Abstract
We disrupted the reverse gyrase gene from a hyperthermophilic archaeon, Thermococcus kodakaraensis KOD1. An apparent positive supercoiling activity that was observed in the host strain was not found in the disruptant strain. We found that a lack of reverse gyrase led to a retardation in growth that was more striking at higher temperatures. However, the disruption of the reverse gyrase gene did not lead to a lethal phenotype at 90°C. This study provides experimental evidence that reverse gyrase is not a prerequisite for hyperthermophilic life.
Reverse gyrase is an enzyme that introduces positive supercoils into covalently closed DNA (12). The enzyme is comprised of two domains: the C-terminal domain of the protein resembles DNA topoisomerases of the type IA family, and the N-terminal domain harbors the motifs conserved in helicases of superfamilies 1 and 2 (3, 6). Reverse gyrase has attracted much attention due to the fact that the gene that codes for it is the only gene that is present in all hyperthermophilic genomes and absent from all mesophilic and thermophilic genomes, indicating that the enzyme is the one and only hyperthermophile-specific protein (5, 7).
Thermococcus kodakaraensis KOD1 is a hyperthermophilic archaeon isolated from a solfatara on Kodakara Island, Kagoshima, Japan (1, 11). The strain is a strict anaerobe and grows heterotrophically on a variety of organic substrates, including starch, pyruvate, and amino acids (1). We have recently determined the entire genome sequence of this strain (unpublished data). Moreover, a gene disruption system has been developed for this strain, allowing us to take a genetic approach to studying reverse gyrase in T. kodakaraensis (17). In order to provide experimental evidence as to whether reverse gyrase is a prerequisite for hyperthermophilic life, we disrupted the reverse gyrase gene of T. kodakaraensis (rgyTk) and examined the growth characteristics of the mutant strain at various temperatures.
A detailed search of the T. kodakaraensis genome sequence revealed the presence of only one reverse gyrase gene on the genome (GenBank accession no. AB117612). The deduced amino acid sequence consisted of 1,222 amino acid residues and resembled those of enzymes from the hyperthermophiles Pyrococcus horikoshii (accession no. NP_142736; 73.2% identity with the T. kodakaraensis enzyme), Pyrococcus furiosus (accession no. AAB49283; 72.8% identity), “Pyrococcus abyssi” (accession no. CAB50173; 72.7% identity), Methanococcus jannaschii (accession no. AAB99531; 42.6% identity), and Sulfolobus acidocaldarius (accession no. A47445; 38.1% identity). An intein of 489 residues was present in exactly the same position as those found in the enzymes from P. horikoshii and M. jannaschii. Phylogenetic analysis revealed that the reverse gyrase from T. kodakaraensis (RgyTk) was positioned in the same branch as the enzymes from the three Pyrococcus strains and M. jannaschii (7), branching off between them with bootstrap values of 100 (data not shown). We also searched for genes that might encode the individual helicase-like and topoisomerase-like domains of reverse gyrase, as the two domains were found to be encoded by two separate genes in “Nanoarchaeum equitans” (10, 21). Besides the single reverse gyrase gene, we detected one topoisomerase I gene and two genes encoding the two subunits of topoisomerase VI. The topoisomerase I gene encoded a protein of 1,229 residues with an intein of 511 residues, resembling the structure of the topoisomerase I from P. furiosus (72.7% identity). In terms of helicases, no other gene product with notable similarity to the helicase-like domain of RgyTk could be detected. The open reading frame that most resembled the domain was an orthologue of the DNA double-strand break repair Rad50 ATPase of P. furiosus (46.8% identity) (9).
