 4 | − | + | − | ND | ND |
Growth on TBAB medium without glucose.
Pink colony on TBABG medium.
Percentage of colonies forming heat-resistant spores.
Ability to oxidize artificial electron donor TMPD.
Accumulation of organic acids on purification agar medium plates.
Anaerobic growth in medium containing nitrate.
Expression of a ctaA-lacZ fusion under phosphate-limiting conditions.
Percentage of wild-type level of alkaline phosphatase induction.
ND, not determined.
Identification of Tn10 insertions linked to the cmp mutation by transformation.
Tn10 mutagenesis of ΔresDE cmp strains was performed by using a Tn10 insertion library generated in JH642. Transformants of the Tn10 library into ΔresDE cmp strains were screened for any Tn10 insertion close to the cmp locus that had cotransformed with a wild-type copy of the cmp gene leading to a phenotypic reversion from that of a cmp ΔresDE strain to a ΔresDE strain. Screening was performed as described in Materials and Methods and identified six candidate Tn10 insertions, which were selected for further study.
Backcross transformation of chromosomal DNA from each Tn10 into a number of ΔresDE cmp strains was performed to determine the proximity of each Tn10 to the cmp locus. These transformations placed the candidate Tn10 insertions in two classes. The first class, MH5882 (Tn10-1), was linked by backcross transformation to the cmp locus in 100% of the transformants screened; while the second class, MH5883 (Tn10-2), was linked in 70% of the transformants screened.
Additionally, we have shown that MH5882 (ΔresDE cmp cydD::Tn10-1) behaves phenotypically as a ΔresDE strain with regards to growth, red pigment formation, sporulation, TMPD oxidation, acid accumulation, anaerobic growth, and alkaline phosphatase induction (Table 3). This indicates that this Tn10 insertion is sufficient to complement the cmp mutation.
Tn10-1 and Tn10-2 are inserted approximately 75° apart on the chromosome.
DNA adjacent to Tn10-2 in MH5883 was amplified by inverse PCR and sequenced. The sequence revealed that Tn10-2 was inserted after bp 922 of a 23S rRNA gene. To determine which of the 10 genomic copies of the 23S rRNA contained Tn10-2, we performed PBS1 transduction mapping, which placed Tn10-2 in rrnE-23S (54.4°) between phoB (64% linked, 53°) and purB (84% linked, 56.9°).
The use of similar techniques with strain MH5882 placed Tn10-1 after bp 316 in cydD. cydD is located at 339° in the B. subtilis genome within an operon that encodes cytochrome bd oxidase (cydAB) and a putative ABC transporter (cydCD) required for the transport of cytochrome bd ubiquinol oxidase (35). These data place Tn10-1 and Tn10-2 approximately 75° apart on the B. subtilis chromosome, indicating that both insertions cannot be cotransformed with the cmp locus. Because Tn10-1 caused a reversion from the ΔresDE cmp phenotype to the ΔresDE phenotype 100% of the time, we reasoned that cydD and cytochrome bd might be essential to the cmp mutant phenotype and that Tn10-2 was likely close to the cmp locus.
cmp maps to the phoB-purB region.
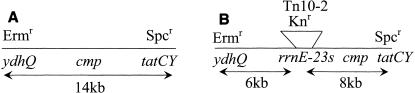
PBS1 transduction mapping showed that phoB (MH1562) and purB (1A601) cotransduced with the cmp locus at frequencies of 66 and 58%, respectively, indicating that the cmp locus was close to Tn10-2. The location of the cmp mutation was further analyzed in a three-factor cross, using MH5881 (ΔtatCY ΔydhQ) as the donor strain and MH5857 (cmp) as the recipient strain, which placed the cmp mutation between tatCY and ydhQ (Table 4 and Fig. 1). This region contains approximately 14 kb of DNA including rrnE-23S (Fig. 1A). To further define the region containing the cmp mutation, a second three-factor cross was performed with MH5874 (ΔtatCY rrnE-23S::Tn10-2) as the donor strain and MH5857 (cmp) as the recipient strain (Table 4 and Fig. 1). These results clearly demonstrated that the cmp mutation was located between tatCY and rrnE-23S (Fig. 1B).
