Significance
Protein lysine methyl transferases and demethylases, previously identified for histone modification, now are known to modify several nonhistone proteins as well. Their deregulation leads to the development and progression of various diseases, including cancer. Thus these enzymes now stand out as attractive therapeutic targets. We present a detailed study of STAT3 posttranslational modification at K49 by the histone methyl transferase, enhancer of zeste homolog 2 (EZH2). Dimethylation of K49 modulates IL-6–responsive transcription, revealing a previously unrecognized important functional modification of STAT3 activity. Because STAT3 is a major oncogene, and EZH2 function is modified in many cancers, this functional connection between the two raises the possibility that modulation of EZH2 activity might be useful in inhibiting the oncogenic activity of STAT3.
Keywords: posttranslational modification, histone methyltransferase, gene expression
Abstract
Several transcription factors, including p53, NF-κB, and STAT3, are modified by the same enzymes that also modify histones, with important functional consequences. We have identified a previously unrecognized dimethylation of K49 of STAT3 that is crucial for the expression of many IL-6–dependent genes, catalyzed by the histone-modifying enzyme enhancer of zeste homolog 2 (EZH2). Loss of EZH2 is protumorigenic in leukemias, but its overexpression is protumorigenic in solid cancers. Connecting EZH2 to a functionally important methylation of STAT3, which is constitutively activated in many tumors, may help reveal the basis of the opposing roles of EZH2 in liquid and solid tumors and also may identify novel therapeutic opportunities.
STAT3 is activated in 70% of all solid and hematological tumors (1, 2), where it stimulates proliferation, survival, angiogenesis, invasion, and tumor-promoting inflammation. Recently, STAT3 also was found to have an important role in maintaining cancer stem cells, both in vitro and in mouse tumor models, indicating that it is integrally involved in tumor initiation, progression, and maintenance (3). IL-6–induced constitutive activation of STAT3 was observed in neoplastic gastric tissue and is positively correlated with tumor progression (4), and the expression of tyrosine-phosphorylated STAT3 is associated with poor prognosis in colorectal cancer, independent of the mutation status of MSI, CIMP, BRAF, or KRAS (5). The STAT3 gene is rarely mutated in cancer but rather is activated by members of the IL-6 family of cytokines, receptor tyrosine kinases, mutated JAKs, or oncogenic cellular tyrosine kinases, such as SRC (6). Because STAT3 is constitutively activated during disease progression and metastasis, it is a promising therapeutic target. Even though transcription factors are difficult drug targets, accumulating evidence for the crucial roles of posttranslational modifications of transcription factors in mediating target gene expression provides an opportunity to develop drugs against the modifying enzymes rather than against the transcription factors themselves.
STAT3 is activated primarily by phosphorylation of Y705 (7) and secondarily by phosphorylation of S727 (8). The acetylation of STAT3 at K685 (9, 10) has been shown recently to have little or no effect on IL-6–dependent gene expression (11). Ray et al. (12) reported that IL-6–induced gene expression requires p300-mediated acetylation of K49 and K87 and that monoubiquitination at K97 is a key mediator of BRD4-dependent gene expression (13). Like NF-κB and p53, STAT3 is reversibly methylated on lysine residues by histone-modifying enzymes, with important functional consequences (14). Furthermore, STAT3 is known to be di- or trimethylated on K140 or K180 by the histone methyltransferase SET9 (SET domain containing lysine methyltransferase 9) or EZH2 (enhancer of zeste homolog 2), respectively (15, 16). Here we show that IL-6–dependent, previously unidentified, dimethylation on K49 is necessary for the expression of a major fraction of STAT3 target genes. Furthermore, the histone methyltransferase EZH2, which is highly expressed in several cancers, catalyzes this modification in response to IL-6.
Results
STAT3 Is Dimethylated on K49.
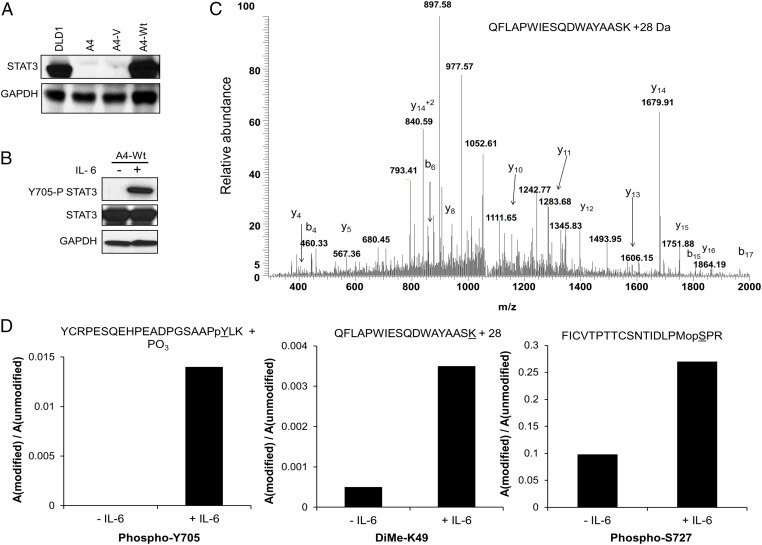
Flag-tagged WT STAT3 (WT-STAT3) was introduced into the STAT3-null human cell line A4 at a level comparable to that in parental DLD1 cells (Fig. 1A). Flag-tagged WT-STAT3, which responds normally to IL-6 (Fig. 1B), was purified from untreated or IL-6–treated cells and was analyzed by MS. In a survey MS experiment, a tryptic peptide containing K49 was observed, mostly in its unmodified form. A targeted liquid chromatography (LC)/MS analysis of the QFLAPWIESQDWAYAASK peptide was performed to look more critically for K49 modification (Fig. 1C). There were several sequence-specific ions in the spectra, including the abundant y14 ion, which confirms the addition of 28 Da of mass to the peptide, and the b17 ion, which is consistent with the unmodified QFLAPWIESQDWAYAAS portion of the peptide. The results indicate the addition of 28 mass units to K49, consistent with dimethylation, and reveal that the relative abundance of the modified peptide is increased in the IL-6–treated sample (Fig. 1D). The previously reported acetylation (12) of STAT3 at K49 was not detected in our samples by MS analysis.