In order to disrupt rgyTk, a ΔtrpE strain was utilized as the host strain. The trpE gene encodes one subunit of anthranilate synthase (20), which is necessary for tryptophan biosynthesis, and therefore the ΔtrpE strain displays strict tryptophan auxotrophy. A gene disruption plasmid, designed to delete the entire rgyTk gene with an intact trpE gene as a marker, was constructed. A DNA fragment containing the entire reverse gyrase gene along with 1,005 and 1,001 bp of the 5′- and 3′-end-flanking regions, respectively, was amplified by PCR from the genomic DNA of T. kodakaraensis. The primers used were 5F1 (5′-GTGGGCCCGCAATGTTCATTACTCTA) and3R1 (5′-TTGCAGCCATGTCCTCAAGGGCGAAG). Thisfragment was inserted into pUC18, and the resulting plasmid was used as a template for inverse PCR in order to amplify all regions other than the reverse gyrase coding region. The primers used were 5R1 (5′-GGCCCCACCCTCTAAGTCTTTCTCTG) and 3F1 (5′-TGGTGTTTTAATCGCTTGCCACTATC). This linear DNA fragment was ligated with a DNA fragment including the trpE gene under the control of the pyrF gene promoter of T. kodakaraensis (17). The plasmid obtained was purified and sequenced in order to confirm the sequence and proper orientation of the following DNA fragments: the 5′-end-flanking region of rgyTk followed by the promoter region, the trpE coding region, and the 3′-end-flanking region of rgyTk (Fig. 1A). Restriction enzymes and other modifying enzymes were purchased from Takara Shuzo (Kyoto, Japan) or Toyobo (Osaka, Japan). The QIAEX gel extraction kit (QIAGEN, Hilden, Germany) was used to recover DNA fragments from the agarose gel. For the isolation of plasmid DNA, a plasmid minikit (QIAGEN) was used. DNA sequencing was carried out with a BigDye terminator cycle sequencing kit and a model 3100 capillary DNA sequencer (Applied Biosystems, Foster City, Calif.).
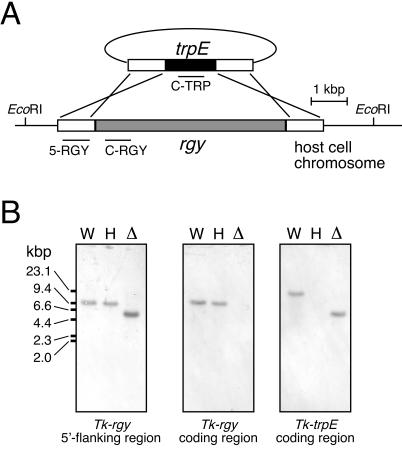
FIG. 1.
Disruption of rgyTk. (A) Strategy for disruption of the rgyTk gene via double crossover recombination. Regions corresponding to the probes used in the analysis described for panel B are indicated. (B) Confirmation of rgyTk gene disruption by Southern blot analyses. A digoxigenin-DNA labeling and detection kit (Roche Diagnostics) was used according to the manufacturer's instructions. The probes used were 5-RGY, a 728-bp fragment in the upstream region of the rgyTk gene (left panel), C-RGY, a 698-bp fragment within the rgyTk coding region (middle panel), and C-TRP, a 700-bp fragment within the T. kodakaraensis trpE coding region (right panel). Genome DNA from the wild type (W), the ΔtrpE host strain (H), and the Δrgy mutant strain (Δ) were digested with EcoRI. Results with 5-RGY show the decrease in length of the EcoRI fragment (from 9,360 to 5,641 bp) due to the replacement of the rgyTk gene with the T. kodakaraensis trpE gene, and those with C-RGY show the absence of the rgyTk gene on the Δrgy mutant strain chromosome.
