Abstract
An analysis of the Pseudomonas aeruginosa genomic sequence revealed three gene clusters, PA1381-1393, PA2231-2240, and PA3552-3558, in addition to the alginate biosynthesis gene cluster, which appeared to encode functions for exopolysaccharide (EPS) biosynthesis. Recent evidence indicates that alginate is not a significant component of the extracellular matrix in biofilms of the sequenced P. aeruginosa strain PAO1. We hypothesized that at least one of the three potential EPS gene clusters revealed by genomic sequencing is an important component of P. aeruginosa PAO1 biofilms. Thus, we constructed mutants with chromosomal insertions in PA1383, PA2231, and PA3552. The mutant with a PA2231 defect formed thin unstructured abnormal biofilms. The PA3552 mutant formed structured biofilms that appeared different from those formed by the parent, and the PA1383 mutant formed structured biofilms that were indistinguishable from those formed by the parent. Consistent with a previous report, we found that polysaccharides were one component of the extracellular matrix, which also contained DNA. We suggest that the genes that were inactivated in our PA2231 mutant are required for the production of an EPS, which, although it may be a minor constituent of the matrix, is critical for the formation of P. aeruginosa PAO1 biofilms.
Pseudomonas aeruginosa is an important opportunistic pathogen that is capable of causing several different types of infections, including chronic biofilm infections. For example, P. aeruginosa infects the lungs of people with the genetic disease cystic fibrosis (CF), and chronic CF lung infections are considered biofilm infections (5, 12, 30). Mature P. aeruginosa biofilms consist of groups of cells growing together in clusters embedded in an extracellular polymeric matrix (4, 5). Quite often, P. aeruginosa isolated from the lungs of CF patients exhibits a mucoid phenotype resulting from the overproduction of the exopolysaccharide (EPS) alginate (12, 34). However, strains that do not produce much alginate are capable of forming mature biofilms in vitro (18). In fact, the commonly studied laboratory strain PAO1 makes little or no alginate (17, 36).
If alginate is not the extracellular matrix for biofilms of strain PAO1, what is? A recent report showed that a major polymer in the extracellular matrix of P. aeruginosa PAO1 biofilms is DNA (35). However, this does not rule out the possibility that P. aeruginosa PAO1 makes an EPS other than alginate and that this polysaccharide is an important constituent of the extracellular polymeric matrix.
A clue that polysaccharides other than alginate might be involved in P. aeruginosa biofilms came from the P. aeruginosa genome sequencing project. The annotation of the genome suggested that there are gene clusters in addition to the alginate biosynthesis gene cluster that may be involved in the synthesis of EPSs. One of these clusters is PA1381-1393, a second is PA2231-2245, and a third is PA3552-3558 (31). The PA1381-1393 cluster has an anomalously low G+C content, and the encoded polypeptides show sequence similarity to polypeptides encoded by a gene cluster which is required for polymyxin resistance and for the aminoarabinose modification of lipid A in Salmonella enterica serovar Typhimurium (9). The PA3552-3558 cluster is adjacent to a cluster of alginate biosynthetic genes. Many of the polypeptides encoded by genes in the PA2231-2245 cluster show similarity to glycosyltransferases, export proteins, or polysaccharide polymerases. The first three genes in this cluster have been studied by Rocchetta et al. (23), who suggested that PA2232 is involved in the biosynthesis of the lipopolysaccharide (LPS) A-band O antigen.
We generated mutations in one gene selected from each cluster (PA1383, PA2231, and PA3552) and studied the influence of the mutations on biofilm development. Our PA2231 mutant showed a severe defect in biofilm formation. Thus, we studied this mutant in more detail. Our results, taken together with those reported in accompanying papers (8, 13), indicate that the PA2231 gene cluster is an important determinant of P. aeruginosa biofilm development. We have adopted the nomenclature of Jackson et al. (13), and we will refer to the genes of the PA2231-2245 cluster as psl (polysaccharide synthesis locus) genes throughout the remainder of this article.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
The strains and plasmids used for this study are listed in Table 1. Escherichia coli and P. aeruginosa were routinely grown at 37°C in Luria-Bertani (LB) broth or on LB agar. For growth curves and microtiter dish biofilm assays, P. aeruginosa was grown in tryptic soy broth (TSB) (Difco). The medium for flow cell biofilms was 1% TSB. Agar plates of Vogel-Bonner minimal medium (33) were used to select exconjugants when pEX18Tc was used as a suicide vector. For EPS purification and for experiments in which double-stranded DNA was visualized by staining the biofilms with PicoGreen (Molecular Probes Inc.), we used M63 minimal medium supplemented with 3% glycerol, 0.4% glutamate, and trace minerals (0.0013% EDTA, 0.0055% ZnSO4 · 7H2O, 0.0025% FeSO4, 0.00078% MnSO4 · H2O, 0.0002% CuSO4 · 5H2O, 0.00012% Co(NO3)2 · 5H2O, and 0.000089% Na2B4O7 · 10 H2O). Media were supplemented with antibiotics for marker selection or plasmid maintenance as follows. For E. coli, we used 100-μg/ml ampicillin, 15-μg/ml gentamicin (GEN), 25-μg/ml chloramphenicol, 100-μg/ml streptomycin (STR), 12-μg/ml tetracycline (TET), and 20-μg/ml nalidixic acid. For P. aeruginosa, we used 150-μg/ml carbenicillin, 100-μg/ml GEN, 500-μg/ml STR, and 50- or 100-μg/ml TET.
