Abstract
By generating prostaglandins, cyclooxygenase-2 (Cox-2/Ptgs2) plays a critical role in regulating inflammatory responses. While several inflammatory stimuli have been shown to increase Ptgs2 expression, less is known about how the transcription of this gene is terminated. Here we show that stimulation of macrophages with yeast zymosan, a TLR2/6 and dectin-1 agonist, causes a transient increase in the expression of Ptgs2 accompanied by a simultaneous increase in the expression of the transcriptional repressor, Activating transcription factor-3 (Atf3). The expression of Ptgs2 was significantly higher in resident peritoneal macrophages isolated from Atf3−/− mice than that from Atf3+/+ mice and was associated with higher prostaglandin production upon stimulation with zymosan. In activated macrophages, Atf3 accumulated in the nucleus and chromatin-immunoprecipitation analysis showed that Atf3 is recruited to the Ptgs2 promoter region. In acute peritonitis and in cutaneous wounds, there was increased leukocyte accumulation and higher levels of prostaglandins (PGE2/PGD2) in inflammatory exudates of Atf3−/− mice compared with WT mice. Collectively, these results demonstrate that during acute inflammation Atf3 negatively regulates Ptgs2 and therefore dysregulation of this axis could potentially contribute to aberrant Ptgs2 expression in chronic inflammatory diseases. Moreover, this axis could be a new therapeutic target for suppressing Ptgs2 expression and the resultant inflammatory responses.
Keywords: Inflammation, prostaglandins, lipid mediators
Introduction
Cyclooxygenases/prostaglandin endoperoxide synthases (Cox/Ptgs 1 & 2) generate prostaglandins that play well-documented roles in regulating vascular tone, thrombosis, inflammation and pain [1–3]. Because of their critical role in regulating inflammation, cyclooxygenase enzymes are therapeutic targets of one of the most widely used classes of drugs, namely, the non-steroidal anti-inflammatory drugs (NSAIDs) [4]. While Ptgs1 is constitutively expressed, Ptgs2 is induced to high levels during inflammation [5, 6]. Previous studies have shown that Ptgs2 transcription is initiated by several transcription factors associated with inflammatory signaling cascades, including NF-κB, AP-1, Sp1 and C/EBP [6]. Less is known about how the transcription of Ptgs2 is repressed, although repression elements in the promoter region of the Ptgs2 gene have been identified, and some negative regulatory factors including nuclear receptor co-repressor (NCoR), the poly (ADP-ribose) polymerase-1 (Parp-1) and sumoylated CEBPB have been shown to repress Ptgs2 expression in cervical cancer cells, pancreatic β-cells and carcinoma cells, respectively [6–9]. Nevertheless, it is not known how during resolution of acute inflammation, Ptgs2 is repressed in tissues and immune cells.
The Activating transcription factor 3 (Atf3) is a member of the ATF/CREB family of basic leucine zipper (bZip) transcription factors. It forms homodimers with itself or heterodimers with other bZip proteins, and the resulting dimers can function as a transcriptional activator or repressor [10]. Atf3 is induced by a variety of cellular stressors, such as ischemia and endoplasmic reticulum stress, and there is accumulating evidence indicating that Atf3 negatively regulates toll-like receptor (TLR)-response genes including interleukin 6 (Il-6) and Il-12b [10–12]. Computational approaches predict that because of proximity of ATF/CRE sites to NF-κB and AP-1 binding sites in the Ptgs2 promoter, Atf3 could also be a potential regulator of Ptgs2. Studies showing that Ptgs2 is one of the genes upregulated in Atf3-deficient cardiomyocytes [13] supports this possibility, although regulation of Ptgs2 by Atf3 per se has not been extensively studied in immune cells. Here, we provide direct evidence that, during the development and resolution of acute inflammation, Atf3 negatively regulates Ptgs2 in leukocytes.
Materials and Methods
Acute peritonitis and cutaneous wounds
To induce peritonitis, male 8–12 week old WT (C57/BL6J; Jackson laboratories) and age-matched Atf3-deficient mice (C57/BL6J background) [14] were administered yeast zymosan (0.04mg/g; i.p.; Sigma) and inflammatory exudates were collected 4h after injection [15]. In some experiments, mice received prostanoid receptor antagonist, AH6809 (10ng; i.p.), or COX-2 inhibitor, NS-398 (100ng; i.p.), 30 minutes prior to zymosan administration. All procedures were approved by the University of Louisville IACUC. Inflammatory exudates were obtained by lavaging the peritoneum with 5ml DPBS−/− and leukocytes were enumerated and identified by flow cytometry using anti-Ly6G (polymorphonuclear neutrophils; PMN) and anti-F4/80 (macrophages) antibodies (Biolegend). Expression of Atf3 and Ptgs2 mRNA was evaluated in cell pellets obtained from inflammatory exudates (see below). Cytokines and chemokines were measured in cell-free exudates (Aushon biosystems and Ebioscience Flowcytomix). Cutaneous wounds were created as described previously [16]. Briefly, the dorsal skin was shaved and treated with depilatory cream after anesthesia. The skin was rinsed and two circular, full thickness wounds (skin and panniculus carnosus) were created using a 5-mm biopsy punch. Wounds were covered by a semipermeable polyurethane dressing and wound tissue was collected 24h post-wounding. Wound tissue was formalin fixed, paraffin embedded and sectioned. Accumulation of PMN was evaluated in deparaffinized sections of wound tissue using anti-myeloperoxidase (MPO) antibodies and an UltraVision detection system (Thermo Scientific, Labvision).
In vitro macrophage incubations
Resident peritoneal macrophages were isolated from WT or Atf3-deficient mice in DPBS−/− and incubated for 1h in 24-well plates at 37°C in DMEM containing 10% FBS. Non-adherent cells were removed and the macrophages were either left untreated or stimulated with yeast zymosan (50µg/ml) for 3 or 6h. Supernatants were collected for LC-MS/MS analysis (see below) and cell pellets were used for gene expression analysis or Western blot. For this, RNA was extracted using the RNeasy mini kit (Qiagen), followed by cDNA synthesis. RT-PCR amplification was performed with SYBR-green qPCR master mix (SA Biosciences) using a 7900HT fast system (Applied Biosystems) and commercially available primers for murine Atf3 (IDT), Ptgs2, Ptgs1, microsomal prostaglandin E synthase 1 (mPges1), hematopoietic prostaglandin D synthase (hPGDS), arachidonate 5 lipoxygenase (Alox5), Alox5 activating protein (ap) and interleukin 1 beta (Il-1β) (SA biosciences). Relative expression was determined by the 2−ΔΔCT method after internal normalization to Hprt. To assess COX-2 protein expression, macrophages treated without or with zymosan for 6h and lysates were prepared. Expression of COX-2 was determined using an anti-COX-2 antibody (Thermo Scientific) and was normalized for loading using an anti-β-actin antibody (Sigma).