Fig. 1.
STAT3 is dimethylated on K49. (A) STAT3-null A4 cells were infected with a retroviral construct expressing WT-STAT3, and stable pools of cells were selected with G418. The level of STAT3 expression, analyzed by the Western method, corresponds to that in the DLD1 parental cells. (B) A4-WT-STAT3 cells were treated with IL-6 (50 ng/mL) and soluble IL-6 receptor (sIL-6R) (62.5 ng/mL) for 1 h, and total cell lysates were analyzed for Y705 phosphorylation. (C) MS analysis of tryptic peptides of STAT3 indicates that K49 is dimethylated. Flag-tagged WT-STAT3 was immunoprecipitated from untreated or IL-6–treated A4-WT-STAT3 cells. The tryptic K49 peptide QFLAPWIESQDWAYAASK was included in the targeted analysis. A peptide consistent with a 28-Da mass modification of K49 was identified. (D) The relative abundance of the modified form of the K49 peptide was greater in the IL-6–treated samples. The levels of the modified forms of the peptides containing Y705, K49, and S727 were analyzed.
Mutation of K49 Impairs STAT3 Activation in Response to IL-6.
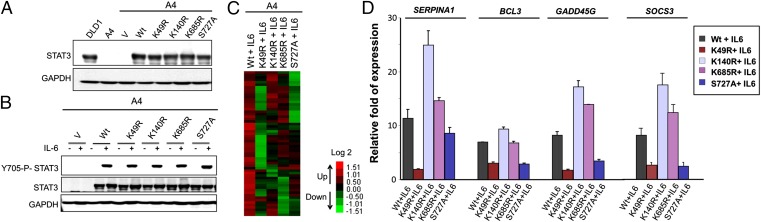
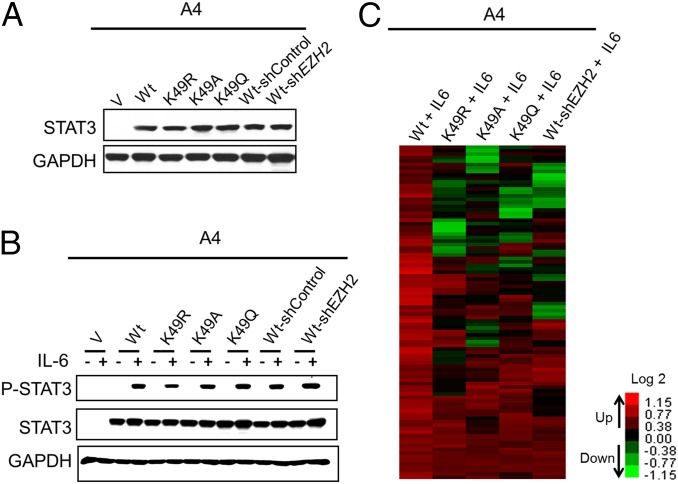
To determine the role of K49 dimethylation of STAT3 on gene expression in response to IL-6, we stably expressed the K49R mutant in A4 cells at a level comparable to that of WT-STAT3 (Fig. 2A). To compare the differential effect of K49 methylation of STAT3 with other previously reported STAT3 posttranslational modifications that affect IL-6–dependent gene expression, we included the K140R, K685R, and S727A mutations (8–11, 15) in the same experiment (Fig. 2A). A4 cells expressing WT-STAT3 or each of the four mutant proteins were treated with IL-6 for 4 h. Western analysis revealed that none of the mutations inhibit the phosphorylation of Y705 (Fig. 2B). A microarray analysis then was performed to identify genes whose expression was increased in response to treatment with IL-6. Of the 47,321 probes on the array, 62 (representing 59 genes) were up-regulated by WT-STAT3 in response IL-6. The expression of 33 of these 59 genes was impaired twofold or more by the K49R mutation, and the S727A, K140R, and K685R mutations reduced the expression of 23, 19, and 18 genes, respectively (Fig. 2C and Table S1). The inhibitory effect of the K49R mutation on the IL-6–induced expression of the BCL3, SERPINA1, GADD45G, and SOCS3 mRNAs was confirmed by real-time PCR (Fig. 2D). We conclude that the K49R mutation impairs the expression of a major subset of IL-6–induced STAT3-dependent genes.
