Abstract
Current models of biofilm formation by Pseudomonas aeruginosa propose that (i) planktonic cells become surface associated in a monolayer, (ii) surface-associated cells form microcolonies by clonal growth and/or aggregation, (iii) microcolonies transition to a mature biofilm comprised of exopolysaccharide-encased macrocolonies, and (iv) cells exit the mature biofilm and reenter the planktonic state. Here we report a new class of P. aeruginosa biofilm mutant that defines the transition from reversible to irreversible attachment and is thus required for monolayer formation. The transposon insertion carried by the sadB199 mutant was mapped to open reading frame PA5346 of P. aeruginosa PA14 and encodes a protein of unknown function. Complementation analysis and phage-mediated transduction demonstrated that the transposon insertion in PA5346 was the cause of the biofilm-defective phenotype. Examination of flow cell-grown biofilms showed that the sadB199 mutant could initiate surface attachment but failed to form microcolonies despite being proficient in both twitching and swimming motility. Closer examination of early attachment revealed an increased number of the sadB199 mutant cells arrested at reversible attachment, functionally defined as adherence via the cell pole. A positive correlation among biofilm formation, irreversible attachment, and SadB level was demonstrated, and furthermore, RpoN and FleR appear to negatively affect SadB levels. Fractionation studies showed that the SadB protein is localized to the cytoplasm, and with the use of GPS-linker scanning mutagenesis, the C-terminal portion of SadB was shown to be dispensable for function, whereas the two putative domains of unknown function and the linker region spanning these domains were required for function. We discuss the results presented here in the context of microbial development as it applies to biofilm formation.
Pseudomonas aeruginosa is a model organism for studying biofilm formation in gram-negative bacteria. Planktonic (free-swimming) P. aeruginosa initiates surface colonization in a flagellum-dependent manner (24), then forms transient (“reversible”) surface interactions, and subsequently becomes firmly (“irreversibly”) attached (15, 18, 43). It has been recently demonstrated for P. fluorescens that an ABC transporter and a large secreted protein are necessary for irreversible attachment by this organism (12); however, the mechanism by which this process occurs in P. aeruginosa has not been explored. The earliest events in the pathway whereby planktonic bacteria form surface-associated microbial communities are unclear; however, it is clear that bacteria sample surface niches via reversible attachment before taking up permanent residence (12, 15, 18, 43). This commitment to irreversible attachment is a crucial step in biofilm formation because initial surface colonizers are likely the foundation upon which the mature biofilm will be built. After irreversibly attaching, P. aeruginosa proceeds to form microcolonies in a type IV pilus- and a GacA-dependent manner (22, 24, 28). As microcolonies become matrix-enclosed macrocolonies, cell-to-cell signaling is thought to become increasingly important (8, 13). It has been proposed that this transition from a planktonic to a biofilm lifestyle is a developmental process (23).
A previous study identified surface attachment-defective (sad) mutants of P. aeruginosa that are unable to form a biofilm (24). Here we report the characterization of one of these strains that carries a mutation in the sadB gene. The sadB locus encodes a protein of unknown function and is required for biofilm formation under all conditions tested. Furthermore, we show that the sadB mutant is defective in transitioning from reversible to irreversible attachment. To our knowledge, this is the first report of a genetic determinant of P. aeruginosa defining the transition from reversible to irreversible attachment.
MATERIALS AND METHODS
Bacterial strains, media, and chemicals.
All bacterial strains and plasmids used in this study are shown in Table 1. P. aeruginosa PA14 was grown on rich lysogeny broth (LB) or minimal medium. M63 minimal salts (27) medium supplemented with MgSO4 (1 mM) was the base medium for static biofilm assays. EPRI minimal medium (8) was used in flow cell experiments. Minimal media were supplemented, as indicated, with the following: glucose, 0.2%; Casamino Acids (CAA), 0.5%; arginine, 0.4%; citrate, 0.4%; and succinate, 0.4%. Unless noted otherwise, antibiotics were used at the indicated concentration: carbenicillin, 500 μg/ml; tetracycline, 150 μg/ml, kanamycin, 500 μg/ml. All studies were performed at 37°C. Unless noted otherwise, all enzymes used for DNA manipulation were purchased from Invitrogen (Carlsbad, Calif.). Cloning was carried out in Escherichia coli JM109 by standard methods (3), and constructs were electroporated into P. aeruginosa as described previously (4).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| P. aeruginosa PA14 | Wild type | 31 |
| PA14 sadB199::Tn5B21; Tcr | This study | |
| PA14 flgK::Tn5B30; Tcr | 24 | |
| PA14 pilB::Tn5B30; Tcr | 24 | |
| PA14 fleR::TnphoA; Knr | F. Ausubel | |
| PA14 ΔrpoN | F. Ausubel | |
| PA14 crc-24::Tn5B30 | 22 | |
| PA14 gacA::kan; Knr | L. Rahme | |
| PA14 ΔlasR | F. Ausubel | |
| Plasmids | ||
| pUCP18 | Cloning vector; Cbr Apr | 35 |
| pNC5 | PA5346 in pUCP18; Cbr Apr | This study |
| pNC72 | GPS-LS construct | This study |
| pNC111 | GPS-LS construct | This study |
| pNC294 | GPS-LS construct | This study |
| pNC227 | GPS-LS construct | This study |
| pNC396 | GPS-LS construct | This study |
| pNC429 | GPS-LS construct | This study |
| pNC447 | GPS-LS construct | This study |
Isolation of P. aeruginosa PA14 sadB199 mutant.
The PA14 sadB199 strain was previously identified in a genetic screen for mutants defective in biofilm initiation (24). For the purposes of this study, the original mutant was reconstructed into the wild-type PA14 strain by phage-mediated transduction (5). The reconstructed strain is used in all the studies presented here.
Molecular techniques.
The DNA sequence flanking transposon insertions was determined and mapped by arbitrary primed PCR as previously described (26). The transposon carried by the sad-199 mutant was mapped to PA5346 by use of the published P. aeruginosa PAO1 genome (37). All other molecular techniques were performed as previously described (3). Plasmid pNC5 contains a wild-type copy of PA5346. Based on the PAO1 sequence, primers were designed ∼500 bp upstream and downstream of PA5346: P226, 5′-GGCGAAGCTTCTGATGATCATCGAGAACCC-3′, and P227, 5′-GGCGGAATTCGGACCTGGTGTTCCAGTTGC-3′. Primers were engineered to create a PCR fragment that could be digested with HindIII and EcoRI and ligated into pUCP18 (35) digested with those same enzymes. This construct presumably carries the native sadB promoter.
Biofilm formation assays. (i) Ninety-six-well microtiter plate assay.
