Abstract
Iron affects the physiology of bacteria in two different ways: as a micronutrient for bacterial growth and as a catalyst for the formation of hydroxyl radicals. In this study, we used DNA microarrays to identify the C. jejuni genes that have their transcript abundance affected by iron availability. The transcript levels of 647 genes were affected after the addition of iron to iron-limited C. jejuni cells. Several classes of affected genes were revealed within 15 min, including immediate-early response genes as well as those specific to iron acquisition and metabolism. In contrast, only 208 genes were differentially expressed during steady-state experiments comparing iron-rich and iron-limited growth conditions. As expected, genes annotated as being involved in either iron acquisition or oxidative stress defense were downregulated during both time course and steady-state experiments, while genes encoding proteins involved in energy metabolism were upregulated. Because the level of protein glycosylation increased with iron limitation, iron may modulate the level of C. jejuni virulence by affecting the degree of protein glycosylation. Since iron homeostasis has been shown to be Fur regulated in C. jejuni, an isogenic fur mutant was used to define the Fur regulon by transcriptome profiling. A total of 53 genes were Fur regulated, including many genes not previously associated with Fur regulation. A putative Fur binding consensus sequence was identified in the promoter region of most iron-repressed and Fur-regulated genes. Interestingly, a fur mutant was found to be significantly affected in its ability to colonize the gastrointestinal tract of chicks, highlighting the importance of iron homeostasis in vivo. Directed mutagenesis of other genes identified by the microarray analyses allowed the characterization of the ferric enterobactin receptor, previously named CfrA. Chick colonization assays indicated that mutants defective in enterobactin-mediated iron acquisition were unable to colonize the gastrointestinal tract. In addition, a mutation in a receptor (Cj0178) for an uncharacterized iron source also resulted in reduced colonization potential. Overall, this work documents the complex response of C. jejuni to iron availability, describes the genetic network between the Fur and iron regulons, and provides insight regarding the role of iron in C. jejuni colonization in vivo.
Iron is known to catalyze a wide range of biochemical reactions essential for most living organisms (1). For example, it plays a crucial role as a cofactor in DNA synthesis as well as in electron transfer reactions. The broad use of this transition metal is due to its physicochemical properties (15). Iron readily forms complexes with common biological donor ligands such as oxygen, nitrogen, and sulfur. This allows its insertion into the active sites of many metabolic proteins. Iron's value resides in the reactivity of the Fe3+/Fe2+ redox couple, which enables it to catalyze enzymatic reactions (15). Paradoxically, this iron reactivity is also responsible for the generation of hydroxyl radical (·OH), which is particularly biotoxic (29). The production of this highly reactive radical is a component of oxidative stress and can damage all biological macromolecules (17, 29).
Iron bioavailability in an aerobic neutral pH environment and in the mammalian host is limited to 10−18 M and 10−24 M, respectively. This level is far below the minimum requirement for bacterial growth (10−7 M) (7). Consequently, microorganisms have evolved complex systems to efficiently capture iron, regulate its acquisition, and detoxify its excess. Many bacteria acquire iron by synthesizing and exporting powerful ferric ion chelators called siderophores (7). Together with cell surface receptors specific for the iron-siderophore complexes, they provide iron to the organisms under the most nutritionally depleted conditions. Other bacterial iron transporter mechanisms include reduction via a surface reductase and subsequent transport of the ferrous iron across the membrane (38), proteolytic degradation of the host iron-binding proteins (9), and surface receptors for mammalian iron carriers such as transferrin, lactoferrin, and heme (1).
The significance of iron acquisition and metabolism for successful microbial proliferation is displayed by numerous examples from medical and environmental biology (33). One example is Pseudomonas aeruginosa, an opportunistic human pathogen. Siderophore-deficient mutants of P. aeruginosa show no virulence when injected into burned mice (the infection model) (24). Remarkably, wild-type virulence is restored by coinjection with a purified siderophore (24). Siderophore production has also been shown to be a virulence-associated factor for several other pathogens, including Escherichia coli (19, 25), Yersinia pestis (4), and Aeromonas hydrophila (22). Undoubtedly, iron is a key environmental signal for the pathogenesis of many, if not all, human pathogens.
Members of the bacterial genus Campylobacter are responsible for an outstanding number of food-borne infections (23). Upon entrance into the mammalian host, Campylobacter must colonize, survive, and replicate in the gastrointestinal tract and should consequently be able to efficiently acquire the essential iron nutrient. Only a few strains of C. jejuni have been shown to produce siderophores (7 strains of 26 tested), and these siderophores are uncharacterized (11). It is not known yet if C. jejuni NCTC 11168 (the strain used in our study) produces a siderophore. However, all strains of Campylobacter tested were able to acquire iron from enterobactin, a siderophore produced by Escherichia coli and other enteric bacteria (11). Analysis of the annotated genome sequence of C. jejuni NCTC 11168 predicts the lack of genes required for enterobactin biosynthesis, suggesting that Campylobacter may acquire iron in the gastrointestinal tract via siderophores (such as enterobactin) produced by the indigenous microflora, even if it does not synthesize its own siderophore.
In order to avoid iron toxicity, microorganisms must achieve an effective iron homeostasis by tightly regulating the expression of genes encoding the proteins involved in iron acquisition and metabolism in response to iron availability (33). In gram-negative bacteria, Fur is the ferric uptake regulator for the transcription of these genes (1, 6). The Fur protein is a homodimer which, upon binding its corepressor Fe2+, binds to a consensus sequence (named the Fur box) at the promoter of Fur-regulated genes, repressing their transcription (1, 6).
The ferric uptake regulator gene (fur) from C. jejuni NCTC 11168 was characterized previously (50, 55). The in vitro protein profile of a fur null mutant was studied and revealed at least three iron-regulated proteins from the outer membrane, four from the periplasm and periplasmic membrane and two from the cytoplasm (50). The N-terminal amino acid sequences of two of the outer membrane proteins, CfrA and ChuA, have high homology with ferric siderophore receptors. A C. jejuni strain lacking ChuA was unable to use hemoglobin or hemin as an iron source; thus, ChuA is likely the receptor for these compounds (54). The two iron-regulated cytoplasmic proteins, catalase (KatA) and alkyl hydroperoxide reductase (AhpC), were identified as oxidative stress defense proteins. The other proteins were not characterized further. Interestingly, in contrast to other gram-negative bacteria, oxidative stress and iron acquisition are regulated separately in C. jejuni (54). While genes encoding proteins involved in iron uptake have been shown to be Fur regulated, genes encoding proteins involved in the oxidative stress defense have been shown to be PerR regulated (54). In fact, previous studies have shown that the expression of katA and ahpC is iron repressed via the PerR regulator (51).
The complete genome sequence of C. jejuni NCTC 11168 suggests the presence of several other potential iron assimilation mechanisms (28). These consist of at least five putative iron-binding proteins, two putative outer membrane ferric siderophore receptors, two putative iron uptake ABC transporter systems (permeases and ATP transporters), a complete putative hemin uptake transporter system, a ferrous iron transporter protein, and three putative TonB/ExbB/ExbD complexes. In addition to its nutritional role, iron availability has also been shown to be a key signal for pathogens to sense that they have invaded the host. Indeed, virulence factors other than iron transport systems are often iron regulated (33). However, to date little is known about the iron regulon in Campylobacter species and whether other Campylobacter virulence determinants are regulated by iron availability.
Here we present a genomewide picture of the C. jejuni NCTC 11168 response to iron availability with DNA microarray technology. Moreover, this study further defines the importance of iron homeostasis and acquisition in vivo.
MATERIALS AND METHODS
Bacterial strain and growth conditions.
The bacterial strains used in this study are listed in Table 1. E. coli DH5α was routinely cultured aerobically at 37°C in Luria-Bertani (LB) broth or on LB agar plates. Plasmid-containing strains were grown in medium supplemented with chloramphenicol at 20 μg/ml. C. jejuni NCTC 11168 was obtained from the National Collection of Type Cultures and routinely maintained at 37°C in a microaerophilic chamber (Don Whitley, West Yorkshire, England) containing 83% N2, 4% H2, 8% O2 and 5% CO2 on Mueller-Hinton (MH) agar plates, MH medium, or minimal essential medium alpha (MEM α) (Invitrogen) with chloramphenicol added as required at a concentration of 20 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli DH5α | endA1 hsdR17(rK− mK−) supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA argF U169 deoR [φ80dlacΔ(lacZΔM15)] | Invitrogen |
| C. jejuni | ||
| AS 144 | C. jejuni NCTC 11168 | National Collection of Type Cultures |
| AS 230 | AS 144 Δfur | This study |
| AS 269 | AS 144 ΔcfrA | This study |
| AS 265 | AS 144 ΔceuE | This study |
| AS 211 | AS 144 ΔCj0178 | This study |
| Plasmids | ||
| pUC19 | Cloning and suicide vector, Ampr | Biolabs |
| pRY 111 | Cmr resistance gene | 57 |
| pAS 226 | pUC19 carrying fur | This study |
| pAS 227 | pUC19 carrying Δfur | This study |
| pAS 229 | pUC19 carrying Δfur::cmr | This study |
| pAS 266 | pUC19 carrying cfrA | This study |
| pAS 267 | pUC19 carrying ΔcfrA | This study |
| pAS 268 | pUC19 carrying ΔcfrA::cmr | This study |
| pAS 261 | pUC19 carrying ceuE | This study |
| pAS 262 | pUC19 carrying ΔceuE | This study |
| pAS 263 | pUC19 carrying ΔceuE::cmr | This study |
| pAS 207 | pUC19 carrying Cj0178 | This study |
| pAS 208 | pUC19 carrying ΔCj0178 | This study |
| pAS 209 | pUC19 carrying ΔCj0178::cmr | This study |
Cmr, chloramphenicol resistance gene; Ampr, ampicillin resistant.
Microarray construction.
The C. jejuni NCTC 11168 microarray was constructed as previously described (40, 42). Briefly, a set of 3,308 oligonucleotides were designed with Primer 3 (code available at http://www.genomewi.mit.edu/genome_software/other/primer3.htm) to amplify, via PCR, an internal fragment from each of the 1,654 predicted open reading frames (ORFs) identified in the annotated genomic sequence of C. jejuni NCTC 11168. Genomic DNA (20 ng) was used as a template in the first round of PCR amplifications with standard methods in a 96-well plate format. The PCR products were analyzed by agarose gel electrophoresis. Successful PCR products were reamplified to reduce the amount of residual genomic DNA carried over from the first PCR. PCRs without product or with incorrectly sized products were performed again by modifying the reaction conditions or by designing new primers.
DNA fragments were obtained for approximately 98% of the ORFs. PCR products were purified with the Millipore PCR96 cleanup kit and quantified with the PicoGreen double-stranded DNA quantitation reagent from Molecular Probes. They were diluted in a 50% dimethyl sulfoxide solution at a concentration of 75 ng/μl and rearrayed into a 384-well format. They were then printed on aminosilane-coated glass microscope slides (CMT GAPS-II; Corning Inc., Corning, N.Y.) with an arrayer robot (Molecular Dynamic) in a repeating 22 by 7 spot pattern. Each block was printed in duplicate. Finally, the DNA fragments were immobilized onto the slides by baking at 80°C for 4 h. The quality of the microarray printing, the efficiency of the DNA binding to the slide, and the spot morphology were assessed by direct labeling of the spotted DNA with a fluorescent nucleic acid stain (POPO-3 {POPO-3: 2,2′-[1,3-propanediylbis[(dimethyliminio)-3,1-propanediyl-1(4H)-pyridinyl-4-ylidene-1-propen-1-yl-3-ylidene]]bis[3-methyl]} iodide; Molecular Probes). In addition, the hybridization capacity of the bound DNA was confirmed with fluorescently labeled genomic DNA.
