Significance
Venom peptides have attracted considerable attention because of their value as pharmacological tools and their potential for development as novel pharmaceuticals and bioinsecticides. There is also a growing interest in venoms as model evolutionary systems, particularly for understanding antagonistic coevolutionary processes. We previously demonstrated that although centipede venoms are rich in novel proteins and peptides, there are considerable differences in venom complexity between high-order taxa. We show that this disparity appears to stem from morphological limitations of the venom gland, and that most centipede venoms likely evolve under constraints imposed by low-complexity toxin production facilities. Thus, the centipede venom apparatus should be a useful model system for gaining insight into the impact of morphological constraints on venom evolution.
Keywords: venom evolution, venom-gland morphology, centipede, mass spectrometry imaging, venom optimization hypothesis
Abstract
Venom represents one of the most extreme manifestations of a chemical arms race. Venoms are complex biochemical arsenals, often containing hundreds to thousands of unique protein toxins. Despite their utility for prey capture, venoms are energetically expensive commodities, and consequently it is hypothesized that venom complexity is inversely related to the capacity of a venomous animal to physically subdue prey. Centipedes, one of the oldest yet least-studied venomous lineages, appear to defy this rule. Although scutigeromorph centipedes produce less complex venom than those secreted by scolopendrid centipedes, they appear to rely heavily on venom for prey capture. We show that the venom glands are large and well developed in both scutigerid and scolopendrid species, but that scutigerid forcipules lack the adaptations that allow scolopendrids to inflict physical damage on prey and predators. Moreover, we reveal that scolopendrid venom glands have evolved to accommodate a much larger number of secretory cells and, by using imaging mass spectrometry, we demonstrate that toxin production is heterogeneous across these secretory units. We propose that the differences in venom complexity between centipede orders are largely a result of morphological restrictions of the venom gland, and consequently there is a strong correlation between the morphological and biochemical complexity of this unique venom system. The current data add to the growing body of evidence that toxins are not expressed in a spatially homogenous manner within venom glands, and they suggest that the link between ecology and toxin evolution is more complex than previously thought.
Venoms are exceptionally complex mixtures of inorganic salts, low molecular weight compounds, peptides, and proteins. Venom peptides and proteins, collectively referred to as toxins, are the primary components of most venoms, and they can be extremely diverse, with a single venom containing up to several thousand unique toxins (1, 2). In animals that rely on venom for prey capture and handling, diet and foraging ecology is thought to be one of the major drivers of toxin evolution (3). In this scenario, diversification of toxins is driven by antagonistic coevolution between predator and prey, and is often evidenced by rapid radiation and sustained diversity of toxin gene families (4–9).
However, venom is also an energetically expensive commodity that tends to be used sparingly and only when physical means of safely overpowering prey are insufficient (10). The degree of coevolution, and hence in many cases toxin-gene diversity, is therefore substantially dependent on the degree to which an animal relies on biochemical (i.e., venom) versus physical means for overpowering prey. Consistent with this scenario, toxin diversity is often inversely correlated with physical strength. For example, the slender-clawed buthid scorpions generally have more complex venoms than their larger-clawed relatives, which sometimes only use venom for predation when they are juveniles (11). Similarly, the evolution of prey constriction in snakes has led to a reduction in, or secondary loss of, venom systems despite these species still feeding on formidable prey (12–15). However, in centipedes (Chilopoda), which represent one of the oldest yet least-studied venomous lineages on the planet, this inverse relationship between venom complexity and physical subdual of prey appears to be absent.
There are ∼3,300 extant centipede species, divided across five orders (16). Perhaps the most conspicuous shared feature of centipedes is the presence of a pair of venom claws, termed forcipules, which are in fact modified walking appendages (17). Forcipules differ substantially between orders, with some of the most striking differences being found between the orders Scolopendromorpha and Scutigeromorpha, which both contain some of the largest species of centipede. Although scutigeromorphs (house centipedes) possess delicate stiletto-like forcipules similar to those of ancestral centipedes from over 430 Mya, the forcipules of scolopendromorphs, and in particular members of the family Scolopendridae, are powerful, heavily sclerotized structures (18). Nevertheless, members of both orders feed primarily on invertebrates and will attack prey exceeding their own body weight; scutigeromorphs, primarily by ambushing and chasing down prey, and scolopendrids seemingly via opportunistic encounters.
Surprisingly, despite lacking the formidable physical weaponry of scolopendrid centipedes, scutigeromorph venoms appear to be relatively noncomplex (19). The venom of a single scolopendrid species can contain over 30 distinct toxin families, often more than half of which are cysteine-rich peptides smaller than 10 kDa. In contrast, venom of the scutigeromorph species Thereuopoda longicornis contains only 18 toxin families of primarily high molecular weight, and it lacks the peptide-toxin diversity found in scolopendrid venoms. This lack of venom complexity is surprising, especially compared with the remarkable toxin diversity seen in other venomous ambush predators, such as spiders and scorpions, and it seems inconsistent with the seemingly high dependence of T. longicornis on its venom for prey capture.
The answer to this conundrum may lie in the organization of the centipede venom gland, which is thought to have evolved from an invagination of the cuticle into a duct and associated weaponization of the cuticular dermal glands (20–23). The venom gland is composed of discrete subglands, or secretory units, each consisting of only three to four cells. These secretory units are individually connected to the lumen through a one-way valve formed by the distal canal cell that penetrates the chitinous duct through a pore. This simple architecture means that the number of secretory units (and secretory cells) present in centipede venom glands depends on the length of the porous part of the venom duct, termed the calyx (24). Interestingly, whereas the calyx only extends for ∼20% of the length of the venom gland in most centipedes, scolopendrids are unique in that they have evolved a calyx that spans nearly the entire length of the venom gland, resulting in a 20-fold increase in the number of secretory units present in a single gland (20, 25). We therefore hypothesized that the difference in toxin diversity between T. longicornis and the scolopendrid species could be because of differences in venom gland complexity resulting in differential morphological constraints on venom evolution.
