Abstract
A high-density oligonucleotide DNA microarray, a genechip, representing the 4.6-Mb genome of the facultative phototrophic proteobacterium, Rhodobacter sphaeroides 2.4.1, was custom-designed and manufactured by Affymetrix, Santa Clara, Calif. The genechip contains probe sets for 4,292 open reading frames (ORFs), 47 rRNA and tRNA genes, and 394 intergenic regions. The probe set sequences were derived from the genome annotation generated by Oak Ridge National Laboratory after extensive revision, which was based primarily upon codon usage characteristic of this GC-rich bacterium. As a result of the revision, numerous missing ORFs were uncovered, nonexistent ORFs were deleted, and misidentified start codons were corrected. To evaluate R. sphaeroides transcriptome flexibility, expression profiles for three diverse growth modes—aerobic respiration, anaerobic respiration in the dark, and anaerobic photosynthesis—were generated. Expression levels of one-fifth to one-third of the R. sphaeroides ORFs were significantly different in cells under any two growth modes. Pathways involved in energy generation and redox balance maintenance under three growth modes were reconstructed. Expression patterns of genes involved in these pathways mirrored known functional changes, suggesting that massive changes in gene expression are the major means used by R. sphaeroides in adaptation to diverse conditions. Differential expression was observed for genes encoding putative new participants in these pathways (additional photosystem genes, duplicate NADH dehydrogenase, ATP synthases), whose functionality has yet to be investigated. The DNA microarray data correlated well with data derived from quantitative reverse transcription-PCR, as well as with data from the literature, thus validating the R. sphaeroides genechip as a powerful and reliable tool for studying unprecedented metabolic versatility of this bacterium.
Rhodobacter sphaeroides belongs to the facultatively phototrophic anoxygenic proteobacteria, which are known for their broad range of energetic and metabolic capabilities. To derive energy for growth, R. sphaeroides uses aerobic respiration in the presence of oxygen, whereas in the absence of oxygen it can either use anoxygenic photosynthesis (in the presence of light), anaerobic respiration (in the dark, in the presence of appropriate electron acceptors), or fermentation (with appropriate substrates) (23, 26, 38). The metabolic capabilities of R. sphaeroides include, but are not limited to, dinitrogen fixation, utilization of single-carbon compounds (carbon dioxide or methanol), and utilization or production of molecular hydrogen, as well as detoxification of metal oxides and oxyanions.
In R. sphaeroides, transitions from one growth mode to another are affected by diverse environmental factors. Oxygen tension is among the most influential factors that dictate the mode of energy generation. At high oxygen tension, R. sphaeroides lacks a photosynthetic apparatus. Synthesis of the photosystem (PS) is controlled by oxygen levels and is accompanied by profound intracellular changes. A decrease in oxygen tension results in upregulated expression of genes encoding various components of the PS. This results in the de novo formation of the intracytoplasmic membrane system that houses the PS. In addition to the PS genes, the oxygen level is known to affect the expression of genes involved in other energy generating and metabolic processes. Some of the regulatory systems involved in the oxygen-dependent regulation of gene expression have been identified (reviewed in references 31 and 38). Under microoxic and anoxic conditions, light plays an important role in the physiology and behavior of R. sphaeroides. Several putative photoreceptors are present in the genome of R. sphaeroides 2.4.1, whose precise roles have yet to be fully elucidated (3, 12, 13, 25).
We have constructed an R. sphaeroides genome-wide DNA microarray, a genechip, in order to study the qualitative and quantitative parameters of transcriptome changes in this metabolically versatile bacterium in response to environmental conditions. The genechip was custom-designed and manufactured by Affymetrix, Santa Clara, Calif. The sequences of the open reading frames (ORFs) and intergenic regions for the genechip were derived from the original annotation of the R. sphaeroides 2.4.1 genome performed at the Oak Ridge National Laboratory (ORNL; http://genome.ornl.gov/microbial/rsph/03nov00_obsolete/) after extensive revision. We present here details of the construction of the R. sphaeroides genechip and evaluate its performance. We also provide an overview of the transcriptome changes observed in R. sphaeroides grown under three diverse growth modes, i.e., aerobic respiration with saturating oxygen, anaerobic respiration in the dark (with dimethyl sulfoxide [DMSO] as a terminal electron acceptor) and anaerobic photosynthesis (under medium light intensity). We focus our discussion on the transcriptome changes pertaining to energy metabolism and redox balance maintenance.
(The preliminary findings reported here were presented at the 102nd General Meeting of American Society for Microbiology in Salt Lake City, Utah, 2002 [abstr. H120].)
MATERIALS AND METHODS
Growth conditions.
R. sphaeroides 2.4.1 was grown in Sistrom's minimal medium A with succinate as carbon source (6). The 60-ml cultures of R. sphaeroides were grown at 30°C in 100-ml glass culture tubes. The following gas mixtures were vigorously sparged during growth: 30% O2, 69% N2, and 1% CO2 (oxic conditions) or 98% N2 and 2% CO2 (anoxic-light conditions; illuminated with white light at 10 W/m2). Anoxic-dark cultures were grown in tightly capped tubes with no sparging; the medium was supplemented with 10% (vol/vol) Luria-Bertani medium (24) and 0.3% (vol/vol) DMSO.
RNA extraction.
R. sphaeroides cells in the early exponential phase (A600 = 0.16 to 0.20 for cultures sparged with oxygen; A600 = 0.4 to 0.5 for cultures grown under anoxic conditions) were collected into centrifugation bottles containing shaved ice. Rifampin was added prior to centrifugation to a final concentration of 200 μg ml−1. Cells were pelleted by a brief centrifugation at high speed, and cell pellets were frozen at −80°C until further processing. RNA was extracted as described earlier (11). Briefly, the cell lysis buffer from the RNeasy midi kit (Qiagen, Chatsworth, Calif.) and an equal volume of sterile zirconium beads were added to the frozen cell pellets. Cells were disrupted by a 1-min vigorous shaking in a Mini-BeadBeater (Biospec Products, Bartlesville, Okla.). RNA was extracted from the supernatants of cell lysates by using the RNeasy midi kit. All RNA preparations were tested for the lack of genomic DNA contamination by quantitative real-time PCR.
Description of the R. sphaeroides genechip.
A high-density oligonucleotide microarray, a genechip, was custom designed and manufactured by Affymetrix based on the submitted sequences of ORFs and intergenic regions (www.uwyo.edu/molecbio/gomelsky/gomelsky.htm/genechip). The oligonucleotide sequences on the R. sphaeroides genechip correspond to the coding strand of the ORFs, and biotin-labeled cDNA was used for hybridizations (an “antisense” genechip). Both strands of the intergenic regions are present on the genechip as separate probe sets. The majority of ORFs and intergenic regions are represented by probe sets comprised of 14 probe pairs. These are scattered across the chip to decrease location-specific effects of hybridization. Each pair contains a “perfect match”, i.e., a 25-nucleotide (nt) oligonucleotide identical to the coding sequence and a “mismatch,” a 25-nt oligonucleotide containing one mismatched nucleotide in the middle of the oligonucleotide (used as control for hybridization specificity). Probe sets for several genes (RSP0258, pufBA; RSP0282, ppsR; RSP0284, bchF; RSP0314, pucBA; RSP0698, fnrL; RSP1518, prrA; RSP1565, appA; RSP4294, 16S rRNA; RSP4295, 23S rRNA) are present in five copies each.
Sample preparation and genechip hybridization conditions.
cDNA synthesis, fragmentation, and terminal labeling with biotin were performed according to the GeneChip Pseudomonas aeruginosa genome array expression analysis protocol (Affymetrix). Approximately 1.6 to 2.0 μg of biotin-labeled cDNA fragmented to a size of 50 to 200 nt was used for hybridizations. The genechips were hybridized by using a Fluidics Station and scanned by using a Genechip Scanner (both from Affymetrix) at the University of Colorado Cancer Center Microarray Core Facility according to specifications provided by the manufacturer.