TABLE 1.
Strains and plasmids used for this study
| Strain or plasmid | Characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAO1 | Parent | B. Iglewski |
| Δ1383Sm | PA1383 mutant of PAO1, Smr Spr | This study |
| ΔpslAGm | PA2231 mutant of PAO1, Gmr | This study |
| ΔpslAnull | PA2231 mutant of PAO1, unmarked | This study |
| Δ3552Gm | PA3552 mutant of PAO1, Gmr | This study |
| E. coli | ||
| DH5-α | 24 | |
| S17-1λpir | thi pro hsdR recA RP4-2 (Tet::Mu)(Km::Tn7) λ | 21 |
| SM10 | thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu Kmr | 28 |
| Plasmids | ||
| pKV69 | Suicide vector, Cmr Tcr | 3 |
| pGmΩ1 | Source of Ω (aacC1 with transcriptional termination signals in both orientations), Ampr | 27 |
| pHRP315 | Source of Ωf (aadA with transcriptional termination signals in both orientations), Smr Spr | 20 |
| pEX18Tc | Suicide vector, Tcr | 11 |
| pMRP9-1 | GFP expression vector, Cbr | 6 |
| Mini-CTX-lacZ | Suicide vector, Tcr | 2 |
| pMM11 | pKV69 carrying 4.7-kb (from bp 1500183 to 1505052) fragment containing PA1383, in BamHI-SphI site | This study |
| pMM12 | pMM1 with aadA Smr Spr (SmaI-EcoRV fragment from pHRP315) cassette between the two BstEII sites of ORF PA1383 | This study |
| pMM21 | pKV69 carrying 4.4-kb (from bp 24521491 to 2456529) fragment containing PA2231, in BamHI-PstI site | This study |
| pMM22 | pMM2 with aacC1 Gmr cassette (SmaI fragment from pGmΩ1) between the two MluI sites of ORF PA2231 | This study |
| pMM31 | pEX18Tc carrying 1.2-kb (from bp 3978642 to 3979870) fragment of PAO1 chromosomal DNA in HindIII- BamHI site | This study |
| pMM32 | pMM3 with 1.2-kb (from bp 3981000 to 3982279) fragment of PAO1 chromosomal DNA in BamHI-EcoRI site | This study |
| pMM33 | pMM31 with aacC1 Gmr cassette (BamHI fragment from pGmΩ1) in BamHI site | This study |
| pMMC2231 | Mini-CTX-lacZ carrying 1.9-kb (from bp 2453177 to 2455101) fragment of PAO1 chromosomal DNA in BamHI-SphI site | This study |
Construction of P. aeruginosa mutants.
We used standard methods for manipulating plasmids and DNA fragments (1). PA1383 was interrupted by the insertion of an aadA gene cassette (Smr Spr). A 4.7-kb P. aeruginosa PAO1 chromosomal DNA fragment (from bp 1500183 to 1505052) containing PA1383 was amplified by use of an Expand long-template PCR kit (Roche Applied Science). The forward primer contained a BamHI site (5′-CGGGATCCGGAGGACGAGAATCGAAACGCCATC-3′) and the reverse primer contained an SphI site (5′-ACATGCATGCGAAGGACCAACCTGATCGAACAGG-3′). The PCR fragment was cloned into BamHI- and SphI-digested pKV69 to give pMM11. A 2.0-kb SmaI-EcoRV fragment of pHRP315 containing the aadA gene was ligated with BstEII-digested pMM11, which had been polished with T4 DNA polymerase, to create pMM12. There is an insertion in pMM12 575 bp downstream of the translational start site of PA1383 and 736 bp upstream of the stop codon. For construction of the mutant, pMM12 was mobilized from E. coli S17-λpir into P. aeruginosa PAO1 by conjugation as follows. Ten microliters of a late-logarithmic-phase culture of E. coli S17-λpir(pMM12) grown at 30°C was mixed with 100 μl of a late-logarithmic-phase culture of P. aeruginosa PAO1 grown at 42°C without shaking. The mixture was spread onto LB agar plates. After 16 to 24 h at 30°C, transconjugants were selected by plating on LB agar containing GEN or STR. We then screened for strains showing sensitivity to TET. Southern blotting was used to confirm that the TET-sensitive strain contained an aadA insertion in PA1383.