Here we show that the discrepancy in venom complexity between scutigeromorph and scolopendrids could indeed be a result of the different morphological attributes of their venom glands. We show that the venom system of T. longicornis, which had not previously been studied in detail, is well developed and occupies a significant portion of the forcipule. Furthermore, the forcipules lack compensating structural morphological adaptations, indicating that venom plays an important role in the ecology of this species despite lacking the complexity of scolopendrid centipedes. Thus, our results suggest that centipede venom evolution may be constrained by the morphology of the venom gland. Moreover, our data support the hypothesis that, in contrast with other venomous arthropods, the high level of toxin diversity found in scolopendrids is a highly derived trait in centipedes.
Results
Comparative Morphology of Forcipules.
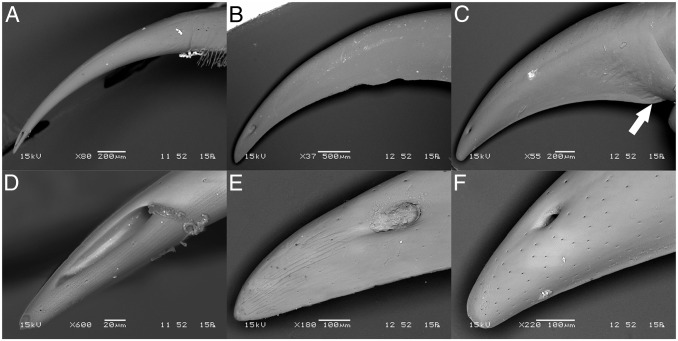
Three centipede species were selected to provide a comparison between Scutigeromorpha (T. longicornis) and Scolopendromorpha, with representatives of the latter chosen from two distinct scolopendrid subfamilies (Scolopendrinae: Scolopendra morsitans and Otostigminae: Ethmostigmus rubripes). Scanning electron microscopy (SEM) revealed marked structural differences between the shape of the forcipules in these three species (Fig. 1). In T. longicornis the apical claw is shaped like a hypodermic needle, bearing no sign of a cutting ridge or any adaptation to lateral movement during prey handling (Fig. 1A). In contrast, both scolopendrids possess a cutting edge reminiscent of a sickle (S. morsitans) or knife (E. rubripes) (Fig. 1 B and C). The evidence for structural adaptation of the cuticle to a cutting function is especially pronounced in the heavily chitinized forcipule of E. rubripes, where a structural transition is particularly evident between the cutting edge and the rest of the forcipular cuticle (Fig. 1C). The physical role of scolopendrid forcipules is also apparent from the position of the pore through which venom is secreted. Although the pore is very close to the tip of the forcipule in T. longicornis, it is located further back in both scolopendrids, possibly to improve the mechanical strength of the tip. Despite having more slender forcipules, the pore is much longer in T. longicornis (∼80 µm) than in the scolopendrid species (∼50 µm) (Fig. 1 D–F). Although the longer pore in T. longicornis could adversely impact the mechanical strength of the forcipular tip, it might also improve diffusion of venom into prey and prevent obstruction of the pore when venom is delivered via a straight stabbing motion.
Fig. 1.
Morphology of the apical claws of E. rubripes, S. morsitans, and T. longicornis. Backscattered electron SEM micrographs of the forcipule apical claw (A–C) and tip of apical claw (D–F) of (A and D) T. longicornis, (B and E) S. morsitans, and (C and F) E. rubripes. Arrow indicates beginning of blade-like structure on the inner curvature of the proximal part of the forcipule apical claw.
Because of morphological differences of the apical claws, we also investigated the presence of other potential structural adaptations. Among these, incorporation of heavy-metal ions is often associated with cuticular structural adaptations to mechanical wear in arthropods (26, 27). Although zinc was previously found in the forcipule of an unknown scolopendromorph centipede (27), no areas of increased brightness indicative of enriched metal concentration were observed in our backscattered SEM micrographs (Fig. 1) (26). Consistent with this observation, energy dispersive spectroscopy (EDS) did not provide conclusive evidence of zinc incorporation in the species we studied (Fig. S1). Thus, metal composition does not appear to be a good indicator of cuticular structure-function relationships in centipedes. EDS did reveal low levels of bromine in the apical claw of the species we studied (Fig. S1). The structural implications of this observation remain unclear as halogen occurrence in the absence of metals does not seem to correlate with harder or stiffer cuticle (28).
Comparative Morphology of Venom Glands.
