Abstract
Background
To provide a point of reference to study the epidemiology and clinical expression of canine babesiosis in China.
Methods
A total of 30 dogs infected with canine babesiosis were evaluated by mean of clinical history, physical examination, hematological, restriction fragment length polymorphism of PCR products (PCR-RFLP) and sequencing analysis.
Result
The most prevalent clinical abnormalities were lethargy (100%), anorexia (100%), pale or icteric mucous membranes (80%), fever (70%) and dark urine (70%). Hematology parameters revealed that anemia and thrombocytopenia were the major abnormalities in blood of dogs infected with canine babesia. The results of PCR-RFLP and sequencing analysis indicated that B. gibsoni was the main species responsible for canine babesiosis cases at the time of the study in Nanjing, China.
Conclusions
The results provide valuable information for better understanding of the epidemiology of canine babesiosis in China.
Keywords: Canine babesiosis, Babesia gibsoni, PCR-RFLP, Epidemiology, Hematology
Introduction
Babesiosis is one of the most common infections of free-living animals worldwide and is gaining increasing interest as an emerging zoonosis in humans. Canine babesiosis is important tick-borne protozoal disease of dogs with a worldwide distribution. The parasites replicate in the red blood cells and initiate a mechanism of antibody-mediated cytotoxic destruction of circulating erythrocytes (1). The severity of babesiosis in dogs ranges from subclinical infection, the development of mild anemia to widespread organ failure and death. The diagnosis of infection with Babesia is usually based on the detection of pathogens in peripheral blood under a microscope.
All large Babesia (3-5 μm) were designated B. canis, whereas all small Babesia were thought to be B. gibsoni (0.5-2.5 μm) (2). Large B. canis was divided into three different species, namely B. canis vogeli, B. canis canis and B. canis rossi (3). B. canis vogeli is the most widespread canine piroplasm which found in tropical, subtropical and Mediterranean regions. B. canis canis was found in central Europe. B. canis rossi was found in Sub-Saharan and South Africa. B. gibsoni occurred in Asia, North American, and Northern and Eastern Africa (4). Canine babesiosis has varying clinical signs which depend on Babesia species, host immunity, age and concurrent diseases. The most common found in canine babesiosis are fever, anemia, jaundice, hemoglobinuria, depression, and weight loss (5).
To provide a point of reference to study the epidemiology and clinical expression of canine babesiosis in China, we evaluated 30 cases of canine babesiosis by mean of clinical history, physical examination, hematological, restriction fragment length polymorphism of PCR products (PCR-RFLP) and sequencing analysis.
Materials and Methods
Sample collection and laboratory analysis
Thirty dogs of different breeds, ages naturally infected with canine Babesia were collected from the Animal Hospital of Nanjing Agricultural University between September 2012 and September 2013. A routine physical examination performed beforehand. Basic information on the breed, age, gender, tick infestation history and access to the outdoors were provided by the owners. EDTA-anticoagulated blood was collected for complete blood counts, smear observations and PCR analysis. Complete blood count (CBC) was assessed with an automatic cell counter (ABX Micros 60, France). The following parameters were assessed: red blood cell count (RBC), hemoglobin concentration (HGB), hematocrit (HCT), mean corpuscular volume (MCV), mean corpuscular hemoglobin (MCH), mean corpuscular hemoglobin concentration (MCHC), red distribution width (RDW), PLT count, white blood cell count (WBC), WBC differential count including neutrophils, lymphocytes, monocytes. Blood smears were prepared, air dried, stained with Giemsa solution. Blood smears were examined by light microscopy at × 100.
DNA extraction and PCR amplification
Genomic DNA was isolated from 200 μL aliquots of EDTA-anticoagulated blood using TIANamp Genomic DNA Kit (TIANGEN, China), according to the manufacturer’s instruction. The concentration of DNA was estimated using optical density (O.D.) readings at 260 nm, and the purity of DNA was checked by calculating the ratio of the O.D. readings at 260 nm and 280 nm, measured using a spectrophotometer (Eppendorf, Germany).