TABLE 4.
Transformation mapping of the cmp locusa
| Transformation recipient | Transformation donor | Transformant phenotype | Frequency (%) |
|---|---|---|---|
| MH5857 (cmp) | |||
| Cross 1 | MH5881 (tatCY::SpcrydhQ::Ermr) | Spcr Ermscmp | 41 |
| Spcr Erms | 38 | ||
| Spcr Ermr | 14 | ||
| Spcr Ermrcmp | 7 | ||
| Cross 2 | MH5874 (tatCY:SpcrrrnE-23s:Knr) | Spcr Kns | 59 |
| Spcr Knr | 21 | ||
| Spcrcmp Kns | 15 | ||
| Spcrcmp Knr | 5 |
The frequency of each transformant obtained is shown. In cross 1, three-factor cross transformation using MH5881 (ΔydhQ ΔtatCY) as the donor strain and MH5857 (cmp) as the recipient strain was selected on Spcr and screened for Ermr and the cmp phenotype. In cross 2, three-factor cross transformation using MH5874 (ΔtatCY rrnE-23s::Tn10-2) as the donor strain and MH5857 (cmp) was selected on Spcr and screened for Neor and for the cmp phenotype.
FIG. 1.
Transformation mapping of the cmp locus. The frequency of each transformant obtained is shown in Table 4. Three-factor cross transformation using MH5881 (ΔydhQ ΔtatCY) as the donor strain and MH5857 (cmp) as the recipient strain was selected on Spcr and screened for Ermr and the cmp phenotype, as shown in Table 4. Three-factor cross transformation using MH5874 (ΔtatCY rrnE-23S::Tn10-2) as the donor strain and MH5857 (cmp) was selected on Spcr and screened for Neor and for the cmp phenotype, as shown in Table 4. (A) The first three-factor cross (cross 1 in Table 4) places cmp in the 14-kb region between ydhQ and tatCY. (B) The second three-factor cross (cross 2 in Table 4) localizes cmp between the Tn10-2 insertion and tatCY.
Each ΔresDE cmp strain has a mutation in YdiH.
To determine the exact location and nature of the cmp mutation, we sequenced the 8-kb region between rrnE-23S and tatCY in MH5857 (ΔresDE cmp). The primers used in this sequencing are found in Table 2, and the procedure for this sequencing is described in Materials and Methods. The only alteration from the published sequence (16) in this 8-kb region of MH5857 was a 13-bp deletion 136 bp into the coding sequence of ydiH (Table 5).
TABLE 5.
Mutations found in ydiH in ΔresDE cmp strains
| Strain | Mutation | Result |
|---|---|---|
| MH5888 | G157 → A | Asp → Asn |
| MH5859 | A insertion after 189 | Frameshift |
| MH5857 | Δ136-149 | Frameshift |
| MH5887 | A7→ T | Nonsense |
Additional isolates of ΔresDE strains containing a cmp mutation were sequenced, yielding three additional alleles of ydiH (Table 5). MH5859 contained an insertion of a single A-T base pair after bp 189 of ydiH. MH5887 contained an A-to-T change 7 bp into ydiH. Finally, MH5888 contained a G-to-A change 157 bp into ydiH.
The mutation found in MH5857 created a frameshift after the 45th codon of ydiH that leads to a stop codon after 79 amino acids (aa). The mutation found in MH5859 created a frameshift after the 63rd codon of ydiH that placed a stop codon 4 codons later. The mutation found in MH5887 created a nonsense codon after the second codon in ydiH, encoding a peptide of just 2 aa. The mutation in MH5888 creates a full-length protein but has a base pair substitution at position 157 that leads to a missense mutation changing aa 53 from aspartate to asparagine. The effect of this cmp mutation will be nonpolar with regards to tatAY and tatCY, the genes downstream of ydiH in the operon. Residue 53 appears to be crucial to the protein function, and based on the results of a BLAST search, is conserved among the 10 additional members of the family of proteins that includes YdiH. This family is composed of prokaryotic DNA-binding proteins and includes p25, a recently characterized DNA-binding protein from T. aquaticus YT-1 (6). Together, these data suggest that the nature of the cmp mutation is a loss-of-function mutation in ydiH, a gene encoding a protein with homology to a family of AT-rich DNA-binding proteins.