We transformed the ΔtrpE strain of T. kodakaraensis by using the plasmid shown in Fig. 1A. All procedures for transformation of T. kodakaraensis and gene disruption have been described in detail elsewhere (17). Transformants were selected using a synthetic medium in the absence of tryptophan. The medium consisted of 0.8× artificial seawater (16) [16 g of NaCl, 2.4 g of MgCl2 · 6H2O, 4.8 g of MgSO4 · 7H2O, 0.8 g of (NH4)2SO4, 160 mg of NaHCO3, 240 mg of CaCl2 · 2H2O, 0.4 g of KCl, 336 mg of KH2PO4, 40 mg of NaBr, 16 mg of SrCl2 · 6H2O, and 8 mg of ammonium iron citrate per liter] supplemented with 5.0 ml of modified Wolfe's trace minerals [0.5 g of MnSO4 · 2H2O, 0.1 g of CoCl2, 0.1 g of ZnSO4, 0.01 g of CuSO4 · 5H2O, 0.01 g of AlK(SO4)2, 0.01 g of H3BO3, and 0.01 g of NaMoO4 · 2H2O per liter] per liter, 5.0 ml of vitamin mixture (10 mg of niacin, 4 mg of biotin, 10 mg of pantothenate, 10 mg of lipoic acid, 4 mg of folic acid, 10 mg of p-aminobenzoic acid, 10 mg of thiamine, 10 mg of riboflavin, 10 mg of pyridoxine, and 10 mg of cobalamin per liter) per liter, 19 amino acids (75 mg of alanine, 125 mg of arginine HCl, 100 mg of asparagine · H2O, 50 mg of aspartic acid, 250 mg of cysteine HCl · H2O, 50 mg of glutamine, 200 mg of glutamic acid, 200 mg of glycine, 100 mg of histidine HCl · H2O, 100 mg of isoleucine, 100 mg of leucine, 100 mg of lysine HCl, 75 mg of methionine, 75 mg of phenylalanine, 125 mg of proline, 75 mg of serine, 100 mg of threonine, 100 mg of tyrosine, and 50 mg of valine per liter), 0.8 mg of resazurin per liter, and 2.0 g of elemental sulfur per liter (pH was adjusted to 6.9 with NaOH). Prior to inoculation, Na2S was added to the medium until the medium became transparent. In the case of plate culture, instead of elemental sulfur and Na2S, 2 ml of a polysulfide solution (10 g of Na2S · 9H2O and 3 g of sulfur flowers in 15 ml of H2O) per liter and 1% Gelrite were added to solidify the medium. All components were purchased from Wako Pure Chemical Industries (Osaka, Japan) or Nacalai Tesque (Kyoto, Japan).
Although we were aware of the possibility that the gene disruption would lead to a lethal phenotype, particularly at high temperatures, we were able to isolate transformants exhibiting tryptophan autotrophy at 85°C. In order to confirm that the reverse gyrase gene was disrupted in the transformants, PCR analyses and Southern blot analyses were performed. The results of PCR indicated that the reverse gyrase gene was removed from its locus and replaced by the trpE gene (data not shown). Southern blot analyses were performed with three probes (Fig. 1A). The probe corresponding to the 5′-end- flanking region of the reverse gyrase gene was amplified with the primers 5F2 (5′-CTCAAAGCTCCTGTCTCCTTTCGTTA) and 5R2 (5′-GCGAACTCCTCAAAGGCCTCTTCCAA), resulting in a probe of 728 bp. The probe (698 bp) within the coding region of the reverse gyrase gene was amplified with the primers CF1 (5′-ACGCTCTAAAGGGCAAGAAGAGCTAC) and CR1 (5′-CAGGCCGCCCTTTCCGAGCCTTGTGA), while the probe (700 bp) within the coding region of the trpE gene was amplified with the primers TF1 (5′-TCCATCATCGGGGGGAAGATCGAAGAGC) and TR1 (5′-CGAACGCGTTTTTCCCCTCATCGAGTT). All DNA fragments were labeled with the digoxigenin-DNA labeling and detection kit (Roche Diagnostics, Mannheim, Germany), and all methods were carried out as recommended by the manufacturer. The results of Southern blot analyses (Fig. 1B) confirmed that the reverse gyrase gene had been replaced by the trpE gene and further indicated that the reverse gyrase gene was not present in any other regions of the genome. We therefore designated this strain as the Δrgy strain.
