Significance
In diploid organisms, trans-allelic interactions control gene expression, providing a tight spatial and temporal level of transcription regulation. Although homologous trans-allelic interactions are quite abundant in various organisms such as Drosophila, plants, and fungi, they have not been widely reported in mammals. This article demonstrates that such a trans-allelic association is evident in mammals and involves the homologous spatial proximity of Tnfα alleles as a prerequisite for the biallelic expression of the Tnfα gene. We believe the phenomenon we describe here provides mechanistic insights for the regulation of gene allelic expression and mRNA dosage control necessary for fine-tuning physiological processes in mammals.
Keywords: homologous spatial proximity, lncRNAs, Tnfα, macrophages
Abstract
Physiological processes rely on the regulation of total mRNA levels in a cell. In diploid organisms, the transcriptional activation of one or both alleles of a gene may involve trans-allelic interactions that provide a tight spatial and temporal level of gene expression regulation. The mechanisms underlying such interactions still remain poorly understood. Here, we demonstrate that lipopolysaccharide stimulation of murine macrophages rapidly resulted in the actin-mediated and transient homologous spatial proximity of Tnfα alleles, which was necessary for the mono- to biallelic switch in gene expression. We identified two new complementary long noncoding RNAs transcribed from the TNFα locus and showed that their knockdown had opposite effects in Tnfα spatial proximity and allelic expression. Moreover, the observed spatial proximity of Tnfα alleles depended on pyruvate kinase muscle isoform 2 (PKM2) and T-helper-inducing POZ-Krüppel-like factor (ThPOK). This study suggests a role for lncRNAs in the regulation of somatic homologous spatial proximity and allelic expression control necessary for fine-tuning mammalian immune responses.
Tight control of total mRNA levels in a cell is essential for cellular homeostasis and normal physiology. The mRNA levels of a gene are regulated at multiple levels, and in addition to mRNA splicing, turnover, and translation, they also involve the epigenetic regulation of gene transcription (1). Transcriptional control in eukaryotic cells involves the tissue-specific activation and binding of transcription factors, which mediate their mode of action on the chromatin fiber, determined by both histone and DNA modifications (2). Another defining principle of transcriptional regulation is provided by the levels of chromatin and genome organization, which are also affected by discrete subnuclear entities, being regions of increased local concentration of protein or RNA molecules (3, 4). On the basis of the tremendous advancement of technologies that depict the genome’s organization in diverse cell types and tissues, we now know that the regulatory mechanisms involved in gene transcription include the formation of transcription networks mediated by long-range chromatin interactions (3, 5–7).
The chromosome conformation capture-based approaches in combination with fluorescence in situ hybridization to DNA (DNA FISH) have shown that a gene locus may be involved in a chromatin network formed by either intrachromosomal or interchromosomal interactions. In mammals, examples of long-range chromatin interactions have been described for the alpha and beta globin loci (8, 9), imprinted loci (10, 11), the two homologous X chromosomes (12–15), the olfactory receptor genes and the H enhancer (16, 17), and the IFNγ and T-helper-type 2 cytokine gene loci expressed in alternate cell fates of CD4+ T cells (18). The public research consortium ENCODE (the Encyclopedia of the DNA Elements) was recently launched in an attempt to identify all functional elements in the human genome (19). Systematic integrated analysis of the genome-wide chromatin interactions, which emerged from the project’s data (20–22), showed that long-range chromatin interactions are more prominent than previously thought.
Most long-range interchromosomal interactions that have been functionally characterized in interphase nuclei so far involved loci localized on nonhomologous chromosomes, with some exceptions (13, 23–25). During meiosis, most organisms go under the process of pairing their homologous chromosomes, which is usually restricted in the germ line. Somatic homologous pairing, however, has been extensively observed in dipteran insects, where it is evident in diverse cell types. In 1954, E.B. Lewis introduced the term transvection to describe cases in Drosophila melanogaster in which homologous pairing influenced gene expression involving the action of enhancers in trans (26). These interchromosomal interactions have been studied extensively in several systems, pointing out that such a mechanism regulating gene expression in trans may be a general phenomenon.