LC-MS/MS analysis of lipid mediators
Macrophage supernatants or inflammatory exudates were collected for targeted LC-MS/MS analysis of leukotriene B4 (LTB4) and prostaglandins E2/D2 (PGE2/D2) [15]. Upon sample collection, 2 volumes of cold methanol containing deuterium-labeled (d4) PGE2 was added and samples were placed at −80°C to allow for protein precipitation. Lipid mediators were extracted using solid-phase (C18) columns and methyl formate fractions were collected and taken to dryness under a stream of N2 gas. After resuspending in methanol, samples were analyzed using an HPLC system (Shimadzu prominence) equipped with a C18 reverse-phase column (4.6 mm × 50 mm) coupled to a triple quadrupole mass spectrometer (AB Sciex; API2000). The instrument was operated in negative ionization mode and the mobile phase consisted of methanol:water:acetic acid (60:40:0.01, vol/vol/vol), which was ramped to 80:20:0.01 over 3 min and to 95:5:0.01 in the next 14 min at a constant flow rate of 400µL per min. Lipid mediators were identified and quantified using multiple reaction monitoring (MRM) and transitions for LTB4 (335>195) and PGE2/PGD2 (351>189). We note that under these LC conditions, PGE2 and PGD2 are not readily separated and are therefore presented as “PGE2/D2”. Extraction recovery was determined using internal d4-PGE2, while all lipid mediators were quantified based on external calibration curves using authentic standards (Cayman chemical).
Nuclear localization of Atf3
RAW 264.7 (ATCC) macrophages were incubated with zymosan (50µg/ml) for 6h. The NE-PER Nuclear and Cytoplasmic Extraction kit (Thermo Scientific) was used to obtain nuclear extracts that were analyzed by immunoblotting with anti-Atf3 (Santa Cruz) and anti-Hdac1 (Cell Signaling) antibodies. Immunoblots were developed using Luminata Forte Western HRP Substrate (Millipore), detected using a Typhoon 9400 variable mode imager (Amersham Biosciences), and quantified with ImageQuant TL.
Chromatin immunoprecipitation (ChIP)
ChIP was carried out according to the manufacturers’ instructions (Santa Cruz) and essentially as described in [17]. Briefly, RAW 264.7 macrophages were stimulated with zymosan (50µg/ml) for 6h and the cells were fixed with 1% formaldehyde for 10 min at room temperature. Cross-linking was terminated with glycine (0.125M). Cell lysis was carried out in ice-cold lysis buffer and crude nuclear extracts were obtained by centrifugation. Pellets were resuspended in high salt lysis buffer on ice and DNA was sheared by sonication. The resulting chromatin solution was precleared with salmon sperm/protein-A agarose and a sample of input DNA (3%) was collected. Immunoprecipitation was carried out overnight at 4°C with anti-Atf3 antibody (200 µg/0.1 ml) or normal rabbit serum (NRS). Antibody–protein–DNA complexes were captured, beads were resuspended in elution buffer and cross-links were reversed. The supernatant was incubated at 67°C overnight and DNA was extracted with a QIAquick PCR Purification kit (QIAGEN). PCR was conducted using promoter specific primers (see Fig. 2) for murine Ptgs-2; sense 5′-CGCAACTCACTGAAGCAGAG -3′ and antisense 5′- TCCTTCGTGAGCAGAGTCCT-3′. PCR products were separated on 2% agarose gels and bands captured under UV illumination.
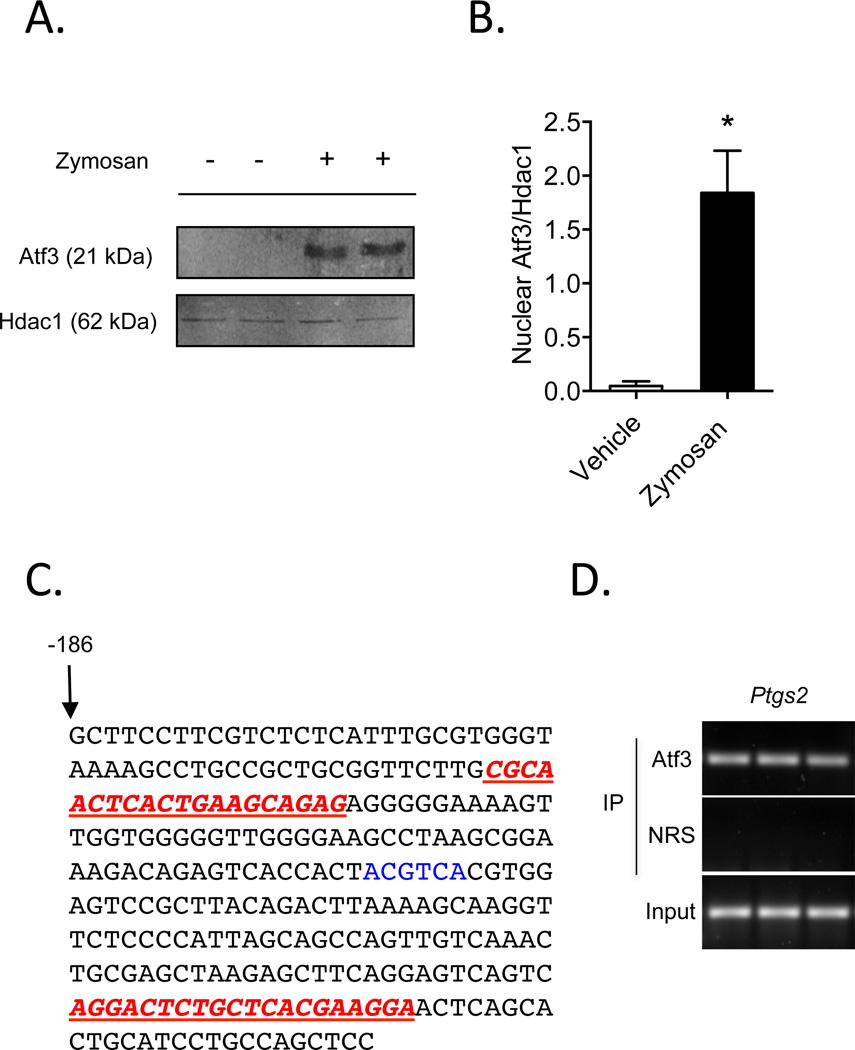
Figure 2. Atf3 is recruited to the murine Ptgs2 promoter in activated macrophages.
(A) Nuclear localization of Atf3 in zymosan-stimulated RAW 264.7 macrophages. Histone deacetylase (Hdac)-1 is shown as a control for nuclear isolation and quantification of band intensities is shown in panel B. (C) The abbreviated sequence of the Ptgs2 promoter, with primers used for the ChIP assay (underlined in red) and an ATF/CRE binding site (highlighted in blue) shown. (D) ChIP analysis of Atf3 bound to the Ptgs2 promoter in macrophages stimulated with zymosan, with non-immune rabbit serum (NRS) control and total DNA input (3%) shown. Results are mean ± SEM, n=3/group. *P<0.05
Statistical analysis
Experimental results are mean ± SEM. Statistical significance (P<0.05) was determined using an unpaired two-tailed Student’s t test or one-way ANOVA, followed by Tukey’s multiple comparisons post-test, as appropriate.