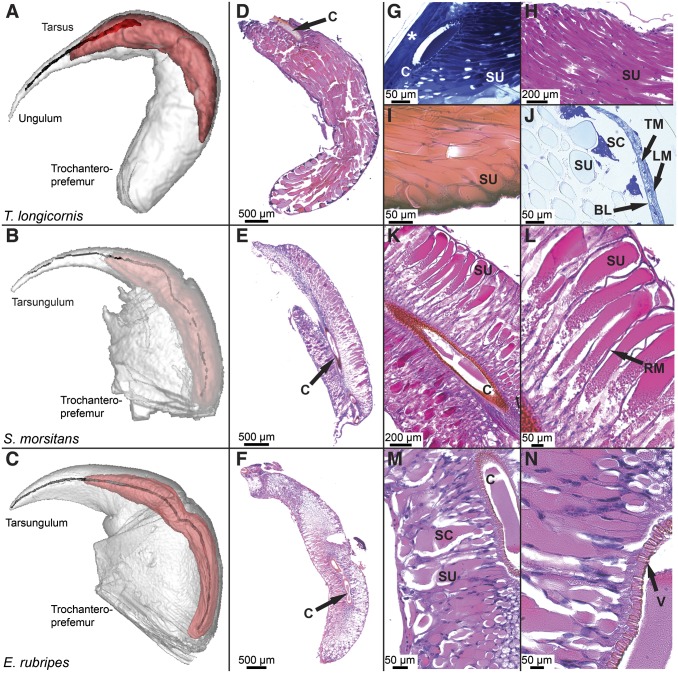
MRI reconstruction of forcipules (Fig. 2 A–C) showed that the venom glands of all species are elongate structures that run from the apical claw, along the outer curvature of the forcipule, and terminate in the proximal segment of the forcipule, the trochanteroprefemur. Although the venom glands of both scolopendrids terminate near the coxal segment of the forcipule, the venom gland in T. longicornis terminates approximately midway along the trochanteroprefemur. Nevertheless, estimating the volume occupied by each reconstructed layer revealed that the venom gland occupies about 21% of the total volume of the forcipule in T. longicornis. Similarly, the venom gland makes up about 20% of the forcipule in S. morsitans, but only about 7% in E. rubripes. Along with the morphological and structural adaptations of the apical claw, it therefore appears that E. rubripes is more adapted to physical use of its venom apparatus than either T. longicornis or S. morsitans.
Fig. 2.
Comparative morphology of the venom apparatus of T. longicornis (Top), S. morsitans (Middle), and E. rubripes (Bottom). (A–C) Three-dimensional MRI reconstruction showing forcipule (white), venom gland (red), and chitinous duct and calyx. (D–F) Low-magnification overview of transverse sections of venom glands stained with H&E (D and F) or PAS (E) showing differences in the arrangement of secretory units in relation to the calyx. (G–J) High-magnification image showing organization of secretory units in the venom gland of T. longicornis: (G) Toluidine blue-stained diagonal section of intact forcipule showing attachment of the calyx to the cuticle (asterisk) and tightly packed secretory units (SU) connected to the lumen through pores in the calyx (C). (H) Jones-PAS stained transverse section displaying secretory units lined with darkly stained nuclei of intermediary cells. (I) Eosin-stained section showing point of termination at the basal lamina and wide diameter of each secretory unit. (J) Alcian yellow-stained section of same region with darkly stained secretory cells (SC), basal lamina (BL), transverse muscle (TM), and longitudinal muscle (LM) layers. (K–N) Magnified sections of E and F showing details of venom gland organization in S. morsitans and E. rubripes along with (L) the presence of radial muscle fibers (RM) and (N) the valve-like structure (V) connecting the secretory unit to the lumen.
Although each secretory unit in the venom glands of all three centipede species are comprised of the same type of cells (20), the organization of these units in the glands of the three species is noticeably different (Fig. 2 D–N). This difference results from the calyx occupying only ∼20% of the length of the venom gland in T. longicornis, compared with ∼90% in both scolopendrids. Consequently, the glands of E. rubripes and S. morsitans are composed of numerous short secretory units that project almost perpendicularly out from the calyx to the basal lamina. Secretory units are interspersed by striated muscle spanning between the calyx and the basal lamina, which is also wrapped in striated muscle. In T. longicornis, secretory units are highly elongate, pear-shaped structures that span up to the entire length of the gland and can be up to several millimeters long. Each unit is lined with intermediary cells and appears to terminate with a cell in which the nucleus is rich in heterochromatin, likely a secretory cell, at the basal lamina. Although the basal lamina surrounding the gland is wrapped in both transverse and longitudinal striated muscle fibers probably involved in venom secretion, there appears to be little supporting structure within the gland. We could not detect any differentiation between secretory units apart from a difference in length depending on the point of termination along the basal lamina.
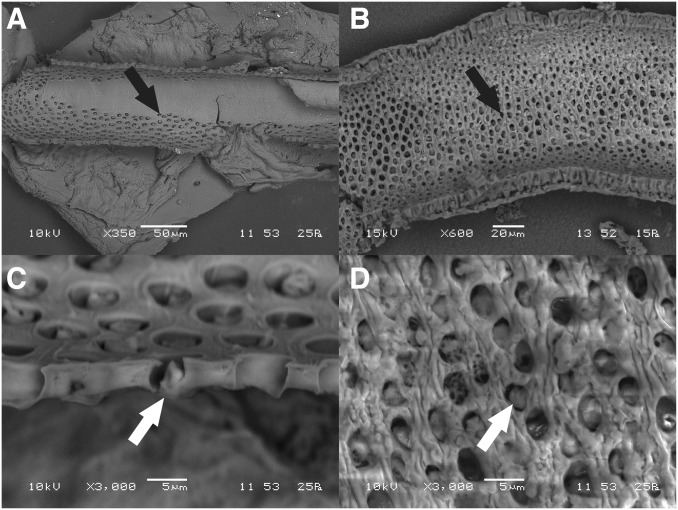
The differences in organization of the venom glands in the scutigerid and scolopendrid species are also evident in the morphologies of their calyxes. Because of the larger-sized secretory units of scutigerids, the pores leading into the lumen from each secretory unit are ∼5 µm in diameter in both S. morsitans and T. longicornis, despite the latter specimen being a smaller animal with a smaller venom gland and narrower calyx (Fig. 3). The density of pores is also higher in S. morsitans, resulting in a much larger overall number of secretory units in the gland. These results agree with those from the comparative study by Dugon and Arthur (20), and support the hypothesis that the high morphological complexity of scolopendrid venom glands may be unique among centipedes (19).