All these transsensing regulatory mechanisms ultimately point to the complex regulation of physiological processes in a cell. Innate immune responses, although tightly regulated, lack such mechanistic insight regarding the dynamic regulation of chromatin and genome organization. Macrophages, as crucial mediators of an innate immune response, can be activated by lipopolysaccharide (LPS) of Gram-negative bacteria via Toll-like receptor 4 that ultimately leads to the activation of several classes or responsive genes, such as the cytokine tumor necrosis factor alpha (TNFα) (27). TNFα is a proinflammatory cytokine with a critical role in the initiation of innate and adaptive immune responses. Although TNFα deficiency causes increased susceptibility to infection, resulting in complete lack of B-cell follicles or causing tuberculosis, prolonged high concentrations of TNFα can result in severe tissue damage, autoimmunity, and cancer (28–30). It is evident that a tightly regulated balance of TNFα levels is of critical importance. Tnfα gene transcription is controlled in a cell type-specific and stimulus-specific manner. Nonetheless, it also requires a tight spatial and temporal level of regulation (31, 32).
In our study, we explored the subnuclear interacting events that regulate Tnfα gene expression. We focused on the identification of factors (proteins or RNA molecules) capable of mediating its subnuclear localization pattern and investigated the mechanisms involved in the fine-tuning of Tnfα mRNA levels.
Results
Homologous Spatial Proximity of Tnfα Alleles Is Detected Early upon LPS Stimulation of Macrophages.
To study the subnuclear localization of the two Tnfα alleles, we performed experiments in thioglycollate-elicited and LPS-stimulated murine peritoneal macrophages. The lymphotoxin/TNFα (LT/TNF) locus includes three genes; namely, the LT beta (Ltb), TNFα (Tnfα), and LTα (Lta) genes (Fig. 1A). Quantitative reverse transcription coupled to PCR (qRT-PCR) experiments performed in primary macrophages revealed that the Tnfα gene is expressed on a time course of LPS stimulation, whereas no mRNA was detected for the Lta and Ltb genes (Fig. 1B). Subsequently, we performed DNA FISH, using fluorescently labeled bacterial artificial chromosome-DNA probes encompassing the LT/TNF locus. Interallelic distances were measured and normalized to the cell nuclear volume [normalized distance (ND) defined in Fig. 1C] to control for differences in the volume between individual cells. We found a considerable decrease of the Tnfα allele mean ND after 1 h of LPS stimulation (ND = 0.265), representing shorter Tnfα alleles distance in average. The homologous spatial proximity of the Tnfα alleles that was detected in wild-type macrophages was abolished in macrophages isolated from MyD88−/− mice, indicating the necessity for LPS signaling. More importantly, we found that in Delta3−/− macrophages, harboring an 8.4-kb deletion in the LT/TNF locus (Fig. 1A), the Tnfα alleles do not undergo homologous association, indicating the specificity of association to the LT/TNF locus (Fig. 1 D and E).
Fig. 1.
LT/TNF locus subnuclear relocalization upon LPS stimulation of macrophages. (A) The LT/TNF locus on murine chromosome 17. (B) qRT-PCR analysis of the total Tnfα mRNA levels. (C) Calculation formula for the ND of Tnfα alleles. (D) Single z-sections of DNA FISH analysis for the LT/TNF locus in LPS-stimulated primary macrophages. The Tnfα alleles distance was normalized for the volume of each cell, and the frequencies of cells with a normalized distance from 0 to 1 were plotted. Mean ND, blue triangle. (Scale bar, 2 μm.) Sample size, 2,931 cells. (E) Mean ND of total number of cells. The lowest mean ND was detected for the wild-type macrophages.