Software for genome annotation, curation, and genechip data analysis.
R. sphaeroides 2.4.1 genome annotations were generated by the Genome Analysis Group at the Oak Ridge National Laboratory. Artemis software (34; http://www.sanger.ac.uk/Software/Artemis/) was used for visual genome inspection and ORF assignments. BLAST, PSI-BLAST (1; http://www.ncbi.nlm.nih.gov/BLAST/index.html), COG (36; http://www.ncbi.nlm.nih.gov/COG/), KEGG (20; http://www.genome.ad.jp/kegg/), and SMART (35; http://smart.embl-heidelberg.de) were used for ORF analysis.
The following software packages were used for genechip data analysis: Microarray Analysis Suite (MAS 5.0; Affymetrix), Robust Multi-Array Analysis (RMA) (19; http://www.stat.berkeley.edu/users/bolstad/RMAExpress/RMAExpress.html), and GeneSpring 4.2 (Silicon Genetics). MAS 5.0 was used for estimation of the number of genes detected on a genechip (scored as “present,” “marginal,” and “absent”). Gene expression values for subsequent analysis were recalculated by using RMA with quantile normalization, which provided a better correlation for expression of genes belonging to operons and regulons (data not shown). The expression data obtained here were deposited in the Gene Expression Omnibus database of the National Center for Biotechnology Information (www.ncbi.nih.gov/geo) in the “three growth modes” series, GSE532, under the following accession numbers: oxic conditions (30% sparged oxygen) (GSM1670, GSM1671, and GSM8108), anoxic-light conditions (GSM2416, GSM2417, and GSM8107), and anoxic-dark-DMSO conditions (GSM2418, GSM2419, and GSM8109). Original probe intensity images (“cel” files) are available upon request.
qPCR.
Aliquots of the cDNA samples used for genechip hybridization, prior to fragmentation, were used for quantitative reverse transcription-mediated real-time PCR (qPCR). The iCycler iQ real-time PCR detection system (Bio-Rad, Richmond, Calif.) with SYBR Green chemistry was used to monitor amplification and to quantify the amount of PCR products. The primers and conditions used for probe amplification were described earlier (11). The rpoZ gene encoding the ω subunit of RNA polymerase was used to normalize expression values for all other genes. Each qPCR reaction was performed in triplicate. Average data from at least two experimental replicates are reported.
RESULTS AND DISCUSSION
Curation of R. sphaeroides genome annotation.
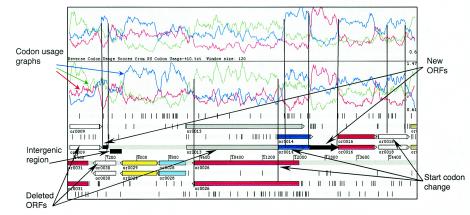
The R. sphaeroides 2.4.1 genome was sequenced by the U.S. Department of Energy (DOE) Joint Genome Institute and the laboratory of S. Kaplan at the University of Texas Medical School (23). To select the sequences for the probe sets to be represented on the DNA microarray, we analyzed 4152 ORFs from the original genome annotation generated by the ORNL (http://genome.ornl.gov/microbial/rsph/03nov00_obsolete/). In that version of the annotation, several hundred ORFs were found to overlap with their same-strand neighbors at either the 3′ or 5′ ends or to overlap with possible ORFs on the opposite strand. Examples of such overlaps are shown in Fig. 1. Since overlapping ORFs are not characteristic of microbial genomes (2), we assumed that the majority of overlaps resulted from either incorrect ORF assignment or from misidentified start codons.
FIG. 1.
Curation of the R. sphaeroides genome annotation. A representative 9-kb DNA fragment of the R. sphaeroides genome annotated by ORNL (http://genome.ornl.gov/microbial/rsph/03nov00/obsolete) is displayed in Artemis software (34). ORFs in each of the six frames identified by the original annotation are shown as colored horizontal arrows. Codon usage graphs for three reading frames (red, green, and blue) on both DNA strands (top panel, forward strand; lower panel, reverse strand) are shown. A profile of a putative ORF that has codon usage characteristic of R. sphaeroides appears alleviated compared to profiles in alternative reading frames. A start and end of an ORF are characterized by a sharp rise and fall, respectively, of a codon usage curve. “Deleted ORFs,” “New ORFs” (shown as solid black arrows), and “Start codon change” indicate changes introduced to the original ORNL annotation. “Intergenic region” (shown as a black box) indicates a sequence between or0039 and a new ORF used for probe set construction. Black vertical lines correspond to revised borders of the ORFs. or0009 was deleted because it extensively overlapped with or0031 and had codon usage uncharacteristic of R. sphaeroides. or0013 was deleted because it extensively overlapped with or0026 and or0014, both of which had proper codon usage and showed similarity to other proteins from the databases. or0030 was deleted and replaced with a new ORF on the opposite strand showing codon usage characteristic of R. sphaeroides. A new ORF was predicted between or0014 and or0016 based upon codon usage. Start codons were corrected for the overlapping ORFs, or0026 and or0014, based on their codon usage profiles, thus eliminating the overlap between them.
Because the R. sphaeroides 2.4.1 genome is GC-rich (ca. 69% G+C), we explored the possibility of improving ORF assignment by relying on the highly biased codon usage characteristic of R. sphaeroides. We constructed a codon usage table based upon the amino acid sequences of those R. sphaeroides proteins from the SwissProt database, which were identified and studied prior to the genome sequencing effort. This codon usage table was applied, by using Artemis software, to obtain codon usage profiles of the entire genome (Fig. 1). We manually inspected regions from the ORNL annotation where ORF overlaps existed. As illustrated in Fig. 1, in many cases, only one putative ORF at a given locus possessed the codon usage characteristic of R. sphaeroides. However, opposite-strand ORFs, representing the “mirror” or “shadow” images of the putative ORFs derived from the GC-biased codon usage of the sense strand, were also found (Fig. 1). Putative ORFs whose existence and/or borders were predicted based upon codon usage alone were submitted to the databases COG, KEGG, and Pfam and further analyzed by using PSI-BLAST. Approximately 89% of the redefined ORFs had at least one hit, i.e., they had significant similarity to proteins or protein domains from other organisms. Relying on the R. sphaeroides codon usage, we deleted 379 presumed nonexistent ORFs and changed the start codons for about 800 ORFs (Fig. 1).
To reveal potentially missed ORFs, we manually examined all DNA regions that were larger than 0.35 to 0.50 kb and located between the redefined ORFs. This inspection revealed a number of ORFs, for which either codon usage profiles or BLAST results warranted an ORF assignment. When these ORF assignments were made, the genome of 2.4.1 was not yet fully assembled and existed in the format of 160 contigs separated by 0- to 1.5-kb gaps (unpublished data). To limit the loss of the ORFs located within the intercontig gaps, we included putative 3′- or 5′-truncated ORFs located at the contig ends if they conformed to the R. sphaeroides codon usage. In more than 30 instances we identified 5′ ends of the putative ORFs with known function at the end of one contig and their 3′ ends at the beginning of the following contig. The DNA sequences of the 5′ and 3′ ends of apparently the same ORFs were combined to design a single probe set per ORF.