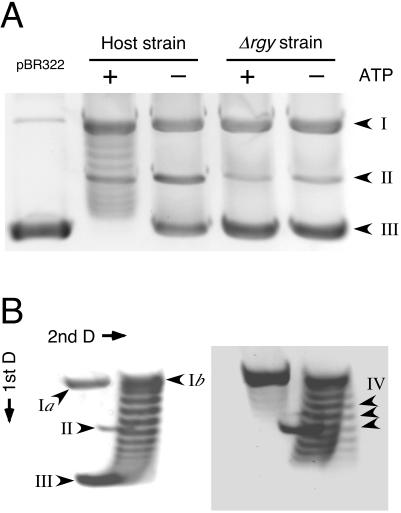
We examined the reverse gyrase activity in the host strain and the Δrgy strain. The host strain and the Δrgy strain were grown at 85°C in the same medium used for growth comparisons (see below). As many competing activities due to various nucleases and topoisomerases are observed in the cell extracts of various hyperthermophiles (4, 14, 15, 18), partial cell fractionation was carried out with both strains as described elsewhere (14, 15) with slight modifications. Cells were suspended in buffer A (50 mM NaH2PO4-Na2HPO4 [pH 7.0], 1 mM dithiothreitol, 1 mM EDTA) containing 1.2 M NH4Cl, 1 mM EGTA, 1 mM sodium bisulfite, 1 mM phenylmethylsulfonyl fluoride, 3 μg of leupeptin per ml, and 3 μg of pepstatin A per ml and lysed by ultrasonication. After centrifugation (20,000 × g, 1 h), the supernatant was collected, and polyethyleneimine (Sigma, St. Louis, Mo.) was added to obtain a final concentration of 0.36%. After gentle mixing for 15 min, the mixture was centrifuged at 20,000 × g for 1 h. The supernatant was further clarified by ultracentrifugation at 90,000 × g for 1 h. Ammonium sulfate was added to the supernatant to achieve 70% saturation, and the mixture was centrifuged at 20,000 × g for 1 h. The pellet was dissolved in buffer A with ammonium sulfate at 35% saturation and centrifuged at 20,000 × g for 1 h. The supernatant was used as the partially purified extract. The standard assay mixture (final volume, 20 μl) contained 35 mM Tris-HCl (pH 8.0), 72 mM KCl, 5 mM MgCl2, 5 mM dithiothreitol, 5 mM spermidine, and 1 μg of negatively supercoiled pBR322. When necessary, ATP was added at a final concentration of 2 mM. After 4 μg of the partially purified extract was added, the reaction mixture was incubated for 3 min at 75°C, and the reaction was stopped by the addition of 10 μl of stop buffer (3% sodium dodecyl sulfate, 0.03% bromophenol blue, 30 mM EDTA, 30% glycerol). In order to detect DNA-relaxing activity, the reaction product was analyzed by 1% agarose gel electrophoresis with Tris-borate-EDTA buffer (90 mM Tris, 90 mM boric acid, and 2 mM EDTA [pH 8.0]). The gel was stained with SYBR Gold (Invitrogen, Carlsbad, Calif.) and visualized under UV illumination. As shown in Fig. 2A, significant DNA-relaxing activity was observed in the case of the host strain. This activity was ATP dependent, suggesting the presence of reverse gyrase in this partially purified fraction. In order to examine the positive supercoiling activity of this fraction, the reaction product was analyzed by two-dimensional agarose gel electrophoresis. After the first dimension, the gel was soaked for 3 h in Tris-borate-EDTA buffer containing 4.5 μg of chloroquine per ml, which was also used as the electrophoresis buffer in the second dimension. Chloroquine decreases the mobility of negatively coiled DNA during agarose gel electrophoresis (8). We could clearly observe the formation of positively supercoiled DNA in the presence of ATP (Fig. 2B). In contrast, we did not observe significant DNA-relaxing activity in the fractions obtained from the Δrgy strain (Fig. 2A).
FIG. 2.