We constructed two PA2231 (pslA) mutants. One contained an aacC1 cassette (Gmr) with a transcriptional terminator inserted in pslA. The other was a deletion mutant. The insertion mutant, P. aeruginosa ΔpslAGm, was constructed as follows. A 4.3-kb PAO1 PA2231 chromosomal DNA fragment (bp 2452149 to 2456529) was amplified by Expand long-template PCR (Roche Applied Science). The 5′ primer contained a BamHI site (5′-CGGGATCCGCGATATCCTGCAACTGGTCGAAC-3′) and the 3′ primer contained a PstI site (5′-AACTGCAGATGTCGTTGAAGCGGACGATGTC-3′). The PCR product was cloned into BamHI- and PstI-digested pKV69 to give pMM21. A 1.6-kb SmaI-EcoRV fragment of pGmΩ1 that contained the aacC1 gene was ligated with MluI-digested pMM21, which had been polished with T4 DNA polymerase, to create pMM22. This construct has the aacC1 gene inserted 808 bp downstream of the translational start site of PA2231 and 208 bp upstream of the stop codon. The aacC1 insertion mutation was confirmed by Southern blotting. The generation of a PA2231 mutation in P. aeruginosa PAO1 with pMM22 involved a procedure similar to that described for the construction of the PA1383 mutant (see above). To generate the pslA deletion mutant, we amplified ΔpslAnull, a 1.2-kb PAO1 chromosomal DNA fragment extending from 1,051 bp upstream through 51 bp downstream of the predicted pslA start codon, by Expand long-template PCR. The 5′ primer contained a HindIII site (5′-CCCAAGCTTGACAACCTCTGCGAGATCT-3′) and the 3′ primer contained an XbaI site (5′-GCTCTAGAGAAGTACTCGATGAATCCAG-3′). The PCR product was cloned into HindIII- and XbaI-digested pEX18Tc to give pMM201. A 1.2-kb PAO1 chromosomal DNA fragment extending from 23 bp upstream to 1,176 bp downstream of the PA2231 stop codon was amplified with a 5′ primer containing an XbaI site (5′-GCTCTAGACTCCTGACCAAGGAAGTCTA-3′) and a 3′ primer containing an EcoRI site (5′-CGGAATTCCGATGCGCTTGATCTTGAAG-3′). The PCR product was cloned into XbaI- and EcoRI-digested pMM201 to generate pMM202. For the generation of P. aeruginosa ΔpslAnull, E. coli SM10(pMM201) was mated with P. aeruginosa PAO1. Equal amounts of late-logarithmic-phase cultures of E. coli grown at 30°C with shaking and of strain PAO1 grown at 42°C without shaking were mixed and grown on LB agar. Cells from the agar plates were suspended in phosphate-buffered saline, and a strain with a single crossover was selected on minimal medium plus TET. Exconjugants that were resistant to TET were counterselected by using LB agar plus 5% sucrose. The resulting strain, ΔpslAnull, was shown to have a PA2231 deletion by PCR.
The generation of the PA3552 mutant was done by the insertion of an aacC1 cassette. A 1.2-kb PAO1 chromosomal DNA fragment upstream of PA3552 (bp 3978642 to 3979870) was amplified by Expand long-template PCR with a forward primer containing a HindIII site (5′-CCCAAGCTTCTGGAAGAACTGAAGAAGCACGACGCC-3′) and a reverse primer with a BamHI site (5′-CGCGGATCCGTCCAGTGACATGTAATGAAGCCTCGG-3′). The PCR product was cloned into HindIII- and BamHI-digested pEX18Tc to yield pMM31. A 1.3-kb PAO1 chromosomal DNA fragment (bp 3981000 to 3982279) downstream of PA3552 was amplified in a similar fashion by using the forward primer 5′-CGGGATCCGTCGATGAAGCCCTATCCGATCGACCT-3′ and the reverse primer 5′-CGGAATTCGGTAGTAGAAGGAGAACAGGAAGTCC-3′. The PCR fragment was cloned into BamHI- and EcoRI-digested pMM31 to create pMM32. A 1.6-kb aacC1-containing BamHI fragment of pGmΩ was ligated with BamHI-digested pMM32 to create pMM33. This construct has the insertion 15 bp downstream of the translational start site of open reading frame (ORF) PA3552 and 10 bp upstream of the stop codon. To generate a P. aeruginosa PA3552 mutant strain, we used a procedure similar to that described above for the generation of the PA2231 aacC1 insertion mutant.
Biofilm experiments.
Biofilm formation in a static system was measured as described by O'Toole and Kolter (19), with some modifications. Briefly, TSB (100 μl/well in 96-well polystyrene microtiter dishes) was inoculated with a mid-exponential-phase culture to give an optical density at 600 nm of 0.005. After 24 h at 37°C in a humidified chamber, the liquid culture was removed by aspiration and the biofilm attached to the microtiter dish wells was stained with 0.1% crystal violet for 15 min. The crystal violet solution was removed by aspiration, and the wells were rinsed three times by submerging the plates in distilled water. The crystal violet remaining in the wells was dissolved in 200 μl of 95% ethanol-5% acetic acid and measured as the absorbance at 590 nm. We also grew biofilms in flow cell microscopic observation cells. For these experiments, P. aeruginosa contained pMRP9-1, which codes for green fluorescent protein (GFP) (6). This plasmid did not influence the planktonic growth of the P. aeruginosa strains used for our studies (data not shown). The flow cells and conditions have been described elsewhere (29). Biofilm architecture was monitored by scanning confocal laser microscopy (SCLM). For experiments with the DNA stain PicoGreen, we used a 1:1,000 dilution of PicoGreen gently injected into flow cells in which biofilms of P. aeruginosa without the GFP plasmid were grown.