A loss-of-function mutation in ydiH is the sole mutation responsible for the cmp phenotype.
To determine if the loss-of-function mutation in ydiH found in our ΔresDE cmp strains is the sole mutation responsible for the cmp phenotype in that background, we created an independent disruption of ydiH (MH5891) by cloning a 516-bp internal fragment of ydiH into pDG1727 and transforming the plasmid to JH642 to yield MH5891. Transformation of chromosomal DNA from MH5202 (ΔresDE) into MH5891 (ΩydiH) yielded a resDE ydiH double mutant strain (MH5893). The growth of MH5893 (ΔresDE ΩydiH) was similar to that of a ΔresDE cmp strain on both TBAB and TBABG plates (Table 3). MH5893 (ΩydiH ΔresDE) was phenotypically identical to the ΔresDE cmp strain with regard to anaerobic growth and acid accumulation, sporulation, TMPD oxidase activity, and levels of Pho induction (Table 3). The phenotypes of strains bearing a single-crossover insertion in ydiH (MH5891) were unchanged from that of the parent strain, JH642 (Table 3). These data confirm that the nature of the cmp mutation is that of a loss-of-function mutation in ydiH.
Cytochrome bd is aberrantly expressed in a cmp mutant strain.
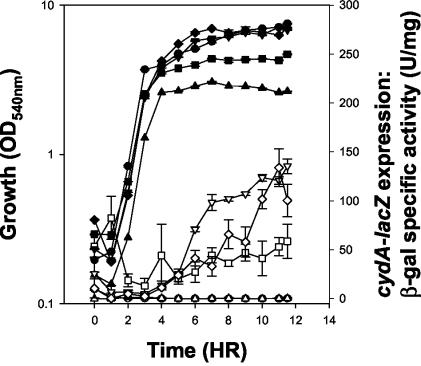
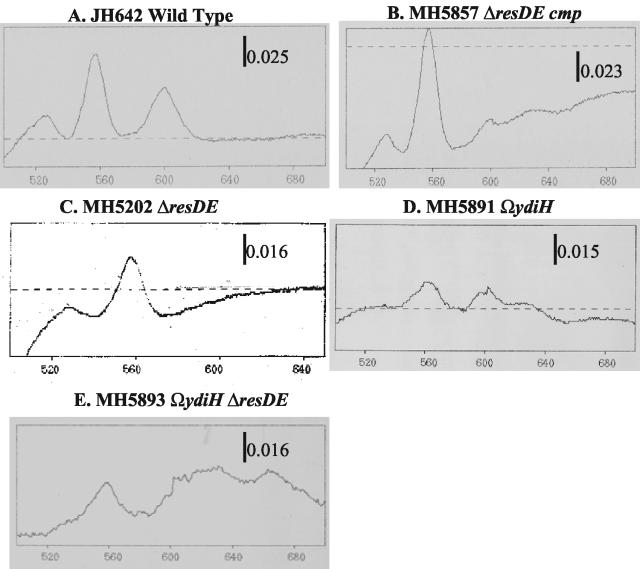
Based on the fact that a Tn10 insertion in cydD caused a suppression of the ΔresDE cmp phenotype to the ΔresDE phenotype 100% of the time, we hypothesized that cytochrome bd played an important role in the cmp phenotype. Aberrant expression of cytochrome bd in strains bearing mutations in ydiH was observed in cydA-lacZ fusion studies (Fig. 2). We found that a wild-type strain and a strain bearing a single mutation in resDE failed to express the cydA-lacZ fusion during stationary phase in cells grown in LB medium with 0.5% glucose. In contrast, strains bearing mutations in ydiH, MH5891 (ΩydiH cydA-lacZ), MH5879 (ΔresDE cmp cydA-lacZ), and MH5893 (ΔresDE ΩydiH cydA-lacZ), all express the cydA-lacZ fusion under these growth conditions. In order to confirm this aberrant expression, we examined light absorption difference (dithionite reduced minus ferricyanide oxidized) spectra from these strains grown under the same conditions to stationary phase (Fig. 3). The aberrant expression of cytochrome bd in strains bearing loss-of-function mutations in ydiH was observed in spectra from a strain bearing the cmp ΔresDE mutation (Fig. 3B), in the parental strain containing a disruption mutation in ydiH (Fig. 3D), and in the ydiH resDE double mutant strain (Fig. 3E). Figure 3B, D, and E show the spectral pattern associated with cytochrome bd: that is a characteristic trough at 650 nm, a peak at 622 nm, and a peak at 595 nm that we would normally not observe in a strain bearing a ΔresDE mutation (Fig. 3C). This expression pattern has previously been established for a strain producing cytochrome bd (35). Cytochrome bd is not present in membranes from a wild-type strain or a ΔresDE strain grown under the same conditions (Fig. 3A and C, respectively). These data suggest that cytochrome bd is being aberrantly expressed in a cmp strain which bears a loss-of-function mutation in YdiH, a member of a family of DNA-binding proteins, suggesting that YdiH might function as a repressor of the cydABCD operon.