Fig. 3.
Inner structure of the calyx. SEM micrographs of the inner wall of the calyx of (A and C) T. longicornis and (B and D) S. morsitans, showing the density of pores (black arrow) and hence number of individual secretory units. Secretory units open into the lumen through these pores by a distal canal cell forming a one-way valve (white arrow), remnants of which can be seen in each of the high-magnification images (C and D). The transition from the nonporous duct to the porous calyx can be observed in A.
Imaging Mass Spectrometry of Venom Glands.
Although the morphological complexity is greater in scolopendrids than other centipedes, this only equates to greater biochemical venom complexity if there is heterogeneous production of venom components across secretory units. We therefore investigated the distribution of molecular ions in the venom glands by matrix-assisted laser desorption ionization (MALDI)-imaging mass spectrometry (IMS), which allows visualization of the spatial distribution of low molecular weight biomolecules across tissues without the need to extract or label samples. The IMS data revealed that peptide toxins are indeed nonuniformly distributed within the venom glands of centipedes (Figs. 4 and 5).
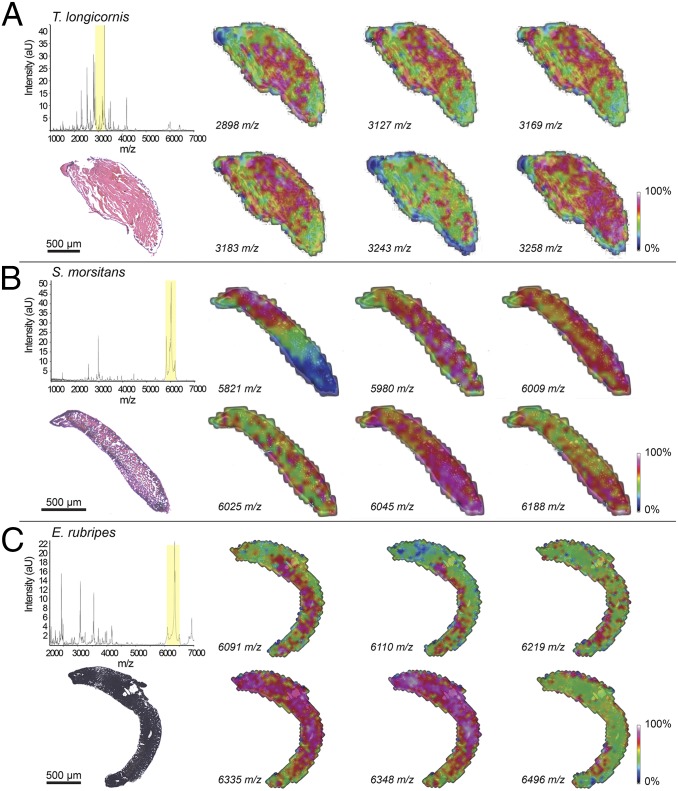
Fig. 4.
MALDI imaging of transverse sections of centipede venom glands. MALDI imaging of venom glands from (A) T. longicornis, (B) S. morsitans, and (C) E. rubripes oriented with top left pointing toward the claw of the forcipule. Normalized spectra averaged across all MS experiments for each section are shown in the left-most panels, above histological images of the sections that were used for MALDI imaging experiments (stained with H&E stain postimaging). The section displayed for E. rubripes was not stained and therefore appears black. Intensities of extracted ions (±1 m/z) from normalized spectra corresponding to peaks within an ∼400 m/z range (indicated by yellow shading in the spectra) are shown as heat-maps across each section.
Fig. 5.
Distribution of secretory unit types in centipede venom glands. Groups of secretory units with representative MS spectra identified by AIC/pLSA calculations performed on IMS data obtained from linear positive analysis of T. longicornis (A) and S. morsitans (B) venom-gland sections.
In T. longicornis, peptide ions could generally be detected in secretory units across the gland but often with highly variable intensities in different regions (Fig. 4A). In addition, several peptide masses were observed with high intensity in regions corresponding to single secretory units surrounded by low-intensity regions (e.g., 3,243 m/z in Fig. 4A). Although the limited spatial resolution of MALDI-IMS (∼50 μm) does not allow comparison between single secretory units, it nevertheless appears likely that there are substantial differences in venom composition even between adjacent secretory units.
A heterogeneous distribution of toxins was also observed in the venom glands of S. morsitans and E. rubripes (Fig. 4 B and C, respectively). Some ions were only found in the distal or proximal region of a section (e.g., 5,821 m/z in Fig. 4B), others were found throughout the section but at higher levels in certain regions (e.g., 6,188 m/z in Fig. 4B), and some ions had almost mutually exclusive distributions within the same region (e.g., 5,980 m/z and 6,025 m/z in Fig. 4B). Although less pronounced, similar evidence for localized toxin production could be observed in IMS experiments performed on E. rubripes sections (e.g., 6,335 m/z and 6,348 m/z in Fig. 4C). Again, there were also patterns of highly localized points of high intensity with some ions showing almost mutually exclusive distributions within the same region of the gland (e.g., 6,091 m/z and 6,219 m/z in Fig. 4C).