Results
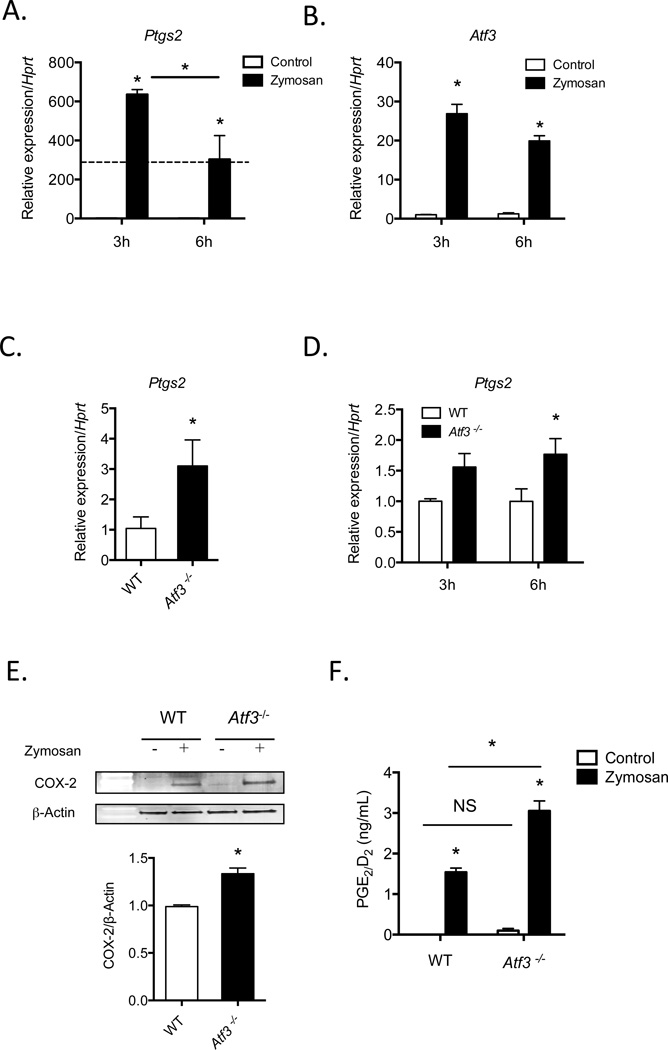
To determine the temporal relationship between induction of Ptgs2 and Atf3, we stimulated macrophages with the TLR2/6 and dectin-1 agonist, zymosan [18, 19]. A robust induction of Ptgs2 was observed after 3h, and its levels declined significantly (~50%) after 6h (Fig. 1A), suggesting termination of transcription or an increase in degradation. In addition, zymosan stimulation of macrophages led to robust induction of Atf3 mRNA, with a time course similar to the induction of Ptgs2 (Fig. 1B). Given this similarity, we assessed whether Atf3 deficiency would alter Ptgs2 expression. We found that in comparison with WT mice, levels of Ptgs2 mRNA were higher in resident peritoneal macrophages isolated from Atf3−/− mice (Fig. 1C). The regulation of Ptgs2 by Atf3 was dynamic, as the difference in Ptgs2 mRNA in unstimulated macrophages was lost after 3h of stimulation with zymosan, whereas Ptgs2 was again elevated in Atf3−/− macrophages after 6h of zymosan stimulation (Fig. 1D). This time course mirrored the induction and repression of Ptgs2 in WT macrophages (Fig. 1A). This increase in Ptgs2 mRNA translated to increased COX-2 protein expression in Atf3-deficient macrophages stimulated with zymosan relative to WT macrophages (Fig. 1E). To determine whether the induced Ptgs2 was catalytically active, we measured the level of its products—prostaglandins E2 and D2 (PGE2/D2), which are generated by downstream synthases in macrophages from the common biosynthetic intermediate, PGH2. Indeed, we found that the higher levels of Ptgs2 in Atf3-deficient macrophages correlated with higher levels of PGE2/D2 in supernatants of macrophages stimulated with zymosan for 6h, while no differences were observed without stimulation (Fig. 1F). Although mRNA levels of Ptgs1 and PG synthases, mPges1 and hPgds, were elevated in Atf3-deficient macrophages at baseline, there were no significant differences in expression of these genes in cells stimulated with zymosan (Supplemental Figure 1). Taken together, these observations suggest that Atf3 negatively regulates Ptgs2 expression in activated macrophages.
Figure 1. Atf3 negatively regulates Ptgs2 expression in macrophages.
(A, B) Levels of Ptgs2 and Atf3 mRNA in resident peritoneal macrophages stimulated without or with zymosan for 3 or 6h. (C) Expression of Ptgs2 mRNA in unstimulated resident peritoneal macrophages isolated from WT or Atf3−/− mice. (D) Levels of Ptgs2 mRNA in resident peritoneal macrophages isolated from WT or Atf3-deficient mice and stimulated with zymosan for 3 or 6h. (E) Western blot of COX-2 protein in WT or Atf3-deficient macrophages stimulated with zymosan for 6h, with quantification shown in the lower panel. (F) PGE2/D2 levels in supernatants of WT and Atf3−/− macrophages stimulated without (control) or with zymosan (6h). Results are mean ± SEM, n=3–7/group. *P<0.05; NS=not significant
We next tested whether Atf3 regulates Ptgs2 gene expression via direct recruitment to the Ptgs2 promoter. In addition to inducing Atf3 transcription (Fig. 1B), zymosan stimulation of macrophages increased nuclear accumulation of Atf3 protein (Fig. 2A & B). As noted, the Ptgs2 promoter contains consensus ATF/CRE binding sites, which are in close proximity to both AP-1 and NF-κB binding sites [11]. Hence, we used chromatin-immunoprecipitation (ChIP) to determine whether Atf3 is recruited to the Ptgs2 promoter in cells stimulated with zymosan. PCR amplification of DNA bound to immunoprecipitated Atf3 (using primers flanking an ATF/CRE binding site −56/−52 upstream of the transcription start site; Fig. 2C) showed that Atf3 is recruited to the Ptgs2 promoter (Fig. 2D). No amplification was observed in samples immunoprecipitated with non-immune rabbit serum (NRS).