Motility assays.
Both swimming and twitching motilities were measured by standard techniques, as described elsewhere (22).
Analysis of extracellular and cell-surface-associated polymers.
We obtained extracellular polymeric matrix material by growing P. aeruginosa strains in 12 large petri dishes (150-mm diameter), each of which contained 75 ml of M63 minimal broth, at 37°C for 48 h without agitation. The gelatinous material loosely attached to the bottom of the petri dishes was harvested by use of a glass spreader. The harvested material was centrifuged at 15,000 × g at 42°C for 1 h. The supernatant fluid was removed and the pellet was suspended in 0.4 M NaCl (final volume, 500 ml). For the extraction of extracellular polymeric substances, the suspension was placed in a 2-liter flask and stirred at 42°C for 30 min. The suspension was centrifuged and the pellet was suspended in 250 ml of 0.4 M NaCl. The supernatant fluid was combined with the supernatant fluid resulting from the first extraction. The EPSs and other polymers were extracted with 95% ethanol at 42°C for 12 h. The precipitated material was collected by centrifugation (4,000 × g at 4°C for 30 min), washed with 95% ethanol, and then washed with absolute ethanol. The washed material was dried and then dissolved in 100 ml of deionized water. Polysaccharides were purified from the resulting viscous solution as follows. One-hundred milliliters of Tris (60 mM, pH 7.5)-MgCl2 (20 mM) and 10 μg of DNase I (Sigma) and RNase A (Qiagen)/ml were added and the solution was incubated at 37°C for 6 h. This resulted in a decrease in viscosity. After the nuclease treatment, 100 μg of proteinase K (Gibco-BRL)/ml was added and the solution was incubated overnight at 55°C. The precipitate formed by this procedure was removed by centrifugation and the supernatant fluid was extracted with hot phenol. Sodium acetate (3 M) was added to bring the acetate concentration to 0.3 M, followed by the addition of ethanol to bring the final ethanol concentration to 80%. After an overnight incubation at 42°C, the precipitate was collected by centrifugation, dissolved in deionized water, and dialyzed against deionized water. The dialysate was lyophilized, and the lyophilized material was analyzed at the University of Georgia Complex Carbohydrate Research Center (37).
To determine the polymer composition of the extracellular matrix, we measured the total carbohydrate, double-stranded DNA, and protein in the crude extracts by a phenol-sulfuric acid assay with glucose as the standard (7), the PicoGreen system with calf thymus DNA as the standard, and a protein assay kit (Sigma) with bovine serum albumin as the standard, respectively. To purify LPS, we used the Hitchcock and Brown method (10). The purified LPS was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (15, 32).
RT-PCR.
The total RNA was isolated from cells grown to an optical density (600 nm) of 2.0 in LB broth by use of a Qiagen RNeasy kit. The DNA was removed as described elsewhere (26). The RNA was then extracted twice with TRIzol (Invitrogen) and once with chloroform-isoamyl alcohol. DNA contamination of the purified RNA was assessed by Expand long-template PCR using rplU primers. Reverse transcription-PCR (RT-PCR) was performed as described elsewhere (16). The cDNA was purified with a Qiagen PCR purification kit and amplified (50 ng) by PCRs (30 cycles) with primers for a series of adjacent genes extending from PA2230 to PA2245 (the sequences of the primers are available upon request). The primers were designed to anneal approximately 250 bp upstream or downstream of the predicted intergenic regions. Genomic DNA was used as a positive control for PCRs.
RESULTS
Influence of mutations in putative EPS synthesis genes on biofilm development.
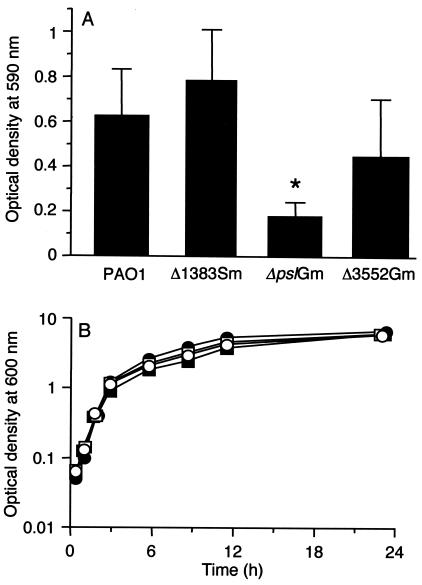
We first examined the biofilm formation of each of the insertion mutants by using a microtiter dish assay (Fig. 1A). In this assay, the parent, the PA1383 mutant, and the PA3552 mutant showed similar levels of biofilm formation. The ΔpslAGm mutant produced substantially smaller amounts of biofilm in microtiter dish wells than did the other strains. The biofilm defect was not the result of a growth defect in ΔpslAGm. All of the strains showed similar growth in batch cultures (Fig. 1B).
FIG. 1.
Growth of the parent and mutant strains in static biofilm cultures in microtiter dishes (A) and in nonbiofilm broth cultures (B). (A) Bars indicate the standard deviations around the means from five independent experiments, with each sample assayed in triplicate. *, P < 0.001 (Mann-Whitney U test). (B) Growth curves for P. aeruginosa PAO1 (open circles), ΔpslAGm (open squares), Δ1383Sm (filled squares), and Δ3552Gm (filled circles).
