Abstract
Background
Naegleria is a free-living amoeba, and pathogenic Naegleria may pose a health risk to people exposed to recreational water. Our objective in this study was to determine if there are pathogenic amoebae in environmental water samples from Changchun, Northeastern China.
Methods
During July to September 2012, a total of 70 water samples were collected from Changchun, Northeastern China, and Naegleria was enriched by in vitro culture and detected by PCR using Naegleria genus-specific primers. Resulting PCR products were sequenced and phylogenetically analyzed to identify Naegleria species.
Results
Naegleria was detected in 65 (92.9%) of 70 water samples. DNA sequence and phylogenetic analyses based on the internal transcribed spacer (ITS) rDNA sequences revealed four Naegleria species, including N. pagei (n = 24) and N. Australiensis (n = 18), N. clarki (n = 13) and N. gruberi (n = 10), in which N. australiensis is pathogenic to mice. But the pathogenic species N. fowleri was not detected.
Conclusion
This is the first report on Naegleria species in Northeastern China, showing that almost all environmental water samples were contaminated with Naegleria, including N. pagei, N. Australiensis, N. clarki and N. gruberi, which should be considered a potential public health threat.
Keywords: Naegleria, PCR, Identification, Changchun, China
Introduction
Free-living amoebae (FLA), feeding on bacteria, fungi, and algae, are ubiquitous in soil and aquatic environments. Of the hundreds of FLA species, Naegleria is one of the predominant FLA found in lakes, rivers, swimming pools, hot spring, geothermal water, discharge from industrial plants (1). More than 30 species of the genus Naegleria, belonging to the family Vahlkampfiidae, have been identified using molecular techniques, in which N. fowleri is the only species pathogenic to humans (2). N. fowleri is transmitted via the nasal mucosa and the olfactory nerve to the brain, causing primary amoebic meningoencephalitis (PAM), an acute and rapidly fatal disease of people following exposure to polluted water (3).
The human case of PAM was first described in Australia in 1965, subsequently found in Florida and Texas in the United States (US). A retrospective study has indicated that the earliest case occurred in Richmond, Virginia from 1951 to 1952 (4). So far approximately 300 cases have been documented worldwide, mostly in USA, Australia and Europe (5, 6). These diseases are almost uniformly fatal with only several survivors. In Asia, the disease has been found in Thailand (7, 8), India (9), Iran (10), and Pakistan (11). In Pakistan, at least 10 people died of the disease in 2009 (12). In China, only 5 cases had been reported in Henan, Hebei and Hainan provinces, respectively, before 2005 (13). However, some cases of PAM may be unreported, as it is difficult to diagnose.
Our objective in this study was to determine if there are pathogenic amoebae in environmental water samples from Changchun, Northeastern China by microbial culture combined with molecular methods.
Materials and Methods
Sample collection
The water samples were collected from Changchun Park (CCP), Zhaoyang Park (ZP), Jingyue Park (JP), Nanhu Park (NP), Yitong River (YR), Xinlicheng River (XR), Children Park (CDP) in Changchun City (125.35°E; 43.88°N), Jilin Province, Northeastern China. The water samples (2 L) were collected within 10 cm of the water surface using a sterile polypropylene bottle and transported to the laboratory for subsequent analyses.
Naegleria detection
To investigate Naegleria in environmental water body, water samples were filtered through 45-mm diameter cellulose nitrate membranes (Pall, USA) with a pore size of 5 μm (14). The water samples were filtered through three to six cellulose nitrate membranes, which were subsequently inverted and placed onto 1.5% non-nutrient agar (NNA) plates containing a lawn of Escherichia coli (15, 16). The number of cellulose nitrate membranes used for each sample depended on the turbidity of the water sample. The plates were sealed and incubated at 37°C for 14 days.