To meet the demands of the subsequent biochemical experiments, we have then used the RAW 264.7 monocyte-derived murine macrophages. We performed DNA FISH experiments in a time course of LPS stimulation. We found a considerable decrease of the Tnfα allele mean ND after 30 min of LPS stimulation (ND = 0.375), representing shorter Tnfα allele distance on average. This decrease in the interallelic distance between the two Tnfα alleles was progressive, comparing the macrophage cell population before LPS stimulation with the cell populations stimulated with LPS for 10, 20, or 30 min. A subsequent increase in the mean ND places the longest interallelic distances after 1 h of LPS stimulation of macrophages, which then decreased in the time course of LPS stimulation (Fig. 2 A and B). Furthermore, cumulative frequency curves for the Tnfα alleles NDs displayed that the macrophage population stimulated with LPS for 30 min was clearly differentiated from the other points during the course of LPS stimulation, as well as the untreated cells (Fig. 2C). This finding was corroborated by statistical analysis for the randomness of the distance distributions for each point of LPS stimulation (Kolmogorov-Smirnov test), portraying a normal distribution for the interallelic distances in untreated cells, but not for the 30-min LPS-stimulated macrophages (P < 0.001; Fig. 2D). This increase in the proximity of the Tnfα alleles could not be attributed to a change in the macrophage cell volume upon LPS stimulation, as the mean nuclear diameter of the cells during the course of activation did not significantly change (SI Appendix, Fig. S1A). The close proximity of the Tnfα alleles in macrophages, after 30 min of LPS stimulation, taken together with the known rarity of somatic homologous pairing in interphase nuclei of mammalian cells, suggested that the homologous spatial proximity of the Tnfα alleles would be temporally transient. Interestingly, we found that this proximity was explained by the fact that the Tnfα alleles paired (ND < 0.1, allele distance shorter than 0.6 μm, given the fact that the mean nuclear diameter is 6 μm; SI Appendix, Fig. S1A) in 18.3 ± 3.3% of the cells after 30 min of LPS stimulation compared with 4.93 ± 3% of untreated macrophages (Fig. 2E).
Fig. 2.
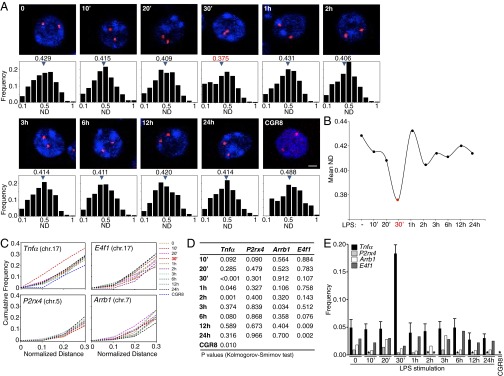
Homologous spatial proximity of the Tnfα locus. (A) Z-sections of DNA FISH analysis. (Scale bar, 2 μm.) Sample size, 441–1104 cells for each time. (B) Mean ND of total number of cells. (C) Cumulative frequency curves for Tnfα, E4f1, P2rx4, and Arrb1 allele pairs. (D) Kolmogorov-Smirnov test. (E) Frequency of cells with an allele ND < 0.1. Bars depict the mean value with SDs from 14 independent experiments. *Value not detected. CGR8, mouse embryonic stem cell line.
To determine whether genomic regions flanking the LT/TNF locus were also drawn into close proximity by the homologous spatial proximity of the Tnfα alleles, we measured the interallelic distances of the E4f1 locus, which is mapped 10.7 Mb upstream of the Tnfα gene on mouse chromosome 17 (SI Appendix, Fig. S2A). As an additional control, we performed DNA FISH experiments and measured the interallelic distances for the gene loci mapped on different mouse chromosomes, such as the P2rx4 locus mapped on chromosome 5 and the Arrb1 locus mapped on chromosome 7 (SI Appendix, Fig. S1 B and C). The cell volume of the mouse macrophage cells did not alter during the course of LPS stimulation for these DNA FISH experiments (SI Appendix, Fig. S1B). The cumulative frequency curves and the frequency of cells with ND < 0.1 for the E4f1 locus, as well as the P2rx4 and Arrb1 gene loci, did not show any evident allelic spatial proximity (Fig. 2 C–E), although they are activated by LPS (SI Appendix, Fig. S3).
In summary, our data highlight a transient homologous spatial proximity event of the Tnfα alleles in mouse macrophages, early upon LPS stimulation, which was specific to Tnfα and was not observed in LT/TNF proximal loci or the tested loci mapped on other mouse chromosomes.
The Tnfα Gene Has a Distinct Pattern of Allelic Gene Expression.