Microtiter plate biofilm assays were performed as previously described (25). Crystal violet (CV)-stained wells were digitally imaged using a Nikon 990 digital camera (Nikon, Melville, N.Y.). The extent of biofilm formation was quantified as previously described (25).
(ii) Microscopy of the air-liquid interface of static biofilms.
Cultures were grown overnight in LB medium and diluted 1:50 into the indicated medium, and 400-μl aliquots were inoculated into 24-well flat-bottomed plates (Corning Inc., Corning, N.Y.). Plates were incubated at a 45° angle (such that the air-liquid interface crossed the center of the flat-bottomed well) for 24 h. Well contents were aspirated and washed two times with fresh M63, and 200 μl of M63 was added to the well. Alternatively (for the experiments presented in Fig. 3) bacteria were pregrown on minimal arginine or glucose medium and allowed to attach for 5 min before the well contents were aspirated and fresh medium was added to the wells without washing. Bacteria attached to the well were then visualized by phase-contrast microscopy with a Leica DM IRB inverted microscope (Leica Microsystems, Wetzlar, Germany) equipped with a charge-coupled device digital camera and a PL Flotar 63× objective lens. Images were captured using a G4 Macintosh computer with the OpenLab software package (Improvision, Coventry, England). Images were processed using Photoshop software (Adobe, Mountain View, Calif.). The quantification of irreversible attachment presented in Fig. 3 was performed by obtaining time-lapse images captured every 2.5 s for 5 min. The functional distinction between reversible and irreversible attachment was made as follows: cells which moved once or not at all over the 5-min interval and attached via the long axis of the cell were scored as “irreversibly attached,” while those cells moving two or more times in this interval and attached via a pole of the cell were scored as “reversibly attached.” The data presented are the averages of results obtained from scoring three sets of movies from two independent cultures. Approximately 200 cells/movie were counted.
FIG. 3.
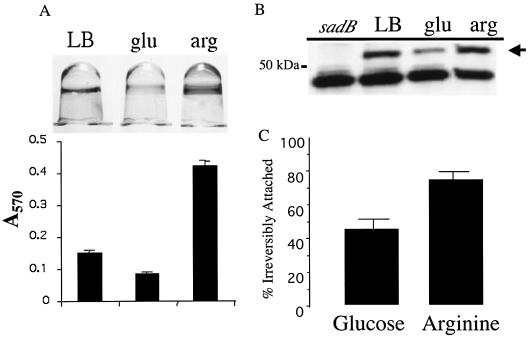
Correlation between SadB level and biofilm formation. (A) Image of 96-well microtiter plate-grown, CV-stained biofilms formed by wild-type cells grown in LB and M63 minimal medium supplemented with glucose or arginine. The medium provided in each case is listed above the well: LB, LB medium; glu, minimal glucose; arg, minimal arginine. Biofilms were allowed to form for 24 h. Below each well is the quantification of CV staining. (B) SadB level in LB or minimal medium supplemented with the carbon sources used in biofilm formation assays presented in panel A. The arrow denotes the SadB cross-reacting band, and the 50-kDa size marker is indicated. The nonspecific band below SadB serves as a convenient loading control. The leftmost lane contains extract derived from the LB medium-grown sadB199 mutant as a control. (C) Percent irreversible attachment of the wild-type strain in minimal medium supplemented with glucose or arginine.
(iii) Biofilm development in flow cells.
Flow cells were assembled and operated as previously described (6, 7), and images were recorded as described above. Initial attachment of cells was determined as follows: upon inoculation into flow chambers, cells were allowed to attach to the glass substratum in the absence of flow for 1 h, and then flow was resumed, removing unattached cells, and images of 10 fields of view were captured (two chambers per strain). Individual cells in each field of view were counted, and the average number of cells attached to the substratum was determined. This experiment was performed in triplicate with one representative data set being shown in Fig. 2B. The reversible attachment data presented in Fig. 2C were obtained as follows: after 24 h of biofilm development, time-lapse movies were made using the microscope and software described above. Movies (see supplemental data at www.dartmouth.edu/∼gotoole/sadBmovies.html) were 1 min long and consisted of images captured every 0.5 s. In a given experiment, the number of reversibly and irreversibly attached cells was determined using the criteria described above for three movies per chamber, and the percentage of reversibly attached cells was calculated.
FIG. 2.
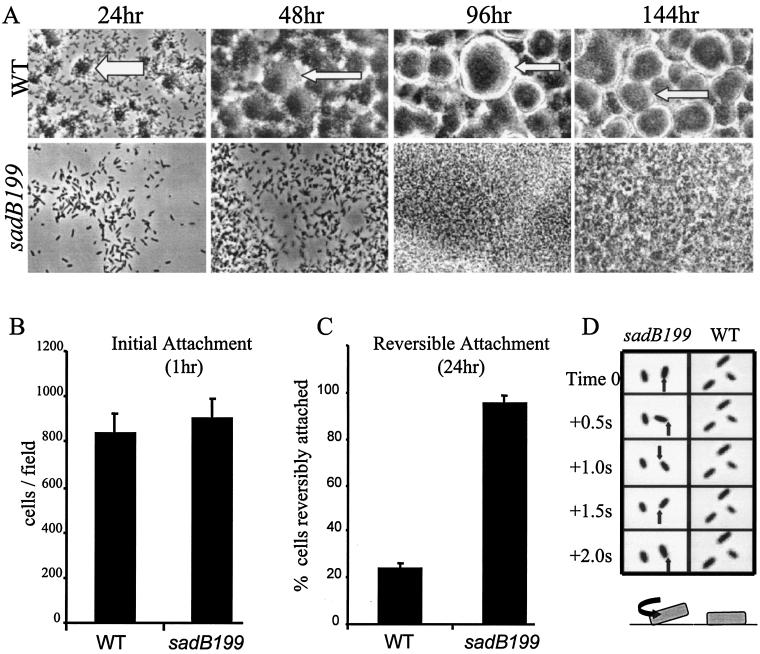
Biofilm formation phenotypes under constant flow conditions. (A) The wild type and the sadB199 mutant were inoculated into flow cells fed with minimal salts EPRI medium supplemented with glucose as the carbon source. Images shown are top-down, phase-contrast micrographs (magnification, ×1,260) recorded over the course of 6 days. At an early time point (24 h), the dark regions (shown by the white arrow) in the wild-type biofilm indicate microcolonies and the gray regions represent the glass surface of the flow chamber. At later time points (48 h and beyond), wild-type macrocolonies are the dark regions surrounded by refractory light-colored halos (indicated by the narrow white arrows). The borders of macrocolonies coincide with these light halos. The glass surface of the chamber cannot be seen in the mature wild-type biofilm (96 and 144 h). The sadB199 mutant biofilm lacks colonial architecture at all time points, and only single cells can be observed on the glass surface at early time points. (B) Quantification of initial attachment of the wild type and the sadB199 mutant to the glass surface of the flow chamber (1 h). (C) Quantification of reversible attachment of the wild type and the sadB199 mutant at 24 h by time-lapse video microscopy. The percentage of reversibly attached cells is shown. See the supplemental data at www.dartmouth.edu/∼gotoole/sadBmovies.html for movies of the reversible attachment phenotype. (D) Image series of initial attachment events. Phase-contrast images were recorded after 24 h of flow cell growth. The black arrow points to the fixed cell pole about which the sadB199 mutant is rotating. The wild-type cells are attached by the long axis of the cell body and do not move over the course of this image series, behaviors typical of irreversible attachment. Below each time course is a cartoon rendition of reversible and irreversible attachment. The black arrow denotes the direction of rotation of the reversibly attached sadB199 mutant bacterium seen in the above micrographs. WT, wild type.