Fig. 2.
K49 mutation of STAT3 impairs its transcriptional activity in response to IL-6. (A) A4 cells were infected with retroviral constructs expressing WT-STAT3 or the STAT3 mutants K49R, K140R, K685R, or S727A, at levels comparable to the level of WT-STAT3 in DLD1 parental cells. Stable pools of cells were selected with G418. Western analyses of total cell lysates are shown. (B) A4-vector control (A4-V), A4-WT, A4-K49R, A4-K140R, A4-K685R, and A4-S727A cells were treated with IL-6 (50 ng/mL) and sIL-6R (62.5 ng/mL) for 4 h, and total cell lysates were analyzed for Y705 phosphorylation. (C) A microarray analysis was performed to identify genes induced in WT-STAT3 and the mutant STAT3 cells upon IL-6 treatment. The heat map shows the induced expression of 59 genes (the full list is given in Table S1). We include only signals that changed by twofold or more, with differential P ≤ 0.05, in IL-6–treated WT-STAT3 cells compared with IL-6–treated vector control cells and untreated WT-STAT3 cells and with average signals >30 in WT cells treated with IL-6. (D) WT-STAT3 cells and A4 cells carrying each of the mutant proteins were treated with IL-6 (50 ng/mL) and sIL-6R (62.5 ng/mL) for 4 h, and the relative expression of the BCL3, SERPINA1, GADD45G, and SOCS3 genes, compared with the untreated cells, was determined by quantitative PCR (qRCR). Values are shown as means, with SDs, from triplicate experiments.
Y705 Phosphorylation Is Required for Dimethylation of STAT3.
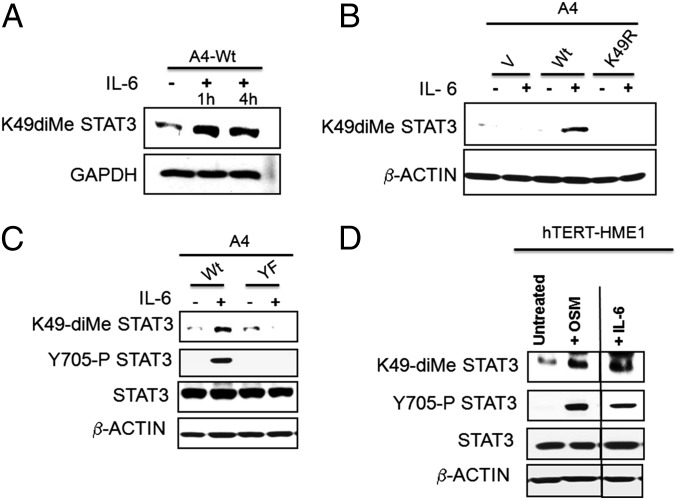
A rabbit polyclonal antibody raised against a STAT3 peptide carrying dimethyl K49 was used to detect K49 dimethylation in response to IL-6. The specificity of the generated polyclonal antibody is confirmed as shown in Fig. S1. Although a basal level of dimethylation was seen in untreated cells (also seen in the MS analysis of Fig. 1C), the level was increased substantially in response to IL-6 (Fig. 3A). As expected, cells expressing the K49R mutant failed to show IL-6–dependent dimethylation of K49 (Fig. 3B). Moreover, Fig. 3C shows that Y705 phosphorylation is necessary for the IL-6–induced increase in K49 dimethylation. To explore the dependence of K49 dimethylation in another cell line and in response to a different STAT3-activating ligand, we treated the nontumorigenic human mammary epithelial cell line hTERT-HME1 with oncostatin M (OSM) or IL-6. Y705 was phosphorylated and K49 was dimethylated in response to either IL-6 or OSM (Fig. 3D).
Fig. 3.
STAT3 Y705 phosphorylation is required for K49 dimethylation of STAT3. (A) WT-STAT3 cells were treated with IL-6 (50 ng/mL) and sIL-6R (62.5 ng/mL) for 1 or 4 h, respectively, and total cell lysates were analyzed for K49 dimethylation by using an antibody that recognizes this specific modification. (B) A4 cells expressing WT or K49R STAT3 were treated with IL-6 (50 ng/mL) and sIL-6R (62.5 ng/mL) for 1 h, and total cell lysates were analyzed for K49 dimethylation. (C) A4 cells expressing WT or Y705F STAT3 were treated with IL-6 (50 ng/mL) and sIL-6R (62.5 ng/mL) for 1 h, and total cell lysates were analyzed for K49 dimethylation and Y705 phosphorylation. (D) hTERT-HME1 cells were treated with OSM (100 ng/mL) or IL-6 (50 ng/mL) and sIL-6R (62.5 ng/mL) for 1 h, and total cell lysates were analyzed for K49 dimethylation, Y705 phosphorylation, and total STAT3.