We did not detect any correlation between the variability in toxin abundance observed via IMS and our histological results. However, the uneven distribution of masses across the glands strongly suggests that peptides are heterogeneously expressed both between and within different regions of the venom gland. An Akaike information criterion (AIC) calculation identified four distinct regions in the glands of all species. Subsequent probabilistic latent semantic analyses (pLSA) revealed that these groups were particularly spatially distinct in the scolopendrid glands, presumably as a result of the extended calyx (Fig. 5). Representative spectra of the four groups in S. morsitans showed that they are all dominated by high-intensity ions corresponding to the mass range of just a few toxin families (Fig. 5B). This finding suggests that any single secretory unit or unit-type only produces a limited number of peptides within a gene family in high abundance. Hence, the diversity within toxin families observed in the venom is likely to result from differential production across multiple secretory units.
It should be noted that although the mass spectra from the venom gland of T. longicornis appears more complex than either of the scolopendrid species, MALDI is prone to ion suppression from competition in the laser source by highly abundant and ionizable components. The relatively higher abundance of detectable ions in the scolopendrid venom glands (19) therefore means that suppression effects are likely to be greater than in T. longicornis, leading to apparently less-diverse mass spectra.
Discussion
Centipedes first appeared over 430 Mya, and they are possibly the oldest venomous terrestrial taxon (23). The split between the scutigeromorph and scolopendromorph ancestors probably occurred soon after appearance of the ancestral form, and hence this split represents one of the most ancient known divergences between animals with homologous venom apparatuses (29). Reflecting the long time span since this ancestral divergence, the venom glands of T. longicornis and the two scolopendrid species are dramatically different. Despite having a forcipule appearing to be primarily adapted for venom delivery, as opposed to inflicting physical damage, the venom gland of T. longicornis is relatively simple in terms of cellular organization. The gland contains only ∼1,000 individual secretory units that each seem to rely on just one secretory cell. In contrast, the venom glands of S. morsitans and E. rubripes are much more complex, containing 10- to 100-fold more secretory units, and hence secretory cells. It is particularly interesting that E. rubripes has a much more complex venom and venom gland than T. longicornis, despite the fact that it appears to have evolved morphological adaptations that facilitate more physical use of the forcipule.
Although histological examination of venom glands did not provide convincing evidence of specialization between secretory units, IMS revealed that low molecular weight (< 10 kDa) toxins are found at different levels of abundance across secretory units in the venom gland of all three species examined. This type of structurally heterogeneous toxin expression supports the hypothesis of Rosenberg and Hilken (22) that the centipede venom gland is essentially a composite gland of semiautonomous subglands. Our results suggest that each of these subglands express only a limited number of venom components in high abundance. The observed complexity of the secreted venom may therefore be because of the combined diversity of expressed toxin genes between secretory units. Thus, evolution of greater gland complexity, and thereby more secretory units, likely represents a morphological adaptation that facilitated biochemical diversification of the venom arsenal of centipedes.
The correlation between calyx length, number of secretory units, and venom complexity has implications for use of centipede venoms as a source of novel bioactive peptides (30, 31). The elongated calyx found in scolopendrids is a highly derived feature among centipedes that appears to be largely limited to this family (23). Furthermore, Dugon and Arthur (20) recently found that the calyx in two geophilomorph and lithobiomorph species contained even fewer pores than the scutigeromorph Scutigera coleoptrata. This finding suggests that relatively low peptide-toxin diversity with a predominance of high molecular weight components is an ancestral feature of centipede venoms. It appears, therefore, that scolopendrids may be unique among centipedes in having evolved venoms rich in disulfide-bridged toxins. Such molecules are potentially attractive for the development of therapeutics and agrochemicals (32, 33).
Interestingly, individual peptide masses were observed not only at variable intensities within distinct regions of the gland but also between different areas along the length of the gland (Figs. 4 and 5). This observation is compatible with compartmentalization of toxin production in venom glands, as has been previously reported for snakes, spiders, scorpions, and cone snails, where it has been proposed to enable biochemical modulation of secreted venom components (10, 34). Recently it was shown that fish- and mollusk-eating members of the venomous marine snail genus Conus use this compartmentalization to produce two pharmacologically and biochemically distinct secretions for defensive and predatory envenomations (35). Intriguingly, the components of each of the two secretion types were produced in distinct “defensive” and “predatory” sections of the venom duct, indicative of deliberate control over the composition of the secreted venom. Moreover, venom composition has been shown to vary during sequential predatory secretions by cone snails, indicating that these venomous animals can control not just the type of secretion (i.e., defensive or predatory) but also the biochemical composition within these venom categories (36).
Although modulation of venom secretion has yet to be demonstrated in centipedes, the identification, via AIC/pLSA analysis of IMS spectra, of distinct toxin-secreting regions along the length of the venom gland in scolopendrids is intriguing. Scolopendra subspinipes mutilans has been shown to change its predatory behavior depending on the amount of venom available in the gland (37). Furthermore, the potential presence of nerve endings in the vicinity of the pore caps through which venom is secreted into the lumen has been reported from scolopendromorph venom glands (20, 38). Coupled with the presence of both radial and peripheral muscle fibers, this suggests that biochemical venom modulation in scolopendrids may be under neuronal control. Although we cannot assign any function to the observed ions comprising the different AIC/pLSA groups, it is tempting to speculate that scolopendrid centipedes in particular may be able to control the secretion of defensive versus offensive venoms, or even the secretion of prey-specific toxins.
In conclusion, centipede venom evolution appears to be constrained by the morphology of the venom gland, with toxin diversity in the venom being a function of the number of secretory units. The observed disparity in venom complexity between scolopendrid and scutigeromorph centipedes likely stems from morphological limitations of the venom gland, with the scolopendrid venom gland having evolved to accommodate a more diverse toxin arsenal. Thus, the centipede venom apparatus might be a useful model system for gaining insight into the limitations placed on venom evolution by morphological constraints. Our results provide further support for the concept that toxins are generally not universally expressed throughout the glands of venomous animals, and they suggest that the link between ecology and toxin evolution is more complex than previously realized.