Sampling and isolation of total RNA.
The iron-restricted Campylobacter cells were grown in 250 ml of MEM α (Invitrogen) microaerobically at 37°C. At early mid-log phase (optical density at 600 nm of approximately 0.1), a 50-ml sample was removed (time zero) and immediately mixed with 5 ml of a cold RNA degradation stop solution (10% buffer-saturated phenol in ethanol), which has been previously shown to keep the bacterial transcriptome intact (5, 42). Ferrous sulfate was added to the remaining 200 ml of broth culture to a concentration of 40 μM. Then, samples of 25 ml were collected at 1, 3, 5, 7, 9, and 15 min after the addition of ferrous ion, rapidly mixed with 2.5 ml of the RNA degradation stop solution, and placed on ice. Cells were immediately collected by centrifugation at 4°C (10 min, 8,000 × g) and resuspended in lysozyme-TE buffer (50 mM Tris-Cl [pH 8], 1 mM EDTA, 0.5 mg/ml lysozyme). Total RNA was isolated with a hot phenol-chloroform protocol (47). After ethanol precipitation, the RNA was resuspended in RNase-free water, and the remaining traces of genomic DNA were removed by two consecutive treatments with DNase I (Invitrogen) and the RNA was further purified with a RNeasy kit (Qiagen, Valencia, Calif.). The absence of genomic DNA was confirmed by PCR with several sets of primers previously used for the microarray construction. The RNA concentration was determined with the RiboGreen RNA quantitation reagent from Molecular Probes, following the manufacturer's protocol. The RNA integrity was evaluated by agarose gel electrophoresis. Purified total RNA was stored at −80°C.
Probe labeling and slide hybridization.
Total RNA from each growth condition was converted to cDNAs in the presence of aminoallyl-dUTP with Superscript II (Invitrogen) at 42°C. The reverse transcription reaction was performed as follows: 16 μg of total RNA was combined with 10 μg of random hexamers in a 34.35-μl reaction mixture containing 8 μl of 5× Superscript II reverse transcriptase buffer and 2 μl of 0.1 M dithiothreitol. After a 5-min incubation at 65°C, the reaction mixture was brought to a final volume of 40 μl by adding (final concentrations): 0.5 mM each of dGTP, ATP, and CTP; 0.16 mM dTTP; 0.34 mM aminoallyl-dUTP; and 2 μl of Superscript II and incubated at 42°C for 120 min.
At the end of the first-strand synthesis, the reaction was stopped and the RNA was hydrolyzed by adding 4 μl of 50 mM EDTA and 2 μl of 10 N NaOH and incubating at 65°C for 20 min. This reaction was neutralized by adding 4 μl of 5 M acetic acid. The aminoallyl-labeled cDNA was purified from free amines and unincorporated aminoallyl-dUTP by adding 450 μl of water and spinning through a Microcon YM-30 filter (Millipore) for 8 min at 8,000 × g. This washing step was repeated three times. After the last wash, the aminoallyl-labeled probes were concentrated to less than 8 μl under vacuum in a SpeedVac and adjusted to a final volume of 10 μl by adding 1 μl of 1 M sodium carbonate (pH 9.0) and water. The resulting aminoallyl-labeled cDNA was coupled to monoreactive fluors (Amersham) by adding 10 μl of dimethyl sulfoxide containing one-sixth of one reaction vial FluoroLink indocarbocyanine or indodicarbocyanine dye and incubating for 45 min at room temperature in the dark. This reaction was quenched by adding 4.5 μl of 4 M hydroxylamine and incubating for 15 min at room temperature in the dark.
Fluorescent indocarbocyanine- and indodicarbocyanine-labeled cDNAs were combined and purified with Qiaquick PCR spin columns according to the manufacturer's instructions (Qiagen, Valencia, Calif.). The fluor-labeled cDNA mix was dried under vacuum with a SpeedVac and resuspended in 15.14 μl of water, to which was added 2.5 μl of salmon sperm DNA (10 mg/ml), 9 μl of 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate, pH 7), 0.36 μl of 10% sodium dodecyl sulfate (SDS), and 9 μl of formamide. Prior to hybridization, microarray slides were prehybridized at 42°C for 45 min in prehybridization buffer (25% formamide, 5× SSC buffer, 0.1% SDS, and 1% bovine serum albumin), rinsed with water, and dried by spinning. The probe was denatured by boiling for 2 min, followed by cooling to 42°C. The probe was then applied to the microarray slide under a coverslip (Grace Bio-labs), placed in a humidified chamber (ArrayIt), and incubated at 42°C overnight. Following hybridization, slides were washed in 2× SSC-0.1% SDS for 5 min at 42°C, 0.1× SSC-0.1% SDS for 10 min at room temperature, and four times in 0.1× SSC for 1 min at room temperature. Slides were then rinsed with distilled water and dried by centrifugation.
Data collection and analysis.
Microarrays were scanned with a ScanArray 3000 confocal scanner (Perkin Elmer) at 10-μm resolution and analyzed with GenePix Pro 4 software (Axon Instruments, Foster City, Calif.). Spots were removed from further analysis if either of two criteria was met: the spots were localized within regions of hybridization or slide abnormalities, or the fluorescent mean intensities in both channels, 1 (indodicarbocyanine) and 2 (indocarbocyanine), were below three times the standard deviation of the local background. By the second criterion, all 192 negative controls were uniformly excluded from the microarray data. Then, the fluorescence intensity in each wavelength was normalized by applying a locally weighted linear regression (Lowess) with the MIDAS software (available from TIGR, http://www.tigr.org/software/) (35). Following normalization, the ratio of channels 2 to 1 was log2 transformed, and the data were statistically analyzed with the empirical Bayes method (21).
The time course experiment was repeated twice (biological replicate), and at least two measurements were generated per experiment (technical replicate). For the comparison of the wild-type and Fur mutant transcriptomes, the microarray experiment was repeated three times (biological replicate). Genes were selected as being differentially expressed if their P value was equal to or below 10−6 and their change in transcript abundance was >2-fold. Finally, genes were grouped by hierarchical clustering analysis with the Genesis software, available from Graz University of Technology (http://genome.tugraz.at) (44).
Real-time quantitative RT-PCR.
Real-time quantitative RT-PCR was performed with the ABI Prism 7700 DNA analyzer (Applied Biosystems, Foster City, Calif.) and the QuantiTect SYBR Green RT-PCR kit (Qiagen, Valencia, Calif.) according to the following protocol. This reverse transcription step was immediately followed by concomitant activation of the HotStart Taq DNA polymerase and deactivation of the reverse transcriptases by heating the reaction at 95°C for 15 min. PCR amplification was composed of 35 cycles of denaturation at 94°C for 15 s, annealing at 55°C for 30 s, and extension at 72°C for 45 s.
In order to confirm the generation of specific PCR products, the PCR was immediately followed by melting curve analysis of the RT-PCR product according to the manufacturer's recommendations (Applied Biosystems). The microarray data for 22 genes were confirmed by real-time RT-PCR analysis (chuC, chuD, Cj1658, exbB2, flaA, flgE2, fliD, fliS, ilvC, kpsE, kpsM, lpxK, p19, peb2, peb3, slyD, tonB2, waaD, waaE, pglE [also named wlaK], pglF [also named wlaL], and pglH [also named wlaC]). The primers used are the same as those utilized for the construction of the microarray. The sequences of these primers are available upon request. The relative expression level of each gene was normalized to either slyD (encoding the peptidyl-prolyl cis-trans isomerase) or ilvC (encoding the ketol-acid reductoisomerase). The expression of both slyD and ilvC was found to be invariant under different growth conditions, allowing the use of their expression level as a reference value for quantification. Quantitative values were obtained with the comparative threshold cycle (ΔΔCT) method, as recommended by Applied Biosystems. The CT value corresponds to the PCR cycle at which there is the first detectable increase in fluorescence associated with the exponential growth of the PCR products. The transcript level from each RNA sample was assayed six times, and the mean CT value was used for further analysis. For data analysis, the induction of the specific genes was calculated as 2−ΔΔCT, where ΔΔCT = ΔCT, tmin − ΔCT, 0 min, and ΔCT, tmin and 0 min are obtained by subtracting the mean CT value of the specific gene from the mean CT value of the reference gene at time t (tmin) or time zero (0 min).
Protein electrophoresis and glycoprotein analysis.
C. jejuni NCTC 11168 grown with and without iron was harvested by centrifugation to yield individual bacterial pellets from each time point equal to an optical density of 0.3 at 600 nm. The harvested pellets were immediately frozen at −70°C. For glycoprotein analysis, frozen bacterial pellets were lysed with modified SDS sample buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, no glycerol, bromophenol blue, or β-mercaptoethanol), and protein levels were assessed by Micro BCA (Pierce). Based on the measured protein levels, dilutions of the samples were prepared in SDS sample buffer (modified sample buffer with 25% glycerol, 720 mM β-mercaptoethanol, and 0.01% bromophenol blue) to contain 4.5 μg of protein/10 μl. SDS-10 and 12% PAGE gels were loaded with 4.5 μg of total protein per lane and electrophoresed until the bromophenol blue was at the bottom of the gel. Initially, gels were silver stained (Bio-Rad Silver Stain Plus) to verify that the protein measurements from Micro BCA were accurate. If necessary, samples were adjusted to give more uniform silver staining through the use of gel imaging equipment and software (UVP Epi Chem II Darkroom and UVP labworks Image Acquisition and Analysis Software).
Once the samples were diluted to provide equal protein loading, duplicate gels were either silver stained or electrotransferred to polyvinylidene difluoride membranes (Millipore Immobilon-P). The protein blots were blocked with 1% bovine serum albumin in phosphate-buffered saline (0.01 sodium phosphate, 0.15 M sodium chloride, pH 7.2) containing 0.05% Tween 20 for 30 min, washed briefly in phosphate-buffered saline with 0.05% Tween 20, and incubated with 5 μg of horseradish peroxidase-labeled Wisteria floribunda lectin (E-YLabs) per ml in phosphate-buffered saline without Tween 20 for 1 h. After lectin incubation, the blots were washed twice for 10 min each with phosphate-buffered saline containing 0.05% Tween 20. Excess Tween 20 was removed by brief rinsing in Tris-HCl (pH 7.6). Blots were then reacted with Tris-HCl (pH 7.6), 0.6 mg of 3,3′-diaminobenzidine per ml, and 0.03% hydrogen peroxide. Data resulting from blots and silver-stained gels were digitally recorded (UVP Biodocit System).
Operon mapping.
Total RNA was extracted from exponentially growing C. jejuni cells after growth in MEM-α as described above. First-strand cDNA synthesis and subsequent PCR amplifications were performed with the Qiagen One-Step RT-PCR system according to the manufacturer's recommendations and with the primer pairs listed in Table 2 and represented in Fig. 2. From 100 to 1,000 ng of total RNA was used for each reaction.
TABLE 2.