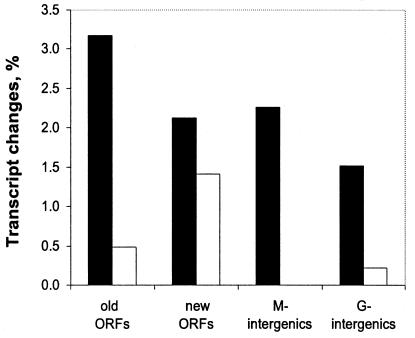
The total number of newly identified ORFs, complete and truncated, was 526. The protein sequences corresponding to these ORFs were submitted to COG, KEGG, Pfam, and BLAST searches and 55% returned at least one entry, suggesting that these sequences encode products with similarity to proteins or protein domains from other organisms. Expression profiles of the newly identified ORFs were analyzed under the three growth conditions and compared to the expression profiles of the redefined ORFs (see below). As is evident from Fig. 2, the percentages of ORFs, which were differentially expressed in response to growth conditions, for the newly identified ORFs were similar to those of the redefined ORFs, suggesting that the majority of the newly identified ORFs were correctly identified. The total number of ORFs that underwent modification was ca. 1,700 (379 + 526 + 800), which accounts for ca. 40% of the 4152 ORFs in the original genome annotation. Our curated annotation contains 4,292 ORFs, each of which is assigned an RSP number. The extensive curation of ORF assignments indicates that annotation of genomes that contain a relatively high percentage of unique ORFs requires significant human involvement. It also suggests that for GC-rich organisms codon usage may be the single most reliable method for ORF identification and for determination of start codons of the ORFs.
FIG. 2.
Expression level changes (anoxic-light versus oxic conditions) in different categories of probe sets presented on the genechip. Bars: ▪, expression increases (>5-fold); □, expression decreases (>5-fold). Old ORFs, ORFs identified in the original ORNL annotation and redefined based on codon usage; new ORFs, newly identified ORFs; M-intergenics, intergenic regions containing putative short ORFs; G-intergenics, intergenic regions with no identifiable ORFs (see the text for details).
R. sphaeroides genechip composition.
The R. sphaeroides high-density oligonucleotide DNA microarray, genechip, was designed and manufactured by Affymetrix based on the DNA sequences provided by us (see Materials and Methods). Four types of sequences are represented on the genechip: (i) 4,292 ORFs from R. sphaeroides 2.4.1 described above; (ii) 47 rRNA and tRNA encoding genes; (iii) 65 selected ORFs from R. sphaeroides strains other than 2.4.1 (to be described elsewhere); and (iv) intergenic regions, i.e., DNA fragments containing no identified ORFs after codon usage-based revision of the genome annotation.
Intergenic regions larger than ca. 0.35 to 0.50 kb that did not contain readily identifiable ORFs (394 in total) were divided into two categories. 171 intergenic regions contained putative ORFs of approximately 90 amino acids or longer that had no significant BLAST hits. The DNA fragments corresponding to these putative ORFs were represented on the genechip by both DNA strands. A significant percentage of these showed differential expression in response to growth conditions (Fig. 2, M- intergenics). It is therefore likely that many of these short ORFs correspond to functional proteins.
The remaining intergenic regions that contained no apparent ORFs were arbitrarily divided into 0.3- to 0.5-kb fragments (Fig. 2, G- intergenics). Sequences of both DNA strands of these fragments are represented on the R. sphaeroides genechip. The differential expression of some representatives from this group suggests that they contain putative genes that were missed in our analysis. Future studies will be required to verify the existence and functions of such putative genes hidden within the intergenic regions. However, the capacity to detect expression of most of the “missed” genes is built into the R. sphaeroides genechip.
Evaluation of the R. sphaeroides genechip performance and experimental reproducibility.
To our knowledge, the R. sphaeroides genechip is based on the genome with the highest GC-content of oligonucleotide-based microarrays available from Affymetrix. We therefore subjected the R. sphaeroides genechip to extensive testing for its representativeness, sensitivity, and reliability.
(i) Genechip representativeness.
Hybridization of the R. sphaeroides genomic DNA to the genechip yielded a measure of genechip representativeness. When fragmented genomic DNA (0.5 to 2.0 μg) was used for hybridization, ca. 97.0% ± 0.2% of the probe sets derived from strain 2.4.1 (including probe set duplicates) were determined to be “present,” 0.85% ± 0.05% “marginally present,” and 2.1% ± 0.2% “absent” after analysis by the MAS 5.0 software. If these numbers were directly applicable to transcriptome analysis, i.e., if all RNAs were present in equimolar levels, one would anticipate the genechip to miss ca. 2 to 3% of the R. sphaeroides genes by using the MAS 5.0 software.
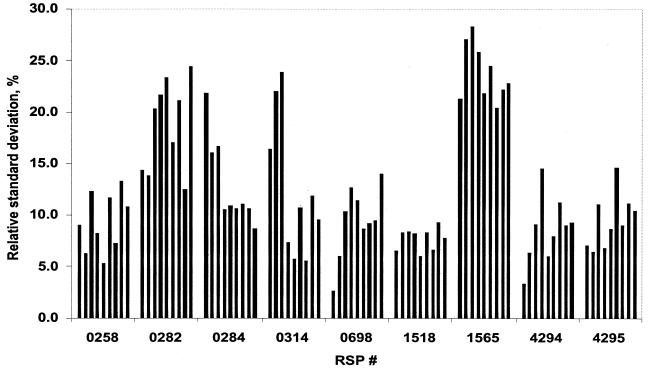
(ii) Intrachip reproducibility.
To evaluate intrachip reproducibility, we analyzed expression values of nine genes that were present in five copies located at various positions on the genechip. The data from nine hybridized genechips are shown in Fig. 3. The average standard deviation calculated for the probe sets of all nine ORFs under three growth conditions was 12.7%. Some copies differed from the average by as much as 28%. Because this variation is independent of growth conditions, it can be taken as a measure of reproducibility. For the majority of ORFs, the standard deviations for each of the five copies did not exceed 15% of the average value on a given chip. This suggested that differences in expression values for the same probe set measured on different genechips that are <15% are likely to be unreliable.
FIG. 3.
Intrachip reproducibility. Each bar represents a standard deviation (as a percentage) from the average signal value (from five copies of a gene) for a given gene on a given genechip. Bars 1 to 3, oxic conditions; bars 4 to 6, anoxic-light conditions; bars 7 to 9, anoxic-dark-DMSO conditions. RSP #, designation of a gene (probe set) on the R. sphaeroides genechip.
(iii) Genechip sensitivity.
The total number of genes whose expression is detectable under any given growth condition is an indication of microarray sensitivity. The percentage of ORFs whose expression was detected as present (MAS 5.0 software) on each of the three replicate genechips used for a given growth condition was 58 to 82% of the total number of ORFs. These numbers are comparable to the percentage of ORFs detected by other bacterial DNA microarrays (5, 21, 22, 29, 33), suggesting that the R. sphaeroides microarray is reasonably sensitive.
(iv) Experimental reproducibility.
We analyzed variation between three biological replicates, i.e., R. sphaeroides cultures grown independently under each of the three growth conditions used in the present study. RMA analysis (19) was adopted as a method of choice because it showed superior correlation of expression of genes in operons (data not shown). Pearson correlation coefficients (R), calculated based on the RMA analysis, were in the range of 0.985 to 0.991 for oxic conditions, 0.982 to 0.990 for anoxic-light and 0.922 to 0.963 for anoxic-dark conditions. The lower experiment-to-experiment reproducibility for anoxic-dark conditions is probably resulted from the fact that these cultures were grown in completely filled tubes, where accumulation of the product of anaerobic respiration with DMSO might have inhibited growth of replicate cultures to somewhat different extents. Both oxic and anoxic-light cultures were grown under much better controlled conditions (see Materials and Methods), which resulted in the higher experiment-to-experiment reproducibility.
Verification of genechip data. (i) Alternative techniques.