FIG. 2.
Growth and cydA-lacZ expression from B. subtilis strains cultured for 11.5 h in LB medium supplemented with 0.5% glucose. Growth, solid symbols; β-galactosidase, open symbols. •, MH5878 (wild-type cydA-lacZ); ▴, MH5880 (ΔresDE cydA-lacZ); ▾, MH5879 (ΔresDE cmp cydA-lacZ); ▪, MH5891 (ΩydiH cydA-lacZ); ♦, MH5893 (ΔresDE ΩydiH cydA-lacZ).
FIG. 3.
Light absorption difference (dithionite reduced minus ferricyanide oxidized) spectra of membranes from strains JH642 (wild type), MH5202 (ΔresDE), MH5887 (ΔresDE cmp), MH5891 (ΩydiH), and MH5893 (ΩydiH ΔresDE). B. subtilis strains were grown in LB medium with 0.5% glucose and harvested during stationary phase. A representative spectrum is shown for each strain. (A) JH642. (B) MH5857. (C) MH5202. (D) MH5891. (E) MH5893.
ydiH binds to the cydA promoter.
Winstedt et al. (35) described primer extension experiments using mRNA harvested from cells grown to the exponential-growth phase, transition phase, or stationary phase that identified a major cydA transcript start site located at −193 bp upstream of a translational start site. A putative SigA-dependent −10 and −35 region (Fig. 4A) was found upstream of the transcriptional start site. An inverted repeat, −114 to −107, was proposed as the binding site for regulatory protein(s) (35) (Fig. 4A). To determine if YdiH's regulation was direct or indirect, binding of YdiH to the cydA promoter was tested by using gel shift assays (Fig. 4B). The YdiH-cydA DNA complex formed at a concentration of 316 nM YdiH. An increasing concentration of YdiH led to the formation of a large-molecular-weight complex, suggesting that YdiH binds to DNA at more than one site and/or YdiH oligomerized upon DNA binding. To further understand the nature of YdiH-DNA complex, we conducted DNase I protection assays (Fig. 4C). YdiH protected three regions on either strand. The regions that were protected were −28 to −54, −60 to −86, and −88 to −122 on the coding strand and −32 to −59, −65 to −91, and −94 to −126 on the noncoding strand. The regions between these three protected regions were more sensitive to DNase I digestion than that of YdiH-free sample, indicating these regions were exposed due to YdiH binding. Further DNA sequence analysis revealed three tandem 22-bp repeats, −120 to −109, −85 to −64, and −53 to −32, with a consensus sequence, 5′-TTTGTGAA(A/T/G)TA(T/C/G)TGA(G/T)CAA(A/T)(A/T/G)T-3′ (Fig. 4D). Additional gel shift assays using a cydA promoter probe (PCR product of primers FMH792 and FMH794, Fig. 4A) that lacks the YdiH binding site, showed no YdiH-cydA complex formation when YdiH concentrations as high as 1 μM were used (data not shown). A graphical description of YdiH binding relative to all transcription and translation start sites is found in Fig. 4E.