Motility assays. (i) Swimming assay.
Bacteria were analyzed for swimming motility as described previously (25). Briefly, a colony of the indicated strain was inoculated into M63 agar (0.3% agar) supplemented with glucose and CAA and incubated for 24 h. The diameter of the movement was measured in millimeters.
(ii) Twitching assay.
Bacteria were analyzed for twitching motility as described previously (41). Briefly, a colony of the indicated strain was inoculated into M63 agar (1.5% agar) supplemented with glucose and CAA and incubated for 24 h. The diameter of the twitching zone at the plate-agar interface was measured in millimeters. Swimming and twitching experiments were performed in triplicate, and the averages and standard deviations of the data are presented.
Cell fractionation.
The fractionation procedure was modified from that described by Lohia et al. (16). Briefly, overnight LB medium-grown cultures were diluted 1:1,000 in fresh LB medium and then incubated for 16 h. Cultures (10 ml) were harvested, washed in PBS, and then resuspended in 1 ml of PBS and chilled on ice. Cells were lysed by passage through a French pressure cell two times. Unbroken cells were removed by centrifugation (7 min at 16,000 × g). The supernatant was centrifuged at 100,000 × g for 1 h to pellet the membrane fraction. The supernatant (soluble cytoplasmic fraction) was transferred to a clean tube and kept on ice. The pellet (total membrane fraction) was resuspended in 500 μl of PBS and stored on ice. Protein concentrations of the fractions were determined using a protein assay kit (Bio-Rad Laboratories, Hercules, Calif.). Five micrograms of total protein was used for Western analysis as described below.
Determining SadB levels.
For the experiment presented in Fig. 3B cells were grown overnight in LB medium, diluted 1:1,000 into the indicated medium, and allowed to grow to stationary phase (∼16 h). The rpoN mutants grew poorly on minimal medium and were supplemented with 0.05% CAA. The cells were pelleted, resuspended in 1 ml of PBS, and lysed in a French pressure cell, and the extract was clarified by centrifugation. A protein assay was performed on the whole-cell lysate, 5 μg of total protein was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and SadB levels were analyzed by Western blotting. For the data presented in Fig. 4 bacterial strains were grown overnight in the indicated medium, normalized to an optical density at 600 nm (OD600) of 0.5, diluted 1:1,000 in the same medium, and allowed to grow to stationary phase (∼16 h). The OD600 values were measured, and 200-μl aliquots normalized to an OD600 of 0.5 were harvested and resuspended in SDS loading buffer containing 1 mM dithiothreitol. Samples were then analyzed by SDS-PAGE and Western analysis.
FIG. 4.
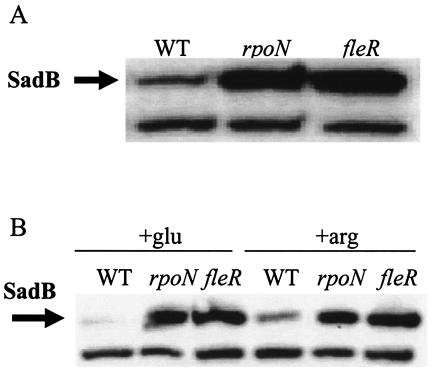
FleR and RpoN affect SadB levels. (A) The indicated strains were grown for ∼16 h in LB medium and normalized to OD600, and then equal amounts of culture were lysed and separated by SDS-PAGE. The arrow indicates the SadB cross-reacting band. (B) The indicated strains were grown overnight in M63 minimal medium supplemented with either glucose (+glu) or arginine (+arg) and prepared and analyzed as described above. WT, wild type.
SDS-PAGE and Western analysis.
Samples were suspended or diluted in SDS loading buffer containing 1 mM dithiothreitol (32), boiled for 5 min, and then separated on a 4 to 15% Tris-HCl gradient gel (Bio-Rad Laboratories). Proteins were transferred to a nitrocellulose membrane and probed with antibodies designed against a peptide corresponding to residues 160 to 180 of the PA5346 open reading frame (ORF; Biosynthesis Inc., Lewisville, Tex.). Western blots were developed using ECL detection reagents per the instructions of the manufacturer (Amersham, Little Chalfont, Buckinghamshire, England).
GPS-LS mutagenesis.
The GPS-LS mutagenesis kit was used to perform linker scanning mutagenesis of the sadB gene. The protocol was performed as described by the manufacturer (New England Biolabs, Beverly, Mass.). Briefly, the complementing construct pNC5 (containing the PA5346 ORF) was used as the target DNA for mutagenesis with the GPS-LS system, which uses TnsABC* transposase to randomly insert a Tn7-based transprimer into the target DNA. Mutagenized plasmids were harbored in E. coli JM109 and screened for transprimer insertions by PCR with primers specific to the PA5346 ORF. The primers used were as follows: P226, 5′-GGCGAAGCTTCTGATGATCATCGAGAACCC-3′, and P227, 5′-GGCGGAATTCGGACCTGGTGTTCCAGTTGC-3′. Transprimers were removed from these clones by restriction digestion with PmeI as described by the manufacturer, resulting in a 15-bp insertion that results in either an insertion of 5 amino acids (in four of six reading frames) or a TAA stop codon in the remaining two frames. Constructs were introduced into E. coli JM109 for DNA sequencing to map the site of insertion and to determine whether an insertion or stop codon was introduced before transfer into P. aeruginosa by electroporation (4). Once moved to P. aeruginosa, constructs were analyzed for their ability to affect biofilm formation in the 96-well microtiter plate assay. The stability of the mutant proteins encoded by the GPS-LS vectors was analyzed by SDS-PAGE and Western analysis. As described above, samples were prepared by harvesting 10 ml of an overnight LB medium-grown culture. Cells were lysed and clarified by centrifugation, and the protein concentration of the whole-cell lysate was determined. Five micrograms of total protein was used for Western analysis.