Materials and Methods
Specimen Collection.
E. rubripes and T. longicornis were purchased from Mini Beast Wildlife (www.minibeastwildlife.com.au), and S. morsitans were collected from the Darling Downs region, Queensland, Australia.
SEM and EDS.
SEM was used to observe the cuticular structures of the centipede venom apparatus. Incorporation of heavy-metal ions is often associated with structural adaptations to mechanical wear in the arthropod cuticle (27), potentially representing cuticular adaptations of forcipules for physical use in overpowering prey. The presence of metal ions can be detected by backscattered electron SEM as regions with high signal intensity, but this is not particularly sensitive and it does not provide elemental composition. We therefore also used EDS to detect incorporation of structural inorganic compounds into the cuticle. Whole forcipules were fixed in 4% (wt/vol) neutrally buffered formalin (NBF) for 48 h, washed in distilled water, and sequentially dehydrated in ethanol. Samples were desiccated under vacuum overnight, mounted on stubs, and examined with a JEOL JSM 6460LA scanning electron microscope operated at 8–15 kV in backscattered electron mode. Spot-based EDS was performed on uncoated samples using the same JEOL JSM 6460LA equipped with a MiniCup EDS detector and X-ray maps analyzed in JEOL Analysis Program. Spots were analyzed at the tip, along the cutting ridge, and the outer curvature of the forcipular (tars)ungulum (Fig. S1). To obtain details of the internal structure of the calyx, venom glands from S. morsitans and T. longicornis were dissected, macerated in 10 M KOH for 2 h, and the calyx cut in half before desiccation. Samples were mounted on stubs and platinum-coated with a Eiko IB-5 sputter coater, before examination using the same JEOL JSM 6460LA operated at 10–15 kV in secondary electron mode.
Magnetic Resonance Imaging.
MRI was used to obtain an accurate estimation of the relative volume of the forcipule occupied by the venom gland in each species. This method also provided a unique opportunity to observe the 3D shape of the venom glands without intrusive dissection or sectioning techniques. For fixation, specimens were anesthetized by CO2 for 30 min and the entire forcipular segment placed in NBF. Before imaging, NBF was removed by four 1-h washing steps in PBS, and then the sample was incubated overnight in 0.1% Magnevist (Bayer) in PBS. After removal of NBF, samples were submersed in perfluoro-ether Fomblin (Solvay Solexis) and placed under vacuum to prevent air artifacts. Imaging was performed on a 16.4 T (700 MHz) vertical 89-mm-bore system (Bruker BioSpin) using a Bruker Micro2.5 gradient system with a gradient sensitivity of 2.5 G⋅cm⋅A. Transmit/receive radiofrequency coils comprised either a 5-mm-diameter solenoid coil or 10-mm-quadrature birdcage resonator (M2M Imaging) depending on sample size. Bruker ParaVision 5.0 software was used for image acquisition, and anatomical images were acquired using a 3D FLASH (Fast Low Angle Shot) gradient echo sequence. Imaging parameters were: TR/TE = 40/8 ms, flip angle 20°, four to eight excitations. The field-of-view and matrix were varied to fit the individual samples with the resulting voxels ranging between 20- and 40-µm isotropic resolution. Total scan time was 8–15 h per sample, depending on size and resolution. MRI data were processed using Medical Imaging Processing, Analysis, and Visualization software v6.0.0 and 3D image segmentation, surface rendering and volumetric measurements of the glands were performed manually using ITK-SNAP.
Imaging Mass Spectrometry.
IMS was guided by published protocols (39, 40), but with sample preparation optimized for centipede venom glands as recently described (41). FlexControl 3.3 (Bruker) was used to operate an UltraFlex III TOF-TOF mass spectrometer (Bruker) in linear positive mode, with the range set to 1,000–9,000 m/z. A small laser size was chosen to achieve a spatial resolution of 50 µm, and matrix ion suppression was enabled up to 980 m/z. Individual IMS experiments were performed using FlexImaging 4.0 (Bruker). FlexImaging was used to establish the geometry and location of the section on the slide based upon the optical image, choose the spatial resolution, and call upon FlexControl to acquire individual spectra, accumulating 200 shots per raster point. FlexImaging was subsequently used to visualize the data in 2D ion-intensity maps, producing an averaged spectrum based upon the normalized individual spectra collected during the experiment. After IMS experiments, 100% methanol was used to wash off the matrix and selected slides were subjected to histological examination.
pLSA, as incorporated in ClinProTools 3.0 (Bruker) and SCiLS Lab (SCiLS), was used to assign spectra to regions. However, histological examination was unable to distinguish morphologically distinct types of secretory units within the venom glands. The number of groups specified for the analyses were therefore derived from an AIC calculation as incorporated in ClinProTools.
Histology.
Histology was used to investigate the general structure of venom glands and potentially account for any differences observed by IMS. Of the fixatives trialed (NBF, Bouin’s fixative, and RCL2) the best results were obtained by fixation in 50% (vol/vol) RCL2/ethanol using the same protocol as per IMS tissue preparation (41). H&E-stained heterochromatin and ribonucleoprotein blue, whereas connective tissue, muscle, cytoplasm, and venom were stained various shades of red. To complement this staining, we used Periodic Acid-Schiff (PAS) with hematoxylin to visualize substances rich in carbohydrates (pink to magenta) and nuclei (dark blue). A Ventana Special Stains automated slide stainer (Ventana Medical Systems, Roche) was also used to stain slides with Alcian Blue PAS pH 2.5 (860-002), Alcian yellow (860-017), Jones H&E (860-019), and Jones PAS (860-014). These stains were used to visualize basement membranes (Jones), mucins (Alcian yellow), as well as acid mucosubstances and sialic acid-containing glycoproteins (Alcian blue PAS).