The positive Naegleria-like samples were detected by PCR using Naegleria genus-specific primers (5'-GAACCTGCGTAGGGATCATTT-3' and 5'-TTTCTTTTCCTCCCCTTATTA-3'), and Naegleria fowleri species-specific primers (5'-GTGAAAACCTTTTTTCCATTTACA-3' and 5'-AAATAAAAGATTGACCATTTGAAA-3') (17, 18). PCR reactions were performed as described as elsewhere (19). Then PCR products were separated on a 1% agarose gel. Products were visualized by ethidium bromide staining and imaged under UV light.
Sequencing and phylogenetic analysis
Resulting PCR products were sequenced and compared with each other and with reference sequences. Phylogenetic analyses based on the ITS and 5.8S sequences were conducted using the software MEGA 4 (http://www.megaso-ftware.net/) (20). The reliability of branches in the tree was assessed by bootstrap analysis with 1,000 replicates.
For comparative phylogenetic analysis, the following sequences were retrieved from the GenBank: N. americana (AJ566623), N. andersoni (X96572), N. australiensis (AY293307), N. canariensis (AJ973124), N. clarki (GU597045), N. fowleri (X96567), N. galeacystis (X96578), N. gruberi (AJ132031), N. indonesiensis (AJ243444), N. italica (X96574), N. jamiesoni (X96570), N. laresi (AJ566630), N. lovaniensis (X96568), N. minor (X96577), N. pagei (AJ566633), N. peruana (AJ785757), N. philippinensis (AM167890), N. pussardi (X96571), N. tihangensis (AJ566631).
Results
A total of 70 environmental water samples were collected from CCP, ZP, JP, NP, YR, XR, and CDP in Changchun, Northern China, 10 samples in each site (Table 1). The water samples were filtered through cellulose nitrate membranes, and cultured on NNA plates with Escherichia coli at 37°C. Naegleria-like trophozoites were normally observed after 48 or 72 hours of culture, and the cyst stage could be seen after at least three days. The flagellate stage could not usually be seen unless induced by sterile distilled water to the surface agar. After 14 days of culture, 65 water samples (92.9%) were found positive for Naegleria-like trophozoites. The 65 isolates were further identified by PCR using N. fowleri species-specific primers and Naegleria genus-specific primers.
Table 1.
Naegleria species in environmental water samples from Changchun, Northeastern China
| Sampling site | No. of sample | Naegleria species (%) | |||
|---|---|---|---|---|---|
| N. australiensis | N. pagei | N. clarki | N. gruberi | ||
| Changchun Park (CCP) | 10 | 4 (40.0) | 5 (50.0) | 0 | 0 |
| Zhaoyang Park (ZP) | 10 | 0 | 10 (100.0) | 0 | 0 |
| Jingyue Park (JP) | 10 | 1 (10.0) | 3 (30.0) | 6 (60.0) | 0 |
| Nanhu Park (NP) | 10 | 4 (40.0) | 3 (30.0) | 0 | 0 |
| Yitong River (YR) | 10 | 9 (90.0) | 0 | 0 | 0 |
| Xinlicheng Reservoir (XR) | 10 | 0 | 3 (30.0) | 7 (70.0) | 0 |
| Children Park (CDP) | 10 | 0 | 0 | 0 | 10 (100.0) |
| Total | 70 | 18 (25.7) | 24 (34.3) | 13 (18.6) | 10 (14.3) |
The N. fowleri species-specific primers did not show any amplicon bands against all 65 isolates. In contrast, Naegleria genus-specific primers produced amplicons against all the isolates, which could be differentiated into 4 groups represented by 379-bp, 495-bp, 460-bp and 395-bp, respectively, (Fig. 1). Resulting PCR products were sequenced and compared with each other and with reference sequences. The unique sequences have been submitted to the GenBank database (JX413059, JX267141, JX294421, and JX861079). DNA sequence showed the presence of four Naegleria species in Changchun, including N. gruberi, N. clarki, N. pagei, and N. Australiensis (Fig. 2). Among the 65 positive samples, 24 (34.3%) were N. pagei, which were the dominant species; 18 (25.7%) were N. Australiensis; 13 (18.6%) were N. clarki; and 10 (14.3%) were N. gruberi.
Fig. 1.