RESULTS
Isolation and initial characterization of the sad-199 mutant.
Previous work reported the results of a genetic screen for surface attachment-defective (sad) mutants of P. aeruginosa PA14 that are defined by their inability to form a biofilm in a microtiter dish (22, 24). Here we describe the analysis of one of those mutants, sad-199 (Table 1). The phenotype of this mutant grown on M63 medium supplemented with glucose and CAA is shown in Fig. 1A and B. This strain is also defective for biofilm formation when grown on minimal medium supplemented with glucose alone, CAA alone, arginine, succinate, or citrate (data not shown). The transposon insertion responsible for the biofilm defect of the sad-199 mutant was mapped, using arbitrary primed PCR, to the PA5346 ORF of the P. aeruginosa PAO1 genome (37). The PA5346 ORF is 1,410 bp encoding a protein 470 residues in length with a predicted molecular mass of 52 kDa. Based on the published annotation, the PA5346 ORF is monocistronic and the protein does not belong to any known functional class (37).
FIG. 1.
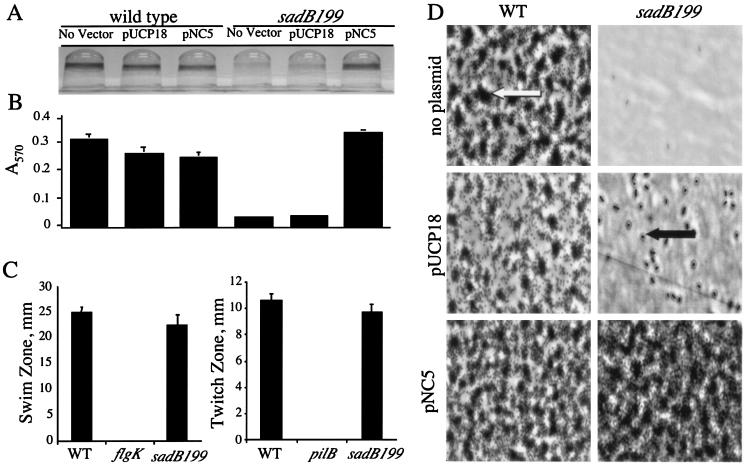
Biofilm formation phenotypes under static conditions. (A) Image of CV-stained biofilms formed by the wild type and the sadB199 mutant carrying either no plasmid, a vector control (pUCP18), or plasmid pUCP18 with the PA5346 (sadB+) ORF (pNC5). Cells were grown in minimal medium containing glucose and CAA for 24 h before being stained with CV. (B) Quantification of CV-stained wells. Ethanol was used to solubilize the CV associated with the biofilm, and the A570 of the resulting solution was measured. (C) Swimming and twitching zones were determined for the wild type and the sadB199 mutant. No swimming was observed in a control strain lacking flagella (flgK), and no twitching was observed in a control strain lacking type IV pili (pilB). (D) Direct visualization of biofilm formation on polyvinyl chloride at the air-liquid interface by phase-contrast microscopy (magnification, approximately ×1,400; see Materials and Methods for details). The large dark regions (indicated by the white arrow) represent microcolonies, the small dark rods (indicated by the black arrow) represent individual cells, and the light gray areas are the polyvinyl chloride surface. Cells were grown in minimal medium containing glucose and CAA for 24 h before washing and visualization. WT, wild type.
Several lines of evidence support the conclusion that PA5346 is required for biofilm formation. First, in the screen for biofilm mutants, three independent transposon insertions in the PA5346 ORF (including sad-199) were isolated and mapped to this locus by arbitrary primed PCR. All strains with mutations in this ORF were rendered biofilm deficient (data not shown). In addition, the sad-199 mutant used in the experiments described below was reconstructed by phage-mediated transduction into a fresh genetic background and genetic linkage was demonstrated between the antibiotic resistance marker carried by the transposon insertion and the observed biofilm-deficient phenotype for six of six transductants tested.
The sad-199 mutant does not differ from the wild type with respect to growth in shaking or static cultures, in rich (LB) medium or the minimal medium tested (data not shown). The sad-199 mutant also has no discernible defects in flagellum-mediated swimming and type IV-mediated twitching motility (Fig. 1C); therefore, the sad-199 strain did not have a defect in these known biofilm functions.
Complementation of the sad-199 mutation.
To confirm that a functional copy of PA5346 is required for biofilm formation, complementation analysis was performed. The PA5346 ORF and promoter region were cloned, resulting in the construction of plasmid pNC5 (see Materials and Methods for details). This construct and the vector control were introduced into wild-type and sad-199 backgrounds. The results of the complementation analysis are shown in Fig. 1A and B. The sad-199 strain is incapable of forming a biofilm in minimal medium supplemented with glucose and CAA as determined by absence of a CV-stained ring, whereas the wild-type strain stains positively (Fig. 1A). Quantification of the staining is presented in Fig. 1B. However, when the PA5346 ORF is introduced extrachromosomally on pNC5 to the sad-199 mutant, full restoration of biofilm formation is observed. Quantification of CV staining shows the level of biofilm formation by the complemented sad-199 strain to be equal to that of the wild-type strain (Fig. 1B). The vector control (pUCP18) has no effect on biofilm formation in either the wild-type or the mutant background, indicating that the presence of a high-copy-number plasmid such as pUCP18 does not induce biofilm formation, nor does it rescue the sad-199 mutant. In addition, having multiple copies of the PA5346 ORF does not have a deleterious effect on biofilm formation as determined by the introduction of pNC5 into the wild-type background. Thus, the introduction of multiple copies of the PA5346 ORF on plasmid pNC5 does not obviously alter biofilm formation, and the small decrease in the level of biofilm formation determined by CV staining is equivalent to that seen with the vector control (Fig. 1B).
Biofilms of the wild-type and mutant strains carrying the vector and pNC5 were also examined microscopically (Fig. 1D). The sad-199 mutant was first examined under static conditions by phase-contrast microscopy. This method allows for examination of the biofilm from the initiation of cell surface contacts through microcolony formation. The data show that the wild-type cells form a biofilm comprised of microcolonies, whereas the sad-199 mutant cells are largely absent from the surface. However, introduction of pNC5 into the sad-199 mutant rescues the biofilm deficiency and restores wild-type microcolony formation. The vector control had no effect on biofilm formation. Therefore, the microscopic data support the observations made for the 96-well plate assay.