Given that part of the venom duct of T. longicornis is fused to the cuticle at the point where the calyx begins, it is difficult to dissect the gland and retain the intact structure of this region of the gland. We therefore also embedded whole forcipules of T. longicornis in LR White resin. The 4% NBF-fixed samples were dehydrated with ethanol in a series of three 1-h steps before immersion in 50% (vol/vol) resin/ethanol overnight. Samples were then infiltrated twice with 100% resin for 4 h before curing in fresh resin at 60 °C for 24 h. Sections were cut at 5-µm thickness and stained with Toluidine blue.
Supplementary Material
Acknowledgments
We thank Hanne H. Thoen, Ryan Hodgson, James Curran, and Ole H. Falck for help with collection of Scolopendra morsitans; Richard and Janine Pechey and Marco Inserra for permission to collect on their properties; the Queensland NMR Network and the National Imaging Facility for instrument access and technical support; and the reviewers of this manuscript for their insightful comments and constructive feedback. This study was supported in part by Australian Research Council Grant DP130103813 (to G.F.K.) and Australian National Health & Medical Research Council Principal Research Fellowship (to G.F.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424068112/-/DCSupplemental.
References
- 1.Escoubas P, Sollod B, King GF. Venom landscapes: Mining the complexity of spider venoms via a combined cDNA and mass spectrometric approach. Toxicon. 2006;47(6):650–663. doi: 10.1016/j.toxicon.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 2.Nascimento DG, et al. Moving pieces in a taxonomic puzzle: Venom 2D-LC/MS and data clustering analyses to infer phylogenetic relationships in some scorpions from the Buthidae family (Scorpiones) Toxicon. 2006;47(6):628–639. doi: 10.1016/j.toxicon.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 3.Casewell NR, Wüster W, Vonk FJ, Harrison RA, Fry BG. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol Evol. 2013;28(4):219–229. doi: 10.1016/j.tree.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 4.Fry BG, et al. The toxicogenomic multiverse: Convergent recruitment of proteins into animal venoms. Annu Rev Genomics Hum Genet. 2009;10:483–511. doi: 10.1146/annurev.genom.9.081307.164356. [DOI] [PubMed] [Google Scholar]
- 5.Brust A, et al. Differential evolution and neofunctionalization of snake venom metalloprotease domains. Mol Cell Proteomics. 2013;12(3):651–663. doi: 10.1074/mcp.M112.023135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fry BG, et al. Molecular evolution and phylogeny of elapid snake venom three-finger toxins. J Mol Evol. 2003;57(1):110–129. doi: 10.1007/s00239-003-2461-2. [DOI] [PubMed] [Google Scholar]
- 7.Kordiš D, Gubenšek F. Adaptive evolution of animal toxin multigene families. Gene. 2000;261(1):43–52. doi: 10.1016/s0378-1119(00)00490-x. [DOI] [PubMed] [Google Scholar]
- 8.Ruder T, et al. Molecular phylogeny and evolution of the proteins encoded by coleoid (cuttlefish, octopus, and squid) posterior venom glands. J Mol Evol. 2013;76(4):192–204. doi: 10.1007/s00239-013-9552-5. [DOI] [PubMed] [Google Scholar]
- 9.Weinberger H, et al. Positions under positive selection—Key for selectivity and potency of scorpion α-toxins. Mol Biol Evol. 2010;27(5):1025–1034. doi: 10.1093/molbev/msp310. [DOI] [PubMed] [Google Scholar]
- 10.Morgenstern D, King GF. The venom optimization hypothesis revisited. Toxicon. 2013;63:120–128. doi: 10.1016/j.toxicon.2012.11.022. [DOI] [PubMed] [Google Scholar]
- 11.Casper GS. Prey capture and stinging behavior in the Emperor Scorpion, Pandinus imperator (Koch) (Scorpiones, Scorpionidae) J Arachnol. 1985;13(3):277–283. [Google Scholar]
- 12.Fry BG, et al. The structural and functional diversification of the Toxicofera reptile venom system. Toxicon. 2012;60(4):434–448. doi: 10.1016/j.toxicon.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Fry BG, et al. Evolution of an arsenal: Structural and functional diversification of the venom system in the advanced snakes (Caenophidia) Mol Cell Proteomics. 2008;7(2):215–246. doi: 10.1074/mcp.M700094-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Fry BG, et al. Squeezers and leaf-cutters: Differential diversification and degeneration of the venom system in toxicoferan reptiles. Mol Cell Proteomics. 2013;12(7):1881–1899. doi: 10.1074/mcp.M112.023143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vidal N. Colubroid systematics: Evidence for an early appearance of the venom apparatus followed by extensive evolutionary tinkering. J Toxicol Toxin Rev. 2002;21(1-2):21–41. [Google Scholar]
- 16.Edgecombe GD, Giribet G. Evolutionary biology of centipedes (Myriapoda: Chilopoda) Annu Rev Entomol. 2007;52(1):151–170. doi: 10.1146/annurev.ento.52.110405.091326. [DOI] [PubMed] [Google Scholar]
- 17.Lewis JGE. The Biology of Centipedes. Cambridge Univ Press; Cambridge, UK: 1981. [Google Scholar]