Primers used in this study
| Primer | DNA sequence (5′-3′) |
|---|---|
| fur-01 | GGAAGATCTGGCTTAAGGGTATTATCAATG |
| fur-02 | GGAAGATCTCATATCAACATAACGTTTACG |
| fur-03 | CGCGGATCCAAGTAAAACATCATATTCCAC |
| fur-04 | CGCGGATCCGGTGTTTGTGGTGATTGTAAT |
| cfrA-01 | ATGCAGATCTGCTTCTGGCTTTACCCAAG |
| cfrA-02 | ATGCAGATCTTCCCAGCCATTTGTAAAGC |
| cfrA-03 | ATGCGGATCCTGCCTTTGTAGGACTTTGAGC |
| cfrA-04 | ATGCGGATCCAACAATGTTTCGCCAAGAGC |
| ceuE-01 | ATGCGAATTCGATACATTAAGCGGGGGACA |
| ceuE-02 | ATGCGCATGCTGTGCTTAGCGTAGGTTTTGG |
| ceuE-03 | ATGCGGATCCTTTTTAGCAGGAACGCCAAC |
| ceuE-04 | ATGCGGATCCGCAACAAAGAACGTGCTCAA |
| Cj0178-01 | ATGCGAATTCTTTGTAACTCCTGGGGCAAC |
| Cj0178-02 | ATGCGAATTCTTGCACCTCTGCTCTCATTG |
| Cj0178-03 | ATGCAGATCTTTTTTCTACCGGCAAGTTCG |
| Cj0178-04 | ATGCAGATCTTTTCAGCTGCCATACACGAG |
| exbB1-F | AATGCAGAAGCAAATGCAAC |
| exbD1-R | GCCATTCTCTTTTAATTTTTGCAT |
| exbD1-F | AATGGCTCACAAAGAAGAGGAG |
| tonB1-R | ATTTCTTTAGGCGGTTTTGG |
| exbB2-F | GGCATTTATAGCTTTTTGGTGTG |
| exbD2-R | AAATTTTCATGTTCTTTGGCTTT |
| exbD2-F | CATTATGCTTGTTTTGCTTGC |
| tonB2-R | TCCATTATAGTGTGGAAAATTTAAAGA |
| exbB3-F | TGGCTTTCTTTATATTTTATTTTAGCA |
| exbD3-R | TTCTTAAGACTGAAATAACATCGTCA |
| exbD3-F | TGCTTGTATTGCTTGCGATT |
| Cj0111-R | TTTTTATTTTGCGGAGGATAAGT |
| fldA-R | CATTGTCAAGAGCAAGTCCAA |
| Cj1383c-F | CCGCAAATTTATCGCTATTT |
| Cj1383c-R | AATTTGATTTGATATTTTATACGGAAC |
| Cj1384c-F | GGTGCAGATGAAATCACTCCT |
| Cj1658-F | ATGCAGATCTGCTCAGCTTTTGCTAGGGTAGA |
| p19-R | ATCAACATGGCGTCCAAAAC |
FIG. 2.
Operon mapping by RT-PCR analysis of iron- and Fur-regulated genes. The template RNA was purified from mid-log-phase bacteria grown in iron-rich and iron-limited MEM-α for iron-induced (exbB3, exbD3, and Cj0111) and iron-repressed genes, respectively. Predicted RT-PCR fragments with gene names are shown at the bottom. The gel lanes match the RT-PCR fragment labels. Lanes M1 and M2 show the 1-kb and 100-bp DNA ladders, respectively.
The cotranscription of exbB1, exbD1, and tonB1 was assayed with a combination of four primers: exbB1-F, exbD1-R, exbD1-F, and tonB1-R. The cotranscription of exbB2, exbD2, and tonB2 was assayed with primers exbB2-F, exbD2-R, exbD2-F, and tonB2-R. The cotranscription of exbB3, exbD3, and Cj0111 was assayed with primers exbB3-F, exbD3-R, exbD3-F, and Cj0111-R. The cotranscription of fldA, Cj1383c, and Cj1384c was assayed with primers fldA-R, Cj1383c-F, Cj1383c-R, and Cj1384c-F. Finally, the cotranscription of Cj1658 and p19 was assayed with primers Cj1658-F and p19-R. As a positive control, each operon was also amplified with the same primers, with chromosomal DNA as a template. As a negative control, PCRs (without previous reverse transcription) were performed on the same RNA templates to confirm the absence of contaminating genomic DNA. RT-PCR products were electrophoresed through 1% agarose gels containing ethidium bromide and visualized with UV light. Ladders of 1 kb and 100 bp (Bayou Biolabs, Harahan, La.) served as size markers.
Construction of the C. jejuni mutants.
fur, cfrA, ceuE, and Cj0178 mutants of C. jejuni NCTC 11168 were constructed with the same inactivation strategy. C. jejuni NCTC 11168 chromosomal DNA was prepared with the Wizard Genomic DNA purification kit (Promega), and the fur gene was PCR amplified with the primers fur-01 and fur-02 (both of which contain a BglII site). The PCR was catalyzed with Pfx DNA polymerase (Invitrogen) and hot-started to ensure high specificity of the products being synthesized. The resulting 1,479-bp fragment was digested with BglII and ligated to the BamHI-restricted pUC19 vector (yielding plasmid pAS226). A deletion of 390 bp was generated within the fur gene by inverse PCR with primers fur-03 and fur-04, both of which contain a BamHI restriction site. The PCR product was cut with BamHI, self-ligated, and introduced into E. coli DH5α, yielding plasmid pAS227. A BamHI-restricted chloramphenicol resistance cassette from plasmid pRY111 (57) was then cloned into pAS227 (previously digested with BamHI), yielding pAS229. The orientation of the chloramphenicol resistance cassette was determined by DNA sequencing. The plasmid construct containing the chloramphenicol resistance cassette in the same orientation as the gene of interest was used to transform C. jejuni NCTC 11168 with a standard protocol (57) to generate the fur mutant by allelic exchange. Transformants were selected on MH agar plates containing 20 μg of chloramphenicol per ml. Finally, the double homologous recombination event was confirmed by analyzing the chloramphenicol-resistant clones by PCR with the corresponding gene primer sets. The primers and plasmids used for mutant construction are listed in Table 2.
The cfrA mutant (AS269), the ceuE mutant (AS265), and the Cj0178 mutant (AS211) were constructed in a similar fashion. Briefly, the cfrA mutant, AS269, was constructed by amplification of a 1,903-bp DNA fragment containing the cfrA gene with primers cfrA-01 and cfrA-02, both of which contain a BglII site. The product was digested with BglII and cloned into the BamHI-restricted pUC19 vector, yielding plasmid pAS266. A 556-bp deletion was created within the cfrA gene by inverse PCR with primers cfrA-03 and cfrA-04 (both of which contain a BamHI site). The chloramphenicol resistance cassette was introduced into the created BamHI site of the cfrA gene, yielding the final construct pAS268.
The ceuE mutant AS265 was constructed following PCR amplification of a 1,743-bp DNA fragment containing the ceuE gene with primers ceuE-01 (which contains an EcoRI restriction site) and ceuE-02 (which contains an SphI restriction site). The product was digested with EcoRI and SphI and cloned into the EcoRI- and SphI-restricted pUC19 vector, yielding pAS261. A 522-bp deletion was created within the ceuE gene by inverse PCR with primers ceuE-03 and ceuE-04 (both of which contain a BamHI site). The chloramphenicol resistance cassette was introduced into the created BamHI site of the ceuE gene, yielding the final construct pAS263.
The Cj0178 mutant AS211 was constructed as follows: a 1,991-bp DNA fragment (harboring the Cj0178 gene) was PCR amplified with primers Cj0178-01 and Cj0178-02, both of which contain an EcoRI site. The product was digested with EcoRI and cloned into an EcoRI-restricted pUC19 vector, yielding plasmid pAS207. A deletion of 511 bp was generated within the Cj0178 gene by inverse PCR with primers Cj0178-03 and Cj0178-04, both of which contain a BglII restriction site. The chloramphenicol resistance cassette was introduced into the created BglII site of the Cj0178 gene, yielding the final construct pAS209.
Growth promotion assays.
The ability of iron sources to promote the growth of C. jejuni NCTC 11168 wild-type and mutant strains was assessed on standard iron-limited assay plates as described by others (2, 12, 13). Briefly, the bacterial strains to be tested were grown to mid-log phase in MH medium, harvested by centrifugation, and resuspended in MH medium to an optical density of 1.0 at 600 nm; 1 ml of this bacterial suspension was added to 24 ml of melted MH agar containing 40 μM desferriferrioxamine mesylate salt (DFO), poured into petri dishes, and allowed to solidify. Sterile disks containing 10 μl of enterobactin (at a concentration of 10 mM) were laid on the surface, and growth zones were measured after 24 h of incubation at 37°C under microaerophilic conditions. Enterobactin was isolated from bacterial growth supernatants by standard protocols as previously described (43).
Chick colonization assays.
One-day-old specific-pathogen-free broiler chicks were obtained from Tyson Farms, Arizona. Chicks were housed in a room maintained at 22°C and equipped with a brooder maintained at 33 to 35°C. Chicks were checked upon arrival by taking cloacal swabs for culture to verify that they did not carry Campylobacter. Chicks were provided water and a commercial chicken starter diet ad libitum. Food and water were withheld for 2 to 3 h prior to the challenge. For this challenge, C. jejuni wild-type and mutant strains were cultured in an MH broth biphasic culture medium at 37°C under microaerophilic conditions. At mid-log phase, the bacterial strains were harvested and resuspended in phosphate-buffered saline buffer. Each chick (3 to 4 days old) was inoculated orally with 1 ml of a bacterial suspension containing between 1 × 105 and 3 × 105 viable C. jejuni. A control group of uninfected birds was also included. Each strain of C. jejuni was inoculated into five birds. The ability of each C. jejuni strain to colonize the chick ceca was evaluated 4 days postchallenge by determining viable counts as described by others (12, 30). Briefly, the chicks were humanely euthanized, and their ceca were collected. The cecal contents were homogenized, serially diluted in phosphate-buffered saline, and plated onto Campylobacter agar base (Oxoid CM935) containing the Campylobacter selective karmali supplements (Oxoid SR167E). Viable counts were expressed as logarithms of CFU per gram of cecum. The data were statistically analyzed with a nonparametric Mann-Whitney rank sum test. P values below 0.05 were regarded as statistically significant.
Computational analysis of the Fur promoter.
Fur-regulated genes were selected according to the transcriptional profiling experiments. Intergenic regions and 500-bp regions extending upstream from the start codon of every gene were retrieved with the regulatory sequence analysis tools at http://rsat.ulb.ac.be/rsat/ (49). These upstream regions were analyzed for the presence of a potential promoter motif with the MEME algorithm at http://meme.sdsc.edu/meme/website/intro.html (T. L. Bailey and C. Elkan, presented at the Second International Conference on Intelligent Systems for Molecular Biology, Menlo Park, Calif., 1994). Finally, a consensus sequence logo was built by compilation of the potential motif sequences of each regulated genes with WebLogo (http://weblogo.berkeley.edu/logo.cgi) (37).
RESULTS AND DISCUSSION
Experimental design, statistical analysis, and validation of the microarray data.
Genes encoding proteins involved in iron metabolism are commonly known to be iron repressed (1, 7, 33). Therefore, in order to determine the global change in gene expression profile elicited in C. jejuni in response to iron availability, we performed two sets of experiments.