PCR was conducted with a set of primers (forward primer B.com 339-F: 5′-GTCTTGTAATTG GAATGATGGTGAC-3′, reverse primer B.com 339-R: 5′-ATGCCCCCAACCGTTCCTATTA-3′) that amplified 340 bp fragment of the 18S rRNA genes from B. canins and B. givsoni but not mammalian DNA (6). Amplification of the full length 18S rRNA genes (1700 bp) were performed using universal primers: the forward primer B.com 1700-F: 5′- AACCTGGTTG ATCCTGCCAGTAGTCAT-3′ and the reverse primer B.com 1700-F 5′- GAATGATCCTTCCGCA GGTTCACCTAC-3′ (7). PCR reaction mixtures contained the following components: 1 μL genomic DNA template (0.5 μg/μL), 12.5 μL of Premix Ex Taq (Takara, Dalian), 0.5 μL of each primer (10 μmol/L), and 10.5 μL water. Touchdown PCR amplification was performed in Veriti 96-well Thermal Cycler (Applied Biosystems, USA). The initial touch down cycle was denaturation at 96 °C for 30 s, annealing at 65 °C for 30 s, and extension at 72 °C for 30 s (90 s for full length 18S rRNA genes). During the touchdown phase, the annealing temperature was decreased at the rate of 1 °C for every cycle of the amplification reaction. After 10 touchdown cycles, 25 standard PCR cycles were performed under the following conditions: denaturation at 96 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 30 s (90 s for full length 18S rRNA genes), and a final extension at 72 °C for an additional 5 min. PCR products were resolved by electrophoresis through a 2% agarose gel for about 30 min at 120 V in Tris-acetate-EDTA buffer.
Restriction digestion and sequencing analysis
PCR products were purified using the TIANgel Midi Purification Kit (TIANGEN, China). The 339 bp PCR product was subjected to restriction enzyme digestion using Hinf I according to manufacturer’s instructions (Takara, Dalian) and visualized by the 12% denaturing polyacrylamide gel electrophoresis. The 1700 bp PCR product was cloned in pMD18-T vector (Takara, Dalian) according to the manufacturer’s instructions. The sequencing of positive plasmid was conducted by Invitrogen, using both the universal forward and reverse primers of pUC plasmid. The sequenced products were analyzed using SeqMan (DNASTAR Inc.) Sequence alignments were performed using BLASTN, which is available on the NCBI server (http://www.ncbi.nlm.nih.gov).
Results
A total of 30 dogs infected with canine babesiosis were diagnosed by clinical examination and observation of intra-erythrocytic inclusions within blood smears. Giemsa-stained blood smears showed the presence of small pear-shaped parasites (Fig. 1). Of the 30 dogs, males (80%) were more affected than females (20%). The median age was 48 moths (range, 2-108). The most prevalent clinical abnormalities were lethargy (100%), anorexia (100%), Pale or icteric mucous membranes (80%), fever (70%) and dark urine (70%). Results of hematology parameters are shown in Table 1. Thirty dogs (100%) had the RBC, HGB and HCT values below the reference values. Twenty-night dogs had the PLT values below the reference values, while only one case (3.3%) presented values of PLT within the normal values. The mean values of the other parameters were normal in dogs infected by canine babesiosis.
Fig. 1.
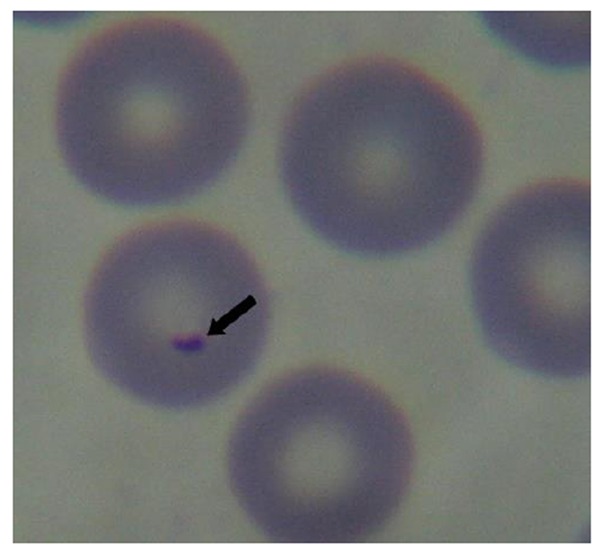
Morphology of B. gibsoni (× 1000)
Table 1.