Complementation experiments were also performed in a minimal medium with arginine as the sole carbon source, and the same results as those reported in Fig. 1 were observed (data not shown). In addition, the complementation experiments were performed in both media mentioned above with a mid- to-low-copy-number plasmid, and the results observed were identical to those for the high-copy-number pUCP18 vector (data not shown). Taken together, these results allow us to firmly conclude that the sad-199 mutant phenotype is caused by a defect in the PA5346 ORF. Based on its role in biofilm development, we have named the PA5346 ORF sadB.
sadB is required for the transition from the reversible to the irreversible attachment stage of biofilm formation.
To determine the role of the SadB protein within the broader context of the current models for biofilm formation, we used a static biofilm assay to examine the stage in biofilm formation in which a sadB199 mutant was blocked. Figure 1D shows that by 24 h the wild-type strain is fully capable of attaching to the substratum and aggregates into microcolonies (white arrow), whereas the sadB199 mutant is almost completely devoid of surface-attached cells, with just occasional individual cells attached (black arrow). Most of the cells present in the sadB199 field of view are out of focus (shown by a halo of light surrounding them), indicating that they are not in the same focal plane as the substratum and are not surface associated. Identical results were observed when a minimal medium supplemented with arginine was used (data not shown). The data in Fig. 1D confirm that sadB199 has a biofilm defect that manifests before the stage of microcolony formation. From these data, it appeared that the sadB199 mutant failed to initiate stable surface interactions. However, due to the rinse step involved in the static assay, we could not conclude whether the sadB199 mutant failed to initiate surface interactions or made loose surface interactions that were overcome by rinsing.
To determine whether the sadB mutant could initiate surface interactions, a once-through flow cell system was used to monitor biofilm formation (6). The flow cell system allows the growth of the biofilm in a small, optically clear chamber that is continuously irrigated with fresh medium (Fig. 2A). By 24 h the wild-type has formed microcolonies of ∼100 cells. By 48 h, macrocolonies are formed, continue to increase in size, and are maintained over the following days observed in this study. In contrast, few of the sadB199 mutant cells are attached at 24 h and no microcolonies have formed, and while cells continue to accumulate over the next 5 days, the structure of the wild-type biofilm is never observed; instead, a mat of cells is formed by the mutant. These data suggest that the sadB199 mutant is not simply delayed in biofilm formation but is blocked from forming a typically structured community.
To further define the defect in attachment, we then examined the biofilm at earlier time points. Flow cells were inoculated with the wild type or the sadB199 mutant and allowed to incubate in the absence of flow for 1 h, after which flow resumed removing the planktonic bacteria, and the surface-attached bacteria were counted immediately. Inspection of the flow cell showed that the sadB199 mutant is able to initiate surface association at a level equal to that of the wild type (Fig. 2B). In this experiment, ∼900 bacterial cells per microscopic field were present for both the wild type and the sadB199 mutant. The ability of the sadB mutant to initiate surface colonization in a flow cell to the same extent as the wild type, combined with the observed lack of surface-attached sadB199 mutant cells after rinsing in the static assay (Fig. 1D), suggested that the sadB199 mutant cells are capable of interacting with the substratum but that this interaction is relatively unstable.
In addition to periodic inspection of the flow cell, time-lapse image series were acquired after ∼24 h of biofilm formation and examined (see supplemental data at www.dartmouth.edu/∼gotoole/sadBmovies.html). We noted that many of the sadB199 mutant cells at this point were attached by a single pole and were observed to spin about that pole, while in contrast a majority of the wild-type cells were adherent via the long axis of the cell and unmoving. These observations were consistent with the historically observed processes of reversible and irreversible attachment (15, 18, 33, 43) and with recent studies from Hinsa et al. (12). Reversible attachment has been functionally defined as a moving cell that interacts with a surface via the cell pole, while irreversible attachment is defined as a cell oriented parallel to the surface and unmoving (12, 15, 18, 34, 43).
By inspecting the time-lapse movies, individual cells in each field of view were scored as reversibly attached (associated via a cell pole and in motion) or irreversibly attached (associated via the long axis of cell and unmoving), and the percentage of reversibly attached cells was calculated for each strain (Fig. 2C). These data show that ∼90% of the cells in a sadB199 mutant biofilm are reversibly attached after 24 h in a flow cell, whereas only ∼25% of the cells of a wild-type biofilm are reversibly attached. Thus, ∼75% of the wild-type cells were scored as irreversibly attached compared to only ∼10% for the sadB199 mutant. Several images from a time-lapse series are shown in Fig. 2D, illustrating reversible and irreversible attachment. Time-lapse movies were also made at 48 h, and at this time point a majority of cells in the wild-type biofilm were associated with a microcolony or macrocolony, whereas ∼90% of cells in the sadB199 mutant cells were still reversibly attached. These data indicate that the sadB199 mutant cells are blocked at the transition from the reversible to the irreversible stage of biofilm formation.
SadB levels correlate with the transition to irreversible attachment.
Based on the phenotype of the sadB199 mutant, we hypothesized that the levels of the SadB protein might be correlated with early events in biofilm formation. We had previously observed that biofilm formation was most robust on minimal arginine medium or LB medium, a relatively weak biofilm was formed on glucose-grown cells (Fig. 3A), and intermediate levels of biofilm formation were observed when cells were grown on minimal medium supplemented with either glucose plus CAA, citrate, or succinate (data not shown).
To determine whether the different levels of biofilm formation observed on various media were correlated with different SadB levels, Western blot assays were performed under these conditions (see Materials and Methods for details) (Fig. 3B). The results show that the highest observed levels of SadB are present when P. aeruginosa is grown in a nutrient-rich medium (LB medium) or when arginine is provided as the sole carbon and energy source. Thus, media that resulted in the highest intensity of CV staining also resulted in the highest level of SadB protein. In contrast, relatively low levels of SadB protein were observed in glucose-grown cells (Fig. 3B) despite the fact that cells grew significantly better on glucose than on arginine. Intermediate levels of SadB were observed when P. aeruginosa was grown on glucose plus CAA, citrate, or succinate (data not shown). While these data do not suggest a causal link between SadB level and biofilm formation, they do provide, to our knowledge, the first example of a protein whose level correlates positively with the extent of biofilm formation.
Based on the expression studies above and the observed role of SadB in the transition to irreversible attachment, we predicted that SadB levels might also correlate with the cells' commitment to irreversible attachment. To test this hypothesis, wild-type cells were grown under conditions that lead to relatively high (+Arg) or low (+Glu) SadB levels and the frequency of cells irreversibly attaching was determined using the static air-liquid interface assay described in Materials and Methods. As shown in Fig. 3C, there was an approximately twofold-greater commitment to irreversible attachment with arginine-grown cells than with glucose-grown cells. These data suggest a possible functional consequence of the increased SadB levels under these conditions.