STAT3 Is Methylated at K49 by the Histone Methyltransferase EZH2.
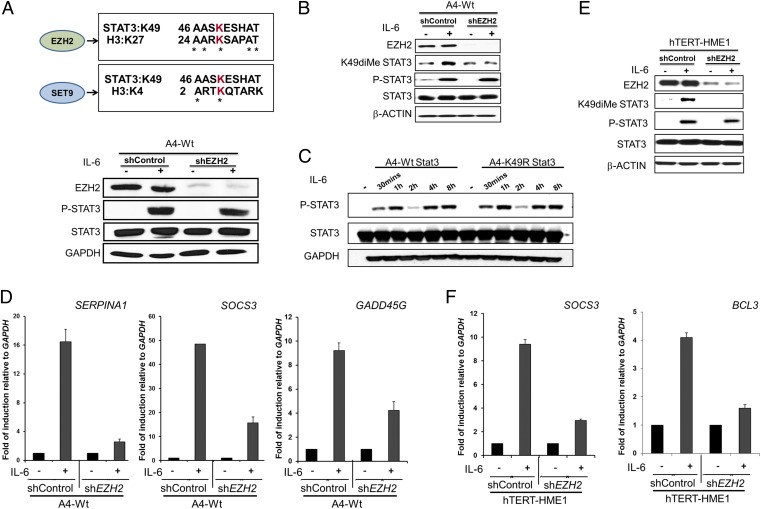
Several histone methyltransferases have been shown to modify transcription factors and thereby significantly affect their ability to induce gene expression. We investigated the possible involvement of two enzymes that previously have been shown to methylate different residues of STAT3, namely SET9, which methylates K140 (15), and EZH2, which methylates K180 (16), in K49 dimethylation. A comparison of histone target sequences for EZH2 revealed homology with the STAT3 sequence surrounding K49, in which five of nine residues are identical with the EZH2 site on histone 3, but no such homology was seen for SET9 (Fig. 4A). When we knocked down the expression of EZH2 in A4 cells expressing WT-STAT3, there was no effect on the tyrosine phosphorylation of STAT3 in response to IL-6 (Fig. 4A), but EZH2 knockdown prevented the dimethylation of K49 of STAT3 in response to IL-6 (Fig. 4B). Similarly, it is evident from a kinetic analysis (Fig. 4C) that K49 mutation does not affect Y705 phosphorylation of STAT3 following IL-6 treatment. The biphasic pattern of Y705 phosphorylation of STAT3 in Fig. 4C is in accord with our previous observations (17). The IL-6–induced expression of three genes whose expression is inhibited by the K49R mutation, SERPINA1, SOCS3, and GADD45G, also was inhibited when EZH2 was knocked down (compare Fig. 2D and Fig. 4D). Kim et al. (16) reported that EZH2 is required for the trimethylation of K180 of STAT3 in glioblastoma stem cells, and they also showed that mutation of K180 strongly inhibited the phosphorylation of Y705. However, in the colon tumor A4 cells that we have studied, MS analysis did not reveal any methylation of K180, and, as shown in Fig. 4B, EZH2 knockdown did not inhibit Y705 phosphorylation following treatment with IL-6. We attribute these different results to differences in the cells analyzed. To confirm the involvement of EZH2 in K49 dimethylation in a nontumorigenic cell line, we knocked down its expression in hTERT-HME1 cells, treated the cells with IL-6, and assayed the induction of two STAT3-dependent genes. In this experiment, EZH2 knockdown impaired both the dimethylation of K49 in response to IL-6 (Fig. 4E) and the induced expression of SOCS3 and BCL3 (Fig. 4F). In summary, we demonstrate that EZH2 is responsible for the IL-6–dependent dimethylation of K49 of STAT3 and that the loss of EZH2 greatly impairs IL-6–dependent STAT3-mediated gene expression.
Fig. 4.
STAT3 is dimethylated on K49 by EZH2. (A) The sequence of STAT3 surrounding K49 is aligned against the EZH2 target sequence surrounding H3 K27 and the SET9 target sequence surrounding H3 K4. Knockdown of EZH2 in A4 cells expressing WT-STAT3 was confirmed by Western analysis. To determine whether EZH2 knockdown impairs STAT3 Y705 phosphorylation, cells with or without EZH2 knockdown were treated with IL-6 (50 ng/mL) and sIL-6R (62.5 ng/mL) for 1 h and were analyzed for Y705 phosphorylation. (B) EZH2-knockdown and control WT-STAT3 cells were treated with IL-6 (50 ng/mL) and sIL-6R (62.5 ng/mL) for 4 h and were analyzed for K49 dimethylation, Y705 phosphorylation, and total STAT3. (C) WT-STAT3 and K49R-STAT3 cells were treated with IL-6 (50 ng/mL) and sIL-6R (62.5 ng/mL), and Y705 phosphorylation of STAT3 was determined. (D) WT-STAT3 cells, with or without EZH2 knockdown, were treated with IL-6 (50 ng/mL) and sIL-6R (62.5 ng/mL) for 4 h, and the relative expression of the SERPINA1, SOCS3, and GADD45G genes was determined by qPCR. Values are shown as the means, with SDs, from triplicate experiments. (E) EZH2-knockdown and control hTERT-HME1 cells were treated with IL-6 (50 ng/mL) and sIL-6R (62.5 ng/mL) for 4 h and were analyzed for EZH2 knockdown, K49 dimethylation, Y705 phosphorylation, and total STAT3 expression. (F) hTERT-HME1 cells, with or without EZH2 knockdown, were treated with IL-6 (50 ng/mL) and sIL-6R (62.5 ng/mL) for 4 h, and the relative expression of the SOCS3 and BCL3 genes was determined by qPCR. Values are shown as the means, with SDs, from triplicate experiments.