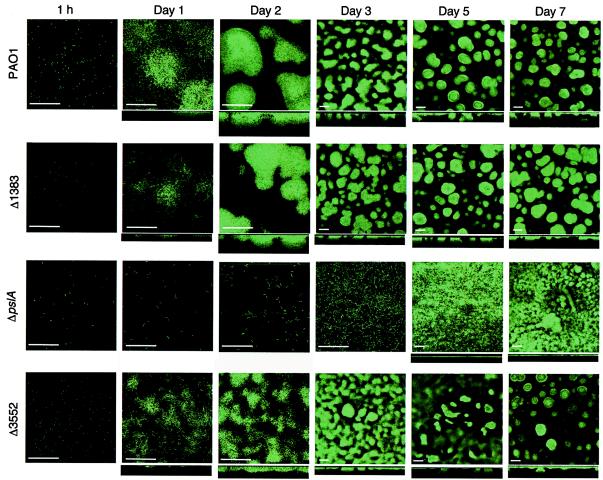
To further assess the influence of the mutations on biofilm development, we transformed the parent and the mutants with a plasmid that codes for the constitutive expression of GFP (6) and monitored the development of biofilms in flow cells over a period of 7 days by using SCLM (Fig. 2). Bacteria attached to the surface within 1 h. There was little difference in the attachment of the parent and that of any of the mutants at this early stage in biofilm formation. After 24 h, it was apparent that microcolonies were forming in the case of all strains but P. aeruginosa ΔpslAGm. After 2 days, all of the strains except ΔpslAGm had formed substantial microcolonies that were over 20 μm deep. The PA3552 mutant appeared to be thinner and somewhat less structured than the parent strain. We had the impression that the biofilms of the PA3552 mutant were more fragile than those of the parent, but we do not have a method to measure fragility. The ΔpslAGm mutant remained a thin sparse layer of cells at day 2 and even after 3 days. By day 3, mushroom-like structures were apparent in all strains but P. aeruginosa ΔpslAGm. Upon further incubation in the flow cells, the mushroom-like structures of all strains but the pslA mutant were maintained. The pslA mutant eventually formed a multilayered growth on the surface, but it was thin and unstructured, with a depth of <10 μm, compared to the depths of the mushroom-like structures formed by the other strains (>25 μm).
FIG. 2.
Influence of putative EPS gene mutations on P. aeruginosa biofilm development. The parent and mutants (indicated on the left) were grown in flow chambers, and SCLM images were acquired at the times shown at the top. Horizontal sections are shown for each time, and vertical reconstructions are shown underneath many of the horizontal images. Bars, 50 μm (note that the magnification for images acquired on days 3 to 7 was different from that for images acquired during earlier stages of development).
PA2231, or pslA, is the first gene in a cluster of genes suspected to be involved in the synthesis of an EPS. The insertion of a GEN resistance marker may have had a polar effect on the expression of downstream genes in the cluster. Thus, we constructed a deletion mutant, ΔpslAnull, which showed a pattern of biofilm development in flow cells that was indistinguishable from that of ΔpslAGm (data not shown). However, complementation with a pslA-containing plasmid did not restore biofilm development to the parental pattern. The pslA-complementing plasmid contained the PA2231-2233 cluster. Note that in an accompanying article, Jackson et al. (13) show that PA2231-2232 mutants form abnormal biofilms with characteristics similar to those of our PA2231 mutants and that the PA2231-2232 mutant biofilm defect can be complemented with a cosmid containing a 22-kb insert including PA2231-2236. Taken together, the data from both investigations suggest that a gene or genes in this cluster are critical for biofilm development.
Flagella are involved in the initial stages of P. aeruginosa biofilm development, and type IV pilus-mediated twitching motility is involved in biofilm formation (19), particularly in the maturation of microcolonies to mature mushroom-like structures on a glass surface (14). Thus, we determined whether the ΔpslAGm strain had a defect in either swimming or twitching motility that could explain the abnormal biofilm development. We discerned no significant differences in swimming or twitching among any of the strains (Table 2).
TABLE 2.
Influence of mutations in putative EPS synthesis genes on twitching and swimming motilities of P. aeruginosa
| Strain | Motility (mm of migration from point of inoculation)a
|
|
|---|---|---|
| Swimming | Twitching | |
| PAO1 | 16 ± 1 | 14 ± 1 |
| Δ1383Sm | 20 ± 0.5 | 14 ± 1 |
| ΔpslAGm | 17 ± 1 | 13 ± 0.5 |
| Δ3552Gm | 20 ± 0.5 | 16 ± 1 |
The results are means ± standard deviations from six independent experiments for swimming motility and from 12 independent experiments for twitching motility.
The extracellular matrix of P. aeruginosa biofilms.