Strains lacking FleR and RpoN have increased SadB levels.
Our microscopic analysis indicated a role for SadB early in the transition from planktonic growth to forming a biofilm; therefore, we hypothesized that SadB expression might be under the control of known biofilm regulators, in particular a regulatory system (or systems) that controls an early step in biofilm formation, but not under the control of regulators required in later steps of this process. Specifically, SadB levels were examined in strains containing single mutations in fleR or rpoN, which controls the expression of pili and flagella, both of which are required early in biofilm formation, as well as crc, gacA, and lasR, which are involved in steps downstream of monolayer formation. We also examined whether SadB levels were affected in a flagellar structural gene mutant (flgK).
Compared to the wild-type strain, SadB levels were unchanged in all of the above mutants, with the notable exception of the fleR and rpoN mutants. SadB levels were monitored in both logarithmic (data not shown) and stationary-phase (Fig. 4A) cells, and in both growth phases, SadB levels were markedly elevated in the fleR and rpoN backgrounds.
To determine whether expression levels of SadB are dependent on RpoN and FleR under other growth conditions, minimal medium supplemented with glucose or arginine was inoculated with wild-type, rpoN, and fleR strains and processed for SDS-PAGE and Western analysis. As expected for the wild-type strain, SadB levels were lower in glucose-grown than in arginine-grown bacteria, while in the rpoN and fleR strains SadB levels were elevated under both conditions (Fig. 4B).
Subcellular localization of SadB.
Various computational tools and algorithms (PSORT, PSIPRED, and TMpred) predicted that SadB might be an integral inner membrane protein. However, there was wide variability among the algorithms in terms of predicting the location and number of transmembrane domains. On the basis of the various prediction programs, we made educated guesses regarding putative periplasmic regions which served as the basis for the construction of sadB::phoA fusion proteins. The PhoA protein was fused downstream of amino acids 20, 88, 156, and 360 of SadB; however, none of the fusions appeared active, judged by the lack of blue color development on agar plates containing the alkaline phosphatase substrate 5-bromo-4-chloro-3-indolylphosphate (data not shown). In a second strategy, random phoA insertions were generated using both an exonuclease digestion strategy and TnphoA mutagenesis (9, 17). Once again, neither technique yielded clones that possessed alkaline phosphatase activity (data not shown). These data are consistent with the conclusion that SadB does not have any domains located in the periplasm and is unlikely to be a transmembrane protein.
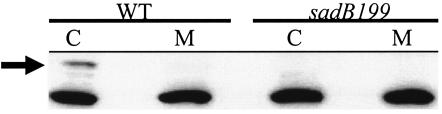
Fractionation studies were performed with antibodies raised to SadB to determine the localization of this protein. Cells were fractionated into cytoplasm and total membrane fractions as described previously (16), and these fractions were analyzed by Western blotting. As shown in Fig. 5, a band corresponding to the predicted size of SadB (52 kDa) is detected in the cytoplasmic fraction of wild-type cells. This band is absent in the membrane fraction of the wild type and both fractions of the sadB199 mutant strain.
FIG. 5.
SadB localization. Soluble cytoplasmic (C) and membrane (M) fractions of the wild type (WT) and the sadB199 mutant were generated and separated by SDS-PAGE. Western analysis was performed with a SadB peptide antibody. The arrow indicates the SadB band.
While the absence of SadB from the membrane fraction is at odds with some computer predictions, the fractionation data are in good agreement with the lack of alkaline phosphatase-positive clones generated in the experiments described above. Taken together, these data are consistent with the conclusion that SadB is a cytoplasmically localized protein.
Structure-function analysis of SadB.
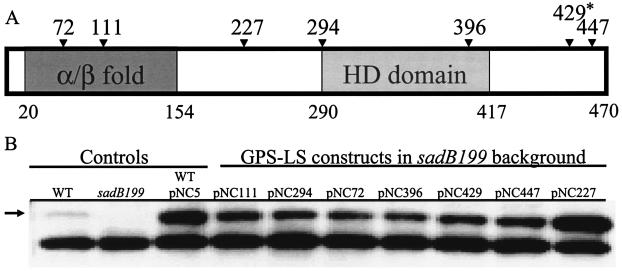
Linker scanning mutagenesis revealed that both predicted domains of the SadB protein, as well as the linker region joining these domains, are required for biofilm formation. Four insertions mapped to the α/β domain, eight insertions mapped to the HD domain, two insertions were isolated between these two domains, and two insertions mapped to the C terminus of the protein. A diagram of representative insertions and truncations is shown in Fig. 6A, and the corresponding Western blot assay performed on strains carrying each construct indicated whether the mutant proteins were expressed and stable (Fig. 6B). The level of the SadB protein in all strains harboring the GPS-LS mutagenized vectors is comparable to that of strains harboring the unmutagenized vector (pNC5) and significantly higher than the wild-type level of the protein. The increased SadB protein level in these strains is most likely an effect of plasmid copy number. All but the two C-terminal-most insertions (429 and 447) rendered the SadB protein nonfunctional with respect to biofilm formation in complementation experiments in the microtiter plate assay. Furthermore, none of these mutant constructs were dominant to the wild-type SadB protein (data not shown).
FIG. 6.
Structure-function analysis of SadB. (A) Diagram of SadB. Numbers below the diagram represent residues that define the borders of the putative domains. The α/β fold extends from residue 20 to 154, and the HD domain extends from residue 290 to 417. Numbers above the diagram represent residues at which 5-amino-acid insertions occurred. The asterisk denotes a GPS-LS mutation that resulted in a truncated protein. The figure is not to scale. (B) Stability of proteins resulting from GPS-LS mutagenesis. Protein from whole-cell lysates (5 μg) was resolved by SDS-PAGE and analyzed by Western analysis with a SadB peptide antibody. The arrow denotes the SadB cross-reacting band. All GPS-LS constructs were named pNC followed by a numerical designation that represents the residue of insertion or truncation. Plasmid pNC5 (sadB+) was the target for GPS-LS mutagenesis. WT, wild type.