The first set of experiments addressed the immediate response of C. jejuni gene expression to the addition of ferrous sulfate. The changes in transcript levels were determined as a function of time after the addition of iron. Briefly, cultures were grown to mid-log phase in the iron-limited medium MEM-α, in which the genes encoding proteins involved in iron metabolism are highly expressed. This medium contains a trace amount of iron and has been used previously to study C. jejuni iron metabolism (50). At the initial time point (0 min), a sample of the growth culture was removed and the total RNA was extracted. Then, ferrous sulfate was added to the remaining bacterial culture at a concentration of 40 μM. Under this condition, the genes encoding proteins involved in iron metabolism are repressed.
Samples from the growth culture were then removed at 1, 3, 5, 7, 9, and 15 min, and the total RNA was purified. All total RNA samples were reverse transcribed and fluorescently labeled as previously described (40, 42). Finally, the relative abundance of gene transcripts at each time point after the addition of ferrous sulfate was compared with the level of transcripts at the 0-min time point (corresponding to the transcriptome of C. jejuni grown in iron-limited condition) by the C. jejuni NCTC 11168 microarray. Each hybridization was repeated twice, yielding two technical replicates for each time point. In addition, two independent time course experiments were carried out, constituting two biological replicates. A total of 24 measurements were performed per gene. The data were normalized, merged, and reported as the log2 ratios of the transcript abundance of C. jejuni grown in iron-starved conditions (time zero) to that of C. jejuni after the addition of ferrous sulfate to the growth medium. The significance of the differential abundance of transcripts was analyzed with a regularized t test based on a Bayesian statistical analysis of variance (22). This statistical method has been shown to be more reliable than a simple t test by reducing the false-positive rate. Genes were selected as being differentially expressed, with a P value below 10−6 (which corresponds to a significance level below 0.01 after Bonferroni correction) and a minimum of a twofold change in transcript abundance in at least one of the six time points.
The bacterial response to the sudden addition of a nutrient is not necessarily identical to the transcriptome profile during balanced growth. Indeed, the physiological response of E. coli to the sudden addition of acetate differs significantly from the transcriptome profile of E. coli during balanced growth in an acetate-containing medium (27). Therefore, the second set of experiments served to compare the transcriptome profiles of C. jejuni grown to mid-log phase in both iron-limited (MEM-α) and iron-rich medium (MEM-α containing 40 μM ferrous sulfate). This experiment was repeated three times (biological replicates) and analyzed as for the time course experiment.
To support the reliability of our microarray data, the expression of 22 genes was confirmed by real-time RT-PCR (data not shown). A high level of concordance was observed between the microarray results and the real-time RT-PCR data (with a correlation coefficient of 0.98). However, as in our previous gene expression study (in which we used the same microarray platform), microarray data analysis underestimates changes compared to real-time RT-PCR measurements (40). This difference between these two technologies has been reported previously by others and suggests a smaller dynamic range for microarray analysis (56). Nevertheless, the high level of correlation between the two technologies confirms the reliability of the data produced in this study. Moreover, our microarray data are in agreement with previous results from others who studied iron regulation in C. jejuni at the level of one or a few genes (50).
Global analysis and kinetics of C. jejuni gene expression in response to iron availability.
Table 3 highlights the number of up- and downregulated genes grouped by hierarchical clusters and functional categories, respectively. The complete statistical analysis can be found online at http://www.cvm.okstate.edu/research/Facilities/CampyLab/Index.htm. Three main points can be extracted from these data: up to 500 genes are transiently differentially expressed in the time course experiment; differentially expressed genes are equally distributed between up- and downregulated genes; and the number of genes differentially expressed during balanced growth (mid-log-phase experiment; 208 genes) is considerably smaller than the number of genes differentially expressed at 5, 7, 9, and 15 min after the addition of iron (278, 353, 462, and 370 genes, respectively). It is apparent from these data that the immediate response of C. jejuni to the sudden addition of iron differs significantly from the bacterial transcriptome during steady-state growth in the presence of iron at mid-log phase. Consequently, the observed transcriptome profile of C. jejuni at 1, 3, 5, 7, 9, and 15 min after the addition of ferrous sulfate likely represents the bacterial adaptation to the new growth medium, while the transcriptome profile at mid-log phase represents the steady-state transcriptome.
TABLE 3.
Up- and downregulated genesa
| Functional category | No. of genes
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 min
|
3 min
|
5 min
|
7 min
|
9 min
|
15 min
|
Mid-log
|
||||||||
| + | − | + | − | + | − | + | − | + | − | + | − | + | − | |
| Small-molecule degradation | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 3 | 0 | 3 | 1 | 0 |
| Energy metabolism | 0 | 0 | 6 | 2 | 14 | 9 | 14 | 17 | 18 | 22 | 11 | 17 | 23 | 2 |
| General intermediary metabolism | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 3 | 1 | 4 | 0 | 5 | 1 | 1 |
| Amino acid biosynthesis | 1 | 0 | 2 | 1 | 3 | 3 | 1 | 3 | 4 | 5 | 10 | 5 | 2 | 5 |
| Polyamine synthesis | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Purines, pyrimidines, nucleosides, and nucleotides | 0 | 0 | 1 | 1 | 2 | 1 | 4 | 2 | 5 | 2 | 7 | 2 | 3 | 0 |
| Biosynthesis of cofactors, prosthetic groups, and carriers | 0 | 0 | 2 | 1 | 3 | 4 | 2 | 6 | 2 | 9 | 2 | 9 | 1 | 8 |
| Fatty acid biosynthesis | 0 | 0 | 3 | 1 | 5 | 2 | 5 | 3 | 5 | 3 | 5 | 0 | 1 | 0 |
| Broad regulatory functions | 0 | 1 | 0 | 1 | 0 | 2 | 1 | 2 | 1 | 3 | 0 | 3 | 0 | 1 |
| Signal transduction | 0 | 0 | 2 | 2 | 2 | 6 | 2 | 7 | 4 | 5 | 1 | 5 | 0 | 2 |
| Ribosomal protein synthesis and modification | 0 | 0 | 9 | 0 | 15 | 0 | 31 | 0 | 39 | 0 | 43 | 1 | 1 | 8 |
| Ribosome maturation and modification | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 2 | 0 | 2 | 0 | 0 | 1 |
| Aminoacyl tRNA synthetases and their modification | 0 | 0 | 1 | 1 | 1 | 2 | 3 | 2 | 4 | 3 | 3 | 1 | 1 | 1 |
| Nucleoproteins | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| DNA replication, restriction/modification, repair | 0 | 0 | 0 | 0 | 2 | 1 | 3 | 2 | 8 | 3 | 5 | 1 | 2 | 2 |
| Protein translation and modification | 0 | 0 | 1 | 1 | 1 | 1 | 2 | 1 | 7 | 1 | 6 | 1 | 0 | 0 |
| RNA synthesis, RNA modification, and DNA transcription | 0 | 0 | 1 | 0 | 2 | 1 | 4 | 1 | 4 | 1 | 5 | 1 | 1 | 1 |
| Phospholipids | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Degradation of macromolecules | 0 | 0 | 0 | 0 | 2 | 0 | 3 | 0 | 2 | 0 | 1 | 2 | 1 | 2 |
| Membranes, lipoproteins, and porins | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 0 | 2 |
| Surface polysaccharides, lipopolysaccharides, and antigens | 0 | 0 | 0 | 1 | 2 | 1 | 6 | 1 | 8 | 3 | 3 | 1 | 4 | 0 |
| Surface structures | 0 | 0 | 2 | 6 | 2 | 9 | 2 | 12 | 2 | 12 | 1 | 7 | 5 | 0 |
| Murein sacculus and peptidoglycan | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 2 | 1 | 3 | 1 | 1 | 1 |
| Miscellaneous periplasmic proteins | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 2 | 1 | 2 |
| Amino acid and amine transport/binding proteins | 0 | 0 | 0 | 0 | 3 | 1 | 3 | 0 | 6 | 5 | 2 | 5 | 4 | 0 |
| Cation transport/binding proteins | 0 | 1 | 1 | 5 | 2 | 10 | 2 | 14 | 1 | 13 | 1 | 11 | 1 | 14 |
| Carbohydrate and organic acid transport/binding proteins | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 3 | 1 | 4 | 1 | 5 | 0 | 0 |
| Anion transport/binding proteins | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 2 | 0 | 2 | 0 | 2 | 0 | 0 |
| Other transport/binding proteins | 0 | 0 | 4 | 5 | 7 | 10 | 6 | 11 | 6 | 14 | 4 | 12 | 6 | 13 |
| Chaperones, chaperonines, and heat shock proteins | 0 | 0 | 0 | 1 | 3 | 3 | 3 | 1 | 3 | 1 | 2 | 2 | 0 | 2 |
| Cell division | 0 | 0 | 1 | 0 | 2 | 0 | 1 | 0 | 2 | 0 | 1 | 0 | 0 | 0 |
| Chemotaxis and mobility | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 2 | 0 | 0 | 0 |
| Protein and peptide secretion | 1 | 0 | 3 | 0 | 5 | 0 | 6 | 0 | 8 | 0 | 5 | 0 | 1 | 1 |
| Detoxification | 0 | 0 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 3 | 1 | 2 | 1 | 4 |
| Pathogenicity | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| Insertion sequence, plasmid-related function | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Drug/analogue sensitivity and antibiotic resistance | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 3 | 1 | 3 | 0 | 2 | 1 |
| Conserved hypothetical proteins | 0 | 0 | 1 | 2 | 4 | 2 | 4 | 3 | 4 | 2 | 1 | 0 | 1 | 2 |
| Unknown | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 |
| Miscellaneous | 1 | 1 | 6 | 5 | 9 | 9 | 10 | 9 | 12 | 13 | 7 | 11 | 8 | 6 |
| Hypothetical unknown proteins | 4 | 2 | 27 | 23 | 40 | 49 | 58 | 55 | 84 | 65 | 61 | 49 | 33 | 20 |
| Total | 8 | 5 | 79 | 62 | 139 | 139 | 186 | 167 | 254 | 208 | 202 | 168 | 106 | 102 |
The number of genes upregulated (+ columns) and downregulated (− columns) at different time points following the addition of ferrous sulfate to a mid-log-phase iron-limited C. jejuni culture is shown. Genes are grouped by functional categories according to the Sanger Center annotation.
Notably, groups of transcripts whose abundance was affected in the time course experiment and not in the steady-state experiment encode ribosomal proteins, surface structures, and proteins of unknown function (Table 3).
Transcript profiling of C. jejuni at mid-log phase in iron-depleted and iron-rich growth conditions.