The Effects on IL-6–Induced Gene Expression of Mutating K49 of STAT3 or of Knocking Down EZH2 Are Similar.
In addition to the K49R mutant, the K49A and K49Q mutants of STAT3 were analyzed to minimize mutation-specific effects. Arginine is similar in size to lysine and has the same charge, whereas alanine is small and uncharged, and glutamine is similar in size to lysine and also is uncharged. The relative expression of all three mutant proteins in A4 cells was very similar (Fig. 5A), and the phosphorylation of Y705 following IL-6 treatment was virtually the same (Fig. 5B). The results of the array analysis are represented schematically in Table S2 and are illustrated in Figs. S2–S5.
Fig. 5.
EZH2 knockdown and K49 mutation of STAT3 in A4 cells have similar effects on IL-6–dependent, STAT3-mediated gene expression. (A) A4 cells expressing WT-STAT3 or the mutants K49R, K49A, or K49Q were analyzed for STAT3 expression. WT-STAT3 cells with or without EZH2 knockdown also were included in the analysis. (B) A4-vector control (V), WT, K49R, K49A, K49Q, WT-shControl, or WT-shEZH2 cells were treated with IL-6 (50 ng/mL) and sIL-6R (62.5 ng/mL) for 4 h, and total cell lysates were analyzed for Y705 phosphorylation. (C) Genes induced by IL-6 in WT-STAT3 cells, in A4 cells carrying the K49R, A, or Q mutants, and in WT-shEZH2 cells were compared by microarray analysis. The heat map shows the induced expression of 97 genes (the full list in given in Table S3). We include only signals that changed by twofold or more, with differential P ≤ 0.05, in IL-6–treated WT-STAT3 cells compared with IL-6–treated vector control cells and untreated WT-STAT3 cells and with average signals >30 in IL-6–treated WT-STAT3 cells.
Of the 34,694 genes on the arrays, 97 were up-regulated in response to IL-6 in cells expressing WT-STAT3 (Fig. 5C and Table S3). The expression of 57, 56, or 48 of these 97 genes was impaired by twofold or more for the K49R, K49A, or K49Q mutations, respectively (Fig. S2). Furthermore, the expression of 58 of the 97 genes induced by WT-STAT3 on IL-6 treatment was inhibited twofold or more when EZH2 was knocked down in these WT-STAT3 cells (Fig. S3). The expression of more than half of the 58 genes impaired in WT-STAT3 EZH2 knockdown cells also was impaired by the mutation of STAT3 K49 to arginine, alanine, or glutamine (Fig. S4). Pairwise comparisons of genes affected by each of the mutations are shown in Fig. S5. Importantly, the expression of 15 of the 20 genes whose expression was impaired by all three K49 mutants also was inhibited when EZH2 was knocked down.
STAT3 Binds to EZH2.
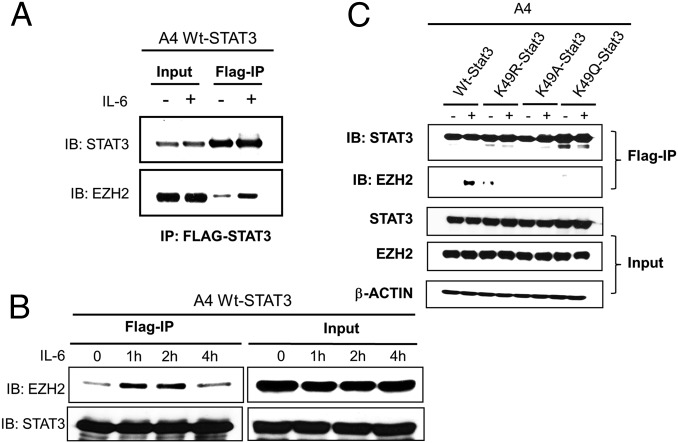
Kim et al. (16) showed that that EZH2 binds to STAT3 in glioblastoma cells. A coimmunoprecipitation experiment (Fig. 6A) demonstrates that WT-STAT3 also binds to EZH2 in A4 cells and that this interaction is increased substantially following treatment with IL-6. Furthermore, a kinetic analysis (Fig. 6B) shows that the binding of EZH2 to STAT3 increases by 1 h, is sustained at 2 h, but decreases to basal levels 4 h after treatment. We did not observe IL-6–induced binding of EZH2 to any of the K49 mutant proteins (Fig. 6C).