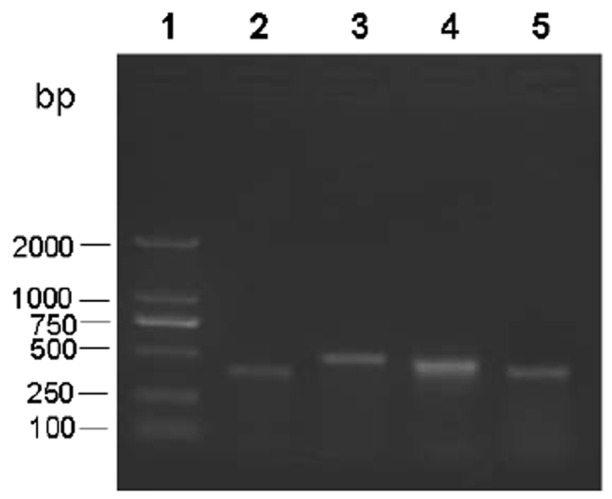
Amplicon bands revealed by genus-specific primers against Naegleria-like isolates. Lane 1 represents the 2,000-bp DNA ladder (Takara); Lanes 2–5 represent the amplicon bands with 379-bp, 495-bp, 460-bp and 395-bp against the different water samples using the Naegleria genus-specific primers, which were grouped into N. gruberi, N. clarki, N. pagei, and N. Australiensis, respectively
Fig. 2.

Phylogenetic relationships of Naegleria identified in this study. A neighbor-joining analysis of the internal transcribed spacer (ITS) rDNA sequences was conducted using the software MEGA 4. Numbers on branches are percent bootstrapping values (>50) using 1,000 replicates. The triangles represent the Naegleria species in the present study
Discussion
In this study, four Naegleria species, including N. gruberi, N. clarki, N. pagei, and N. Australiensis, were detected from 65 (92.9%) water samples in Changchun, Northeastern China. Based on the sequence of the internal transcribed spacer (ITS) rDNA sequences, the N. gruberi isolate was most similar (99%) to N. gruberi (AB298288) in Japan, the N. clarki and N. australiensis isolates were 99% similar to N. clarki (GU597046) and N. australiensis (GU597027) in Taiwan, and the N. pagei isolate showed 100% identity to N. pagei (AJ566633) in USA. Phylogenetic analysis demonstrated that the four Naegleria species formed a clade with N. gruberi, N. clarki, N. pagei, and N. Australiensis,, respectively, and the bootstrap values for their inclusions were high (Fig. 2).
The most pathogenic Naegleria species, N. fowleri, was not identified in the detected water samples from Changchun, China. This may explain why PAME cases have not been reported in the studied areas. The Naegleria species found in the present study are also identified in other countries and regions. For example, N. australiensis is distributed in Europe, North America, and Australia. N. pagei is present in Europe, North America, Africa, and Asia. N. clarki is found in Australia, Africa, N. gruberi is found in Europe, Asia, North America (21).
N. australiensis has been shown to be responsible for the majority of pathogenic isolates in Oklahoma, which can also found in fish brains (22, 23). Mice inoculated intranasally with N. australiensis at 1 × 105 cell exhibits an average mortality rate of 20% (24). N. australiensis is regarded as a potentially pathogenic species by ATCC, although it has never been found in a human (25). However, N. australiensis has been identified in fish brains.
N. pagei, N. clarki, and N. gruberi were non-pathogenic. However, some studies report that Legionella pneumophila, which causes Legionnaire’s disease, can replicate in amoeba host, leading to infection and subsequent intracellular replication (26, 27). Thus, N. gruberi hosting L. pneumophila in aquatic environments may have serious health consequences. N. clarki can also act as vehicle for bacteria, which can harbor two different bacterial populations in the cytoplasm and in the nucleus, respectively, (28). In addition, it has demonstrated that the pathogenic N. fowleri is evolved from the non-pathogenic N. lovaniensis (6).
Conclusion
This is the first report on Naegleria species in Northeastern China, showing that almost all environmental water samples were contaminated with Naegleria, including N. pagei, N. Australiensis, N. clarki and N. gruberi, which should be considered a potential public health threat, as N. australiensis is pathogenic to mice, and N. gruberi may host L. pneumophila.