DISCUSSION
Previous observations and data from this study have shown that initial surface attachment in P. aeruginosa proceeds from transient, cell pole-mediated interactions (reversible attachment) to stable surface interactions that occur via the long axis of the cell body (irreversible attachment) (12, 15, 18, 34, 43). Hinsa et al. demonstrated that for Pseudomonas fluorescens the lapBCE-encoded ABC transporter and the secreted LapA protein are required for the transition from reversible to irreversible attachment (12). A homologous system had not been identified in P. aeruginosa, yet the stages of reversible and irreversible attachment have been observed (34), indicating that factors governing this transition should exist in this organism as well. In this study we have shown that the sadB locus is required for the transition from reversible to irreversible attachment in P. aeruginosa (Fig. 2). We have also demonstrated that SadB levels positively correlate with the extent of biofilm formation and the transition to irreversible attachment observed under static conditions on different sole carbon-energy sources (Fig. 3). To the best of our knowledge this is the first example of a protein with levels that positively correlate with biofilm formation. This observation makes SadB a possible choice as a marker for the cells' commitment to biofilm formation. We hypothesize that the robust biofilm formation observed in certain media, for example, arginine-based minimal medium, is at least in part due to elevated SadB levels that aid in rapid progression to irreversible attachment. However, the exact mechanism by which SadB promotes the transition from reversible to irreversible attachment is unknown.
One of the goals of this study was to determine the role of SadB in biofilm formation within the larger context of present models for biofilm formation. The microscopy studies outlined above suggested that the sadB199 mutant was blocked early in the transition to a biofilm lifestyle. Based on this putative early role in biofilm formation, we predicted that SadB expression might be controlled only by the subset of biofilm regulators required for early (but not late) steps in biofilm formation (Fig. 7). We observed that SadB levels were elevated in both rpoN and fleR mutants (Fig. 4) but were unchanged in all other mutants tested. RpoN is an alternate sigma factor (σ54) required for fleR transcription (2), and FleR is a response regulator required for σ54-dependent transcription of middle flagellar genes (30). FleR has not been demonstrated previously to act as a repressor; however, its activity has not been examined outside of the hierarchy of flagellar gene control. The increased level of SadB in the rpoN mutant may result from the lack of FleR in this strain, as RpoN is required for the expression of fleR. Therefore, FleR may be affecting SadB levels by directly repressing sadB or by activating an unknown (possibly σ54-dependent) repressor of sadB. RpoN may also directly control SadB based on the putative RpoN binding site identified upstream of the sadB ORF—a consensus GC sequence centered at −12 is present and a consensus GG sequence, typically centered at −24, is found at the −26 position relative to the predicted start site of transcription (39). A recent report showed that in P. aeruginosa PAO1 an rpoN mutant forms an abnormal biofilm comprised only of tightly packed microcolonies and proposed that RpoN regulates genes involved in biofilm maturation, such as rhlI (38). However, because rpoN mutants also do not make functional flagella or pili it is possible that RpoN plays a role in multiple stages of biofilm formation. In contrast to the results observed with the rpoN and fleR mutants, SadB levels were unchanged in the crc, gacA, and lasR mutants. This latter group of mutants has defects in biofilm formation events downstream of irreversible attachment (Fig. 7). The flgK mutant (which is unable to make the flagellar hook protein) was also tested for its effect on SadB level because of the link between flagella and exopolysaccharide production in Vibrio cholerae (40) and surface sensing in Vibrio parahaemolyticus (19-21). SadB levels were not altered in the flgK mutant. These regulation studies are consistent with the microscopy experiments in that they implicate SadB in the earliest step(s) of biofilm formation.
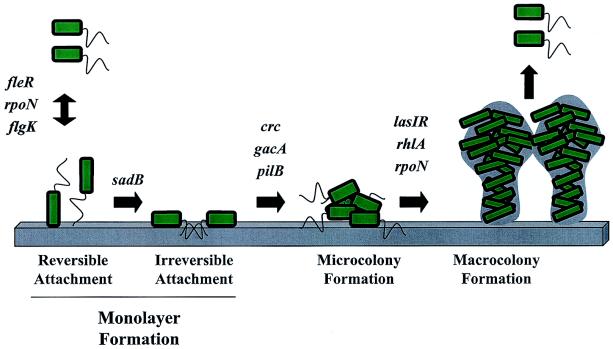
FIG. 7.
A model for biofilm development by P. aeruginosa. Shown is a model for the biofilm developmental pathway of P. aeruginosa that includes known structural and regulatory genes and the corresponding step at which they are predicted to be involved in biofilm formation. Black arrows represent the transitions between steps, and the name of each step is indicated below the figure.
The means by which SadB mediates the transition to irreversible attachment is still not understood, and although SadB contains two conserved domains, neither of these domains has an established function. Structure-function studies demonstrated that both the putative α/β (amino acids 20 to 154) and HD (amino acids 290 to 417) domains are required for biofilm formation; however, the C-terminal portion of SadB appears to be dispensable for function. We also localized SadB to the cytoplasm of P. aeruginosa, suggesting that SadB does not act as a cell surface adhesin. The cytoplasmic localization of SadB is consistent with the predicted functions of the α/β and HD domains in nucleotide binding and phosphohydrolase activity, respectively (1, 10, 11, 42).
O'Toole, Kaplan, and Kolter had proposed previously that biofilm formation represents a new example of a bacterial developmental pathway (23). In the cases of spore formation in Bacillus subtilis, fruiting body formation in Mxyococcus xanthus, and the swarmer-to-stalk transition in Caulobacter crescentus, development has been both historically and functionally characterized by a temporal series of events (e.g., changes in gene expression, protein function, or morphology) that can be blocked at distinct steps by mutation (36). Here we present two lines of evidence that are consistent with this view of microbial development as applied to biofilm formation. First, we have shown that a functional copy of sadB is essential for the transition from reversible to irreversible attachment and that disrupting this early SadB-dependent step does not simply delay but blocks the formation of a typically structured mature biofilm even after incubation in a flow cell for 6 days (Fig. 2A). It is also important that strains lacking a functional flagellum or pilus (structures required at early steps in formation), like the sadB mutant, will eventually form a biofilm in flowing conditions, but the structure of the community formed is markedly different from that observed for the wild-type strain (14). Other examples of mutations that block biofilm formation at various steps during the formation of the mature community are shown in Fig. 7. Second, we demonstrated that the SadB level is regulated by RpoN and FleR, which control production of the flagellum, a factor important very early in the transition to a surface existence (24, 29, 33), but the SadB level is not impacted by regulators shown to be important for downstream events in biofilm formation. While the regulatory factors important for the formation of microcolonies or a mature biofilm, steps presumably downstream of irreversible attachment, do not impact the SadB level, it is yet to be determined whether SadB regulates the expression or activity of these downstream functions. The data presented here are consistent with the idea that SadB can be placed within an ordered hierarchy of functions required to form a biofilm and with the notion that the formation of a biofilm is indeed an ordered and regulated microbial developmental process.
Acknowledgments
We thank D. A. Hogan for helpful experimental input and reading the manuscript and F. Ausubel and L. Rahme for strains. The fleR mutation in this study was obtained from MGH-Parabiosys:NHLBI Program for Genomic Applications, Massachusetts General Hospital and Harvard Medical School, Boston, Mass. (http://pga.mgh.harvard.edu/cgi-bin/pa14/mutants/retrieve.cgi).