Fig. 6.
EZH2 binding to STAT3 is enhanced by IL-6 treatment in a time-dependent manner. (A) WT-STAT3 cells were treated with IL-6 (50 ng/mL) and sIL-6R (62.5 ng/mL) for 1 h, followed by immunoprecipitation of STAT3 from whole-cell extracts with anti-FLAG M2 beads. Western analyses were performed to detect EZH2. (B) WT-STAT3 cells were treated with IL-6 (50 ng/mL) and sIL-6R (62.5 ng/mL) for the indicated times, followed by immunoprecipitation of STAT3 from whole-cell extracts with anti-FLAG M2 beads. Western analyses were performed to detect the time dependence of EZH2 binding. (C) A4 cells expressing WT-STAT3 or one of the three K49 mutants were treated with IL-6 (50 ng/mL) and sIL-6R (62.5 ng/mL) for 1 h, followed by immunoprecipitation of STAT3 from whole-cell extracts with anti-FLAG M2 beads. Western analyses were performed to detect EZH2 binding.
Knockdown of EZH2 Has Little Effect on the Binding of STAT3 to the SOCS3 Promoter.
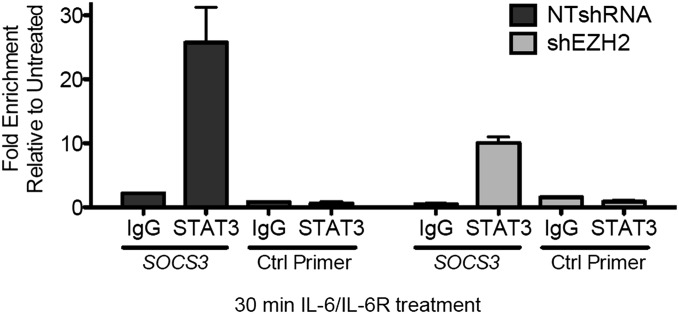
Stark et al. (14, 15) show that gene-specific inducible modifications of transcription factors occur at promoters, where the local chromatin machinery is active, and that these modifications can modulate the local functions of the transcription factors by altering their stability, transactivation potency, and affinity for DNA, in turn affecting the strength and duration of inducible gene expression. Therefore we tested whether EZH2, the enzyme responsible for K49 dimethylation of STAT3, inhibits STAT3 binding to the SOCS3 promoter. A ChIP analysis was performed with anti-STAT3 in IL-6–treated A4-WT-STAT3/shControl and A4-WT-STAT3/shEZH2 cells. The data in Fig. 7 show that, although about 2.5 times more STAT3 was bound to the SOCS3 promoter in the presence of EZH2 than in its absence, there still was substantial binding without EZH2. Because STAT3 occupancy of the SOCS3 promoter following treatment with IL-6 is impaired only slightly in the absence of EZH2, we hypothesize that the stimulatory effect of EZH2 on STAT3-dependent gene expression at susceptible promoters may be caused by events that follow STAT3 binding, for example, the recruitment of positively acting accessory factors.
Fig. 7.
Knockdown of EZH2 has little effect on the binding of STAT3 to the SOCS3 promoter. A4-WT-STAT3/shControl (NTshRNA) and A4-WT-STAT3/shEZH2 (shEXH2) cells were treated with IL-6 (50 ng/mL) and sIL-6R (62.5 ng/mL) for 30 min or were untreated. A ChIP assay then was performed, using an antibody against total STAT3. The immunoprecipitated DNA was amplified by qPCR, using primers specific for STAT3-binding sites on the SOCS3 promoter. Values represent means ± SD of fold enrichments of the IL-6–treated versus untreated samples following analysis by qPCR.
Discussion
Two previous studies, including one from our laboratory, indicate that STAT3 can be methylated by histone methyltransferases. K140 is dimethylated by SET9 and is demethylated by LSD1 (15), and K180 is trimethylated by phosphorylated EZH2 (16). Our MS results (Fig. 1A) and Western data (Fig. 3 A and B) reveal a third site of methylation. In response to IL-6, K49 of STAT3 is dimethylated by EZH2. The amino acid sequences surrounding the K49 methylation site of STAT3 and the K27 EZH2-dependent methylation site of histone H3 are quite similar (Fig. 4A). Y705 phosphorylation of STAT3 following ligand stimulation precedes K49 dimethylation (Fig. 3 C and D), and EZH2 is bound preferentially to tyrosine-phosphorylated STAT3 (Fig. 6 A and B). We also find that none of the K49 mutants tested binds to EZH2 (Fig. 6C). The affinity of tyrosine-phosphorylated STAT3 dimers for EZH2 may be greater than that of unphosphorylated STAT3. Alternatively, or in addition, EZH2 may bind to promoter-bound STAT3 phosphorylated dimers rather than to STAT3 dimers that are not associated with promoters.