Hematological parameters in 30 dogs infected with Canine babesiosis
| Parameters | Mean ± SD | Minimum | Maximum | Reference | |
|---|---|---|---|---|---|
| RBC | × 106/μl | 2.2 ± 1.1 | 0.75 | 4.3 | 5.5-8.5 |
| HGB | g/dl | 4.8 ± 2.2 | 1.8 | 9.6 | 12.3-18 |
| HCT | % | 15.3 ± 6.7 | 4.6 | 27.9 | 37-55 |
| MCV | fl | 72.2 ± 10.0 | 56.6 | 97 | 60-77 |
| MCH | pg | 22.61 ± 2.5 | 19.3 | 28.4 | 18.5-30 |
| MCHC | g/dl | 30.8 ± 2.9 | 22.9 | 36 | 30-37.5 |
| RDW | % | 17.7 ± 3.5 | 12.4 | 24.7 | 14.7-17.9 |
| WBC | ×103/μl | 14.2 ± 7.9 | 4.6 | 38.8 | 5.5-16.9 |
| %NEU | % | 63.5 ± 26.7 | 2.72 | 92.1 | 60-80 |
| %LYM | % | 25.1 ± 17.7 | 6 | 88.5 | 12-30 |
| %MONO | % | 8.4 ± 10.1 | 0.6 | 43.5 | 3-10 |
| NEU | ×103/μl | 9.6 ± 6.7 | 0.6 | 31.8 | 2-12 |
| LYM | ×103/μl | 3.2 ± 2.8 | 0.8 | 15.2 | 0.5-4.9 |
| MONO | ×103/μl | 1.4 ± 2.6 | 0.05 | 13.1 | 0.3-2 |
| PLT | ×103/μl | 63.5 ± 60.2 | 2 | 271 | 175-500 |
As shown in Fig. 2, the primer set B.com 339-F and B.com 339-R successfully amplified an approximately 339 bp DNA fragment from all 30 dogs infected with canine babesiosis. The Babesia species can be distinguished based on the polymorphism of Hinf I restriction site within the 339 bp fragment of the 18S rRNA genes from B. canins (no Hinf I restriction site), and B. gibsoni (Hinf I restriction site). All the 339 bp DNA fragment from30 dogs infected with canine babesiosis could be digested into two DNA fragment (185 bp and 154bp) with the Hinf I, which indicated that the parasites were B. gibsoni.
Fig. 2.
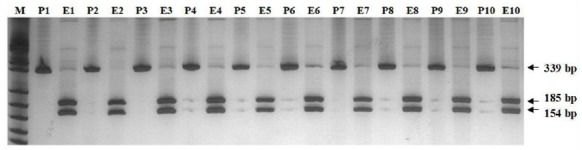
PCR-RFLP analysis with Hinf I for 339 bp PCR products of 18S rRNA/ M:marker;P:PCR products;E:Digestion products with the Hinf I
The 1700 bp PCR products amplified from 3 dogs were purified and cloned in the pMD18-T vector. The sequences of three full length 18S rRNA genes had the same sequences and had been submitted to EMBL sequence databases (Accession No. HG328235, HG328236 and HG328237).
Phylogenetic analysis of the three full length 18S rRNA genes showed that the sequences exhibited a high level of homology (99-100%) with the other B. gibsoni isolated from Japan (Accession No. AB118032, 271082) and USA (Accession No. EU08467, AF396748). The results of sequences analysis were shown that those dogs were infected with B. gibsoni.