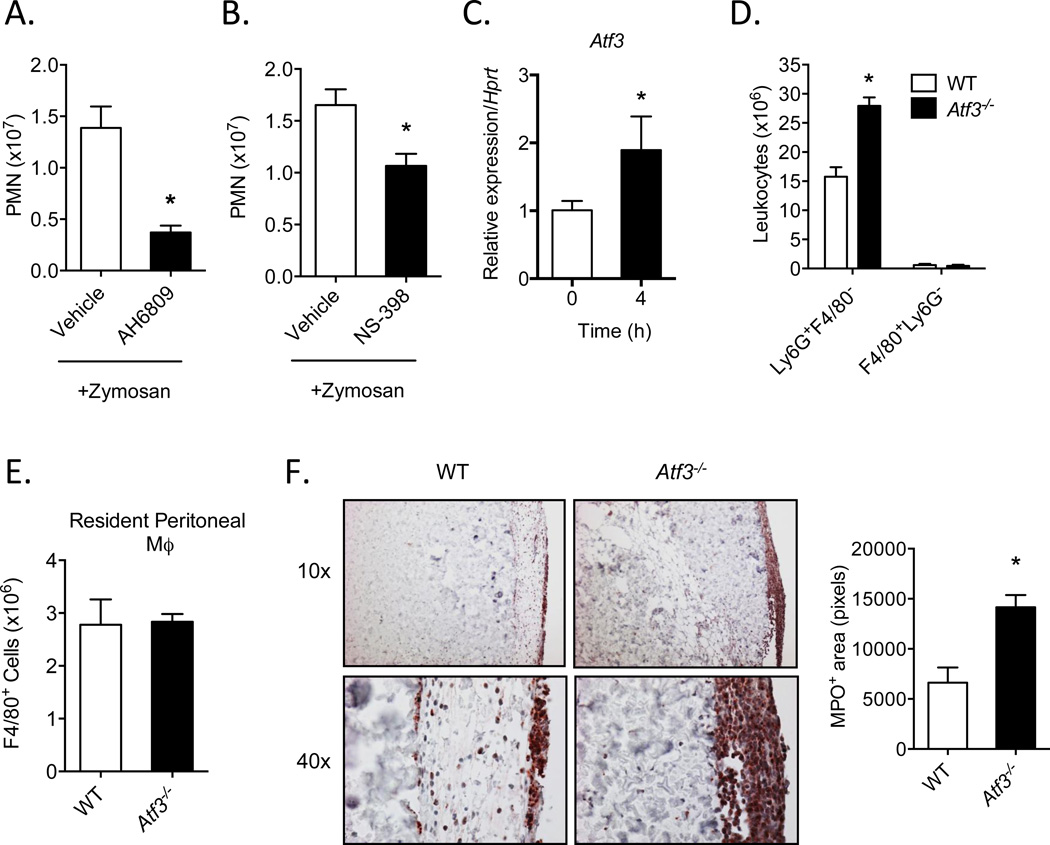
To test the in vivo relevance of our findings, we used an acute model of peritonitis with zymosan as a model stimulus [15]. As expected, i.p. administration of zymosan led to a robust leukocyte infiltrate, most of which were identified as PMN (Ly6G+F4/80−) at this 4h time point (Fig. 3A). Co-administration of an E and D-prostanoid receptor antagonist, AH6809 [20], led to a significant decrease in PMN infiltration in response to zymosan, (Fig. 3A), confirming the role of prostanoids in mediating leukocyte trafficking [1, 5, 21]. In addition, the selective COX-2 inhibitor, NS-398, also significantly decreased PMN infiltration in response to zymosan (Fig. 3B). Measurement of Atf3 showed that there was a significant increase in the gene expression of this transcription factor in exudate leukocytes 4h post-zymosan, when compared with resident, unstimulated leukocytes (Fig. 3C). To assess the significance of this increase in Atf3 and to determine its role in leukocyte trafficking, we measured the levels of PMN and macrophages (F4/80+/Ly6G−) in WT and Atf3-deficient mice. We found that PMN recruitment was significantly higher in Atf3-deficient mice than their WT controls (Fig. 3D). Importantly, levels of peritoneal macrophages in WT and Atf3-deficient mice were similar; this is true for both the resident macrophages from naïve mice (Fig. 3E) and recruited macrophages from zymosan-treated mice (Fig. 3D). Because resident macrophages play an important role in PMN recruitment in this acute peritoneal inflammation model [22], the increased PMN in Atf3-deficient mice (at 4 hours after zymosan injection) is not due to the differences in resident macrophage numbers. However, it is possible that the bioactivity of these macrophages (such as expression of genes important for PMN recruitment) is different (see below Fig. 4), contributing to the increased PMN recruitment. To test whether the increased PMN recruitment in Atf3-deficient mice is limited to this peritonitis model or not, we examined PMN recruitment in a different inflammation model: cutaneous wounds. As shown in Fig 3F, PMN levels were consistently higher in the Atf3-deficient wounds than WT wounds, indicating that Atf3 negatively regulates PMN infiltration in the context of sterile tissue injury. Collectively, these data suggest that Atf3 plays an important role in negatively regulating leukocyte trafficking during acute inflammation.
Figure 3. Atf3 negatively regulates leukocyte trafficking during acute inflammation.
(A, B) Neutrophil (PMN) levels in WT mice administered zymosan (4h) and treated with vehicle (sterile saline), E-prostanoid/D-prostanoid (EP2 and DP1) receptor antagonist, AH6809 (A), or COX-2 inhibitor, NS-398 (B). (C) Expression of Atf3 mRNA in peritoneal leukocytes isolated from naïve mice or during acute peritonitis stimulated by zymosan. (D) Neutrophil (Ly6G+F4/80−) and macrophage (F4/80+Ly6G−) levels in WT and Atf3-deficient mice 4h after induction of peritonitis. (E) Total resident F4/80+ macrophage levels in the peritoneum of naïve WT and Atf3-deficient mice. (F) Histological analysis of myeloperoxidase (MPO)-positive cells in cutaneous wounds of WT and Atf3-deficient mice 24h after wounding, with quantitation of the total MPO+ area in the right panel (4 fields per animal). Results are mean ± SEM, n=3–8/group. *P<0.05
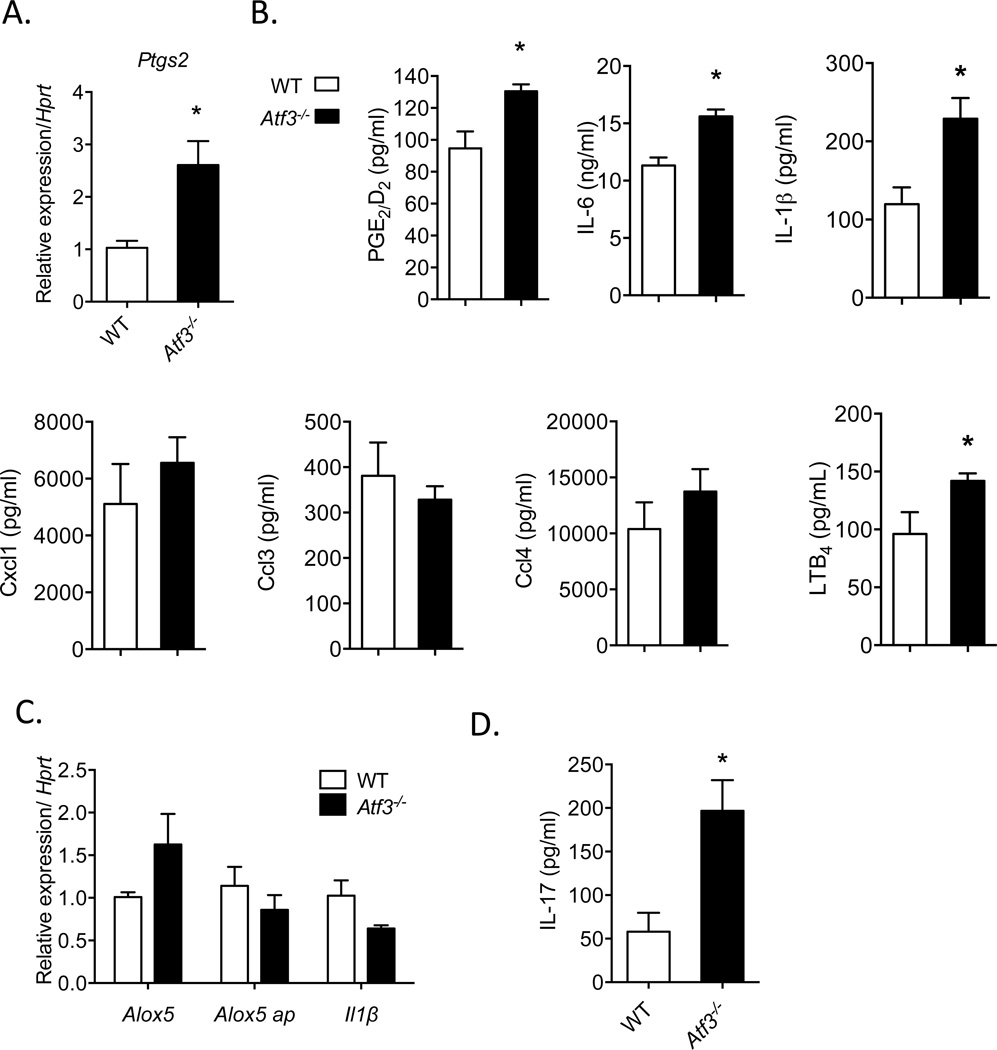
Figure 4. Increased leukocyte accumulation in Atf3-deficient mice is associated with the Ptgs2/IL-17 axis.