Analysis of IL-6–stimulated gene expression reveals that the expression of more than half of the genes induced by WT-STAT3 is impaired when K49 is mutated (Fig. 2). When we knocked down EZH2 expression, IL-6–dependent K49 dimethylation was abrogated (Fig. 4 B and E). IL-6–dependent gene expression also was severely impaired in cells in which EZH2 was knocked down (Fig. 4 D and F), but Y705 phosphorylation was not (Fig. 4C). EZH2 knockdown and three different mutations of K49 impaired the expression of similar sets of dependent genes. Previously, Kim et al. (16) showed that EZH2 binds to STAT3 in glioblastoma stem cells. In addition, the ChIP data of Fig. 7 show that EZH2 has only a minor effect on IL-6–dependent STAT3 binding to the SOCS3 promoter. Our results show that knockdown of EZH2 reduces but does not eliminate SOCS3 expression (unlike mutation of K49), perhaps because the knockdown was incomplete. Because, for IL-6–mediated STAT3-dependent gene expression, the reductions in gene expression and in chromatin association detected by ChIP are not completely different (fivefold versus 2.5-fold), STAT3 K49 methylation may affect both chromatin association and cofactor recruitment. In the future, it will be of interest to compare STAT3 association at additional target promoters (for both methylation-sensitive and methylation-resistant genes) and to expand the correlation between sensitivity to the methylation mark and chromatin recruitment.
In summary, we identify K49 as a previously unidentified dimethylation site on STAT3 and show that this modification has a substantial effect on IL-6–dependent STAT3-mediated gene expression. As reviewed by Stark et al. (14), the lysine methylation of several different transcription factors, including STAT3, has been shown to occur after they have bound to specific promoters. In each case, the methylation affects the expression of some genes but not others. The modifications might alter the affinity of the transcription factor for some promoter sequences but not others, or specific proteins might be recruited to the modified residues (as has been amply observed for methylated lysine residues of histones), with functional consequences for some promoters but not others.
EZH2, the catalytic subunit of polycomb repressive complex 2, methylates K27 of histone H3, leading to chromatin condensation and suppression of gene expression (18–22). EZH2 is overexpressed in a wide range of cancers, including those of breast, prostate, and bladder (23, 24). However, in contrast, Ntziachristos et al. (25) and Simon et al. (26) showed that EZH2 disruption is sufficient to cause T-cell acute lymphoblastic leukemia in mice and that its disruption also favors the development of human T-cell acute lymphocytic leukemia. Furthermore, the ability of EZH2 to function as a crucial modifier of several signaling pathways, independent of its histone methyltransferase activity, adds to the complexity of EZH2 biology. A study by Shi et al. (27) showed that EZH2 can serve as an adaptor protein, linking estrogen receptor to β-catenin to enhance their transcriptional activities, and recent findings by Lee et al. (28) demonstrated that EZH2 enhances NF-κB activity by promoting NF-κB binding to a set of target promoters through direct physical interaction with RelA and RelB in basal-like breast cancer cells. In addition to these roles, EZH2 recently has been implicated in the modification of several transcription factors through its methyltransferase activity, independent of its ability to modify histones. RORα, an orphan nuclear receptor that is involved in several signaling pathways and that inhibits cancer progression in a context-dependent manner, is monomethylated by EZH2 and thus is marked for ubiquitin-mediated degradation (29). EZH2 has been reported to methylate the transcription factor GATA4, thereby repressing its activity (30).
Deregulated expression of IL-6 assists tumorigenesis by inhibiting apoptosis, stimulating angiogenesis, and increasing drug resistance (31). Lysine methylation by histone-modifying enzymes activates several different transcription factors, including STAT3, p53, and NF-κB, that have well-recognized roles in tumorigenesis. Therefore, mutations, amplifications, or deletions of histone lysine methyltransferases and demethylases that also modify transcription factors may affect cancer development and progression significantly (14). For this reason, these enzymes are promising targets for antitumor drugs. Both STAT3 and EZH2 have been found to be hyperactive in numerous cancers (22, 32). Our present data indicate that EZH2 has an important role in IL-6–mediated, STAT3-dependent gene expression, and therefore EZH2 inhibitors are promising therapeutic targets for solid tumors. However, the loss of EZH2 expression is characteristic of myelodysplastic syndrome, myeloproliferative disorders, and acute myeloid leukemia (32, 33), supporting the need to identify alternative targets in different tumor types. However, it is worthwhile considering the use of established EZH2 inhibitors to down-regulate STAT3-dependent gene expression in cancers in which EZH2 is overexpressed.
Materials and Methods
STAT3-null A4 cells were derived from human DLD1 colon cancer cells by homologous recombination (15). Both DLD1 and the STAT3-null A4 cells were grown in McCoy’s 5A medium with l-glutamine (HyClone), supplemented with 5% (vol/vol) FBS, penicillin, and streptomycin (100 μg/mL) (Gibco-BRL). Immortalized hTERT-HME1 cells were grown in mammary epithelial growth medium supplemented with bovine pituitary extract, gentamycin, hydrocortisone, insulin, and human epidermal growth factor (bullet kit), all from Lonza.