Discussion
The primary goal of this research was to investigate the epidemiology and clinical expression of canine babesiosis in Nanjing, China. In the present study, 30 cases infected with Babesia gibsoni were confirmed by mean of clinical history, physical examination, hematological, restriction fragment length polymorphism of PCR products (PCR-RFLP) and sequencing analysis. The cases of canine babesiosis reported in China have been considered to be caused by B. canis vogeli and B. gibsoni. B. gibsoni is the majority parasites infected dog in north of China, such as Jiangsu, Jilin, Henan province. Prevalence of B. canis vogeli in dogs was only reported in Guangdong province, south of China (8). B. gibsoni has a wide geographical distribution and has been reported from Asia, Africa, Europe, the Middle East and Northern America. Dogs are reservoir for B. gibsoni, specific fighting dogs in particular, and not an endemic species of tick. It is speculated that B. gibsoni will be reported eventually from all countries where dogs fighting is practiced (4).
Microscopical test is the simplest and most accessible diagnostic test for detecting intraerythrocytic parasite. But the similarity between species and subspecies has been a limiting factor (9). Serological test is a useful assay for detection of infected dogs, especially if combined with PCR. However, poor specificity, inability to differentiate acute from chronic infection were major limitation of serological tests (10). PCR method represents a powerful tool not only for genotype identification but also in cases when blood smears do not provide a certain diagnosis. Recent advances in molecular methodology (PCR-based methods and subsequent DNA sequencing) have made it possible to detect and identify piroplasms with greater sensitivity and specificity than with traditional methods (11). In this study, microscopical test, PCR-RFLP and sequencing analysis were used for B. gibsoni detection.
Most of the dogs infected with B. gibsoni showed abnormalities such as lethargy, anorexia, fever, anemia, and thrombocytopenia, which were in accordance with other researches (5). The most common abnormalities of hematological parameters were RBC, HGB and HCT, which were observed in all affected dogs. The most consistent and severe change in blood picture observed in the course of canine babesiosis is thrombocytopenia (1, 12). Thrombocytopenia was present in 96.7% of dogs in this study. Leukocyte counts are dependent on more various factors than just Babesia infection. These values are the most inconsistent and the less reliable in terms of characterizing the organism’s reaction in canine babesiosis (12).
The morphological similarity between species and subspecies of Babesia has led to much confusion over accurate diagnosis using light microscopy (13). Moreover, the detection of Babesia parasites is difficult in dogs with unapparent or chronic infection since the level of parasitemia is very low (14). PCR, in combination with restriction fragment length polymorphism (RFLP) analysis offers an effective and rapid means of discriminating between species (13, 15). In this study, all 339 bp DNA fragment from 30 dogs infected with canine babesiosis could be digested with the Hinf I, which indicated that the parasites were B. gibsoni. Sequencing of the full length 18S rRNA genes provides a reliable method for both species identification and phylogentic analysis and allows new sequences to be compared with the sequences of other Babesia (7, 16). The results of this study revealed that canine babesiosis in Nanjing, China were due to infection of B. gibsoni.
Conclusion
In the present study, the most common abnormalities detected in blood of dogs infected with B. gibsoni were anemia and thrombocytopenia. The results of PCR-RFLP and sequencing analysis indicated that B. gibsoni was the main species responsible for canine babesiosis cases at the time of the study in Nanjing, China.
Acknowledgments
This study was supported by a grant from A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) and Nature Science Foundation of Jiangsu Province (BK20130686). The authors declare that there is no conflict of interests.