(A) Levels of Ptgs2 mRNA in leukocytes obtained from inflammatory exudates of WT and Atf3-deficient mice undergoing peritonitis (4h). (B) Levels of PGE2/D2, inflammatory cytokines and chemokines IL-6, IL-1β, Cxcl1, Ccl3, and Ccl4, and leukotriene B4 (LTB4) in peritoneal exudates of WT and Atf3-deficient mice collected 4h after zymosan administration. (C) Expression of Alox5, Alox5 activating protein (ap) and Il-1β mRNA in WT or Atf3-deficient resident peritoneal macrophages stimulated with zymosan for 6h. (D) Levels of IL-17 in peritoneal exudates of WT and Atf3-deficient mice undergoing peritonitis (4h). Results are mean ± SEM, n=3–8/group. *P<0.05
As results showed that inhibition of prostanoid signaling significantly dampens the development of peritonitis and that loss of Atf3 has the opposite effect, we sought to determine how the Ptgs2 pathway was modulated in Atf3-deficient mice during peritonitis. Consistent with our in vitro studies showing that loss of Atf3 increases Ptgs2 levels, we found a significant increase in Ptgs2 mRNA expression in exudate leukocytes obtained from Atf3-deficient mice compared with WT mice (Fig. 4A). This increase in Ptgs2 translated into a significant elevation of PGE2/D2 levels in exudates isolated from Atf3-deficient mice (Fig. 4B). As enhanced acute leukocyte trafficking in Atf3-deficient mice could involve other lipid mediators or chemokines, we measured pro-inflammatory cytokines and chemokines that have either been previously shown to be regulated by Atf3 (i.e., IL-6), or those which have well-documented roles in regulating PMN chemotaxis during acute inflammation. Consistent with previous reports [11], we found that the loss of Atf3 increased levels of IL-6 in inflammatory exudates of mice undergoing peritonitis (Fig. 4B). Levels of IL-1β were also significantly increased. In contrast, levels of PMN chemokine, Cxcl1, were not significantly affected by Atf3 deficiency. Similarly, other leukocyte chemokines, including Ccl3 and Ccl4, were also not different between WT and Atf3-deficient mice, demonstrating selective regulation of inflammatory signaling by Atf3 (Fig. 4B). We also measured exudate levels of leukotriene B4 (LTB4), as this lipid mediator is rapidly generated during acute inflammation and plays a critical role in PMN chemotaxis [23]. As shown in Fig. 4B, levels of LTB4 were significantly elevated in inflammatory exudates of Atf3-deficient mice compared with WT mice. Overall, these results confirm our in vitro studies demonstrating that loss of Atf3 increases Ptgs2 and its downstream products and that Atf3-deficiency increases leukocyte infiltration without altering the generation of classic PMN chemokine, Cxcl1.
Based on our results showing that Atf3-deficient mice have elevated PMN infiltration during acute inflammation and that this is correlated with increased levels of IL-6, IL-1β, LTB4 and PGE2/D2, we next sought to determine how Atf3 regulates the production of these mediators. Previous studies demonstrated that Atf3 down-regulates Il-6 transcription [24]. Our results here indicate that Atf3 is recruited to the Ptgs2 promoter (Fig. 2D); its correlation with the reduced Ptgs2 steady-state mRNA level also suggests that Atf3 down-regulates Ptgs2 gene. To determine whether levels of IL-1β and LTB4 were elevated because of their regulation by Atf3 in macrophages upon exposure to zymosan, or whether they were simply increased as a consequence of increased PMN levels in the peritoneal cavity (which can produce both IL-1β, and LTB4), we assessed the expression of Il-1β and the enzymes required for LTB4 biosynthesis in isolated resident peritoneal macrophages. This analysis showed that expression of arachidonate-5 lipoxygenase (Alox5) and Alox5 activating protein (alox5 ap) was not significantly different in resident peritoneal macrophages isolated from WT or Atf3-deficient mice and stimulated with zymosan (Fig. 4C). Moreover, expression of Il-1β was similar between WT and Atf3-deficient macrophages, suggesting that increased levels of IL-1β and LTB4 in vivo were likely a result of increased PMN levels. Collectively, these results suggest that loss of Atf3 increases PMN infiltration during acute inflammation and that, consistent with the in vitro studies described above, Atf3 deficiency is associated with increased prostanoid production.
Several previous studies have shown that prostaglandins, particularly PGE2, increase PMN infiltration during acute inflammation and that these effects are dependent in part by the downstream production of IL-17, which can promote PMN recruitment to sites of inflammation [21, 25–29]. Indeed, congruent with elevated levels of prostanoids in inflammatory exudates of Atf3-deficient mice (Fig. 4B), there was an increase in exudate levels of IL-17 in Atf3-deficient mice compared with WT mice (Fig. 4D). Overall, the enhanced recruitment of PMN during acute inflammation in Atf3-deficient mice is consistent with amplification of the prostaglandin/IL-17 axis.
Discussion
The results of this study demonstrate that Atf3 negatively regulates Ptgs2 expression during acute inflammation through recruitment to the Ptgs2 promoter. This role of Atf3 is supported by the observation that mice lacking Atf3 had higher PMN infiltration in two distinct models of acute inflammation concurrent with amplification of the Ptgs2 pathway. Our results thus provide novel insights into the dynamic regulation of Ptgs2 expression during acute inflammation.
It is now widely appreciated that temporal regulation of lipid mediator production governs the duration of acute inflammatory responses. For instance, leukotrienes and prostaglandins regulate early phases of inflammation including vascular permeability, vascular caliber, leukocyte trafficking and phagocytosis [30]. Their levels decline rapidly as inflammation begins to resolve; a phase that is regulated by distinct families of lipid mediators that terminate leukocyte trafficking and promote macrophage-mediated clearance of apoptotic cells [30, 31]. Thus, a comprehensive understanding of how lipid mediator signaling is triggered and terminated is important for understanding why in some cases inflammation resolves appropriately while under other conditions it persists chronically at a low levels of activation. Previous studies have shown that Ptgs2 is dynamically regulated during the initiation and resolution of acute inflammation [5]. The increase in Ptgs2 during the initiation of inflammation leads to the generation of PGE2 and is associated with increased edema and leukocyte trafficking, while a later wave of Ptgs2 expression gives rise to PGD2 and its degradation products (i.e., 15-deoxy PGJ2). Inhibition of Ptgs2 during the early phase of inflammation blocks leukocyte infiltration, whereas inhibiting Ptgs2 during the later phase enhances leukocyte accumulation[5]. The findings of the present study add new mechanistic insight into this temporal regulation of Ptgs2 by elucidating that Atf3 plays a role in blunting the first wave of Ptgs2 transcription to control the magnitude of the inflammatory response. As Atf3 itself is induced by the same stimuli that induce Ptgs2 expression, these results add to a growing body of literature demonstrating that Atf3 is a central hub that controls the tone of the inflammatory response to a level appropriate to the extant pathogen load [32].