Cytochrome bd is sufficient to create the cmp mutant phenotype.
In order to confirm the role of cytochrome bd in the cmp phenotype, we created a strain in which the cydABCD operon was under the control of an inducible promoter (Pspac) in a ΔresDE background (MH5885). We then asked if expression of cytochrome bd in this background would be sufficient to create the phenotypes associated with a cmp ΔresDE strain. We found that induction of cytochrome bd was sufficient to compensate for the loss of ResD for all phenotypes normally associated with the cmp mutation (Table 3). The same strain grown without induction of cytochrome bd mimicked the phenotype of a ΔresDE strain (Table 3).
DISCUSSION
In this report, we have shown that spontaneous mutations arise in B. subtilis resDE strains that compensate for the loss of ResD for all known phenotypes that are not associated with ResD's role as a transcriptional activator. Previous reports (7) have demonstrated that strains deficient in CcdA, which is proposed to play a role in keeping two critical cysteinyls in apocytochrome C reduced, can pick up secondary mutations in either bdbC or bdbD which code for thiol-disulfide oxidoreductases. These secondary mutations complement the loss of CcdA for a number of phenotypes. An additional report (25) demonstrated that Bacillus stearothermophilus strains deficient in cytochrome caa3 have the ability to spontaneously produce cytochrome bd. It was proposed that this takes place due to a mutation in a repressor of the operon that produces cytochrome bd. However, the nature of this mutation has not been described to date.
The cmp mutation in a B. subtilis ΔresDE background is a loss-of-function mutation in ydiH. YdiH has been assigned to a family of AT-rich DNA-binding proteins based on sequence homology (16). Included in this family is p25, a protein from T. aquaticus YT-1 that has been recently characterized (6) and was shown to bind AT-rich DNA segments within its regulatory regions. During the course of characterizing the cmp mutation, we determined that cytochrome bd was aberrantly expressed in a ΔresDE cmp background (Fig. 2 and 3B), suggesting that YdiH may serve as a negative regulator of the cydABCD operon. We propose that this regulation is direct based on the results of our gel-shift and DNase I protection data (Fig. 4).
The 22-bp conserved repeats that were protected by YdiH binding (Fig. 4D) suggested that YdiH may bind DNA as a dimer to protect this relatively long region, equal to two turns of the helix. The YdiH homologue protein, p25, an AT-rich DNA-binding protein from T. aquaticus YT-1, is a dimer in solution (6).
The gel-shift pattern of the cydA promoter with increasing concentrations of YdiH suggested that multiple complexes were formed, a fact that was supported by DNase I protection at three sites on the cydA promoter by YdiH (Fig. 4B and C). The 10 to 13 bp between the direct repeats (each approximately equal to one turn of the helix) were apparently bent due to YdiH binding as they were hypersensitive to DNase I digestion. Low concentrations of YdiH initially bound to the −88 to −122 region (Fig. 4C) followed by simultaneous protection of two downstream binding sites at increasing concentrations. This may suggest sequential and/or cooperative binding of YdiH to the cydA promoter, which may be important for YdiH's function as a repressor. The cydA promoter region contains a long untranslated region of 193 bp, a length similar to the leader transcript for the trpE operon (11, 27). The data presented here are consistent with a repressor function for YdiH, which binds at three sites 71 bp downstream of the transcriptional start site. Transcription from the cydA promoter likely proceeds through the initial 71 bp until RNA polymerase encounters YdiH, leading to a loss of production of functional cytochrome bd. Thus, one of the functions of this untranslated region is to allow for repression of transcription of the operon.