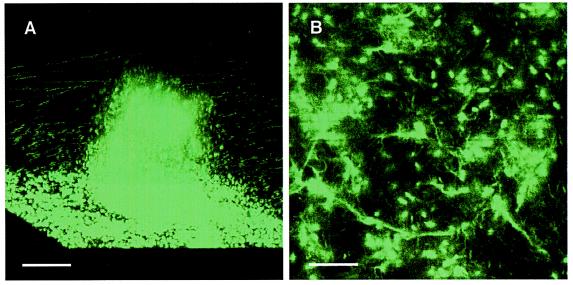
The defect in biofilm development shown by pslA mutants does not appear to result from a general growth defect (Fig. 1B) or from a motility defect (Table 2). This and the fact that PA2231 is the first gene in a cluster of genes annotated as EPS synthesis genes led us to analyze the extracellular matrix of the parent and the ΔpslAGm strain. To obtain enough polysaccharide for our analyses, we grew attached bacteria in relatively large volumes in thin layers under static conditions. We do not know how the biofilms that develop under these conditions compare to those grown in flow cells. The matrix from either strain grown under static conditions consisted primarily of DNA, which was about five-fold more abundant than protein, which was about three or four times more abundant than carbohydrates (Table 3). The finding of abundant DNA in the purified extracellular matrix was consistent with a previous report that DNA is an important polymer in the matrix of P. aeruginosa PAO1 biofilms (35). To extend and confirm this observation, we grew biofilms of the parent in a defined DNA-free medium and stained these biofilms with PicoGreen, a specific fluorescent double-stranded DNA stain. The biofilm matrix was brightly fluorescent, and both the cells and the intercellular matrix were stained (Fig. 3). It is not possible to tell from images such as those shown in Fig. 3 whether the matrix of the entire biofilm is fluorescent or whether there is a specific region encased in DNA. As a control, a PicoGreen-stained planktonic culture fluid showed no extracellular fluorescence.
TABLE 3.
Composition of extracellular matrix of P. aeruginosa PAO1 and ΔpslAGm
| Strain | Total carbohydrate (mg/100 mg or protein)a | Total double-stranded DNA (mg/100 mg of protein)a |
|---|---|---|
| PAO1 | 27.6 ± 4.6 | 529.1 ± 103.7 |
| Δ2231Gm | 30.0 ± 3.4 | 608.9 ± 53.9 |
Results are means and standard deviations from three independent experiments.
FIG. 3.
Images from SCLM of a 7-day P. aeruginosa biofilm stained with the double-stranded DNA stain PicoGreen. (A) Reconstructed three-dimensional image of a mushroom-like structure. Bar, 50 μm. (B) Horizontal section of the biofilm at the glass surface showing that the material between cells is stained as well as the cells in the biofilm. Bar, 10 μm.
Although carbohydrates constituted only a small part of the matrix by weight, an EPS produced by the parent, but not by the ΔpslAGm strain, might yet play an important role in biofilm development (the EPS might also constitute a larger fraction of the matrix in flow cell biofilms). Thus, we removed DNA and protein from the purified matrix material and analyzed the remaining soluble carbohydrate. For the parent, the most abundant sugar monomers were glucose, rhamnose, and mannose (a ratio of about 4 glucose to 3 rhamnose to 1 mannose). For the ΔpslAGm mutant, the polysaccharide was enriched in glucose (glucose constituted about 50% of the parent EPS and 75 to 80% of the ΔpslAGm EPS); however, in separate analyses the levels of rhamnose and mannose were low but variable. Friedman and Kolter (8) report in an accompanying paper that mutations in the psl gene cluster in a strain of P. aeruginosa other than PAO1 result in a mannose-depleted polysaccharide.
In P. aeruginosa, there can be an overlap between EPS synthesis gene functions and LPS functions (23). Thus, pslA could be involved in LPS synthesis and an LPS modification could be responsible for the abnormal biofilm phenotype of pslA mutants. To address this question, we purified LPS from the mutant and the wild type and showed that the two preparations were indistinguishable in a silver-stained sodium dodecyl sulfate-polyacrylamide gel (data not shown). Although this does not exclude the possibility of a subtle alteration in the LPS structure, it is consistent with our conclusion that the mutant produces a defective EPS that is important for normal biofilm development.
Evidence that pslA is the first gene in a 15-gene operon.
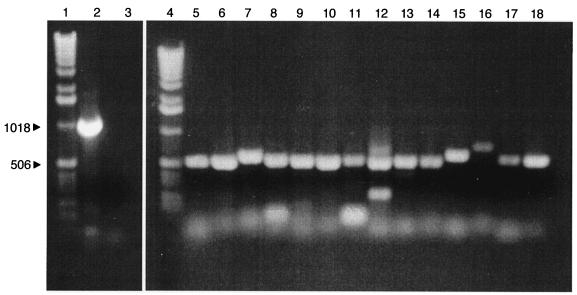
In the P. aeruginosa genome, pslA (or PA2231) is the first gene in a cluster that appears to encode polysaccharide synthesis functions. This cluster extends through PA2245. The next gene, PA2246, is oriented in the opposite direction. To assess whether the genes in the PA2231-2245 cluster are cotranscribed, we first interrogated the existing P. aeruginosa microarray database generated in our laboratory over the past several years (25, 26). The analysis revealed only that the transcripts of these genes are present in a very low abundance in a variety of planktonic cultures. Thus, we used RT-PCR with the RNA isolated from the parent strain, PAO1, and primer pairs spanning the intergenic regions through the PA2231 gene cluster (Fig. 4). The results indicate that PA2230 is not cotranscribed with PA2231. The analysis suggested that each gene in the cluster from PA2231 to PA2245 is cotranscribed with the adjacent gene. This is consistent with the conclusion that the cluster constitutes a large operon in P. aeruginosa. Furthermore, in an independent study, Friedman and Kolter (8) have identified multiple mutations in this gene cluster that result in a specific biofilm phenotype, the lack of pellicle formation. They found that mutations in the PA2231-2245 cluster downstream of pslA result in the production of an EPS consisting primarily of glucose and rhamnose, but not mannose. Thus, we propose that these genes be called pslA to -O.