Several inflammatory stimuli have been shown to increase Ptgs2 expression, including bacterial lipopolysaccharides (LPS), fungal pathogens, and inflammatory cytokines, such as TNF-α and IL-1β [6]. Interestingly, prostaglandins, such as PGE2, can also stimulate Ptgs2 expression as part of a positive feedback loop that amplifies the inflammatory response [6]. Transcription factors linked to these inflammatory signaling pathways, including AP-1, NF-κB (p65/p50), Sp1 and C/EBP, have been implicated in the induction of Ptgs2 and several of these transcription factors bind to the transcriptional coactivator, p300, which is essential for Ptgs2 transcription [6, 33]. Notably, the Ptgs2 promoter contains two cyclic AMP responsive elements (CRE) that have been shown to be important in Ptgs2 induction in response to a diverse array of stimuli, including phorbol esters, the v-src oncogene, and LPS[6, 34–36]. Once generated, Ptgs2 protein is subject to degradation via the endoplasmic reticulum-associated degradation pathway (ERAD), a process that is delayed by NSAIDs[36]. In addition to degradation, transcriptional repression of Ptgs2 may represent another critical control point in regulating the levels of Ptgs2. Indeed, mutation of the E-box region of the Ptgs2 promoter, which is adjacent to one of the ATF/CRE sites (i.e., CRE-1), increases Ptgs2 expression in response to LPS, indicating that an endogenous repressor acts as a negative regulator of Ptgs2 transcription [36]. Moreover, other factors, such as PARP-1, have been shown to repress Ptgs2 transcription by binding to previously unrecognized repression elements in the Ptgs2 promoter far upstream of the transcription start site (−655/−632) [7]. Our results strengthen the view that transcriptional repression of Ptgs2 is an important regulatory mechanism controlling the expression of Ptgs2 during acute inflammation by demonstrating that Atf3 is rapidly recruited to the Ptgs2 promoter and negatively regulates Ptgs2 expression.
It has been previously reported that Atf3 negatively regulates gene transcription by recruiting histone deacetylates (HDACs), such as HDAC1, to alter chromatin structure and thus access to transcription factors [11]. In addition to HDAC1, recent studies have also demonstrated that a sumoylated form of CEBPB, denoted LAP1, recruits HDAC4 to blunt human PTGS2 expression [8]. Thus, it is likely that binding of Atf3 blunts Ptgs2 transcription by regulating chromatin remodeling, given the proximity of ATF/CRE binding sites to both NF-kB and AP-1 binding sites in the Ptgs2 promoter. Future studies will be required to fully elucidate whether this mechanism underlies the ability of Atf3 to modulate Ptgs2 expression, although our results support the notion that loss of Atf3 increases Ptgs2 expression via binding to the promoter region. Interestingly, a splice variant of Atf3 lacking the DNA binding domain (i.e., Atf3ΔZip2), has been shown to regulate NF-κB-dependent transcription of anti-apoptotic genes by displacing CBP/p300 binding to activated NF-κB [37]. Given the critical role of p300 in the transcriptional regulation of Ptgs2 induction, it is intriguing to speculate that Atf3 and its alternatively spliced forms may have multiple distinct roles in regulating Ptgs2 transcription.
Atf3 is induced by several stress signals, including ischemia, ER stress, and oxidative stress [10]. Importantly, Atf3 has emerged as a central negative regulator of TLR signaling and Atf3 itself is induced by the inflammatory signaling pathways (e.g., NF-κB) engaged by ligation of TLR receptors [11, 12]. Although Atf3 was initially studied in the context of signaling through TLR4 [11], it is now clear that Atf3 negatively regulates gene expression induced by several TLRs, including TLR3, 5, 7 and 9 [12]. In addition, activation of the TLR2/6 heterodimer by yeast zymosan induces rapid induction of Atf3 expression in both macrophages and dendritic cells [12]. In our study, we used fungal zymosan as a model stimulus because it robustly stimulates lipid mediator biosynthesis in leukocytes [38]. We found that stimulation of macrophages with zymosan leads to a robust induction of both Ptgs2 and Atf3. The temporal relationship between Atf3 and Ptgs2 is similar to the established negative relationship between Atf3 and other pro-inflammatory cytokines, such as IL-6 and IL-12b [11]. This relationship allows for the rapid induction and repression of Ptgs2, such that its levels start declining by 6h post-stimulation. This time course is similar to the development of inflammation in vivo, which in this model reaches a maximum at 4–6 h and begins to resolve by 24h [15]. Indeed, our results demonstrate increased Ptgs2 levels and increased production of downstream prostanoids in the context of Atf3 deficiency both in vitro and in vivo. This dynamic regulation is likely to be important for enabling the resolution of inflammation, as prostanoids, such as PGE2, are known to suppress macrophage phagocytosis of apoptotic cells [15, 39].
It is notable that loss of Atf3 increased recruitment of PMN in peritonitis, which we attribute in part to a dysregulation of prostanoid biosynthesis. This conclusion is based on the observation that both Ptgs2 and PGE2/D2 levels were increased in inflammatory exudates of Atf3-deficient mice and that inhibition of PGE2/D2 receptors or COX-2, largely blunted PMN infiltration. These findings are consistent with prior work showing that inhibition of COX-2 with NSAIDs diminishes the onset of acute inflammation in several distinct models [5, 15, 40, 41]. Interestingly, a recent study by Boespflug et al. demonstrated that Atf3 has a dual effect on PMN recruitment [42]. They showed that, in a murine model of airway inflammation driven by LPS, levels of Cxcl1 were elevated in the Atf3-deficient airway. However, no changes in PMN infiltration were observed, presumably due to an inherent chemotaxis defect of the Atf3-deficient PMN. Thus, Atf3-deficiency increases Cxcl1, a chemotaxic factor for PMN, in the airway but decreases the inherent ability of PMN to migrate, resulting in no net change in PMN infiltration in the lung. In our study, we did not observe any differences in exudate levels of Cxcl1 at this time point and PMN levels were actually increased in Atf3-deficient mice. The discrepancy between our results and those of Beospflug et al. could be attributed potentially to different model stimuli (TLR4 vs. TLR2/6), or the timing of exudate collection. Alternatively, this difference could be due to the fact that PMN recruitment in our peritonitis model is driven largely by resident tissue macrophages [22], whereas the study by Boespflug et al. found that Atf3-driven changes in Cxcl1 production were due primarily to lung epithelial cells [42]. Nonetheless, our observations are unlikely to be model-specific because, in addition to the acute peritonitis model, we also observed that Atf3-deficient mice have elevated PMN infiltration into skin wounds. Moreover, a similar increase in PMN recruitment has been reported in Atf3-deficient mice during renal ischemia/reperfusion injury [43]. We note that one limitation of our study was the inability to selectively interfere with Atf3 expression in tissue macrophages given that the mice used here were globally deficient in Atf3. In addition, the increase in PMN recruitment in Atf3-deficient mice is likely multifactorial and cannot be solely attributed to COX-2. Clearly, future studies are warranted to interrogate fully the role of Atf3 in leukocyte trafficking and how it relates to the suppressive effects of Atf3 on Ptsg2 transcription in determining the overall inflammatory response.