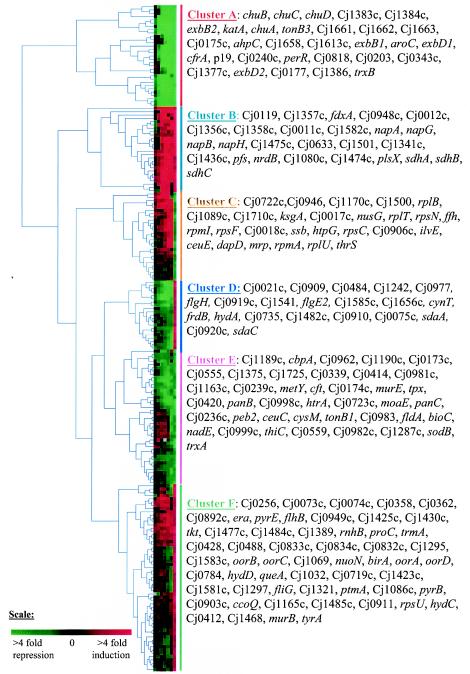
The major objective of this work was to identify the genes associated with the mechanism of iron acquisition and metabolism. Because these genes should be differentially expressed at mid-log phase in response to iron availability, we further analyzed the genes which satisfied this criterion. Globally, the transcript abundance of 208 genes was found to be significantly altered between iron-limited and iron-rich growth conditions during steady state. In order to visualize the temporal expression of these genes in response to the addition of iron, we subjected our microarray data to hierarchical clustering analysis (10, 45). This cluster analysis identified six major clusters, named A, B, C, D, E, and F (Fig. 1).
FIG.1.
Hierarchical cluster analysis of genes found to be significantly up- or downregulated at mid-log phase. Going from left to right, the columns represent the transcriptome change at 1, 3, 5, 7, 9, and 15 min after the addition of ferrous sulfate and at mid-log phase. The intensity of the color is proportional to the change, as represented by the scale at the bottom. Detailed gene names are shown for each cluster.
Clusters A and E are composed of transcripts that decreased in level in response to the addition of iron. Genes from cluster A are downregulated earlier than genes from cluster E. Genes from clusters A and E are downregulated at the steady state between 4- and 256-fold and 2- and 13-fold, respectively. As expected, genes that have been reported or annotated as playing a role in iron acquisition or detoxification belong to these two clusters (Table 4). With the exception of ferritin, which is involved in iron storage, the other proteins are likely involved in the transport of iron or iron complexes through the bacterial membrane. Our microarray data confirm the previous findings showing that the expression of chuA, cfrA, and p19 is iron regulated (50) and also identify new members of the iron regulon. While ChuA appears to be required for hemin transport in C. jejuni (34, 54), the functional annotations of the other iron uptake systems are strictly based on homologies with known proteins from other bacteria. Interestingly, genes from the third ExbB-ExbD energy-transducing complex (exbB3 and exbD3) were found to be iron induced, suggesting that this set of genes are unlikely to be involved in iron acquisition.
TABLE 4.
Some of the genes from clusters A and E that show substantial reduction in expression after the addition of ferrous sulfate
| Category and protein | Proposed functions |
|---|---|
| Iron transport and storage | |
| ChuABCD | Putative hemin uptake system |
| CfrA | Putative ferric siderophore receptor protein |
| Cj1661-1663 and Cj0173c-0175c | Putative ABC transporter systems |
| TonB3, ExbB2, ExbD2, TonB1, ExbB1, and ExbD1 | Members of the three putative TonB-ExbB-ExbD energy-transducing complexes |
| p19 and Cj1658 | Iron-regulated proteins with identity to components of iron transporters |
| Cj0177 | Putative lipoprotein sharing homology with a protein involved in iron acquisition |
| CeuC | Putative enterobactin uptake permease |
| Cft | Ferritin |
| Cj0203, Cj0891c, Cj1163c, and Cj0339 | Putative transmembrane tranporter proteins |
| Oxidative stress responses | |
| PerR | Peroxide stress regulator |
| KatA | Catalase |
| AhpC | Alkyl hydroperoxide reductase |
| Tpx | Thiol peroxidase |
| SodB | Iron-containing superoxide dismutase |
| Iron not required as cofactor, physiologically important under iron-limited conditions? | |
| Cj0414 | Putative oxidoreductase |
| Cj0723c | Zinc metalloprotease involved in macromolecule degradation |
| PanC, PanB, BioC, MoaE, ThiC, NadE, TrxA, and TrxB | Cofactor biosynthesis |
| FldA | Flavodoxin |
| Cj0559 | Oxidoreductase |
| Cj1287c | Malate oxidoreductase |
Both clusters A and E contain genes encoding proteins involved in oxidative stress defense. These proteins are the peroxide stress regulator (PerR), the catalase (KatA), the alkyl hydroperoxide reductase (AhpC), a probable thiol peroxidase (Tpx), and the iron-containing superoxide dismutase (SodB) (Table 4). SodB mediates the dismutation of superoxide anion into water and hydrogen peroxide, which is subsequently converted to water and oxygen by KatA (30). The AhpC protein catalyzes the reduction of alkyl hydroperoxide to alcohols (30). Previous reports have shown that AhpC and SodB are required for C. jejuni survival in aerobic conditions and that a C. jejuni katA mutant is affected in its ability to survive within macrophages (3, 9, 30, 39). The Tpx protein exhibits high homology with antioxidant enzymes from other bacteria, but its role in oxidative stress defense in C. jejuni requires further investigation.
Interestingly, several genes from the A and E clusters encode enzymes that do not require iron as a cofactor and are not expected to play a role in iron acquisition or detoxification and thus may play an essential physiological role under iron-limited conditions (Table 4). It will be interesting to determine what role, if any, these enzymes play in vivo.
Finally, it should be noticed that the expression of the gene annotated as encoding the ferrous iron uptake protein, feoB, was not identified as being significantly repressed by the addition of ferrous iron by the gene selection algorithm described in the Materials and Methods section. Recently, FeoB has been proposed not to be required for ferrous iron uptake in C. jejuni (32). Thus, although we expected an immediate repression of the feoB gene after ferrous iron addition, based on the sequence annotation, recent findings suggest that this gene is not likely to be critical for iron uptake and thus not necessarily iron-regulated. While feoB expression was not affected during the time course experiment, 1.9-fold repression of feoB expression (P = 1.2 × 10−5) was seen at mid-log phase. Our findings suggest that if ferrous iron does regulate the expression of feoB, the effect is long-term rather than immediate.
Clusters B and F contain genes that have their transcript abundance increased in response to the addition of ferrous sulfate. Genes from cluster B are more rapidly and highly induced than genes from cluster F. Changes at mid-log phase vary from 2.6- to 92-fold and from to 2- to 13-fold for clusters B and F, respectively. These two clusters are dominated by genes encoding proteins involved in energy metabolism. Interestingly, many of these genes encode enzymes that require iron for their function. Consequently, these enzymes may provide the most efficient cellular functions in the presence of iron (Table 5). Of interest, the oxidoreductase OorABCD may have a function complementary to that of several of the putative iron-independent oxidoreductase enzymes listed in Table 4 (Cj0414, Cj0559, and Cj1287c) under iron-rich growth conditions. From cluster B, the ferredoxin FdxA (with 14-fold induction at mid-log phase), is worth noting because these results confirm the previously reported induction of fdxA expression by iron (53). An fdxA mutant has been shown to be affected in its aerotolerance ability, suggesting a role for FdxA in oxidative stress defense (53).
TABLE 5.
Some of the genes from clusters B and F that are substantially upregulated in response to iron addition
| Protein(s) | Proposed functions related to energy metabolism |
|---|---|
| SdhABC | Succinate dehydrogenase complex |
| NapABGH | Periplasmic nitrate reductase |
| Cj1357c, Cj1358c | Putative periplasmic cytochrome c's |
| NrdB | Ribonucleoside-diphosphate reductase |
| OorABCD | 2-Oxoglutarate:acceptor oxidoreductase |
| HydC | Ni/Fe-hydrogenase B-type cytochrome subunit |
| Cj0074c | Putative iron-sulfur protein |
| Cj0358 | Putative cytochrome C551 peroxidase |
| Cj0012c | Nonheme iron protein |
Clusters C and D contain genes that are antagonistically expressed between the two sets of experiments (the time course and the mid-log experiment). Genes from cluster C have their transcript abundance increased during the first 15 min and decreased at mid-log phase. Genes from this cluster encode essentially proteins involved in macromolecule biosynthesis and modification (ribosomal proteins, proteins involved in ribosome maturation and modification, DNA replication, restriction, and modification, and RNA synthesis). Genes from cluster D have their transcript abundance decreased during the first 15 min and increased at mid-log phase. These encode proteins involved in energy metabolism, surface structures, and amino acid transport as well as many proteins of unknown function. The significance of the expression profile of the genes from these two clusters is unclear and requires further investigation. However, these results clearly highlight the impact of microarray experimental design on gene expression analysis and demonstrate that time course experiments yield data that are significantly different from those obtained by steady-state experiments.
Characterization of several iron-regulated operons.
The gene clustering analysis suggested that a number of differentially expressed genes might be cotranscribed. Therefore, we used RT-PCR to assess the cotranscription of several sets of genes that were found to be either iron repressed (exbB1-exbD1-tonB1, exbB2-exbD2-tonB2, fldA-Cj1383c-Cj1384c, and Cj1658-p19) or iron induced (exbB3-exbD3-Cj0111) by microarray analysis. None of these genes has been previously described to be cotranscribed. The transcripts were mapped from C. jejuni cells grown to mid-log phase in an iron-limited or iron-rich medium. The RT-PCRs were performed with primers that anneal across pairs of genes (Fig. 2 and Table 2). Each RT-PCR gave a product of the expected size, as shown in Fig. 2. These results confirm the transcriptional organization of these genes in operons.
The operon fldA-Cj1383c-Cj1384c is divergently transcribed from katA and in the opposite direction from its downstream gene and thus likely constitutes a single transcriptional unit. While Cj1383c and Cj1384c encode proteins of unknown function, fldA codes for a flavodoxin. The expression of these three genes was found to be repressed upon addition of iron, similar to the expression of the catalase gene katA, indicating that these other genes might also play a role in oxidative stress defense. The operon exbB2-exbD2-tonB2 is divergently trancribed from Cj1627c and is in the opposite orientation from its downstream gene Cj1631c. Therefore, this set of genes likely constitutes an independent transcriptional unit. Finally, Cj1658-p19, ExbB1-exbD1-tonB1, and exbB3-exbD3-Cj0111 appear to be transcribed together, based on our RT-PCR data.
Effect of iron on Campylobacter glycosylation profile.
Recently, C. jejuni has been shown to possess a system of “general” protein glycosylation encoded by the pgl gene cluster (the same genes were annotated wla in the genome of C. jejuni NCTC 11168) (46). In C. jejuni NCTC 11168, this cluster contains 12 genes, consecutively named wlaB, pglH (wlaC), pglI (wlaD), pglJ (wlaE), pglB (wlaF), pglA (wlaG), pglC (wlaH), pglD (wlaI), wlaJ, pglE (wlaK), pglF (wlaL), and pglG (wlaM). The mutation of several genes from this locus demonstrated the participation of their products in the general glycosylation pathway (20, 46, 58).
Up to 30 genes have been identified to encode potential glycoproteins in C. jejuni (58), of which 15 were found to be iron regulated at mid-log phase by our microarray data analysis (trxA, sodB, Cj0175c, Cj0238, ahpC, Cj0276, Cj0415, Cj0420, tpx, Cj0906c, Cj0998c, Cj1032, tsf, p19, and cgpA). Interestingly, our microarray data suggested that the transcript abundance of the pgl genes is affected by iron availability. Only wlaB, pglH, pglA, and pglC were selected as significantly upregulated (up to threefold) at 7 and 9 min. In addition, the induction of pglH (wlaC) was confirmed by real-time RT-PCR. While pglE and pglF were not selected as differentially expressed with our selection algorithm, RT-PCR analysis revealed a twofold repression (3 min after the addition of ferrous ion). The observed differential expression between the pgl genes which participate in the same pathway for protein glycosylation is unclear and requires further investigation.