Comparison of DNA-relaxing activity and positive supercoiling activity in partially purified extracts of the host strain and the Δrgy strain. (A) DNA-relaxing activity in the partially purified extracts of the host strain and the Δrgy strain were analyzed as described in the text. The substrate was negatively supercoiled pBR322 (1 μg) purified from Escherichia coli (left lane). The presence (+) and absence (−) of ATP (2 mM) are indicated. (B) Positive supercoiling activity in the partially purified extracts of the host strain. Reaction conditions are described in the text, and ATP was added at a concentration of 2 mM. The second-dimension electrophoresis was performed in the presence of 4.5 μg of chloroquine per ml. Two stages of the reaction are shown. The left panel displays the progression of the reaction from negatively supercoiled DNA to relaxed DNA. The right panel displays the formation of positively supercoiled DNA. Different forms of pBR322 are indicated by the following arrowhead labels: I, nicked (Ia) or relaxed (Ib) DNA; II, linear DNA; III, negatively supercoiled DNA; IV, positively supercoiled DNA. D, dimension.
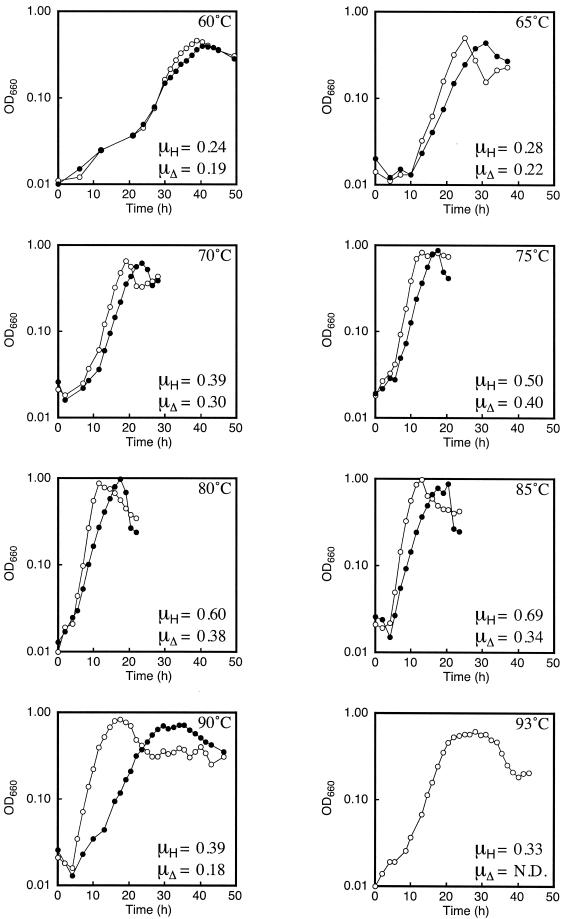
Growth rate comparisons between the host strain and the Δrgy strain were performed at various temperatures in a nutrient-rich medium supplemented with pyruvate and elemental sulfur. The medium contained the following, per liter: 0.8× artificial seawater, 5 g of yeast extract, 5 g of tryptone, 10 g of sodium pyruvate, 0.8 mg of resazurin, and 2 g of elemental sulfur. Prior to inoculation, Na2S was added to the medium until the medium became transparent. This medium leads to high growth rates of the host strain in batch culture, and we used it because we did not want the medium composition to be a rate-limiting factor in our growth experiments. Under these conditions, the host strain displays growth between 60 and 93°C, with the highest growth rates at approximately 85°C (see below). Growth rates of the host strain and the Δrgy strain were examined at various temperatures by measuring the optical density at 660 nm. Growth curves at 60, 65, 70, 75, 80, 85, 90, and 93°C are shown in Fig. 3. We observed that the growth of the Δrgy strain was slower than that of the host strain, a tendency that became more striking at higher temperatures. In contrast to the growth of the host cells, we did not observe growth of the Δrgy strain during the examined time period (49 h) at 93°C. At 93°C, we observed significant browning of the medium. The browning initiated after 20 h, and after 49 h, the medium was a dark brown color. Therefore, we cannot rule out the possibility that the Δrgy strain may be able to grow at 93°C under more-suitable conditions.
FIG. 3.
Growth curves and specific growth rates of the host strain and the Δrgy strain at various temperatures. Representative growth curves of the host strain and the Δrgy strain are shown. Temperatures are indicated in each panel. The specific growth rates of the host strain (μH) and those of the Δrgy strain (μΔ) were calculated from multiple growth curves. Open circles represent the host strain, and filled circles represent the Δrgy strain. N.D., not determined: OD660, optical density at 660 nm.