In characterizing the ΔresDE cmp strain, we found that all phenotypes associated with a resDE mutation are suppressed by the cmp mutation except for ctaA expression and anaerobic growth, two phenotypes associated with ResD's role as a transcriptional activator (Table 3). This suggests that the role of the cmp mutation is not to simply create a mechanism to bypass the activity of ResD in the cell as a transcriptional activator. Rather, it is the expression of cytochrome bd that is responsible for the phenotypic changes associated with a cmp mutation. This hypothesis is strongly supported in our studies with MH5885 (ΔresDE Pspac-cydABCD). In the presence of IPTG, this strain expressed the cydABCD operon and behaved phenotypically as a strain bearing a cmp mutation. In the absence of IPTG, this strain behaves in a manner similar to a ΔresDE strain (Table 3). In the case of sporulation, previous work (34) has suggested that a heme-copper oxidase (either aa3 or caa3) is required for efficient sporulation. However, these sporulation assays were performed in NSMP medium, a medium in which cytochrome bd is poorly expressed (35). Our ΔresDE strains, which cannot make cytochrome aa3 or caa3, are sporulation defective; however, the cmp ΔresDE double mutants that aberrantly express cytochrome bd sporulate at wild-type levels, suggesting that either a heme-copper oxidase or cytochrome bd is sufficient to allow for efficient sporulation.
In an effort to characterize the gene expression patterns of B. subtilis under various respiratory conditions, Ye et al. performed microarray analysis on a strain bearing a resDE mutation grown under aerobic respiratory conditions in 2× YT medium supplemented with 1% glucose and 20 mM K3PO4 (36). They found that the cydABCD operon was induced in a strain bearing a resDE mutation compared to the wild-type strain. A possible explanation for this discrepancy is that during the course of study, their resDE strain acquired a cmp mutation (a frequent phenomenon), leading to aberrant expression of the cydABCD operon. Our studies using 2× YT medium supplemented with 1% glucose and 20 mM K3PO4 demonstrated that cydA-lacZ expression was reduced in a ΔresDE strain in comparison to that in the wild-type strain (data not shown).
Our analysis of B. subtilis strains bearing cmp mutations has provided insight into the function of YdiH. Based on the fact that the cmp mutation appears to be a loss-of-function mutation in ydiH (putative DNA-binding protein) and that strains bearing the cmp mutation aberrantly express cytochrome bd, we proposed that YdiH functions as a negative regulator for cydABCD expression. DNA-binding assays have demonstrated that YdiH directly binds the cydABCD promoter region. This binding takes place at three tandem repeats which are separated by 10 to 13 bp. These direct repeats are located in the long untranslated region of the cydABCD promoter. The aberrant expression of cytochrome bd in a ΔresDE background is sufficient to suppress the phenotypes associated with the loss of cyotochromes aa3 and caa3 in this background. Future studies will determine how cmp mutations bypass the requirement for ResD for full alkaline phosphatase induction (30) and what insight we can gain from our understanding of the cmp mechanism into the role of ResD in alkaline phosphatase induction, where it is normally required for 80% of the wild-type alkaline phosphatase response.
Acknowledgments
This work was supported by National Institutes of Health grant GM-33471 to F.M.H.
We thank Jörg Müller, Nick Kapp, Ruth Chestnut, and Gufou Sun for strains. We thank the Bacillus genetic stock center for PBS1 phage.
REFERENCES
- 1.Bisschop, A., and W. N. Konings. 1976. Reconstitution of reduced nicotinamide adenine dinucleotide oxidase activity with menadione in membrane vesicles from the menaquinone-deficient Bacillus subtilis aroD. Relation between electron transfer and active transport. Eur. J. Biochem. 67:357-365. [DOI] [PubMed] [Google Scholar]
- 2.Carls, R. A., and R. S. Hanson. 1971. Isolation and characterization of tricarboxylic acid cycle mutants of Bacillus subtilis. J. Bacteriol. 106:848-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chary, V. K., E. I. Amaya, and P. J. Piggot. 1997. Neomycin- and spectinomycin-resistance replacement vectors for Bacillus subtilis. FEMS Microbiol. Lett. 153:135-139. [DOI] [PubMed] [Google Scholar]
- 4.Cutting, S. M., and P. B. Vander Horn. 1990. Genetic analysis, p. 24-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Inc., New York, N.Y.