This work was supported by grants from the NIH (1 R01 AI051360-01A1) and the Pew Charitable Trusts to G.A.O. G.A.O. is a Pew Scholar in the Biomedical Sciences.
REFERENCES
- 1.Aravind, L., and E. V. Koonin. 1998. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 23:469-472. [DOI] [PubMed] [Google Scholar]
- 2.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1997. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacteriol. 179:5574-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. A., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1990. Current protocols in molecular biology. Wiley Interscience, New York, N.Y.
- 4.Bloemberg, G. V., G. A. O'Toole, B. J. J. Lugtenberg, and R. Kolter. 1997. Green fluorescent protein as a marker for Pseudomonas spp. Appl. Environ. Microbiol. 63:4543-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Budzik, J. M., W. A. Rosche, A. Rietsch, and G. A. O'Toole. 2004. Isolation and characterization of a generalized transducing phage for Pseudomonas aeruginosa strains PAO1 and PA14. J. Bacteriol. 186:3270-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen, B. B., C. Sternberg, J. B. Andersen, R. J. Palmer, Jr., A. T. Nielsen, M. Givskov, and S. Molin. 1999. Molecular tools for study of biofilm physiology. Methods Enzymol. 310:20-42. [DOI] [PubMed] [Google Scholar]
- 7.Davey, M. E., N. C. Caiazza, and G. A. O'Toole. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:1027-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 9.Franklin, M. J., and D. E. Ohman. 2002. Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O acetylation of alginate in Pseudomonas aeruginosa. J. Bacteriol. 184:3000-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galperin, M. Y., D. A. Natale, L. Aravind, and E. V. Koonin. 1999. A specialized version of the HD hydrolase domain implicated in signal transduction. J. Mol. Microbiol. Biotechnol. 1:303-305. [PMC free article] [PubMed] [Google Scholar]
- 11.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 12.Hinsa, S. M., M. Espinosa-Urgel, J. L. Ramos, and G. A. O'Toole. 2003. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 49:905-918. [DOI] [PubMed] [Google Scholar]
- 13.Kjelleberg, S., and S. Molin. 2002. Is there a role for quorum sensing signals in bacterial biofilms? Curr. Opin. Microbiol. 5:254-258. [DOI] [PubMed] [Google Scholar]
- 14.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48:1511-1524. [DOI] [PubMed] [Google Scholar]
- 15.Lawrence, J. R., P. J. Delaquis, D. R. Korber, and D. E. Caldwell. 1987. Behavior of Pseudomonas fluorescens within the hydrodynamic boundary layers of surface microenvironments. Microb. Ecol. 14:1-14. [DOI] [PubMed] [Google Scholar]
- 16.Lohia, A., A. N. Chaterjee, and J. Das. 1984. Lysis of Vibrio cholerae cells: direct isolation of the outer membrane from whole cells by treatment with urea. J. Gen. Microbiol. 130:2027-2033. [DOI] [PubMed] [Google Scholar]
- 17.Manoil, C., and J. Beckwith. 1986. A genetic approach to analyzing membrane protein topology. Science 233:1403-1408. [DOI] [PubMed] [Google Scholar]
- 18.Marshall, K. C., R. Stout, and R. Mitchell. 1971. Mechanism of the initial events in the sorbtion of marine bacteria to surfaces. J. Gen. Microbiol. 68:337-348. [Google Scholar]
- 19.McCarter, L., M. Hilmen, and M. Silverman. 1988. Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell 54:345-351. [DOI] [PubMed] [Google Scholar]
- 20.McCarter, L., and M. Silverman. 1990. Surface-induced swarmer cell differentiation of Vibrio parahaemolyticus. Mol. Microbiol. 4:1057-1062. [DOI] [PubMed] [Google Scholar]
- 21.McCarter, L. L. 1995. Genetic and molecular characterization of the polar flagellum of Vibrio parahaemolyticus. J. Bacteriol. 177:1595-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Toole, G. A., K. A. Gibbs, P. W. Hager, P. V. Phibbs, Jr., and R. Kolter. 2000. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 182:425-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Toole, G. A., H. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 24.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 25.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 26.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to the study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 27.Pardee, A. B., F. Jacob, and J. Monod. 1959. The genetic control and cytoplasmic expression of “inducibility” in the synthesis of β-galactosidase in E. coli. J. Mol. Biol. 1:165-178. [Google Scholar]
- 28.Parkins, M. D., H. Ceri, and D. G. Storey. 2001. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol. Microbiol. 40:1215-1226. [DOI] [PubMed] [Google Scholar]
- 29.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: defining the roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-294. [DOI] [PubMed] [Google Scholar]
- 30.Prouty, M. G., N. E. Correa, and K. E. Klose. 2001. The novel sigma54- and sigma28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39:1595-1609. [DOI] [PubMed] [Google Scholar]
- 31.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Sauer, K., and A. K. Camper. 2001. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 183:6579-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schweizer, H. P. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 103:109-112. [DOI] [PubMed] [Google Scholar]
- 36.Shimkets, L. J., and Y. V. Brun. 1999. Prokaryotic development: strategies to enhance survival, p. 1-7. In L. J. Shimkets and Y. V. Brun (ed.), Prokaryotic development. ASM Press, Washington, D.C.
- 37.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, and I. T. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 38.Thompson, L. S., J. S. Webb, S. A. Rice, and S. Kjelleberg. 2003. The alternative sigma factor RpoN regulates the quorum sensing gene rhlI in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 220:187-195. [DOI] [PubMed] [Google Scholar]
- 39.Totten, P. A., J. C. Lara, and S. Lory. 1990. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J. Bacteriol. 172:389-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watnick, P. I., C. M. Lauriano, K. E. Klose, L. Croal, and R. Kolter. 2001. The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol. Microbiol. 39:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitchurch, C. B., M. Hobbs, S. P. Livingston, V. Krishnapillai, and J. S. Mattick. 1990. Characterization of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialized protein export system widespread in eubacteria. Gene 101:33-44. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, H., K. Huang, Z. Li, L. Banerjei, K. E. Fisher, N. V. Grishin, E. Eisenstein, and O. Herzberg. 2000. Crystal structure of YbaK protein from Haemophilus influenzae (HI1434) at 1.8 Å resolution: functional implications. Proteins 40:86-97. [DOI] [PubMed] [Google Scholar]
- 43.Zobell, C. E. 1943. The effects of solid surfaces upon bacterial activity. J. Bacteriol. 46:39-56. [DOI] [PMC free article] [PubMed] [Google Scholar]