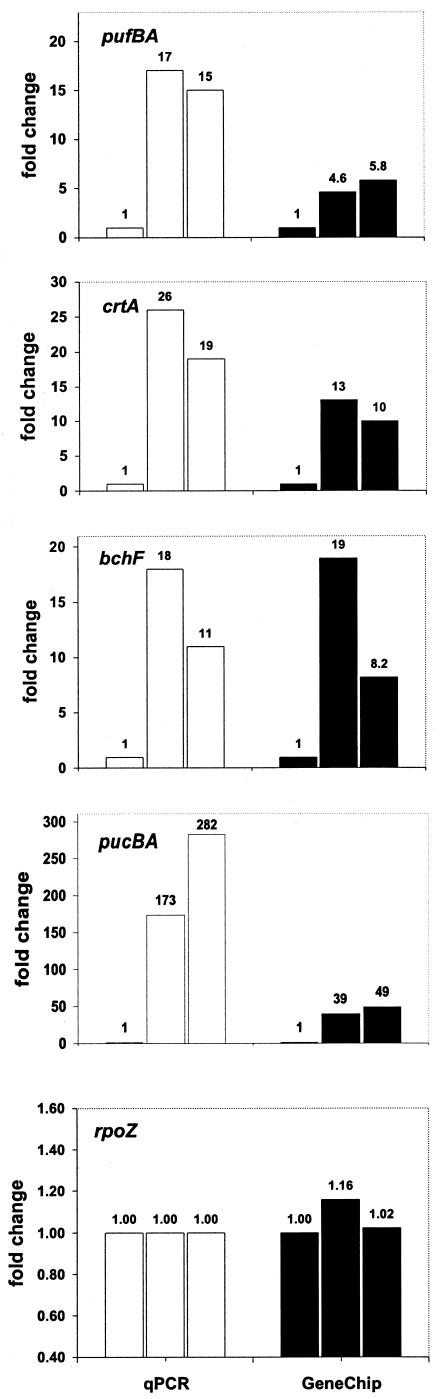
The genechip data were verified by qPCR performed on a selected group of genes with the same cDNA samples that were used for genechip hybridizations. Figure 4 shows changes in transcript levels detected by qPCR compared to those from the genechips. It is evident that the patterns of transcript changes detected by qPCR and the genechips were remarkably consistent, although the observed fold changes were somewhat different. In most cases, genechip underestimated differences compared to qPCR (Fig. 4). This result is not unusual; similar discrepancies in the fold differences between DNA microarrays and qPCR were observed for a variety of DNA microarray platforms (18, 22, 37).
FIG. 4.
Correlation between transcript abundance determined by genechips (▪) and qPCR (□). Transcript level of each gene under oxic conditions (left bars) is assigned a value of 1. The fold change ratios for each gene (compared to oxic conditions) for anoxic-light (center bars) and anoxic-dark (right bars) conditions are shown.
(ii) Expression profiles of selected expression units.
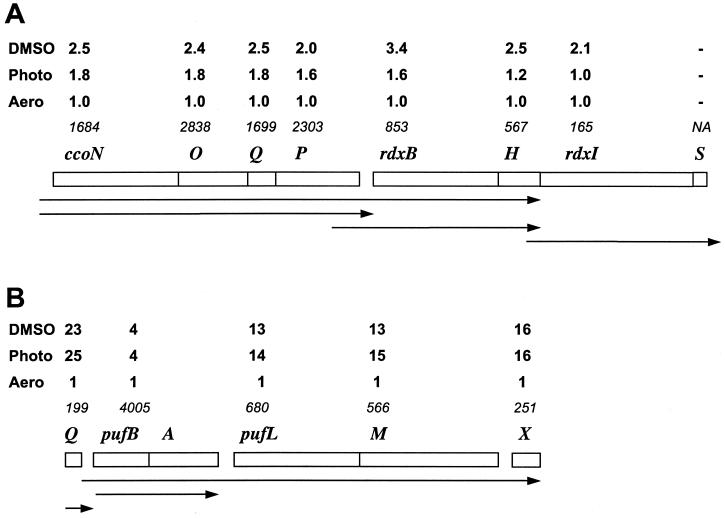
We quantitatively evaluated how the genechip data correlate with the expression profiles of selected operons that have been investigated in detail by traditional techniques. Two genetic loci were chosen, ccoNOQP-rdxBH and Q-pufBALMX. Figure 5 shows the absolute expression values under oxic conditions, as well as fold changes in expression (compared to oxic conditions) of each gene under the different conditions used here.
FIG. 5.
Expression profiles of selected genetic loci. (A) ccoNOQP-rdxBH-rdxIS; (B) Q-pufBALMX. The average gene expression levels from three genechip replicates under oxic conditions (Aero) are shown in italics. Each of these is assigned an arbitrary value of 1 for comparison purposes. The fold changes (compared to oxic conditions) under anoxic-light (Photo) or anoxic-dark (DMSO) conditions are shown. Horizontal arrows correspond to major transcripts (14, 32; reference 8 and references therein).
The ccoNOQP locus is known to form an operon, and the operon structure is evident from the genechip data (Fig. 5A). The absolute expression levels of all four cco genes under oxic conditions were similar, 1.7 to 2.8 × 103 relative fluorescent units, but different from the expression levels of the downstream genes. Expression levels of each of the four cco genes were increased 1.6- to 1.8-fold under anoxic-light conditions and ca. 2.0- to 2.5-fold under anoxic-dark-DMSO conditions compared to their expression levels under oxic conditions. Based upon genechip data, the rdxBH genes apparently form a different expression unit. The absolute expression levels of the rdxB and rdxH genes were similar to each other, 5.7 × 102 to 8.5 × 102 under oxic conditions, but significantly lower than those observed for genes of the ccoNOQP operon. Most importantly, the patterns of expression of ccoNOQP and rdxBH are not identical, indicating the presence of a separate transcript for rdxBH. This is consistent with the data by Roh and Kaplan (32), who showed that rdxBH expression is derived from readthrough from the promoter upstream of ccoN, as well as from a promoter located within the ccoP gene. rdxI is expressed at significantly lower levels, 1.7 × 102, under oxic conditions compared to rdxBH and is apparently independent of rdxBH (Fig. 5A). This is also consistent with the findings by Roh and Kaplan (32).
The Q-pufBALMX locus is known to have a complex expression pattern (14; reference 8 and references therein). The R. sphaeroides Q gene has its own promoter independent of the puf promoters. In agreement, Q gene expression under oxic conditions, 2.0 × 102, is significantly lower than that observed for the pufBA genes, 4.0 × 103 (pufB and pufA are represented by a single probe set on the genechip), and the fold changes in expression levels for the Q gene and pufBA are different (Fig. 5B). The pufL, pufM, and pufX transcript levels change synchronously in response to environmental conditions (Fig. 5B), indicating that these genes belong to the same transcript, as was shown earlier (8, 14). The reason for the lower expression levels of pufX compared to pufLM is not understood; however, lower pufX abundance was observed earlier by direct mRNA measurements (7). The fold induction levels for pufBA and pufLMX are different, suggesting that these genes belong to differentially regulated transcription units (Fig. 5B). This is in agreement with two differentially regulated puf promoters, one for the pufBA transcript and another for the pufBALMX transcript, as well as with the existence of transcription terminators between pufBA and pufLMX (8).
In summary, the expression profiles revealed by the R. sphaeroides genechip are consistent with the known structure of transcription units and transcript abundance derived by alternative techniques.
Overview of R. sphaeroides transcriptome flexibility.