- 5.de Vrij, W., B. van den Burg, and W. N. Konings. 1987. Spectral and potentiometric analysis of cytochromes from Bacillus subtilis. Eur. J. Biochem. 166:589-595. [DOI] [PubMed] [Google Scholar]
- 6.Du, X., and J. J. Pene. 1999. Identification, cloning and expression of p25, an AT-rich DNA-binding protein from the extreme thermophile, Thermus aquaticus YT-1. Nucleic Acids Res. 27:1690-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erlendsson, L. S., and L. Hederstedt. 2002. Mutations in the thiol-disulfide oxidoreductases BdbC and BdbD can suppress cytochrome c deficiency of CcdA-defective Bacillus subtilis cells. J. Bacteriol. 184:1423-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrari, E., S. M. H. Howard, and J. A. Hoch. 1986. Effect of stage 0 sporulation mutations on subtilisin expression. J. Bacteriol. 166:173-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guerout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan, D. 1985. Techniques for transformation of E. coli, p. 109-135. In D. M. Glover (ed.), DNA cloning II. A practical approach. IRL Press, Washington, D.C.
- 11.Henner, D. J., L. Band, and H. Shimotsu. 1985. Nucleotide sequence of the Bacillus subtilis tryptophan operon. Gene 34:169-177. [DOI] [PubMed] [Google Scholar]
- 12.Homuth, G., A. Rompf, W. Schumann, and D. Jahn. 1999. Transcriptional control of Bacillus subtilis hemN and hemZ. J. Bacteriol. 181:5922-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hulett, F. M., C. Bookstein, and K. Jensen. 1990. Evidence for two structural genes for alkaline phosphatase in Bacillus subtilis. J. Bacteriol. 172:735-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jongbloed, J. D., U. Martin, H. Antelmann, M. Hecker, H. Tjalsma, G. Venema, S. Bron, J. M. van Dijl, and J. Muller. 2000. TatC is a specificity determinant for protein secretion via the twin-arginine translocation pathway. J. Biol. Chem. 275:41350-41357. [DOI] [PubMed] [Google Scholar]
- 15.Kapp, N. 1992. Analysis of the phosphate stimulon of Bacillus subtilis. Ph.D. thesis. University of Illinois at Chicago, Chicago.
- 16.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 17.Liu, X., and H. W. Taber. 1998. Catabolite regulation of the Bacillus subtilis ctaBCDEF gene cluster. J. Bacteriol. 180:6154-6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mueller, J. P., and H. W. Taber. 1989. Isolation and sequence of ctaA, a gene required for cytochrome aa3 biosynthesis and sporulation in Bacillus subtilis. J. Bacteriol. 171:4967-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakano, M. M., T. Hoffmann, Y. Zhu, and D. Jahn. 1998. Nitrogen and oxygen regulation of Bacillus subtilis nasDEF encoding NADH-dependent nitrite reductase by TnrA and ResDE. J. Bacteriol. 180:5344-5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakano, M. M., G. Zheng, and P. Zuber. 2000. Dual control of sbo-alb operon expression by the Spo0 and ResDE systems of signal transduction under anaerobic conditions in Bacillus subtilis. J. Bacteriol. 182:3274-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakano, M. M., Y. Zhu, M. Lacelle, X. Zhang, and F. M. Hulett. 2000. Interaction of ResD with regulatory regions of anaerobically induced genes in Bacillus subtilis. Mol. Microbiol. 37:1198-1207. [DOI] [PubMed] [Google Scholar]
- 22.Nicholson, W., and P. Setlow. 1990. Sporulation, germination, and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Inc., New York, N.Y.