- 18.Dugon MM, Black A, Arthur W. Variation and specialisation of the forcipular apparatus of centipedes (Arthropoda: Chilopoda): A comparative morphometric and microscopic investigation of an evolutionary novelty. Arthropod Struct Dev. 2012;41(3):231–243. doi: 10.1016/j.asd.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 19.Undheim EAB, et al. Clawing through evolution: Toxin diversification and convergence in the ancient lineage Chilopoda (centipedes) Mol Biol Evol. 2014;31(8):2124–2148. doi: 10.1093/molbev/msu162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dugon MM, Arthur W. Comparative studies on the structure and development of the venom-delivery system of centipedes, and a hypothesis on the origin of this evolutionary novelty. Evol Dev. 2012;14(1):128–137. doi: 10.1111/j.1525-142X.2011.00527.x. [DOI] [PubMed] [Google Scholar]
- 21.Dugon MM, Hayden L, Black A, Arthur W. Development of the venom ducts in the centipede Scolopendra: An example of recapitulation. Evol Dev. 2012;14(6):515–521. doi: 10.1111/ede.12004. [DOI] [PubMed] [Google Scholar]
- 22.Rosenberg J, Hilken G. Fine structural organization of the poison gland of Lithobius forficatus (Chilopoda, Lithobiomorpha) Nor J Entomol. 2006;53(2):119–127. [Google Scholar]
- 23.Undheim EAB, King GF. On the venom system of centipedes (Chilopoda), a neglected group of venomous animals. Toxicon. 2011;57(4):512–524. doi: 10.1016/j.toxicon.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Bonato L, et al. A common terminology for the external anatomy of centipedes (Chilopoda) Zookeys. 2010;69(69):17–51. doi: 10.3897/zookeys.69.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chao JL, Chang HW. Variation of the poison duct in Chilopoda centipedes from Taiwan. Nor J Entomol. 2006;53(2):139–151. [Google Scholar]
- 26.Cribb BW, et al. Insect mandibles—Comparative mechanical properties and links with metal incorporation. Naturwissenschaften. 2008;95(1):17–23. doi: 10.1007/s00114-007-0288-1. [DOI] [PubMed] [Google Scholar]
- 27.Schofield RSM. Metals in cuticular structures. In: Brownell P, Polis G, editors. Scorpion Biology and Research. Oxford Univ Press; New York: 2001. pp. 234–256. [Google Scholar]
- 28.Cribb BW, et al. Structure, composition and properties of naturally occurring non-calcified crustacean cuticle. Arthropod Struct Dev. 2009;38(3):173–178. doi: 10.1016/j.asd.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Fernández R, et al. Evaluating topological conflict in centipede phylogeny using transcriptomic data sets. Mol Biol Evol. 2014;31(6):1500–1513. doi: 10.1093/molbev/msu108. [DOI] [PubMed] [Google Scholar]
- 30.Yang S, et al. Chemical punch packed in venoms makes centipedes excellent predators. Mol Cell Proteomics. 2012;11(9):640–650. doi: 10.1074/mcp.M112.018853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang S, et al. Discovery of a selective NaV1.7 inhibitor from centipede venom with analgesic efficacy exceeding morphine in rodent pain models. Proc Natl Acad Sci USA. 2013;110(43):17534–17539. doi: 10.1073/pnas.1306285110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King GF. Venoms as a platform for human drugs: Translating toxins into therapeutics. Expert Opin Biol Ther. 2011;11(11):1469–1484. doi: 10.1517/14712598.2011.621940. [DOI] [PubMed] [Google Scholar]
- 33.King GF, Hardy MC. Spider-venom peptides: Structure, pharmacology, and potential for control of insect pests. Annu Rev Entomol. 2013;58:475–496. doi: 10.1146/annurev-ento-120811-153650. [DOI] [PubMed] [Google Scholar]
- 34.Hu H, Bandyopadhyay PK, Olivera BM, Yandell M. Elucidation of the molecular envenomation strategy of the cone snail Conus geographus through transcriptome sequencing of its venom duct. BMC Genomics. 2012;13(1):284. doi: 10.1186/1471-2164-13-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dutertre S, et al. Evolution of separate predation- and defence-evoked venoms in carnivorous cone snails. Nat Commun. 2014;5:3521. doi: 10.1038/ncomms4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prator CA, Murayama KM, Schulz JR. Venom variation during prey capture by the cone snail, Conus textile. PLoS ONE. 2014;9(6):e98991. doi: 10.1371/journal.pone.0098991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dugon MM, Arthur W. Prey orientation and the role of venom availability in the predatory behaviour of the centipede Scolopendra subspinipes mutilans (Arthropoda: Chilopoda) J Insect Physiol. 2012;58(6):874–880. doi: 10.1016/j.jinsphys.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 38.Antoniazzi MM, et al. Comparative morphological study of the venom glands of the centipede Cryptops iheringi, Otostigmus pradoi and Scolopendra viridicornis. Toxicon. 2009;53(3):367–374. doi: 10.1016/j.toxicon.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 39.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: Localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69(23):4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 40.Yarnold JE, et al. High resolution spatial mapping of brominated pyrrole-2-aminoimidazole alkaloids distributions in the marine sponge Stylissa flabellata via MALDI-mass spectrometry imaging. Mol Biosyst. 2012;8(9):2249–2259. doi: 10.1039/c2mb25152c. [DOI] [PubMed] [Google Scholar]
- 41.Undheim EAB, et al. Multifunctional warheads: Diversification of the toxin arsenal of centipedes via novel multidomain transcripts. J Proteomics. 2014;102:1–10. doi: 10.1016/j.jprot.2014.02.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.