The conditions used in the present study correspond to three diverse growth modes of R. sphaeroides, i.e., aerobic respiration in the dark at saturating oxygen tension, anaerobic respiration in the dark with DMSO as a terminal electron acceptor, and anaerobic photosynthesis at medium light intensity. We qualitatively and quantitatively examined the transcriptome changes between these growth modes. The average standard deviation for replicate copies of a given probe set present on a single chip was <15% (see above). When we compared expression values of a given probe set between the genechips, the differences of <15% were regarded as insignificant. To increase the reliability of the comparisons, we applied the ≥15% criterion to all genechips corresponding to a given growth condition. We considered “changed” only genes whose expression values from each of the three replicates under one growth condition (ai) differed by at least 15% from values from each of the three replicates under a different growth condition (aj), i.e., ai ≥ 1.15 aj or ai ≤ 1.15 aj.
Genes encoding tRNAs and rRNAs were excluded from the analysis because their expression levels were not reliably measured for the following reasons. Variations in the abundance of tRNAs stems from the RNA purification protocol used in the present study, which disfavors very small RNA species. Variations in the abundance of rRNAs stems from the fact that their abundance appears to be beyond a linear range of measurements.
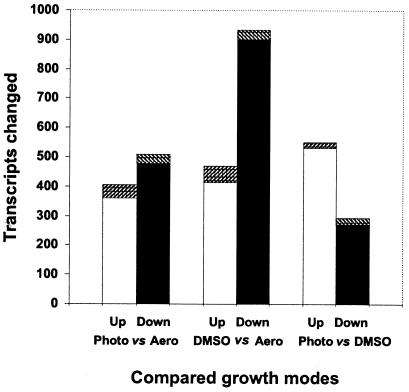
When anoxic-light conditions (photosynthesis) were compared to oxic conditions (aerobic respiration), the abundance of transcripts from 913 probe sets was changed (404 increased and 509 decreased). Of these, expression of 360 ORFs was increased and 479 decreased, which comprises ca. 20% of all of the ORFs represented on the genechip. The abundance of transcripts from 74 intergenic regions was also changed (44 increased and 30 decreased) (Fig. 6). When anoxic-dark conditions (anaerobic respiration with DMSO) were compared to oxic conditions (aerobic respiration), the abundance of transcripts from 1,403 probe sets was changed (469 increased and 934 decreased). Of these, the expression of 1,313 ORFs was changed (413 increased and 900 decreased). This comprises ca. 31% of all ORFs. The abundance of transcripts from 90 intergenic regions was changed (56 increased and 34 decreased) (Fig. 6). When anoxic-light conditions were compared to anoxic-dark conditions, the abundance of transcripts from ca. 845 probe sets was changed (551 increased and 294 decreased). Of these, expression of 530 ORFs was increased and 271 decreased, which comprises ca. 19% of all ORFs. The abundance of transcripts from 44 intergenic regions was also changed (21 increased and 23 decreased) (Fig. 6).
FIG. 6.
Pairwise comparisons of transcriptome changes under diverse growth modes. The numbers of transcripts whose abundance changed are shown. Up, upregulated genes (□, ORFs; ▨, intergenic regions); Down, downregulated genes (▪, ORFs; ▧, intergenic regions); Aero, oxic conditions; Photo, anoxic-light conditions; DMSO, anoxic-dark conditions.
These data show that expression of one-fifth to one-third of the R. sphaeroides ORFs are significantly changed in cells grown under the diverse conditions used here and suggest that massive changes in expression is the major means for adapting to diverse conditions.
Major patterns of gene expression in R. sphaeroides related to energy generation and maintenance of redox balance.
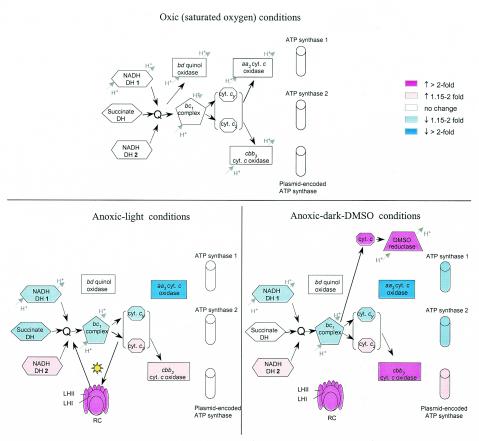
Growth under the oxic, anoxic-dark, and anoxic-light conditions used here requires different pathways of energy generation, i.e., aerobic respiration, anaerobic respiration (with DMSO as terminal electron acceptor) and photosynthesis, respectively. These growth modes also pose different constraints on the redox poise of the cell. Here we focus on expression of genes related to energy generation and maintenance of redox balance under the three growth conditions. Our analysis revealed that most functional changes in these pathways known to occur during adaptation of cells to these growth conditions are reflected in corresponding changes in gene expression. However, several unexpected and currently unexplainable expression patterns exist. Figure 7 summarizes the most significant changes in gene expression, which are discussed below.
FIG. 7.
R. sphaeroides electron transport chain complexes and ATP synthases involved in energy generation under oxic, anoxic-light, and anoxic-dark-DMSO conditions. Arrows indicate electron flow; dashed line arrows indicate anticipated direction of electron flow. The known sites for generation of proton motive force are shown. The expression of genes under oxic conditions is set as the background for comparisons (not colored). Blue corresponds to decreased gene expression of the corresponding proteins compared to expression under oxic conditions as follows: light blue, ≤2-fold decrease; dark blue, >2-fold decrease. Pink corresponds to increased gene expression compared to the expression under oxic conditions as follows: light pink, ≤2-fold increase; dark pink, >2-fold increase. No color corresponds to expression that is not significantly different from the expression under oxic conditions. Proteins whose genes are expressed below reliable detection are not shown. DH, dehydrogenase; Q, quinone-quinol pool; LH, light-harvesting complex; RC, reaction center complex. The reaction center and light-harvesting complexes are not shown under oxic conditions because several bch genes involved in bacteriochlorophyll biosynthesis are not expressed under these conditions and no PS is made.
Based upon the expression patterns (upregulated, downregulated, and unchanged) under three growth conditions, all genes were placed into nine classes. Class 1 includes 266 probe sets, 233 ORFs, and 33 intergenic regions, whose expression was upregulated under both anoxic-dark and anoxic-light conditions compared to oxic conditions. Class 1 genes are concluded to be responsive to the lack of oxygen, irrespective of the presence or absence of light. Several functional categories related to energy generation and redox maintenance are evident in this class.
A large group of genes includes PS genes that encode components of the photosynthetic apparatus. Most of these genes are clustered in the large PS gene cluster (RSP0255-0292), whereas two operons are separate from this cluster (RSP0314-0315 [puc1BAC] and RSP1556-1557 [puc2BA]) (39). Oxygen is known to be the major environmental factor that controls expression of the PS genes (reviewed in references 31 and 38). In this regard, the fold change increases for a given PS gene in response to anoxic conditions are very similar, irrespective of the presence or absence of light. The PS genes that belong to class 1 encode enzymes of bacteriochlorophyll biosynthesis, bch (RSP0260-0263, RSP0273-0274, RSP0277, RSP0279-0281, and RSP0284-0289); enzymes of carotenoid biosynthesis, crt (RSP0264-0268 and RSP0270-0272); enzymes involved in the biosynthesis of isoprenoids, which give rise to precursors for bacteriochlorophyll, carotenoids, and quinols (RSP0254, dxsA; RSP0276, idi); and structural polypeptides (RSP0255-0258, puf; RSP0290, puhA; RSP0314, puc1BA [represented by one probe set]; RSP1556-155, puc2BA), as well as assembly factors for the reaction center and light-harvesting complexes (RSP0259, Q protein; RSP0278, orf428; RSP0291, orf1447; RSP0315, puc1C); and regulatory factors involved in PS formation (RSP0269, tspO; RSP0283, ppaA).