Although these genes are located in a cluster within the C. jejuni genome, no current evidence about the operonic structure(s) for this region has been reported. Despite the complexity of the gene expression data for genes related to protein glycosylation and glycoprotein production, the apparent differential abundance of the transcripts encoding some of the glycoproteins and some of the Wla-Pgl proteins during the time course experiments predicted variation in the glycosylation level and therefore in lectin reactivity. In order to test this hypothesis, the lectin binding properties of whole-cell lysates were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) (Fig. 3A) and revealed (following electrotransfer of the SDS-PAGE gel to a polyvinylidene difluoride membrane) with horseradish peroxidase-labeled Wisteria floribunda lectin (Fig. 3B), which interacts with oligosaccharides terminating with N-acetylgalactosamine (36). The whole-cell lysates were obtained from the same C. jejuni growth cultures as those used to purify the total RNA for the microarray analysis.
FIG. 3.
Whole-cell lysates of C. jejuni proteins analyzed on SDS-12.5% PAGE. (A) Silver staining; (B) lectin blotting. Total proteins were prepared from C. jejuni grown to mid-log phase in iron-limited medium (MEM-α; lanes labeled 0 min and mid-log) or iron-rich medium (MEM-α with 40 μM FeSO4). Lanes labeled 5 min, 9 min, and 15 min correspond to the protein profiles of C. jejuni grown in iron-limited medium at 5, 9, and 15 min after the addition of ferrous sulfate, respectively. The lane labeled mid-log (+Fe) corresponds to the protein profile of C. jejuni grown to mid-log phase in the iron-rich medium.
The lectin blot analysis revealed reduced lectin reactivity with proteins extracted from C. jejuni grown in iron-rich medium (Fig. 3, lanes 5, 9, and 15 min and mid-log [+Fe]) compared to iron-limited medium (Fig. 3, 0 min). Given the lack of data on the functional role of each pgl protein, the correlation between the decreased lectin reactivity and gene expression is difficult to address. Nevertheless, our data clearly demonstrate that proteins from C. jejuni grown in iron-limited medium are hyperglycosylated compared to proteins from C. jejuni grown in iron-rich medium.
Additional supporting evidence regarding the importance of iron as a potential regulator of protein glycosylation comes from ongoing studies involving C. jejuni strains grown on different media and under different temperatures to determine the effects on protein glycosylation. Consistently, strains grown on MH medium (lower iron) showed greater lectin reactivity per amount of protein analyzed than those grown on 5% sheep blood plates (higher iron); this was consistent whether the base was tryptic soy agar or MH agar (D. S. Threadgill, unpublished data). Although the glycosylation profiles cannot be directly compared because of the use of solid versus liquid media, these results further support the potential role of iron in the regulation of protein glycosylation in C. jejuni. Recently, Szymanski and colleagues demonstrated that a C. jejuni pgl mutant is affected in its ability to adhere to and invade INT407 cells as well as to colonize the mouse gastrointestinal tract (45). Because invasion has been shown to be an important component of Campylobacter pathogenesis, the degree of glycosylation may play a role in modulating the level of C. jejuni virulence. This hypothesis is supported by the hyperglycosylated phenotype of C. jejuni grown in iron-limited medium, a growth condition which mimics the iron availability within the host gastrointestinal tract.
Fur regulon.
The ferric uptake regulator Fur has been shown to repress the transcription of at least seven iron-regulated proteins in C. jejuni. Five were characterized and identified as CeuE, ChuD, p19, ChuA, and CfrA (50). The other two were not characterized further. In order to identify other genes transcribed under the control of Fur, the C. jejuni fur gene was disrupted, and two complementary genomewide expression profiling experiments were performed.
In the first experiment, RNAs were extracted from the C. jejuni wild-type stain and its fur mutant at mid-log phase in iron-limited condition. Both RNA pools were reverse transcribed to cDNA, fluorescently labeled, and cohybridized to the microarray slides. Under this growth condition, all the genes that are Fur regulated in the absence of its coeffector Fe2+ (apo-Fur) will be identified. It should be noticed that the transcript level of lysS, which is located downstream of fur, was not affected by the fur mutation, as shown by our microarray data, indicating that the constructed mutation is nonpolar.
In the second experiment, RNAs were extracted from the C. jejuni wild-type strain and its fur mutant 15 min after the addition of ferrous sulfate (at a concentration of 40 μM) to an iron-limited mid-log-phase bacterial culture. The total RNAs were processed as described earlier. Under this growth condition, the genes that are Fur regulated in the presence of Fe2+ will be identified. The microarray data are presented as the ratio of the transcript level of the wild-type strain to that of the fur mutant. Both microarray experiments were repeated three times (biological replicates) with two technical replicates each, yielding six measurements per gene. The data were statistically analyzed as described in the Materials and Methods section and merged to the microarray data from the time course and mid-log experiments. Figure 4 represents the hierarchical clustering analysis of the Fur-regulated genes (with a Pearson correlation distance). In total, 53 genes were found to be Fur regulated and can be grouped into four major hierarchical clusters, named A, B, C, and D (Fig. 4). Of note, unlike other bacterial fur genes, C. jejuni fur expression is characterized by the absence of iron-responsive autoregulation (52). Indeed, fur was not selected as an iron-regulated gene by our microarray analysis of time course and mid-log phase experiments (Fig. 1).
FIG. 4.
Hierarchical cluster analysis of Fur-regulated genes. Columns 1 to 7 correspond to C. jejuni gene expression changes in response to the addition of iron to an iron-limited medium at 1, 3, 5, 7, 9, and 15 min and at mid-log phase, respectively. Columns 8 and 9 represent the change in transcript level of the wild-type C. jejuni strain compared to the fur mutant grown to mid-log phase in iron-limited medium and 15 min after the addition of FeSO4, respectively. The shade of red and green indicates the level of change. Genes are subgrouped into four clusters, named A, B, C, and D.
Cluster A contains eight genes that are iron induced and apo-Fur repressed. Interestingly, Fur repression decreased between 1.2- and 2.1-fold upon the addition of iron but was not completely abolished. This decrease in Fur repression could only partially explain the increase in transcript level of these eight genes following the addition of iron. Indeed, the genes from this cluster displayed an iron induction in their mRNA levels up to fourfold. Consequently, the expression of these genes is likely under the control of an additional regulator. In addition, it should be noted that these microarray experiments cannot distinguish between a direct or indirect effect of Fur on gene expression. The role of these eight genes and why they are regulated by iron could not be deduced from their annotation and thus remains to be fully determined.
Cluster B contains six genes that have their transcript level increased in response to the addition of iron (in the time course experiment) and are either Fur activated or Fur repressed. Of note, the expression of fdxA (encoding a ferredoxin) is apo-Fur repressed. As discussed above, FdxA is probably involved in oxidative stress defense. This protein has been proposed to reduce the oxidized state of the alkyl hydroxyperoxide reductase (AhpC), similar to AhpF in other gram-negative bacterium (53, 54).
Cluster C contains 10 genes that are iron repressed during the time course experiment and are apo-Fur activated. Of interest among these are the five proteins involved in flagellum biogenesis (FlaD, FlgE2, FlgG2, FlgH, and FlgI). While the reduction of the flagellum-related transcripts upon the addition of iron and formation of Fe2+-Fur complex might explain their iron repression during the time course experiment, it is inconsistent with their overexpression at mid-log phase in iron-rich medium. Consequently, the expression of these genes is undoubtedly under the control of other regulators and/or indirectly controlled by Fur. Indeed, three regulators, RpoN, FlgR, and FliA, have been previously shown to regulate flagellar expression (18).
Cluster D contains 29 genes that have their expression repressed by iron and Fe2+-Fur. The expression profile of these genes is in agreement with the well-established model of Fur as a repressor of genes encoding proteins involved in iron acquisition and assimilation. Indeed, 17 genes from this cluster encode proteins involved in heme transport (ChuABCD), iron transporter systems (CfrA, CeuBC, Cj0173c-175c, p19, and Cj1658), and several components of the energy-transducing TonB-ExbB-ExbD complex. Interestingly, another gene from this cluster encodes a thioredoxin (TrxB), which has recently been shown to be PerR regulated and involved in oxidative stress defense in Staphylococcus aureus (16). As an efficient thiol donor, thioredoxin likely reduces the oxidized cellular proteins and therefore contributes to the maintenance of the intracellular redox status. The expression profile of the other genes from this cluster and the function of their products in iron metabolism are unclear and require further investigation.
fur mutant is affected in chick colonization.
It is important to mention that the NCTC 11168 strain used in our study is phenotypically different from the sequenced C. jejuni NCTC 11168 (11168-GS) strain described by Gaynor et al. (13). The NCTC 11168-GS strain was shown to be rod-shaped, nonmotile, and unable to colonize the gastrointestinal tract of chicks. In contrast, the C. jejuni NCTC 11168 strain used in our study is helical and fully motile and able to colonize chicks at a level of 8 × 107 CFU per g of cecal content with an inoculum of 103 CFU (A. Stintzi, unpublished data). Consequently, it is a variant of the genome-sequenced C. jejuni NCTC 11168 strain and was therefore effectively used to investigate the mechanism of chick colonization. Of note, the 11168 strain used in our study, although clearly distinct from NCTC 11168-GS, appears to be similarly poorly invasive in human intestinal INT407 cells (A. Stintzi, unpublished data), suggesting that the invasion defect noted by Gaynor et al. may exist separately from the other defects.
C. jejuni NCTC 11168 and the derivative fur mutant (AS230) were orally inoculated into groups of 2-day-old chicks. The ability of both strains to colonize the chicks was assessed by enumeration of bacteria in the ceca 4 days postinoculation. As shown in Fig. 5, the fur mutant exhibited a significant reduction in colonization compared to the wild-type strain (P < 0.05, nonparametric Mann-Whitney rank sum test). These data clearly demonstrate the importance of iron homeostasis in vivo. The effect of the fur mutation on the ability of C. jejuni to colonize chicks is likely multifactorial. The fur mutation may increase oxidative stress due to the overaccumulation of iron. Additionally, several of the Fur-activated genes might be required for chick colonization. For example, several genes encoding proteins involved in flagellum biosynthesis, which is known to be required for chick colonization, were found to be Fur activated (Fig. 4). Alternatively, the constitutive expression of iron-regulated proteins in the fur mutant could provide attractive targets for chick antibodies, thus reducing the colonization level of this mutant through enhanced immune clearance.
FIG. 5.
Colonization properties of the C. jejuni mutant strains in the chick model. Groups of five chicks were inoculated with the C. jejuni wild-type (wt) strain NCTC 11168 or with the fur, cfrA, ceuE, and Cj0178 mutants (as indicated) at a dose of 1 × 105 to 3 × 105 CFU. The columns represent the means, and the error bars indicate the standard deviations.
Computational analysis of a putative Fur binding sequence.