Inhibition of COX enzymes through NSAIDs has become the mainstay of therapy for chronic inflammation [4, 44], although little is understood about transcriptional repression of Ptgs2. Therefore, dysregulation of the Atf3-Ptgs2 axis may be an important contributing factor underlying increased Ptgs2 expression in chronic diseases and this axis may be a new target for novel pharmacologic intervention.
Supplementary Material
Expression of Ptgs1, microsomal prostaglandin E synthase 1 (mPges1) and hematopoietic prostaglandin D synthase (hPgds) in macrophages isolated from WT or Atf3-deficient mice and left untreated or stimulated with zymosan for 6h. Results are mean ± SEM, n=5–6/group. *P<0.05; NS: not significant.
-
-
Ptgs2 and Atf3 are induced by inflammatory stimuli in macrophages.
-
-
Atf3 binds to the promoter region of the Ptgs2 gene.
-
-
Loss of Atf3 in macrophages increases Ptgs2 levels.
-
-
Atf3 KO mice have increased leukocytes and Ptgs2 levels during acute inflammation.
Acknowledgements
We thank Nalinie Wickramasinghe for expert technical assistance.
This work was supported in part by grants from the National Institutes of Health HL106173 and GM103492). J.H. is the recipient of a National Research Service Award from the National Heart, Lung and Blood Institute (HL116186). These funding sources played no role design or execution of these studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authorship contributions
J.H., Y.T., and M.J.Z. carried out experiments, analyzed data and contributed to the writing of the manuscript; A.B. and S.S. contributed to the planning of the project and writing the manuscript; T. H. provided the Atf3-deficient mice and contributed to the writing of the manuscript. M.S. planned the project, analyzed data and wrote the manuscript.
Disclosure of Conflicts of Interest
None
References
- 1.Hata AN, Breyer RM. Pharmacology and signaling of prostaglandin receptors: multiple roles in inflammation and immune modulation. Pharmacol Ther. 2004;103:147–166. doi: 10.1016/j.pharmthera.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Samuelsson B. Role of basic science in the development of new medicines: examples from the eicosanoid field. J Biol Chem. 2012;287:10070–10080. doi: 10.1074/jbc.X112.351437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smyth EM, Grosser T, Wang M, Yu Y, FitzGerald GA. Prostanoids in health and disease. J Lipid Res. 2009;50(Suppl):S423–S428. doi: 10.1194/jlr.R800094-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ricciotti E, FitzGerald GA. Prostaglandins and inflammation. Arterioscler Thromb Vasc Biol. 2011;31:986–1000. doi: 10.1161/ATVBAHA.110.207449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilroy DW, Colville-Nash PR, Willis D, Chivers J, Paul-Clark MJ, Willoughby DA. Inducible cyclooxygenase may have anti-inflammatory properties. Nat Med. 1999;5:698–701. doi: 10.1038/9550. [DOI] [PubMed] [Google Scholar]
- 6.Kang YJ, Mbonye UR, DeLong CJ, Wada M, Smith WL. Regulation of intracellular cyclooxygenase levels by gene transcription and protein degradation. Prog Lipid Res. 2007;46:108–125. doi: 10.1016/j.plipres.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin Y, Tang X, Zhu Y, Shu T, Han X. Identification of PARP-1 as one of the transcription factors binding to the repressor element in the promoter region of COX-2. Arch Biochem Biophys. 2011;505:123–129. doi: 10.1016/j.abb.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 8.Wang WL, Lee YC, Yang WM, Chang WC, Wang JM. Sumoylation of LAP1 is involved in the HDAC4-mediated repression of COX-2 transcription. Nucleic Acids Res. 2008;36:6066–6079. doi: 10.1093/nar/gkn607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Subbaramaiah K, Dannenberg AJ. Cyclooxygenase-2 transcription is regulated by human papillomavirus 16 E6 and E7 oncoproteins: evidence of a corepressor/coactivator exchange. Cancer Res. 2007;67:3976–3985. doi: 10.1158/0008-5472.CAN-06-4273. [DOI] [PubMed] [Google Scholar]
- 10.Hai T, Hartman MG. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene. 2001;273:1–11. doi: 10.1016/s0378-1119(01)00551-0. [DOI] [PubMed] [Google Scholar]
- 11.Gilchrist M, Thorsson V, Li B, Rust AG, Korb M, Roach JC, et al. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006;441:173–178. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- 12.Whitmore MM, Iparraguirre A, Kubelka L, Weninger W, Hai T, Williams BR. Negative regulation of TLR-signaling pathways by activating transcription factor-3. J Immunol. 2007;179:3622–3630. doi: 10.4049/jimmunol.179.6.3622. [DOI] [PubMed] [Google Scholar]
- 13.Giraldo A, Barrett OP, Tindall MJ, Fuller SJ, Amirak E, Bhattacharya BS, et al. Feedback regulation by Atf3 in the endothelin-1-responsive transcriptome of cardiomyocytes: Egr1 is a principal Atf3 target. Biochem J. 2012;444:343–355. doi: 10.1042/BJ20120125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartman MG, Lu D, Kim ML, Kociba GJ, Shukri T, Buteau J, et al. Role for activating transcription factor 3 in stress-induced beta-cell apoptosis. Mol Cell Biol. 2004;24:5721–5732. doi: 10.1128/MCB.24.13.5721-5732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hellmann J, Zhang MJ, Tang Y, Rane M, Bhatnagar A, Spite M. Increased saturated fatty acids in obesity alter resolution of inflammation in part by stimulating prostaglandin production. J Immunol. 2013;191:1383–1392. doi: 10.4049/jimmunol.1203369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Y, Zhang MJ, Hellmann J, Kosuri M, Bhatnagar A, Spite M. Proresolution therapy for the treatment of delayed healing of diabetic wounds. Diabetes. 2013;62:618–627. doi: 10.2337/db12-0684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diaz-Munoz MD, Osma-Garcia IC, Cacheiro-Llaguno C, Fresno M, Iniguez MA. Coordinated upregulation of cyclooxygenase-2 and microsomal prostaglandin E synthase 1 transcription by nuclear factor kappa B and early growth response-1 in macrophages. Cell Signal. 2010;22:1427–1436. doi: 10.1016/j.cellsig.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 18.Brown GD, Taylor PR, Reid DM, Willment JA, Williams DL, Martinez-Pomares L, et al. Dectin-1 is a major beta-glucan receptor on macrophages. J Exp Med. 2002;196:407–412. doi: 10.1084/jem.20020470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Underhill DM. Macrophage recognition of zymosan particles. J Endotoxin Res. 2003;9:176–180. doi: 10.1179/096805103125001586. [DOI] [PubMed] [Google Scholar]