Several genes of unknown function are located downstream of RSP0292 at the end of the PS gene cluster (RSP0293-0295). The expression pattern of these genes is similar to that of the PS genes. We therefore predict that these genes are involved in PS formation in R. sphaeroides. Similarly, dxsA (RSP0254) located at the other end of the PS cluster, downstream of the puf operon, must be included in the PS gene cluster. dxsA encodes deoxyxylulose-5-phosphate synthase (15), whose product is a common precursor to isoprenoids in the methylerythritol phosphate pathway. Expression of dxsA is six- to ninefold higher under anoxic compared to oxic conditions, which is readily explainable since isoprenoids are needed for bacteriochlorophyll and carotenoid biosynthesis.
A notable exception to the expression pattern of the PS genes is ppsR (RSP0282), which is located in the middle of the PS gene cluster. ppsR encodes a major repressor of PS gene expression. Expression of ppsR is essentially independent of growth conditions, which is consistent with our earlier data based on ppsR-lacZ fusion (13) and PpsR immunoblot (9) analyses. Interestingly, expression of the appA gene (RSP1565), which does not belong to the PS gene cluster, is moderately upregulated (1.7- to 2.0-fold) under anoxic conditions. AppA functions as an antirepressor of PpsR (3, 13, 25). We hypothesize that the increased abundance of the appA transcript contributes to the derepressed expression of the PS genes under anoxic conditions.
Several genes, which do not belong to the PS cluster but are involved in PS formation, are upregulated under both anoxic conditions. These genes encode enzymes involved in the biosynthesis of protoporphyrin IX, a common precursor of heme and bacteriochlorophyll. Expression of hemA (RSP2984) encoding 5-aminolevulinic acid synthase, the first enzyme in protoporphyrin IX biosynthesis, is increased approximately three- to fourfold under anoxic conditions, a finding that is consistent with earlier observations (28). Increased hemA expression is required to satisfy the elevated demand for protoporphyrin IX under anoxic conditions due to bacteriochlorophyll biosynthesis. Consistent with this explanation, expression of the genes encoding most of the subsequent enzymes of the protoporphyrin IX biosynthetic pathway—hemB (RSP2848) for δ-aminolevulinate dehydratase, hemC (RSP0679) for porphobilinogen deaminase, hemN (RSP0317), and hemZ (RSP0699) for both oxygen-independent coproporphyrinogen III oxidases—is increased severalfold under anoxic conditions.
The cco operon (RSP0692-0696) encoding the cytochrome c oxidase of the cbb3 type, belongs to class 1 (see above). It has been shown that the cbb3 cytochrome c oxidase, which has high affinity to oxygen, is apparently active under anoxic conditions (30); however, its precise function has yet to be determined. This expression pattern is opposite to the expression pattern of genes encoding the cytochrome c oxidase of the aa3 type (see class 2).
The cytochrome c2 encoding gene (RSP0296, cycA) is upregulated ∼1.7-fold under both anoxic conditions, a finding that is consistent with earlier observations (4). Higher cycA expression is required to ensure adequate amounts of cytochrome c2 for electron transfer between the reaction center and the bc1 complex under anoxic conditions. It is worth mentioning that expression of genes encoding bc1 complex (RSP1394-1396, pet) is slightly upregulated (1.3- to 1.7-fold on average); however, these genes did not pass the reliability criterion used here.
Genes encoding a putative multisubunit NADH-ubiquinone oxidoreductase (RSP0100-0112) are upregulated two- to fourfold. The R. sphaeroides genome encodes a second set of genes for NADH-ubiquinone oxidoreductase (nuo, RSP2512-2531), which are more similar to the NADH-ubiquinone oxidoreductases from related bacteria, Rhodobacter capsulatus and Paracoccus denitrificans. Expression of the nuo genes is significantly higher in absolute values, than that of the putative enzyme, RSP0100-0112, and is oppositely regulated under anoxic conditions (see class 4). The function of the putative NADH-ubiquinone oxidoreductase (RSP0100-0112) and physiological reasons for upregulation of its expression under both anoxic conditions are not known and require further investigation.
Two clusters of genes involved in carbon dioxide fixation (RSP1280-1285[cbbI operon] and RSP3266-3271 [cbbII operon]) are upregulated under both anoxic conditions. In heterotrophically grown Rhodobacter spp., a major function of carbon dioxide fixation under anoxic conditions is to serve as an “electron sink”, i.e., to balance redox poise of the cell (10, 16, 26, 40). Not surprisingly, expression of the cbb operons is increased 8- to 15-fold (cbbI) and 19- to 55-fold (cbbII) under anoxic-light conditions, where “excessive” reducing power is produced by photosynthesis. Increase in expression of these operons under anoxic-dark conditions is much more moderate, i.e., two- to fivefold, since DMSO reduction functions as the main “electron sink” (see class 3). The presence of carbon dioxide is required for induction of the cbb operons. Whereas the anoxic-light-grown cultures were sparged with the gas mixture containing carbon dioxide, anoxic-dark-grown cultures must have endogenously produced carbon dioxide.
Class 2 includes 335 probe sets, 315 ORFs, and 20 intergenic regions, whose expression was significantly downregulated under both anoxic-dark and anoxic-light conditions compared to oxic conditions. This class includes genes that are repressed in the absence of oxygen, irrespective of light.
The expression levels of several genes involved in oxidative stress response, i.e., catalase (RSP2779, cat), superoxide dismutase (RSP2693, sox), peroxiredoxin (RSP0899), and glutathione peroxidase (RSP2389), are 1.5- to 4.0-fold higher under oxic conditions than under anoxic conditions. Apparently, growth at saturating oxygen concentration results in significant levels of reactive oxygen species in R. sphaeroides, which are detoxified by the products of these genes.
Several groups of genes involved in energy generation are downregulated under both anoxic conditions. Expression of the cta genes encoding a multisubunit cytochrome c oxidase of the aa3 type (RSP1826-1829 and RSP1877) is approximately 5.0- to 7.5-fold lower under anoxic conditions than under oxic conditions. The aa3 cytochrome c oxidase is the primary terminal oxidase operating under high oxygen tension. Consistent with our observations, the amount of the aa3 oxidase is greatly diminished under anoxic conditions (9). Note that the expression pattern of the second cytochrome c oxidase, of the cbb3 type, is opposite to that of the aa3 oxidase (see class 1).
Expression of the NADH-ubiquinone dehydrogenase genes (RSP2512-2531, nuo) is 1.6- to 2.6-fold downregulated under anoxic conditions. It is interesting that this expression pattern is opposite to that of the other putative NADH-ubiquinone dehydrogenase, RSP0100-0112, described above. Neither physiological reasons nor molecular mechanisms responsible for the opposite expression patterns of the two presumed NADH-ubiquinone dehydrogenase complexes in R. sphaeroides are known.
Class 3 includes 202 probe sets, 175 ORFs, and 27 intergenic regions, whose expression was significantly upregulated under anoxic-dark conditions but unchanged under anoxic-light conditions. DMSO reduction under anoxic-dark conditions serves as a major energy-generating pathway. The most profound changes are observed in genes involved in DMSO reduction, i.e., genes encoding DMSO reductase and its cognate cytochrome c (RSP3046-3048, dorCAB), as well as genes involved in the biosynthesis of the molybdopterin cofactor of DMSO reductase (RSP3049-3050, moaAB). Expression of these genes under oxic conditions is at or below detection. It is increased at least 69- to 314-fold under anoxic-dark conditions. In agreement with earlier observations, expression of these genes requires both the lack of oxygen and the presence of DMSO (27).