FIG. 4.
Analysis of the PA2231-2245 gene cluster by RT-PCR of intergenic regions. Lane 1, size markers; lane 2, PCR amplification product of PA2230-2231 cluster with genomic DNA as the template (this serves as a control for RT-PCR of this region); lane 3, RT-PCR of PA2230 and -2231 (the lack of product indicates that PA2230 and -2231 are not cotranscribed); lane 4, size marker; lane 5, RT-PCR of PA2231 and -2232; lane 6, PA2232 and -2233; lane 7, PA2233 and -2234; lane 8, PA2234 and -2235; lane 9, PA2235 and -2236; lane 10, PA2236 and -2237; lane 11, PA2237 and -2238; lane 12, PA2238 and -2239; lane 13, PA2239 and -2240; lane 14, PA2240 and -2241; lane 15, PA2241 and -2242; lane 16, PA2242 and -2243; lane 17, PA2243 and -2244; lane 18, PA2244 and -2245. Control experiments without reverse transcriptase reactions prior to PCR were negative.
DISCUSSION
We have presented evidence that of three gene clusters that may encode functions for nonalginate EPS synthesis in P. aeruginosa PAO1, the PA2231-2245 (pslA to -O) cluster plays an important role in biofilm development. PA2231 (pslA) mutants showed a biofilm defect in both static microtiter dish experiments (Fig. 1) and flow cell experiments (Fig. 2). This is consistent with the findings reported in the two accompanying papers (8, 13). We call the genes in this cluster psl genes by agreement among the authors of all three papers. We also showed that mutants with Ω insertions in the other two gene clusters (PA1383 and PA3552) developed biofilms similar to those of the parent, although the PA3552 mutant biofilms seemed somewhat thinner (Fig. 2) and we had the impression that they were more fragile than those of the parent. Although we introduced the Ω cassette near the beginning of each cluster, we have not investigated whether any of the genes in these clusters are cotranscribed. For this and other reasons, we caution against concluding that the clusters other than the psl cluster are not important for biofilm development. We can only say that a gene or genes in the psl cluster are important for biofilm development.
There are many possible reasons why genes in the psl gene cluster may be required for normal biofilm development. We know that motility is involved in biofilm formation, but both the twitching and swimming motilities of a pslA mutant were comparable to those of the parent (Table 2). There is evidence that LPS influences biofilm architecture (23). One might imagine that the psl gene cluster affects LPS synthesis, but at least by comparing LPSs in silver-stained sodium dodecyl sulfate-polyacrylamide gels, the mutant and the parent were indistinguishable.
The most obvious explanation for an involvement of psl genes in biofilm formation is that they direct the synthesis of an essential EPS. Our finding that the purified EPS from ΔpslAGm is different from that of the parent and the findings of Friedman and Kolter (8) support this explanation. However, in both the parent and the mutant, the EPS may be a minor constituent of the total extracellular material. We judged DNA to be the most abundant polymer in the matrix (Table 3). This was consistent with a previous report that DNA was a major component of the P. aeruginosa PAO1 biofilm extracellular matrix (35). The DNA could have resulted from contamination of the matrix preparation with whole P. aeruginosa cells or there may have been substantial cell lysis as a result of the specific conditions in which we grew the biofilms for matrix analysis. Although this could account for some of the DNA in these preparations, we visualized DNA in both the cells and the matrix of a biofilm growing in a flow cell by staining with PicoGreen (Fig. 3). This provides evidence consistent with, but not in proof of, the hypothesis that there is abundant DNA in the extracellular matrix of flow-cell-grown biofilms. Regardless of the abundance of polysaccharide in the extracellular matrix, both we and Friedman and Kolter (8) have found that mutations in different genes in this locus alter the monomeric constituents of the polysaccharide from extracellular matrix fractions. Thus, the psl (polysaccharide synthesis locus) terminology used to describe the gene cluster seems appropriate. Our evidence indicates that PA2231 is the first of 15 genes in an operon (Fig. 4) and thus we called it pslA. For the parent strain, the most abundant sugars in the EPS were glucose, rhamnose, and mannose. The psl mutants produced an EPS that was predominantly glucose and rhamnose (8). We noted that the sugars that constitute alginate were not detected. This was consistent with previous findings that strain PAO1 biofilms make little or no alginate, at least under certain conditions (36).