- 20.Abramovitz M, Adam M, Boie Y, Carriere M, Denis D, Godbout C, et al. The utilization of recombinant prostanoid receptors to determine the affinities and selectivities of prostaglandins and related analogs. Biochim Biophys Acta. 2000;1483:285–293. doi: 10.1016/s1388-1981(99)00164-x. [DOI] [PubMed] [Google Scholar]
- 21.Lemos HP, Grespan R, Vieira SM, Cunha TM, Verri WA, Jr, Fernandes KS, et al. Prostaglandin mediates IL-23/IL-17-induced neutrophil migration in inflammation by inhibiting IL-12 and IFNgamma production. Proc Natl Acad Sci U S A. 2009;106:5954–5959. doi: 10.1073/pnas.0812782106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cailhier JF, Partolina M, Vuthoori S, Wu S, Ko K, Watson S, et al. Conditional macrophage ablation demonstrates that resident macrophages initiate acute peritoneal inflammation. J Immunol. 2005;174:2336–2342. doi: 10.4049/jimmunol.174.4.2336. [DOI] [PubMed] [Google Scholar]
- 23.Palmblad J, Malmsten CL, Uden AM, Radmark O, Engstedt L, Samuelsson B. Leukotriene B4 is a potent and stereospecific stimulator of neutrophil chemotaxis and adherence. Blood. 1981;58:658–661. [PubMed] [Google Scholar]
- 24.Hai T, Wolford CC, Chang YS. ATF3, a hub of the cellular adaptive-response network, in the pathogenesis of diseases: is modulation of inflammation a unifying component? Gene Expr. 2010;15:1–11. doi: 10.3727/105221610x12819686555015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kihara Y, Matsushita T, Kita Y, Uematsu S, Akira S, Kira J, et al. Targeted lipidomics reveals mPGES-1-PGE2 as a therapeutic target for multiple sclerosis. Proc Natl Acad Sci U S A. 2009;106:21807–21812. doi: 10.1073/pnas.0906891106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolls JK, Linden A. Interleukin-17 family members and inflammation. Immunity. 2004;21:467–476. doi: 10.1016/j.immuni.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 27.Pelletier M, Maggi L, Micheletti A, Lazzeri E, Tamassia N, Costantini C, et al. Evidence for a cross-talk between human neutrophils and Th17 cells. Blood. 2010;115:335–343. doi: 10.1182/blood-2009-04-216085. [DOI] [PubMed] [Google Scholar]
- 28.Yao C, Sakata D, Esaki Y, Li Y, Matsuoka T, Kuroiwa K, et al. Prostaglandin E2-EP4 signaling promotes immune inflammation through Th1 cell differentiation and Th17 cell expansion. Nat Med. 2009;15:633–640. doi: 10.1038/nm.1968. [DOI] [PubMed] [Google Scholar]
- 29.Boniface K, Bak-Jensen KS, Li Y, Blumenschein WM, McGeachy MJ, McClanahan TK, et al. Prostaglandin E2 regulates Th17 cell differentiation and function through cyclic AMP and EP2/EP4 receptor signaling. J Exp Med. 2009;206:535–548. doi: 10.1084/jem.20082293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buckley CD, Gilroy DW, Serhan CN. Proresolving lipid mediators and mechanisms in the resolution of acute inflammation. Immunity. 2014;40:315–327. doi: 10.1016/j.immuni.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levy BD, Clish CB, Schmidt B, Gronert K, Serhan CN. Lipid mediator class switching during acute inflammation: signals in resolution. Nat Immunol. 2001;2:612–619. doi: 10.1038/89759. [DOI] [PubMed] [Google Scholar]
- 32.Litvak V, Ramsey SA, Rust AG, Zak DE, Kennedy KA, Lampano AE, et al. Function of C/EBPdelta in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals. Nat Immunol. 2009;10:437–443. doi: 10.1038/ni.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng WG, Zhu Y, Wu KK. Role of p300 and PCAF in regulating cyclooxygenase-2 promoter activation by inflammatory mediators. Blood. 2004;103:2135–2142. doi: 10.1182/blood-2003-09-3131. [DOI] [PubMed] [Google Scholar]
- 34.Xie W, Fletcher BS, Andersen RD, Herschman HR. v-src induction of the TIS10/PGS2 prostaglandin synthase gene is mediated by an ATF/CRE transcription response element. Mol Cell Biol. 1994;14:6531–6539. doi: 10.1128/mcb.14.10.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schroer K, Zhu Y, Saunders MA, Deng WG, Xu XM, Meyer-Kirchrath J, et al. Obligatory role of cyclic adenosine monophosphate response element in cyclooxygenase-2 promoter induction and feedback regulation by inflammatory mediators. Circulation. 2002;105:2760–2765. doi: 10.1161/01.cir.0000018127.10968.34. [DOI] [PubMed] [Google Scholar]
- 36.Kang YJ, Wingerd BA, Arakawa T, Smith WL. Cyclooxygenase-2 gene transcription in a macrophage model of inflammation. J Immunol. 2006;177:8111–8122. doi: 10.4049/jimmunol.177.11.8111. [DOI] [PubMed] [Google Scholar]
- 37.Hua B, Tamamori-Adachi M, Luo Y, Tamura K, Morioka M, Fukuda M, et al. A splice variant of stress response gene ATF3 counteracts NF-kappaB-dependent anti-apoptosis through inhibiting recruitment of CREB-binding protein/p300 coactivator. J Biol Chem. 2006;281:1620–1629. doi: 10.1074/jbc.M508471200. [DOI] [PubMed] [Google Scholar]
- 38.Dalli J, Serhan CN. Specific lipid mediator signatures of human phagocytes: microparticles stimulate macrophage efferocytosis and pro-resolving mediators. Blood. 2012;120:e60–e72. doi: 10.1182/blood-2012-04-423525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Serezani CH, Kane S, Medeiros AI, Cornett AM, Kim SH, Marques MM, et al. PTEN directly activates the actin depolymerization factor cofilin-1 during PGE2-mediated inhibition of phagocytosis of fungi. Sci Signal. 2012;5:ra12. doi: 10.1126/scisignal.2002448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morris T, Stables M, Hobbs A, de Souza P, Colville-Nash P, Warner T, et al. Effects of low-dose aspirin on acute inflammatory responses in humans. J Immunol. 2009;183:2089–2096. doi: 10.4049/jimmunol.0900477. [DOI] [PubMed] [Google Scholar]
- 41.Schwab JM, Chiang N, Arita M, Serhan CN. Resolvin E1 and protectin D1 activate inflammation-resolution programmes. Nature. 2007;447:869–874. doi: 10.1038/nature05877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boespflug ND, Kumar S, McAlees JW, Phelan JD, Grimes HL, Hoebe K, et al. ATF3 is a novel regulator of mouse neutrophil migration. Blood. 2014;123:2084–2093. doi: 10.1182/blood-2013-06-510909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li HF, Cheng CF, Liao WJ, Lin H, Yang RB. ATF3-mediated epigenetic regulation protects against acute kidney injury. J Am Soc Nephrol. 2010;21:1003–1013. doi: 10.1681/ASN.2009070690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.FitzGerald GA, Patrono C. The coxibs, selective inhibitors of cyclooxygenase-2. N Engl J Med. 2001;345:433–442. doi: 10.1056/NEJM200108093450607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of Ptgs1, microsomal prostaglandin E synthase 1 (mPges1) and hematopoietic prostaglandin D synthase (hPgds) in macrophages isolated from WT or Atf3-deficient mice and left untreated or stimulated with zymosan for 6h. Results are mean ± SEM, n=5–6/group. *P<0.05; NS: not significant.