Interestingly, a cluster of genes, RSP3929-3936, located on one of the R. sphaeroides plasmids which may encode an F0F1-type ATP synthase, is upregulated 1.4- to 4.5-fold under anoxic-dark conditions. Whether this plasmid-encoded ATP synthase is functional and, if so, whether its upregulation affects overall ATP synthesis is unknown. Genes for two additional F0F1-type ATP synthases are present in the genome: RSP1035-1039 and RSP2296-2300. These enzymes are expressed at much higher levels compared to the putative plasmid-encoded ATP synthase, and their expression is downregulated under anoxic-dark conditions, i.e., opposite to the plasmid-encoded genes (see class 5).
Class 4 includes 171 probe sets, 159 ORFs, and 12 intergenic regions, whose expression was significantly downregulated under anoxic-light conditions but was essentially unchanged under anoxic-dark conditions compared to oxic conditions. The genes encoding succinate dehydrogenase (RSP0974-0979, sdh) fall into this category. Succinate serves a dual role in the growth medium used here, i.e., primary carbon source and primary electron donor to the electron transport chains. Decreased expression of the sdh genes might indicate a lower requirement for succinate dehydrogenase under anoxic-light conditions. This could be explained by the fact that, under these conditions, carbon dioxide fixation provides an additional source of carbon. It is noteworthy that although expression of the sdh genes decreased by 1.8- to 2.2-fold it still remained high. A much steeper decline in the activity of succinate dehydrogenase could have been anticipated under these conditions. It is currently unknown how the abundance and activity of the enzymatic complex itself corresponds to the decrease in sdh gene expression. However, our assumption of the lower demand for succinate under anoxic-light conditions is backed up by the observed twofold decrease in expression of the dicarboxylate-binding periplasmic transport protein (RSP0910, dctP).
Class 5 includes 593 probe sets, 580 ORFs, and 13 intergenic regions, whose expression was significantly downregulated under anoxic-dark conditions but was essentially unchanged under anoxic-light conditions. It is worth noting that the expression of numerous genes encoding ribosomal proteins and other proteins involved in translation was downregulated, which is probably indicative of the severalfold-slower growth under anoxic-dark conditions compared to either oxic or anoxic-light conditions.
Expression of the putative chromosome encoded F0F1-type ATP synthases (RSP2296-2300 and RSP1035-1039) was downregulated by approximately twofold. The decreased expression of the ATP synthases may signify the less-efficient electron transport chain which generates a lower proton motive force.
Expression of NAD/NADP transhydrogenase (pntAB, RSP0239-0240) that catalyzes NADPH formation at the expense of proton gradient and NADH was downregulated by ca. 2.0- to 2.5-fold under anoxic-dark conditions. NAD/NADP transhydrogenase plays a role in redox balancing in R. sphaeroides (17). It is possible that decreased expression of this enzyme corresponds to a lower need for NADPH for biosynthetic purposes under anoxic-dark conditions.
Class 6 includes six probe sets, five ORFs, and one intergenic region, whose expression was significantly upregulated under anoxic-dark conditions but was downregulated under anoxic-light conditions. Class 7 includes 137 probe sets, 122 ORFs, and 15 intergenic regions, whose expression was significantly upregulated under anoxic-light conditions but was essentially unchanged under anoxic-dark conditions. Class 8 includes six probe sets, five ORFs, and one intergenic region, whose expression was significantly upregulated under anoxic-light conditions but was downregulated under anoxic-dark conditions. No genes related to energy generation or maintenance of redox balance belong to classes 6 to 8. Class 9 contained genes whose expression was essentially unchanged under the three growth conditions studied.
In conclusion, we have curated the genome annotation of the metabolically versatile proteobacterium R. sphaeroides 2.4.1. In this bacterium, codon usage proved to be the most reliable tool for ORF identification, as well as start codon determination, a finding that probably is true for other GC-rich organisms. We have constructed a DNA microarray for the genome-wide transcriptome analyses and provided evidence for its utility in accurate assessment of transcript abundance. The transcriptome profiles of R. sphaeroides grown under a diverse set of conditions, i.e., aerobic respiration, anaerobic respiration and anaerobic photosynthesis, were analyzed. According to our analysis, expression levels of one-fifth to one-third of all R. sphaeroides ORFs change upon transitions between the three diverse growth modes used here. This suggests that R. sphaeroides uses massive and coordinate changes in expression patterns as the major means of restructuring its metabolic capabilities.
The largest group of differentially expressed genes was comprised of genes whose expression was responsive to the lack of oxygen, regardless of the presence or absence of light (more than 600 genes, classes 1 and 2). The size of this cluster confirms the existing notion that oxygen availability plays a major role in R. sphaeroides metabolism and physiology. The second largest cluster of genes (class 5) corresponded to the genes downregulated under anoxic-dark conditions. Many of these genes are apparently downregulated because of significantly slower growth under these conditions.
We have reconstructed energy generation and redox balance pathways in R. sphaeroides grown under three diverse growth modes and linked these to gene expression. We found that most functional changes involved in these pathways, which were known from R. sphaeroides physiology, are reflected in expression patterns of the corresponding genes. In addition, the present study generated a number of predictions for future investigations. For example, according to the expression data, the PS gene cluster needs to be extended by several genes downstream of the puf and puhA operons. The involvement of these genes in PS formation has yet to be tested. Further, we uncovered the second set of genes encoding a putative NADH dehydrogenase and found that expression of these genes is opposite to expression of the main NADH dehydrogenase. We also identified three differentially expressed sets of genes encoding putative ATP synthases. The underlying reasons and mechanisms of differential expression of these apparently homologous protein complexes are not known. In fact, expression data are the first indication of their functionality. Numerous intergenic regions were found to be differentially expressed. These are candidates for missed ORFs and/or RNAs. Overall, the constructed genechip has proven to be a powerful tool for studying the unprecedented metabolic and energetic versatility of R. sphaeroides.
Acknowledgments
This study was supported by NIH grants NCRR (COBRE) P20 RR15640 (M.G.) and PHS GM15590 (S.K.), DOE grant BER DE-FG02-01ER63232 (M.G. and S.K.), the Cullen Foundation at the University of Texas Health Science Center (UTHSC) (S.K.), and startup funds from the University of Wyoming (M.G.).