If DNA and protein are abundant polymers in the P. aeruginosa PAO1 biofilm, why does an EPS have such a strong influence on the ability of the bacteria to form a biofilm? There are several possible explanations. Perhaps the EPS is synthesized early in biofilm formation and serves as a base upon which the mushroom-like structures are built. Another possibility that we favor is that the EPS functions as a scaffold on which other polymers like DNA become entangled. One might imagine that bacterial cells in a biofilm exist in a cloud of matrix materials, including DNA and protein, and that the EPS serves as a backbone that defines the area in which the cloud exists.
Acknowledgments
Support for this study was provided by a grant from the National Institute of General Medicine (GM59026) and from the W. M. Keck Foundation.
The contents of this study do not necessarily represent the official views of the National Institute of General Medicine.
REFERENCES
- 1.Ausubel, F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1997. Short protocols in molecular biology, 3rd ed. John Wiley & Sons, Inc., New York, N.Y.
- 2.Becher, A., and H. P. Schweizer. 2000. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. BioTechniques 29:948-950, 952. [DOI] [PubMed] [Google Scholar]
- 3.Chugani, S. A., M. Whiteley, K. M. Lee, D. D'Argenio, C. Manoil, and E. P. Greenberg. 2001. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 98:2752-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 5.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 6.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 7.Fox, J. D., and J. F. Robyt. 1991. Miniaturization of three carbohydrate analyses using a microsample plate reader. Anal. Biochem. 195:93-96. [DOI] [PubMed] [Google Scholar]
- 8.Friedman, L., and R. Kolter. 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gunn, J. S., S. S. Ryan, J. C. Van Velkinburgh, R. K. Ernst, and S. I. Miller. 2000. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 68:6139-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 12.Hoiby, N. 1993. Antibiotic therapy for chronic infection of Pseudomonas in the lung. Annu. Rev. Med. 44:1-10. [DOI] [PubMed] [Google Scholar]
- 13.Jackson, K. D., M. Starkey, S. Kremer, M. R. Parsek, and D. J. Wozniak. 2004. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J. Bacteriol. 186:4466-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klausen, M., A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 50:61-68. [DOI] [PubMed] [Google Scholar]
- 15.Lesse, A. J., A. A. Campagnari, W. E. Bittner, and M. A. Apicella. 1990. Increased resolution of lipopolysaccharides and lipooligosaccharides utilizing Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J. Immunol. Methods 126:109-117. [DOI] [PubMed] [Google Scholar]
- 16.Martin, P. R., A. A. Watson, T. F. McCaul, and J. S. Mattick. 1995. Characterization of a five-gene cluster required for the biogenesis of type 4 fimbriae in Pseudomonas aeruginosa. Mol. Microbiol. 16:497-508. [DOI] [PubMed] [Google Scholar]
- 17.Mathee, K., O. Ciofu, C. Sternberg, P. W. Lindum, J. I. Campbell, P. Jensen, A. H. Johnsen, M. Givskov, D. E. Ohman, S. Molin, N. Hoiby, and A. Kharazmi. 1999. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology 145:1349-1357. [DOI] [PubMed] [Google Scholar]
- 18.Nivens, D. E., D. E. Ohman, J. Williams, and M. J. Franklin. 2001. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J. Bacteriol. 183:1047-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motilities are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 20.Parales, R. E., and C. S. Harwood. 1993. Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for gram-negative bacteria. Gene 133:23-30. [DOI] [PubMed] [Google Scholar]
- 21.Pearson, J. P., C. Van Delden, and B. H. Iglewski. 1999. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 181:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rashid, M. H., and A. Kornberg. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rocchetta, H. L., J. C. Pacan, and J. S. Lam. 1998. Synthesis of the A-band polysaccharide sugar d-rhamnose requires Rmd and WbpW: identification of multiple AlgA homologues, WbpW and ORF488, in Pseudomonas aeruginosa. Mol. Microbiol. 29:1419-1434. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Schuster, M., A. Hawkins, C. S. Harwood, and E. P. Greenberg. 2004. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol. Microbiol. 51:973-985. [DOI] [PubMed] [Google Scholar]
- 26.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schweizer, H. P. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15:831-833. [PubMed] [Google Scholar]
- 28.Simon, R., U. Priefer, and A. Puhler. 1983. A broad-host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:37-45. [Google Scholar]
- 29.Singh, P. K., M. R. Parsek, E. P. Greenberg, and M. J. Welsh. 2002. A component of innate immunity prevents bacterial biofilm development. Nature 417:552-555. [DOI] [PubMed] [Google Scholar]
- 30.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 31.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K.-S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 32.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 33.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 34.Welsh, M. J., B. W. Ramsey, F. Accurso, and G. R. Cutting. 2001. Cystic fibrosis, p. 5121-5189. In C. L. Scriver, A. L. Beaudet, W. S. Sly, D. Valle, B. Childs, and B. Vogelstein (ed.), The metabolic and molecular basis of inherited disease, 8th ed. McGraw-Hill, New York, N.Y.
- 35.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 36.Wozniak, D. J., T. J. Wyckoff, M. Starkey, R. Keyser, P. Azadi, G. A. O'Toole, and M. R. Parsek. 2003. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. USA 100:7907-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 E1 Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 96:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]