Additional details and experimental procedures are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Earl Poptic of the Hybridoma Core, Cleveland Clinic, for generating the K49-dimethyl STAT3 antibody, and helpful discussions with Dr. Jeongwu Lee, Cleveland Clinic. This work was supported by National Institutes of Health (NIH) Grant PO1 CA062220 (to G.R.S.) and NIH Shared Instrument Grant 1S10RR031537-01 (to B.W.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1503152112/-/DCSupplemental.
References
- 1.Bar-Natan M, Nelson EA, Xiang M, Frank DA. STAT signaling in the pathogenesis and treatment of myeloid malignancies. JAK-STAT. 2012;1(2):55–64. doi: 10.4161/jkst.20006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frank DA. STAT3 as a central mediator of neoplastic cellular transformation. Cancer Lett. 2007;251(2):199–210. doi: 10.1016/j.canlet.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 3.Xiong A, Yang Z, Shen Y, Zhou J, Shen Q. Transcription factor STAT3 as a novel molecular target for cancer prevention. Cancers (Basel) 2014;6(2):926–957. doi: 10.3390/cancers6020926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z, et al. Activation of STAT3 in human gastric cancer cells via interleukin (IL)-6-type cytokine signaling correlates with clinical implications. PLoS ONE. 2013;8(10):e75788. doi: 10.1371/journal.pone.0075788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morikawa T, et al. STAT3 expression, molecular features, inflammation patterns, and prognosis in a database of 724 colorectal cancers. Clin Cancer Res. 2011;17(6):1452–1462. doi: 10.1158/1078-0432.CCR-10-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Jong PR, Mo JH, Harris AR, Lee J, Raz E. STAT3: An anti-invasive factor in colorectal cancer? Cancers (Basel) 2014;6(3):1394–1407. doi: 10.3390/cancers6031394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akira S, et al. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell. 1994;77(1):63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 8.Wen Z, Zhong Z, Darnell JE., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82(2):241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 9.Wang R, Cherukuri P, Luo J. Activation of Stat3 sequence-specific DNA binding and transcription by p300/CREB-binding protein-mediated acetylation. J Biol Chem. 2005;280(12):11528–11534. doi: 10.1074/jbc.M413930200. [DOI] [PubMed] [Google Scholar]
- 10.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307(5707):269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 11.Dasgupta M, et al. Critical role for lysine 685 in gene expression mediated by transcription factor unphosphorylated STAT3. J Biol Chem. 2014;289(44):30763–30771. doi: 10.1074/jbc.M114.603894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ray S, Boldogh I, Brasier AR. STAT3 NH2-terminal acetylation is activated by the hepatic acute-phase response and required for IL-6 induction of angiotensinogen. Gastroenterology. 2005;129(5):1616–1632. doi: 10.1053/j.gastro.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 13.Ray S, et al. Inducible STAT3 NH2 terminal mono-ubiquitination promotes BRD4 complex formation to regulate apoptosis. Cell Signal. 2014;26(7):1445–1455. doi: 10.1016/j.cellsig.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stark GR, Wang Y, Lu T. Lysine methylation of promoter-bound transcription factors and relevance to cancer. Cell Res. 2011;21(3):375–380. doi: 10.1038/cr.2010.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang J, et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci USA. 2010;107(50):21499–21504. doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim E, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23(6):839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, van Boxel-Dezaire AH, Cheon H, Yang J, Stark GR. STAT3 activation in response to IL-6 is prolonged by the binding of IL-6 receptor to EGF receptor. Proc Natl Acad Sci USA. 2013;110(42):16975–16980. doi: 10.1073/pnas.1315862110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 19.Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci. 2010;35(6):323–332. doi: 10.1016/j.tibs.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10(10):697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 21.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647(1-2):21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 22.Tan JZ, Yan Y, Wang XX, Jiang Y, Xu HE. EZH2: Biology, disease, and structure-based drug discovery. Acta Pharmacol Sin. 2014;35(2):161–174. doi: 10.1038/aps.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleer CG, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100(20):11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varambally S, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 25.Ntziachristos P, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med. 2012;18(2):298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon C, et al. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 2012;26(7):651–656. doi: 10.1101/gad.186411.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi B, et al. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol Cell Biol. 2007;27(14):5105–5119. doi: 10.1128/MCB.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee ST, et al. Context-specific regulation of NF-κB target gene expression by EZH2 in breast cancers. Mol Cell. 2011;43(5):798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 29.Lee JM, et al. EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Mol Cell. 2012;48(4):572–586. doi: 10.1016/j.molcel.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 30.He A, et al. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev. 2012;26(1):37–42. doi: 10.1101/gad.173930.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo Y, Xu F, Lu T, Duan Z, Zhang Z. Interleukin-6 signaling pathway in targeted therapy for cancer. Cancer Treat Rev. 2012;38(7):904–910. doi: 10.1016/j.ctrv.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 32.Lund K, Adams PD, Copland M. EZH2 in normal and malignant hematopoiesis. Leukemia. 2014;28(1):44–49. doi: 10.1038/leu.2013.288. [DOI] [PubMed] [Google Scholar]
- 33.Hock H. A complex Polycomb issue: The two faces of EZH2 in cancer. Genes Dev. 2012;26(8):751–755. doi: 10.1101/gad.191163.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.