We are grateful to Mark Gerasi and Bifeng Gao (UCHSC-Denver) for numerous useful discussions regarding genechip technology and data analysis; to B. Todd Woessner and Dana Swoveland (UCHSC-Denver) for excellent genechip hybridizations; to Garry Miyada, Xue Mei Zhou, and Tanya Trilessky (Affymetrix) for advice and expeditious handling of the R. sphaeroides genechip design; and to Jung Hyeob Roh (UTHSC-Houston) for genomic DNA hybridization experiment. We are also thankful to the COBRE group at the University of Wyoming (Bill Flynn, Don Jarvis, Rich McCormick, Mark Stayton, and Paul Thomas) for their support of development of the R. sphaeroides genechip; to the Genomes-to-Life group (Tim Donohue [University of Wisconsin], Jeremy Edwards [University of Delaware], Jon Hosler [University of Mississippi], and Bill Margolin [UTHSC-Houston] for useful discussions.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boldogkoi, Z. 2000. Coding in the noncoding DNA strand: a novel mechanism of gene evolution? J. Mol. Evol. 51:600-606. [DOI] [PubMed] [Google Scholar]
- 3.Braatsch, S., M. Gomelsky, S. Kuphal, and G. Klug. 2002. The single flavoprotein, AppA, from Rhodobacter sphaeroides integrates both redox and light signals. Mol. Microbiol. 45:827-836. [DOI] [PubMed] [Google Scholar]
- 4.Brandner, J. P., A. G. McEwan, S. Kaplan, and T. J. Donohue. 1989. Expression of the Rhodobacter sphaeroides cytochrome c2 structural gene. J. Bacteriol. 171:360-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caldwell, R., R. Sapolsky, W. Weyler, R. R. Maile, S. C. Causey, and E. Ferrari. 2001. Correlation between Bacillus subtilis scoC phenotype and gene expression determined using microarrays for transcriptome analysis. J. Bacteriol. 183:7329-7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen-Bazire, G., W. R. Sistrom, and R. Y. Stanier. 1957. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J. Cellular Comp. Physiol. 49:25-68. [DOI] [PubMed] [Google Scholar]
- 7.DeHoff, B. S., J. K. Lee, T. J. Donohue, R. I. Gumport, and S. Kaplan. 1988. In vivo analysis of puf operon expression in Rhodobacter sphaeroides after deletion of a putative intercistronic transcription terminator. J. Bacteriol. 170:4681-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donohue, T. J., P. J. Kiley, and S. Kaplan. 1988. The puf operon region of Rhodobacter sphaeroides. Photosynthesis Res. 19:39-61. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Horsman, J. A., E. Berry, J. P. Shapleigh, J. O. Alben, and R. B. Gennis. 1994. A novel cytochrome c oxidase from Rhodobacter sphaeroides that lacks CuA. Biochemistry 33:3113-3119. [DOI] [PubMed] [Google Scholar]
- 10.Gibson, J. L., and F. R. Tabita. 1996. The molecular regulation of the reductive pentose phosphate pathway in proteobacteria and cyanobacteria. Arch. Microbiol. 166:141-150. [DOI] [PubMed] [Google Scholar]
- 11.Gomelsky, L., J. Sram, O. V. Moskvin, I. M. Horne, H. N. Dodd, J. M. Pemberton, A. G. McEwan, S. Kaplan, and M. Gomelsky. 2003. Identification and in vivo characterization of PpaA, a regulator of photosystem formation in Rhodobacter sphaeroides. Microbiology 149:377-388. [DOI] [PubMed] [Google Scholar]
- 12.Gomelsky, M., and G. Klug. 2002. BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem. Sci. 27:497-500. [DOI] [PubMed] [Google Scholar]
- 13.Gomelsky, M., and S. Kaplan. 1997. Molecular genetic evidence suggesting interactions between AppA and PpsR in regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 179:128-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong, L., J. K. Lee, and S. Kaplan. 1994. The Q gene of Rhodobacter sphaeroides: its role in puf operon expression and spectral complex assembly. J. Bacteriol. 176:2946-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahn, F. M., L. M. Eubanks, C. A. Testa, B. S. Blagg, J. A. Baker, and C. D. Poulter. 2001. 1-Deoxy-d-xylulose 5-phosphate synthase, the gene product of open reading frame (ORF) 2816 and ORF 2895 in Rhodobacter capsulatus. J. Bacteriol. 183:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallenbeck, P. L., R. Lerchen, P. Hessler, and S. Kaplan. 1990. Roles of CfxA, CfxB, and external electron acceptors in regulation of ribulose 1,5-bisphosphate carboxylase/oxygenase expression in Rhodobacter sphaeroides. J. Bacteriol. 172:1736-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hickman, J. W., R. D. Barber, E. P. Skaar, and T. J. Donohue. 2002. Link between the membrane-bound pyridine nucleotide transhydrogenase and glutathione-dependent processes in Rhodobacter sphaeroides. J. Bacteriol. 184:400-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holland, M. J. 2002. Transcript abundance in yeast varies over six orders of magnitude. J. Biol. Chem. 277:14363-14366. [DOI] [PubMed] [Google Scholar]
- 19.Irizarry, R. A., B. M. Bolstad, F. Collin, L. M. Cope, B. Hobbs, and T. P. Speed. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa, M., and S. Goto. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khil, P. P., and R. D. Camerini-Otero. 2002. Over 1000 genes are involved in the DNA damage response of Escherichia coli. Mol. Microbiol. 44:89-105. [DOI] [PubMed] [Google Scholar]
- 22.Lee, J.-M., S. Zhang, S. Saha, S. Santa Anna, C. Jiang, and J. Perkins. 2001. RNA expression analysis using an antisense Bacillus subtilis genome array. J. Bacteriol. 183:7371-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackenzie, C., M. Choudhary, F. W. Larimer, P. F. Predki, S. Stilwagen, J. P. Armitage, R. D. Barber, T. J. Donohue, J. P. Hosler, J. Newman, J. P. Shapleigh, R. E. Sockett, J. Zeilstra-Ryalls, and S. Kaplan. 2001. The home stretch, a preliminary analysis of the nearly completed genome of Rhodobacter sphaeroides 2.4.1. Photosynthesis Res. 70:19-41. [DOI] [PubMed] [Google Scholar]
- 24.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Masuda, S., and C. Bauer. 2002. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell 110:613-623. [DOI] [PubMed] [Google Scholar]
- 26.McEwan, A. G. 1994. Photosynthetic electron transport and anaerobic metabolism in purple non-sulfur phototrophic bacteria. Antonie Leeuwenhoek 66:151-164. [DOI] [PubMed] [Google Scholar]
- 27.Mouncey, N. J., and S. Kaplan. 1998. Cascade regulation of dimethyl sulfoxide reductase (dor) gene expression in the facultative phototroph Rhodobacter sphaeroides 2.4.1T. J. Bacteriol. 180:2924-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neidle, E. L., and S. Kaplan. 1993. Expression of the Rhodobacter sphaeroides hemA and hemT genes, encoding two 5-aminolevulinic acid synthase isozymes. J. Bacteriol. 175:2292-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ochsner, U. A., P. J. Wilderman, A. I. Vasil, and M. L. Vasil. 2002. GeneChip® expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 45:1277-1287. [DOI] [PubMed] [Google Scholar]
- 30.Oh, J. I., and S. Kaplan. 1999. The cbb3 terminal oxidase of Rhodobacter sphaeroides 2.4.1: structural and functional implications for the regulation of spectral complex formation. Biochemistry 38:2688-2696. [DOI] [PubMed] [Google Scholar]
- 31.Oh, J. I., and S. Kaplan. 2001. Generalized approach to regulation and integration of gene expression. Mol. Microbiol. 39:1116-1123. [DOI] [PubMed] [Google Scholar]
- 32.Roh, J. H., and S. Kaplan. 2002. Interdependent expression of the ccoNOQP-rdxBHIS loci in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 184:5330-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenow, C., R. M. Saxena, M. Durst, and T. R. Gingeras. 2001. Prokaryotic RNA preparation methods useful for high density array analysis: comparison of two approaches. Nucleic Acids Res. 29:E112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M.-A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 35.Schultz, J., R. R. Copley, T. Doerks, C. P. Ponting, and P. Bork. 2000. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28:231-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tatusov, R. L., D. A. Natale, I. V. Garkavtsev, T. A. Tatusova, U. T. Shankavaram, B. S. Rao, B. Kiryutin, M. Y. Galperin, N. D. Fedorova, and E. V. Koonin. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29:22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuen, T., E. Wurmbach, R. L. Pfeffer, B. J. Ebersole, and S. C. Sealfon. 2002. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 30:E48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeilstra-Ryalls, J., M. Gomelsky, J. Eraso, A. Yeliseev, J. O'Gara, and S. Kaplan. 1998. Control of photosystem formation in Rhodobacter sphaeroides. J. Bacteriol. 180:2801-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng, X., M. Choudhary, and S. Kaplan. 2003. A second and unusual pucBA operon of Rhodobacter sphaeroides 2.4.1: genetics and function of the encoded polypeptides. J. Bacteriol. 185:6171-6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu, Y. S., and S. Kaplan. 1985. Effects of light, oxygen, and substrates on steady-state levels of mRNA coding for ribulose-1,5-bisphosphate carboxylase and light-harvesting and reaction center polypeptides in Rhodopseudomonas sphaeroides. J. Bacteriol. 162:925-932. [DOI] [PMC free article] [PubMed] [Google Scholar]