- 23.Paul, S., X. Zhang, and F. M. Hulett. 2001. Two ResD-controlled promoters regulate ctaA expression in Bacillus subtilis. J. Bacteriol. 183:3237-3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perkins, J. B., A. Sloma, T. Hermann, K. Theriault, E. Zachgo, T. Erdenberger, N. Hannett, N. P. Chatterjee, V. Williams II, G. A. Rufo, Jr., R. Hatch, and J. Pero. 1999. Genetic engineering of Bacillus subtilis for the commercial production of riboflavin. J. Ind. Microbiol. Biotechnol. 22:8-18. [Google Scholar]
- 25.Sakamoto, J., A. Matsumoto, K. Oobuchi, and N. Sone. 1996. Cytochrome bd-type quinol oxidase in a mutant of Bacillus stearothermophilus deficient in caa3-type cytochrome c oxidase. FEMS Microbiol. Lett. 143:151-158. [DOI] [PubMed] [Google Scholar]
- 26.Santana, M., F. Kunst, M. F. Hullo, G. Rapoport, A. Danchin, and P. Glaser. 1992. Molecular cloning, sequencing, and physiological characterization of the qox operon from Bacillus subtilis encoding the aa3-600 quinol oxidase. J. Biol. Chem. 267:10225-10231. [PubMed] [Google Scholar]
- 27.Shimotsu, H., and D. J. Henner. 1984. Characterization of the Bacillus subtilis tryptophan promoter region. Proc. Natl. Acad. Sci. USA 81:6315-6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimotsu, H., and D. J. Henner. 1986. Construction of a single-copy integration vector and its use in analysis of regulation of the trp operon of Bacillus subtilis. Gene 43:85-94. [DOI] [PubMed] [Google Scholar]
- 29.Steinmetz, M., and R. Richter. 1994. Easy cloning of mini-Tn10 insertions from the Bacillus subtilis chromosome. J. Bacteriol. 176:1761-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun, G., S. M. Birkey, and F. M. Hulett. 1996. Three two-component signal-transduction systems interact for Pho regulation in Bacillus subtilis. Mol. Microbiol. 19:941-948. [DOI] [PubMed] [Google Scholar]
- 31.Sun, G., E. Sharkova, R. Chesnut, S. Birkey, M. F. Duggan, A. Sorokin, P. Pujic, S. D. Ehrlich, and F. M. Hulett. 1996. Regulators of aerobic and anaerobic respiration in Bacillus subtilis. J. Bacteriol. 178:1374-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trieu-Cuot, P., C. Poyart-Salmeron, C. Carlier, and P. Courvalin. 1990. Nucleotide sequence of the erythromycin resistance gene of the conjugative transposon Tn1545. Nucleic Acids Res. 18:3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandeyar, M. A., and S. A. Zahler. 1986. Chromosomal insertions of Tn917 in Bacillus subtilis. J. Bacteriol. 167:530-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winstedt, L., and C. von Wachenfeldt. 2000. Terminal oxidases of Bacillus subtilis strain 168: one quinol oxidase, cytochrome aa3 or cytochrome bd, is required for aerobic growth. J. Bacteriol. 182:6557-6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winstedt, L., K.-I. Yoshida, Y. Fujita, and C. von Wachenfeldt. 1998. Cytochrome bd biosynthesis in Bacillus subtilis: characterization of the cydABCD operon. J. Bacteriol. 180:6571-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye, R. W., W. Tao, L. Bedzyk, T. Young, M. Chen, and L. Li. 2000. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J. Bacteriol. 182:4458-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zamboni, N., N. Mouncey, H. P. Hohmann, and U. Sauer. 2003. Reducing maintenance metabolism by metabolic engineering of respiration improves riboflavin production by Bacillus subtilis. Metab. Eng. 5:49-55. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, X., and F. M. Hulett. 2000. ResD signal transduction regulator of aerobic respiration in Bacillus subtilis: ctaA promoter regulation. Mol. Microbiol. 37:1208-1219. [DOI] [PubMed] [Google Scholar]