In E. coli, upon association with Fe2+, the Fur protein binds a 19-bp consensus site (named the Fur box) with the sequence 5′-GATAATGATAATCATTATC-3′, and represses the transcription of the downstream gene. This Fur box was determined by DNase I protection assays and footprinting experiments (1). Three successive interpretations of the functional pattern of Fur binding to this sequence have been proposed (1): originally, Fur was assumed to recognize two 9-bp inverted repeats in the palindromic sequence; this initial assumption was then challenged by studies suggesting that Fur recognizes three adjacent hexameric repeats in a head-to-head-to-tail orientation; and finally, a third interpretation proposed that the 19-bp consensus sequence represents overlapping heptamer inverted repeats for the binding of two Fur dimers. Despite the apparent discrepancy of the Fur box sequence interpretation, the consensus sequence is conserved and has been identified in many bacterial species. C. jejuni Fur was shown to recognize the E. coli consensus Fur box, suggesting the presence of a similar sequence upstream of Fur-regulated genes in C. jejuni (55).
Since this Fur box has been shown to be present upstream of iron- and Fur-repressed genes, we searched for the presence of a conserved sequence element upstream of the initiation codon of the genes from cluster D. As a first step, genes were grouped into potential operonic structures based on our operon mapping experiment and physical distances between genes. Genes that were separated by less than 15 bp and putatively transcribed in the same orientation were assumed to lie on the same operon. By this strategy, 16 probable operons were identified. As a second step, the intergenic DNA regions upstream of the initiation codon of the first gene from each operon were searched for conserved sequence elements conforming to a potential Fur box. Specifically, the upstream sequences of the following genes were analyzed with the MEME algorithm: cfrA, Cj1658, chuA, ceuB, Cj0177, Cj0176c, Cj1384c, Cj1613c, Cj1377c, exbB2, tonB3, trxB, aroC, murE, Cj1375, and acs (3). A 19-bp conserved element was identified upstream of 11 of the genes (cfrA, Cj1658, ceuB, Cj0177, Cj0176c, chuA, Cj1384c, Cj1613c, exbB2, tonB3, and Cj0818). The consensus sequence obtained from this search is shown in Fig. 6. The absence of the element upstream of the other genes suggests that they are indirectly regulated by Fur. Interestingly, the predicted C. jejuni consensus sequence poorly matches the E. coli consensus Fur box (1). Confirmation of this putative C. jejuni Fur box will require additional experimentation, including DNA footprinting.
FIG. 6.
Sequence logo of the potential Fur binding site. The height of each letter indicates the relative frequency of that base at that position. The height of each stack of letters corresponds to the sequence conservation at that position.
Identification of the ferric enterobactin receptor.
In gram-negative bacteria, the translocation of the ferric siderophore complex through the bacterial membranes is energy dependent and usually requires a specific outer membrane receptor, a periplasmic binding protein, and an inner membrane ABC transport system. While the analysis of the C. jejuni NCTC 11168 genome reveals the presence of a periplasmic binding protein (CeuE) and an ABC complex (CeuBCD) with high identity to the Campylobacter coli ferric enterobactin transport system, the specific outer membrane receptor has not been identified yet. In agreement with their role in ferric enterobactin uptake, the ceuBCDE genes were found to be iron and Fur regulated by our microarray analysis. In an effort to characterize the ferric enterobactin receptor in C. jejuni NCTC 11168, we confirmed the ability of ferric enterobactin to promote C. jejuni growth in iron-limited medium and constructed mutations in genes identified by our microarray analysis which might fulfill this function.
First, the ability of C. jejuni NCTC 11168 to utilize ferric enterobactin as a sole iron source was confirmed by a standard growth promotion assay (2, 12, 14). This assay consists of analyzing the ability of ferric enterobactin to support the growth of C. jejuni on iron-restricted medium. This is done by supplementing the growth medium with a sufficient amount of the iron chelator desferrioxamine mesylate salt (DFO) to completely inhibit Campylobacter growth in the absence of iron. As shown in Fig. 7, enterobactin mediates iron acquisition in C. jejuni NCTC 11168. Next, a ceuE mutant was constructed in order to confirm the role of the periplasmic binding protein CeuE in ferric enterobactin transport (Fig. 8). The ceuE mutant was slightly impaired (zone of growth, 2.8 ± 0.9 cm versus 3.5 ± 1.4 cm for the wild type) and not fully abolished in its ability to acquire iron from ferric enterobactin (Fig. 7). This observation, while consistent with the previous results in C. coli, suggests that the CeuE protein is not strictly essential for the utilization of iron from ferric enterobactin.
FIG. 7.
Enterobactin growth promotion tests of the wild-type strain C. jejuni NCTC 11168 and the mutants ΔCj0178, ΔceuE, and ΔcfrA. A halo of growth around the filter paper disk containing 10 μl of enterobactin (10 mM) indicates utilization of the siderophore by the tested strain. The diameters of growth promotion zones ± standard deviation are shown in parentheses.
FIG. 8.
Diagram of the genetic organization of the cfrA, ceuE, and Cj0178 mutants described in this study. Each mutant was constructed by site-directed deletion and insertional mutagenesis with the chloramphenicol resistance marker (Cmr). The length of each deletion is shown in parentheses (in base pairs). The solid black arrow represents the position and orientation of the inserted chloramphenicol antibiotic resistance cassette in each gene. Given the absence of a transcriptional terminator downstream of the chloramphenicol resistance gene and the orientation of this gene with respect to the mutated gene, the constructed mutations are likely nonpolar. All genes are drawn approximately to scale.
Based on our microarray data and the genome annotation, C. jejuni NCTC 11168 possesses only three iron-regulated outer membrane proteins with homology to high-affinity outer membrane receptors, CfrA, ChuA, and Cj0178. Since ChuA has been proposed to be required for hemin transport (54) and a C. coli cfrA mutant appeared to be able to acquire iron from ferric enterobactin (13), Cj0178 was the best candidate for the ferric enterobactin outer membrane receptor. However, as shown in Fig. 7, a C. jejuni Cj0178 mutant (Fig. 8) was not affected in its ability to utilize iron from ferric enterobactin, suggesting that Cj0178 is the outer membrane receptor of an unidentified iron source.
Given that our microarray analysis did not identify any other candidate for the ferric enterobactin receptor, we reevaluated the function of CfrA in enterobactin-mediated iron transport. In contrast to the previous study with C. coli, the ability of the C. jejuni cfrA mutant (Fig. 8) to acquire iron from ferric enterobactin was fully abolished (Fig. 7), demonstrating the role of CfrA as the ferric enterobactin receptor in C. jejuni NCTC 11168. Because the C. jejuni and C. coli CfrA proteins are almost identical (98.7% identity over 696 amino acids), suggesting conserved functions, C. coli may possess an additional receptor that is able to mediate iron acquisition from enterobactin. If so, enterobactin would still be able to support the growth of a C. coli cfrA mutant in an iron-restricted medium. Similar findings have been observed with Salmonella enterica, which has two receptors, FepA and IroN, for ferric enterobactin (31).
Alternatively, it should be noted that the growth promotion assay in the previous C. coli study was performed with EDDHA (ethylenediamine bis[2-hydroxyphenyl] acetic acid) as an iron chelator instead of DFO. These two chelators differ significantly in their effectiveness at binding iron, with DFO being the more powerful iron chelator (41, 59). In aqueous solution, enterobactin could spontaneously break down into the trimers, dimers, and monomers of dihydroxybenzoyl serine (DHBS) (14, 26). As a result, the growth promotion assays may have been performed with a mixture of enterobactin and its breakdown products. Since DHBS is a weaker iron chelator than enterobactin, it will compete less efficiently for iron with DFO than with EDDHA (41). In other words, the breakdown products of enterobactin will be more efficient in delivering iron to the bacterium in a medium containing EDDHA than in one containing DFO. Consequently, the growth promotion observed in the C. coli cfrA mutant might have been DHBS mediated. Similar to E. coli, C. coli may possess an additional outer membrane receptor specific for the DHBS iron complex. In addition, because DHBS-mediated iron acquisition would likely not be observed in a DFO-containing medium, it is possible that C. jejuni also possesses a DHBS-specific receptor. We are currently investigating this possibility.
Ferric siderophore transporters are required for C. jejuni NCTC 11168 chick colonization.
The role of CfrA, CeuE, and Cj0178 in colonization was investigated in the chick model. Groups of five chicks were orally inoculated with the wild-type strain and the mutants at a dose between 1 × 105 and 3 × 105 CFU. As shown in Fig. 5, all three mutants, cfrA, ceuE, and Cj0178, were significantly affected in their ability to colonize the gastrointestinal tract of chicks (P < 0.05, nonparametric Mann-Whitney rank sum test). At 4 days postinfection, the number of cfrA and ceuE mutants per gram of cecum was below the detection limit of our assay (500 CFU/g of cecum). This result clearly demonstrates the requirement of the two outer membrane receptors (CfrA and Cj0178) as well as of the ferric enterobactin periplasmic binding protein (CeuE) in chick gut colonization. Overall, these experiments highlight for the first time the importance of iron acquisition in the gut colonization process by C. jejuni. Moreover, the inability of the ceuE mutant to colonize the ceca indicates an essential role for enterobactin in gut colonization. In addition, we cannot exclude a possible role for CfrA and Cj0178 in adhesion. In support of this possibility, CfrA exhibits significant identity to the E. coli O157 Iha adhesin protein (34.2% identity) (48).
Conclusion.
The transcriptional response of C. jejuni to iron availability is pleiotropic, involving the differential expression of genes belonging to several functional groups, ranging from energy metabolism to cell surface structures to iron acquisition and to oxidative stress defense. Several of these genes encode proteins that have been shown to be involved in the mechanism of Campylobacter pathogenesis. Interestingly, the level of Campylobacter glycoproteins and/or the efficiency of glycan addition to those proteins was found to increase with iron limitation. While the biological function of glycosylation remains to be established, a mutant deficient in the glycosylation process has been shown to be affected in its ability to bind and invade INT407 cells as well as to colonize the mouse gastrointestinal tract (45). Consequently, iron availability likely modulates the abilities of Campylobacter to adhere to, invade, and colonize the host gastrointestinal tract. In support of this hypothesis, a fur mutant showed significantly reduced chick cecum colonization, highlighting the importance of iron homeostasis in vivo. Thus, as in other pathogens, iron might constitute a key environmental signal for controlling Campylobacter pathogenesis.
In addition, this work demonstrated the requirement of iron acquisition in the colonization process and survival of C. jejuni in vivo. the CfrA protein was identified as the ferric enterobactin receptor, not previously annotated in the genome sequence. Finally, chick colonization assays with strains mutated in genes encoding components of ferric siderophore transport systems show the importance of enterobactin in gut colonization.
Acknowledgments
We thank I. Turcot and J. Andrus for contributions to the manuscript. Plasmid pRY111 was graciously provided by P. Guerry. We are grateful to all the staff from the OU and OSU microarray core facilities.
This work was supported by National Institutes of Health grants RO1-AI055612 (to A.S.) and RR15564 (to A.S.) and by National Science Foundation grant NSF-POWRE MCB-9973861 (to D.T.).