The specific growth rates (μ) (μX = dX/dt, where t is time and X is the amount of cells at a given time) of the two strains are shown in Fig. 3 along with the growth curves. The host strain displayed a maximum μ of 0.69 h−1 at 85°C, while the maximum μ of the Δrgy strain (0.4 h−1) was observed at 75°C. The ratio of the specific growth rate of the host strain to that of the Δrgy strain (μΔ/μH) was relatively constant (ca. 0.8) at lower temperatures, between 60 and 75°C. However, at higher temperatures, this ratio displayed a decrease with the elevation in temperature: at 80°C, it was 0.63, at 85°C, it was 0.49, and at 90°C, it was 0.46.
This study aimed to address one question: is reverse gyrase a prerequisite for hyperthermophilic life? The interpretation of our results relies heavily on how we define hyperthermophilic life in terms of temperature. If growth at 90°C is considered hyperthermophilic life, the answer is no. We have clearly shown that a hyperthermophilic cell deprived of reverse gyrase is still capable of growth at 90°C. At this stage, we cannot conclude whether reverse gyrase is necessary for life at temperatures higher than 90°C. Among the known hyperthermophiles, T. kodakaraensis grows at temperatures lower than those preferred by various Methanopyrus (13), Pyrodictium (19), and Pyrolobus (2) species. Disruption of the reverse gyrase genes in these organisms should help to clarify whether reverse gyrase is an essential enzyme at temperatures above 90°C.
It has been pointed out that if reverse gyrase were a prerequisite for hyperthermophilic life, life could not have originated from a hyperthermophile (6). This is based on the fact that reverse gyrase exhibits its unique activity through the coordinated function of two domains or components that are members of two completely different protein superfamilies with distinct ancestral proteins (3, 6). Therefore, the two domains must have evolved independently in less-thermophilic organisms. Although our results do not in any way support the hypothesis of life originating in hot environments, we have clearly shown that life can originate at temperatures up to at least 90°C.
Acknowledgments
This study was partly supported by grants-in-aid from the Ministry of Education, Science, Sports, Culture, and Technology to T.I. (14103011) and H.A. (413/13031047).
REFERENCES
- 1.Atomi, H., T. Fukui, T. Kanai, M. Morikawa, and T. Imanaka. 16 April 2004, posting date. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea [Online.] http://archaea.ws/archive/pdf/volume1/issue4/1-Imanaka.pdf. [DOI] [PMC free article] [PubMed]
- 2.Blöchl, E., R. Rachel, S. Burggraf, D. Hafenbradl, H. W. Jannasch, and K. O. Stetter. 1997. Pyrolobus fumarii, gen. and sp. nov., represents a novel group of archaea, extending the upper temperature limit for life to 113°C. Extremophiles 1:14-21. [DOI] [PubMed] [Google Scholar]
- 3.Confalonieri, F., C. Elie, M. Nadal, C. B. de la Tour, P. Forterre, and M. Duguet. 1993. Reverse gyrase: a helicase-like domain and a type I topoisomerase in the same polypeptide. Proc. Natl. Acad. Sci. USA 90:4753-4757. (Erratum, 91:3478, 1994.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de la Tour, C. B., C. Portemer, M. Nadal, K. O. Stetter, P. Forterre, and M. Duguet. 1990. Reverse gyrase, a hallmark of the hyperthermophilic archaebacteria. J. Bacteriol. 172:6803-6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forterre, P. 2002. A hot story from comparative genomics: reverse gyrase is the only hyperthermophile-specific protein. Trends Genet. 18:236-237. [DOI] [PubMed] [Google Scholar]