Acknowledgments
This work was partially supported by the Chinese National Nature Science Foundation (Nos. 31001057, 31072127), and National Science & Technology Pillar Program during the Twelfth Five-year Plan Period in China (2013BAD12B04). The authors declare that there is no conflict of interests.
References
- 1.Huang SW, Hsu BM. Survey of Naegleria from Taiwan recreational waters using culture enrichment combined with PCR. Acta Trop. 2011;119:114–118. doi: 10.1016/j.actatropica.2011.04.016. [DOI] [PubMed] [Google Scholar]
- 2.Kao PM, Tung MC, Hsu BM, Chou MY, Yang HW, She CY, Shen SM. Quantitative detection and identification of Naegleria spp. in various environmental water samples using real-time quantitative PCR assay. Parasitol Res. 2013;112:1467–1474. doi: 10.1007/s00436-013-3290-x. [DOI] [PubMed] [Google Scholar]
- 3.Barnett ND, Kaplan AM, Hopkin RJ, Saubolle MA, Rudinsky MF. Primary amoebic meningoencephalitis with Naegleria fowleri: clinical review. Pediatr Neurol. 1996;15:230–234. doi: 10.1016/s0887-8994(96)00173-7. [DOI] [PubMed] [Google Scholar]
- 4.Anderson K, Jamieson A. Primary amoebic meningoencephalitis. Lancet. 1972;1:902–903. doi: 10.1016/s0140-6736(72)90772-6. [DOI] [PubMed] [Google Scholar]
- 5.Budge PJ, Lazensky B, Van Zile KW, Elliott KE, Dooyema CA, Visvesvara GS, Beach MJ, Yoder JS. Primary amebic meningoencephalitis in Florida: a case report and epidemiological review of Florida cases. J Environ Health. 2013;75:26–31. [PubMed] [Google Scholar]
- 6.De Jonckheere JF. Origin and evolution of the worldwide distributed pathogenic amo-eboflagellate Naegleria fowleri. Infect Genet Evol. 2011;11:1520–1528. doi: 10.1016/j.meegid.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 7.Charoenlarp K, Jariya P, Junyandeegul P, Panyathanya R, Jaroonvesama N. Primary amoebic meningoencephalitis: a second reported case in Thailand. J Med Assoc Thai. 1988;71:581–586. [PubMed] [Google Scholar]
- 8.Jariya P, Makeo S, Jaroonvesama N, Kunara-tanapruk S, Lawhanuwat C, Pongchaikul P. Primary amoebic meningoencephalitis: a first reported case in Thailand. Southeast Asian J Trop Med Public Health. 1983;14:525–527. [PubMed] [Google Scholar]
- 9.Gupta N, Bhaskar H, Duggal S, Ghalaut PS, Kundra S, Arora DR. Primary amoebic meningoencephalitis: first reported case from Rohtak, North India. Braz J Infect Dis. 2009;13:236–237. doi: 10.1590/s1413-86702009000300016. [DOI] [PubMed] [Google Scholar]
- 10.Movahedi Z, Shokrollahi MR, Aghaali M, Heydari H. Primary amoebic meningoencephalitis in an Iranian infant. Case Rep Med. 2012;2012:782854. doi: 10.1155/2012/782854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saleem T, Rabbani M, Jamil B. Primary amoebic meningoencephalitis: two new cases from Pakistan. Trop Doct. 2009;39:242–243. doi: 10.1258/td.2009.090032. [DOI] [PubMed] [Google Scholar]
- 12.Shakoor S, Beg MA, Mahmood SF, Bandea R, Sriram R, Noman F, Visvesvara GS, Zafar A. Primary amebic meningoencephalitis caused by Naegleria fowleri, Karachi, Pakistan. Emerg Infect Dis. 2011;17:258–261. doi: 10.3201/eid1702.100442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mi JP. Parasites and central nervous system diseases. J Med Pest Control. 2005;7:540–542. in Chinese. [Google Scholar]