References
- 1.Zygner W, Gojska O, Rapacka G, Jaros D, Wedrychowicz H. Hematological changes during the course of canine babesiosis caused by large Babesia in domestic dogs in Warsaw (Poland) Vet Parasitol. 2007;145:146–151. doi: 10.1016/j.vetpar.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 2.Boozer AL, Macintire DK. Canine babesiosis. Vet Clin N Am-Small. 2003;33:885–904. doi: 10.1016/s0195-5616(03)00039-1. [DOI] [PubMed] [Google Scholar]
- 3.Costa-Junior LM, Zahler-Rinder M, Ribeiro MFB, et al. Use of a Real Time PCR for detecting subspecies of Babesia canis. Vet Parasitol. 2012;188:160–163. doi: 10.1016/j.vetpar.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 4.Irwin P. Canine babesiosis: from molecular taxonomy to control. Parasite and Vector. 2009;2(Suppl 1):S4. doi: 10.1186/1756-3305-2-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rene-Martellet M, Chene J, Chabanne L, Chalvet-Monfray K, Bourdoiseau G. Clinical signs, seasonal occurrence and causative agents of canine babesiosis in France: Results of a multiregional study. Vet Parasitol. 2013;197:50–58. doi: 10.1016/j.vetpar.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 6.Birkenheuer AJ, Levy MG, Breitschwerdt EB. Development and evaluation of a seminested PCR for detection and differentiation of Babesia gibsoni (Asian genotype) and B-canis DNA in canine blood samples. J Clin Microbiol. 2003;41:4172–4177. doi: 10.1128/JCM.41.9.4172-4177.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caccio SM, Antunovic B, Moretti A, et al. Molecular characterisation of Babesia canis canis and Babesia canis vogeli from naturally infected European dogs. Vet Parasitol. 2002;106:285–292. doi: 10.1016/s0304-4017(02)00112-7. [DOI] [PubMed] [Google Scholar]
- 8.Li GQ, Zhang HJ, Zhang GL, et al. Molecular identification and phylogenetic analysis of Babesia vogeli from dogs. Chinese Veterinary Science. 2011;41:245–249. [Google Scholar]
- 9.Sadeghi Dehkordi Z, Zakeri S, Nabian S, et al. Molecular and biomorphometrical identification of ovine babesiosis in Iran. Iranian J Parasitol. 2010;5(4):21–30. [PMC free article] [PubMed] [Google Scholar]
- 10.Yamane I, Thomford JW, Gardner IA, et al. Evaluation of the indirect fluorescent antibody test for diagnosis of Babesia gibsoni infections in dogs. Am J Vet Res. 1993;54:1579–1584. [PubMed] [Google Scholar]
- 11.Martin AR, Dunstan RH, Roberts TK, et al. Babesia canis vogeli: a novel PCR for its detection in dogs in Australia. Exp Parasitol. 2006;112:63–65. doi: 10.1016/j.exppara.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Fabisiak M, Sapierzynski R, Klucinski W. Analysis of haematological abnormalities observed in dogs infected by a large Babesia. B Vet I Pulawy. 2010;54:167–170. [Google Scholar]
- 13.Jefferies R, Ryan UM, Irwin PJ. PCR-RFLP for the detection and differentiation of the canine piroplasm species and its use with filter paper-based technologies. Vet Parasitol. 2007;144:20–27. doi: 10.1016/j.vetpar.2006.09.022. [DOI] [PubMed] [Google Scholar]
- 14.Ikadai H, Tanaka H, Shibahara N, et al. Molecular evidence of infections with Babesia gibsoni parasites in Japan and evaluation of the diagnostic potential of a loop-mediated isothermal amplification method. J Clin Microbiol. 2004;42:2465–2469. doi: 10.1128/JCM.42.6.2465-2469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gulanber A, Gorenflot A, Schetters TPM, Carcy B. First molecular diagnosis of Babesia vogeli in domestic dogs from Turkey. Vet Parasitol. 2006;139:224–230. doi: 10.1016/j.vetpar.2006.02.035. [DOI] [PubMed] [Google Scholar]
- 16.Inokuma H, Yoshizaki Y, Matsumoto K, et al. Molecular survey of Babesia infection in dogs in Okinawa, Japan. Vet Parasitol. 2004;121:341–46. doi: 10.1016/j.vetpar.2004.03.012. [DOI] [PubMed] [Google Scholar]