REFERENCES
- 1.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 2.Baig, B. H., I. K. Wachsmuth, and G. K. Morris. 1986. Utilization of exogenous siderophores by Campylobacter species. J. Clin. Microbiol. 23:431-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baillon, M. L., A. H. van Vliet, J. M. Ketley, C. Constantinidou, and C. W. Penn. 1999. An iron-regulated alkyl hydroperoxide reductase (AhpC) confers aerotolerance and oxidative stress resistance to the microaerophilic pathogen Campylobacter jejuni. J. Bacteriol. 181:4798-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bearden, S. W., J. D. Fetherston, and R. D. Perry. 1997. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect. Immun. 65:1659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein, J. A., A. B. Khodursky, P. H. Lin, S. Lin-Chao, and S. N. Cohen. 2002. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. USA 99:9697-9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, V. 1997. Avoidance of iron toxicity through regulation of bacterial iron transport. Biol. Chem. 378:779-786. [PubMed] [Google Scholar]
- 7.Braun, V., and K. Hantke. 2002. Mechanisms of bacterial iron transport, p. 289-311. In G. Winkelmann (ed.), Microbial transport systems. Wiley-VCH Verlag GmBH & Co., Berlin, Germany.
- 8.Day, W. A., Jr., J. L. Sajecki, T. M. Pitts, and L. A. Joens. 2000. Role of catalase in Campylobacter jejuni intracellular survival. Infect. Immun. 68:6337-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doring, G., M. Pfestorf, K. Botzenhart, and M. A. Abdallah. 1988. Impact of proteases on iron uptake of Pseudomonas aeruginosa pyoverdin from transferrin and lactoferrin. Infect. Immun. 56:291-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Field, L. H., V. L. Headley, S. M. Payne, and L. J. Berry. 1986. Influence of iron on growth, morphology, outer membrane protein composition, and synthesis of siderophores in Campylobacter jejuni. Infect. Immun. 54:126-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaynor, E. C., S. Cawthraw, G. Manning, J. K. MacKichan, S. Falkow, and D. G. Newell. 2004. The genome-sequenced variant of Campylobacter jejuni NCTC 11168 and the original clonal clinical isolate differ markedly in colonization, gene expression, and virulence-associated phenotypes. J. Bacteriol. 186:503-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerry, P., J. Perez-Casal, R. Yao, A. McVeigh, and T. J. Trust. 1997. A genetic locus involved in iron utilization unique to some Campylobacter strains. J. Bacteriol. 179:3997-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hantke, K. 1990. Dihydroxybenzoylserine—a siderophore for E. coli. FEMS Microbiol. Lett. 55:5-8. [DOI] [PubMed] [Google Scholar]
- 15.Harris, W. R. 2002. Iron chemistry, p. 1-35. In D. M. Templeton (ed.), Molecular and cellular iron transport. Marcel Dekker, New York, N.Y.
- 16.Horsburgh, M. J., M. O. Clements, H. Crossley, E. Ingham, and S. J. Foster. 2001. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 69:3744-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imlay, J. A., and S. Linn. 1988. DNA damage and oxygen radical toxicity. Science 240:1302-1309. [DOI] [PubMed] [Google Scholar]
- 18.Jagannathan, A., C. Constantinidou, and C. W. Penn. 2001. Roles of rpoN, fliA, and flgR in expression of flagella in Campylobacter jejuni. J. Bacteriol. 183:2937-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Konopka, K., A. Bindereif, and J. B. Neilands. 1982. Aerobactin-mediated utilization of transferrin iron. Biochemistry 21:6503-6508. [DOI] [PubMed] [Google Scholar]
- 20.Linton, D., E. Allan, A. V. Karlyshev, A. D. Cronshaw, and B. W. Wren. 2002. Identification of N-acetylgalactosamine-containing glycoproteins PEB3 and CgpA in Campylobacter jejuni. Mol. Microbiol. 43:497-508. [DOI] [PubMed] [Google Scholar]
- 21.Long, A. D., H. J. Mangalam, B. Y. Chan, L. Tolleri, G. W. Hatfield, and P. Baldi. 2001. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J. Biol. Chem. 276:19937-19944. [DOI] [PubMed] [Google Scholar]
- 22.Massad, G., J. E. Arceneaux, and B. R. Byers. 1991. Acquisition of iron from host sources by mesophilic Aeromonas species. J Gen. Microbiol. 137:237-241. [DOI] [PubMed] [Google Scholar]
- 23.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer, J. M., A. Neely, A. Stintzi, C. Georges, and I. A. Holder. 1996. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 64:518-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neilands, J. B. 1993. Siderophores. Arch Biochem Biophys. 302:1-3. [DOI] [PubMed] [Google Scholar]
- 26.O'Brien, I. G., G. B. Cox, and F. Gibson. 1971. Enterochelin hydrolysis and iron metabolism in Escherichia coli. Biochim. Biophys. Acta 237:537-549. [DOI] [PubMed] [Google Scholar]
- 27.Oh, M. K., L. Rohlin, K. C. Kao, and J. C. Liao. 2002. Global expression profiling of acetate-grown Escherichia coli. J. Biol. Chem. 277:13175-13183. [DOI] [PubMed] [Google Scholar]
- 28.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 29.Pomposiello, J. P., and B. Demple. 2002. Global adjustment of microbial physiology during free radical stress. Adv. Microb. Physiol. 46:320-327. [DOI] [PubMed] [Google Scholar]
- 30.Purdy, D., S. Cawthraw, J. H. Dickinson, D. G. Newell, and S. F. Park. 1999. Generation of a superoxide dismutase (SOD)-deficient mutant of Campylobacter coli: evidence for the significance of SOD in Campylobacter survival and colonization. Appl. Environ. Microbiol. 65:2540-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rabsch, W., U. Methner, W. Voigt, H. Tschape, R. Reissbrodt, and P. H. Williams. 2003. Role of receptor proteins for enterobactin and 2,3-dihydroxybenzoylserine in virulence of Salmonella enterica. Infect. Immun. 71:6953-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raphael, B. H., and L. A. Joens. 2003. FeoB is not required for ferrous iron uptake in Campylobacter jejuni. Can J. Microbiol. 49:727-731. [DOI] [PubMed] [Google Scholar]
- 33.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 34.Rock, J. D., A. H. M. van Vliet, and J. M. Ketley. 2001. Haemin uptake in Campylobacter jejuni. Int. J. Med. Microbiol. 291:125. [Google Scholar]
- 35.Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang, N. Bhagabati, J. Braisted, M. Klapa, T. Currier, M. Thiagarajan, A. Sturn, M. Snuffin, A. Rezantsev, D. Popov, A. Ryltsov, E. Kostukovich, I. Borisovsky, Z. Liu, A. Vinsavich, V. Trush, and J. Quackenbush. 2003. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 34:374-378. [DOI] [PubMed] [Google Scholar]
- 36.Sakiyama, T., M. Kabayama, M. Tomita, J. Nakamura, H. Mukai, Y. Tomita, and K. Furukawa. 1998. Distribution of glycoproteins with beta-N-acetylgalactosaminylated N-linked sugar chains among bovine tissues. Biochim. Biophys. Acta 1380:268-274. [DOI] [PubMed] [Google Scholar]
- 37.Schneider, T. D., and R. M. Stephens. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schroder, I., E. Johnson, and S. de Vries. 2003. Microbial ferric iron reductases. FEMS Microbiol. Rev. 27:427-447. [DOI] [PubMed] [Google Scholar]
- 39.Stead, D., and S. F. Park. 2000. Roles of Fe superoxide dismutase and catalase in resistance of Campylobacter coli to freeze-thaw stress. Appl. Environ. Microbiol. 66:3110-3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stintzi, A. 2003. Gene expression profile of Campylobacter jejuni in response to growth temperature variation. J. Bacteriol. 185:2009-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stintzi, A., and K. N. Raymond. 2002. Siderophore chemistry, p. 273-319. In T. D. Moss (ed.), Molecular and cellular iron transport. Marcel Dekker, New York, N.Y.
- 42.Stintzi, A., and L. Whitworth. 2003. Investigation of the Campylobacter jejuni cold-shock response by global transcript profiling. Genome Lett. 2:18-27. [Google Scholar]
- 43.Stintzi, A., C. Barnes, J. Xu, and K. N. Raymond. 2000. Microbial iron transport via a siderophore shuttle: a membrane ion transport paradigm. Proc. Natl. Acad. Sci. USA 97:10691-10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sturn, A., J. Quackenbush, and Z. Trajanoski. 2002. Genesis: cluster analysis of microarray data. Bioinformatics 18:207-208. [DOI] [PubMed] [Google Scholar]
- 45.Szymanski, C. M., D. H. Burr, and P. Guerry. 2002. Campylobacter protein glycosylation affects host cell interactions. Infect. Immun. 70:2242-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szymanski, C. M., R. Yao, C. P. Ewing, T. J. Trust, and P. Guerry. 1999. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32:1022-1030. [DOI] [PubMed] [Google Scholar]
- 47.Tao, H., C. Bausch, C. Richmond, F. R. Blattner, and T. Conway. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181:6425-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarr, P. I., S. S. Bilge, J. C. Vary, Jr., S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Helden, J. 2003. Regulatory sequence analysis tools. Nucleic Acids Res. 31:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Vliet, A. H., K. G. Wooldridge, and J. M. Ketley. 1998. Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J. Bacteriol. 180:5291-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Vliet, A. H., M. L. Baillon, C. W. Penn, and J. M. Ketley. 1999. Campylobacter jejuni contains two fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J. Bacteriol. 181:6371-6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Vliet, A. H., J. D. Rock, L. N. Madeleine, and J. M. Ketley. 2000. The iron-responsive regulator Fur of Campylobacter jejuni is expressed from two separate promoters. FEMS Microbiol. Lett. 188:115-118. [DOI] [PubMed] [Google Scholar]
- 53.van Vliet, A. H., M. A. Baillon, C. W. Penn, and J. M. Ketley. 2001. The iron-induced ferredoxin FdxA of Campylobacter jejuni is involved in aerotolerance. FEMS Microbiol. Lett. 196:189-193. [DOI] [PubMed] [Google Scholar]
- 54.van Vliet, A. H., J. M. Ketley, S. F. Park, and C. W. Penn. 2002. The role of iron in Campylobacter gene regulation, metabolism and oxidative stress defense. FEMS Microbiol. Rev. 26:173-186. [DOI] [PubMed] [Google Scholar]
- 55.Wooldridge, K. G., P. H. Williams, and J. M. Ketley. 1994. Iron-responsive genetic regulation in Campylobacter jejuni: cloning and characterization of a fur homolog. J. Bacteriol. 176:5852-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wurmbach, E., T. Yuen, B. J. Ebersole, and S. C. Sealfon. 2001. Gonadotropin-releasing hormone receptor-coupled gene network organization. J. Biol. Chem. 276:47195-47201. [DOI] [PubMed] [Google Scholar]
- 57.Yao, R., R. A. Alm, T. J. Trust, and P. Guerry. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127-130. [DOI] [PubMed] [Google Scholar]
- 58.Young, N. M., J. R. Brisson, J. Kelly, D. C. Watson, L. Tessier, P. H. Lanthier, H. C. Jarrell, N. Cadotte, F. St Michael, E. Aberg, and C. M. Szymanski. 2002. Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J. Biol. Chem. 277:42530-42539. [DOI] [PubMed] [Google Scholar]
- 59.Yunta, F., S. Garcia-Marco, J. J. Lucena, M. Gomez-Gallego, R. Alcazar, and M. A. Sierra. 2003. Chelating agents related to ethylenediamine bis(2-hydroxyphenyl)acetic acid (EDDHA): synthesis, characterization, and equilibrium studies of the free ligands and their Mg2+, Ca2+, Cu2+, and Fe3+ chelates. Inorg. Chem. 42:5412-5421. [DOI] [PubMed] [Google Scholar]