- 14.Behets J, Declerck P, Delaedt Y, Verelst L, Ollevier F. Survey for the presence of specific free-living amoebae in cooling waters from Belgian power plants. Parasitol Res. 2007;100:1249–1256. doi: 10.1007/s00436-006-0399-1. [DOI] [PubMed] [Google Scholar]
- 15.Edagawa A, Kimura A, Kawabuchi-Kurata T, Kusuhara Y, Karanis P. Isolation and genotyping of potentially pathogenic Acantha-moeba and Naegleria species from tap-water sources in Osaka, Japan. Parasitol Res. 2009;105:1109–1117. doi: 10.1007/s00436-009-1528-4. [DOI] [PubMed] [Google Scholar]
- 16.Schuster FL. Cultivation of pathogenic and opportunistic free-living amoebas. Clin Microbiol Rev. 2002;15:342–354. doi: 10.1128/CMR.15.3.342-354.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pelandakis M, Serre S, Pernin P. Analysis of the 5.8S rRNA gene and the internal transcribed spacers in Naegleria spp. and in N. fowleri. J Eukaryot Microbiol. 2000;47:116–121. doi: 10.1111/j.1550-7408.2000.tb00020.x. [DOI] [PubMed] [Google Scholar]
- 18.Ithoi I, Ahmad AF, Nissapatorn V, Lau YL, Mahmud R, Mak JW. Detection of Naegleria species in environmental samples from Peninsular Malaysia. PLoS One. 2011;6:e24327. doi: 10.1371/journal.pone.0024327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moussa M, De Jonckheere JF, Guerlotte J, Richard V, Bastaraud A, Romana M, Talarmin A. Survey of Naegleria fowleri in geothermal recreational waters of Guadeloupe (French West Indies) PLoS One. 2013;8:e54414. doi: 10.1371/journal.pone.0054414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 21.De Jonckheere JF. Molecular definition and the ubiquity of species in the genus Naegleria. Protist. 2004;155:89–103. doi: 10.1078/1434461000167. [DOI] [PubMed] [Google Scholar]
- 22.Dykova I, Kyselova I, Peckova H, Obornik M, Lukes J. Identity of Naegleria strains isolated from organs of freshwater fishes. Dis Aquat Organ. 2001;46:115–121. doi: 10.3354/dao046115. [DOI] [PubMed] [Google Scholar]
- 23.John DT, De Jonckheere JF. Isolation of Naegleria australiensis from an Oklahoma Lake. J protozool. 1985;32:571–575. doi: 10.1111/j.1550-7408.1985.tb03077.x. [DOI] [PubMed] [Google Scholar]
- 24.John DT, Howard MJ. Seasonal distribution of pathogenic free-living amoebae in Oklahoma waters. Parasitol Res. 1995;81:193–201. doi: 10.1007/BF00937109. [DOI] [PubMed] [Google Scholar]
- 25.Visvesvara GS, Moura H, Schuster FL. Pathogenic and opportunistic free-living amoe-bae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol Med Microbiol. 2007;50:1–26. doi: 10.1111/j.1574-695X.2007.00232.x. [DOI] [PubMed] [Google Scholar]
- 26.Rowbotham TJ. Preliminary report on the pathogenicity of Legionella pneumophila for fresh-water and soil amoebae. J Clin Pathol. 1980;33:1179–1183. doi: 10.1136/jcp.33.12.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Declerck P, Behets J, Delaedt Y, Margineanu A, Lammertyn E, Ollevier F. Impact of non-Legionella bacteria on the uptake and intracellular replication of Legionella pneumophila in Acanthamoeba castellanii and Naegleria lovaniensis. Microb Ecol. 2005;50:536–549. doi: 10.1007/s00248-005-0258-0. [DOI] [PubMed] [Google Scholar]
- 28.Walochnik J, Muller KD, Aspock H, Michel R. An endocytobiont harbouring Naegleria strain identified as N. clarki De Jonckheere, 1994. Acta Protozool. 2005;44:301–310. [Google Scholar]