- 6.Forterre, P. 1996. A hot topic: the origin of hyperthermophiles. Cell 85:789-792. [DOI] [PubMed] [Google Scholar]
- 7.Forterre, P., C. B. de la Tour, H. Philippe, and M. Duguet. 2000. Reverse gyrase from hyperthermophiles: probable transfer of a thermoadaptation trait from Archaea to Bacteria. Trends Genet. 16:152-154. [DOI] [PubMed] [Google Scholar]
- 8.Forterre, P., G. Mirambeau, C. Jaxel, M. Nadal, and M. Duguet. 1985. High positive supercoiling in vitro catalyzed by an ATP and polyethylene glycol-stimulated topoisomerase from Sulfolobus acidocaldarius. EMBO J. 4:2123-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hopfner, K.-P., A. Karcher, D. S. Shin, L. Craig, L. M. Arthur, J. P. Carney, and J. A. Tainer. 2000. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101:789-800. [DOI] [PubMed] [Google Scholar]
- 10.Huber, H., M. J. Hohn, R. Rachel, T. Fuchs, V. C. Wimmer, and K. O. Stetter. 2002. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature 417:63-67. [DOI] [PubMed] [Google Scholar]
- 11.Imanaka, T., and H. Atomi. 2002. Catalyzing “hot” reactions: enzymes from hyperthermophilic Archaea. Chem. Rec. 2:149-163. [DOI] [PubMed] [Google Scholar]
- 12.Kikuchi, A., and K. Asai. 1984. Reverse gyrase—a topoisomerase which introduces positive superhelical turns into DNA. Nature 309:677-681. [DOI] [PubMed] [Google Scholar]
- 13.Kurr, M., R. Huber, H. König, H. W. Jannasch, H. Fricke, A. Trincone, J. K. Kristjansson, and K. O. Stetter. 1991. Methanopyrus kandleri, gen. nov. and sp. nov. represents a novel group of hyperthermophilic methanogens, growing at 110°C. Arch. Microbiol. 156:239-247. [Google Scholar]
- 14.Nadal, M., E. Couderc, M. Duguet, and C. Jaxel. 1994. Purification and characterization of reverse gyrase from Sulfolobus shibatae. Its proteolytic product appears as an ATP-independent topoisomerase. J. Biol. Chem. 269:5255-5263. [PubMed] [Google Scholar]
- 15.Nadal, M., C. Jaxel, C. Portemer, P. Forterre, G. Mirambeau, and M. Duguet. 1988. Reverse gyrase of Sulfolobus: purification to homogeneity and characterization. Biochemistry 27:9102-9108. [DOI] [PubMed] [Google Scholar]
- 16.Robb, F. T., and A. R. Place (ed.). 1995. Archaea: a laboratory manual, vol. 3. Thermophiles, p. 167-168. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 17.Sato, T., T. Fukui, H. Atomi, and T. Imanaka. 2003. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185:210-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slesarev, A. I. 1988. Positive supercoiling catalysed in vitro by ATP-dependent topoisomerase from Desulfurococcus amylolyticus. Eur. J. Biochem. 173:395-399. [DOI] [PubMed] [Google Scholar]
- 19.Stetter, K. O., H. König, and E. Stackebrandt. 1983. Pyrodictium gen. nov., a new genus of submarine disc-shaped sulphur reducing archaebacteria growing optimally at 105°C. Syst. Appl. Microbiol. 4:535-551. [DOI] [PubMed] [Google Scholar]
- 20.Tang, X.-F., S. Ezaki, H. Atomi, and T. Imanaka. 2001. Anthranilate synthase without an LLES motif from a hyperthermophilic archaeon is inhibited by tryptophan. Biochem. Biophys. Res. Commun. 281:858-865. [DOI] [PubMed] [Google Scholar]
- 21.Waters, E., M. J. Hohn, I. Ahel, D. E. Graham, M. D. Adams, M. Barnstead, K. Y. Beeson, L. Bibbs, R. Bolanos, M. Keller, K. Kretz, X. Lin, E. Mathur, J. Ni, M. Podar, T. Richardson, G. G. Sutton, M. Simon, D. Söll, K. O. Stetter, J. M. Short, and M. Noordewier. 2003. The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc. Natl. Acad. Sci. USA 100:12984-12988. [DOI] [PMC free article] [PubMed] [Google Scholar]