Taking into account that the subnuclear localization of gene loci has a direct effect on the gene’s expression kinetics, we wanted to study whether the transient homologous spatial proximity of the Tnfα alleles had any functional effect on Tnfα gene expression. We performed semiquantitative RT-PCR analysis for the Ltb, Tnfα, and Lta genes of the LT/TNF locus and found that only the Tnfα gene was highly expressed in macrophages upon LPS stimulation (Fig. 3A), in agreement with the data obtained for the primary macrophages. We then performed qRT-PCR experiments and found that Tnfα reached maximal mRNA levels after 1 h of LPS stimulation (Fig. 3B). To analyze the allelic expression pattern of the Tnfα gene at the single-cell level, we performed RNA-DNA FISH experiments, with the simultaneous detection of both the newly synthesized Tnfα mRNA and the DNA of the two Tnfα alleles. The analysis of the RNA-DNA FISH experiments we have performed in macrophages on a time course of LPS stimulation revealed that the highest frequency of Tnfα-expressing cells (74 ± 9.6%) was detected after 1 h of LPS stimulation (Fig. 3C), in agreement with the qRT-PCR results (Fig. 3B). We found that nascent Tnfα mRNA was not detected in resting macrophages, but the gene was rapidly activated upon LPS induction and expressed Tnfα from either one or both alleles, unlike the Lta gene, which was moderately activated by LPS and was only expressed in a monoallelic manner (Fig. 3D). Strikingly, the examination of Tnfα transcription at the single-cell level uncovered a unique allelic pattern of expression. Early on in LPS stimulation, during the first 30 min, Tnfα was mainly expressed from one allele. After 1 h of stimulation, however, about 70% of the expressing cells displayed a pattern of transcription from both alleles. After the 1 h LPS biallelic switch, the frequency of expressing cells steadily declined until it reached basal levels after 24 h of stimulation (Fig. 3E). The switch from mono- to biallelic gene expression coincided with the qRT-PCR results of maximal mRNA levels at the 1h LPS time point, portrayed both by total Tnfα transcript levels and expressing cell frequency (Fig. 3 B and C). In short, Tnfα gene expression follows a distinct monoallelic pattern succeeded by a switch to biallelic expression early on in LPS stimulation of macrophages.
Fig. 3.

Tnfα biallelic expression depends on LPS-stimulated and actin-mediated homologous spatial proximity. (A) mRNA expression analysis for the Tnfα, Ltb, and Lta genes. Hprt1 was used as a loading control. (B) qRT-PCR analysis of the total Tnfα mRNA levels in LPS-stimulated macrophages. (C) Percentage of Tnfα expressing macrophages upon LPS stimulation. (D) RNA-DNA FISH analysis of the LT/TNF locus, along with the nascent Tnfα or Lta mRNA transcripts. (Scale bar, 2 μm.) (E) The switch to Tnfα biallelic expression is detected after 1 h of LPS stimulation. RNA-DNA FISH experiments were analyzed to plot the allelic pattern of Tnfα expression. Bars represent the percentage of Tnfα expressing cells with SDs of three independent experiments. Sample sizes/time point (n) = 205–352 cells. *Value not detected. (F) Frequency of cells with paired Tnfα alleles (ND < 0.1) in LPS-stimulated or LTA-pretreated macrophages, with SDs of three independent experiments. Sample size (n) = 3,760 cells. (G) Frequencies of Tnfα expressing cells in LTA-pretreated compared with untreated LPS-stimulated macrophages with SDs of three independent experiments. Sample size (n) = 3,334 cells. (H) Frequencies of Tnfα biallelically expressing cells. (I) Frequencies of cells with Tnfα allele ND < 0.1 with SDs of three independent experiments. Sample size (n) = 3,779 cells. (J). Frequencies of Tnfα expressing macrophages with SDs of three independent experiments. Sample size (n) = 4,288 cells. (K) Frequencies of Tnfα biallelically expressing macrophages. *Value not detected.
The LPS-Induced and Actin-Mediated Homologous Spatial Proximity of the LT/TNF Locus Is Necessary for the Biallelic Switch in Tnfα Gene Expression.
To further explore whether the homologous spatial proximity of the Tnfα alleles played a role in mediating the subsequent biallelic switch in Tnfα gene expression, we blocked actin polymerization using latrunculin A (LTA) and found that the movement of the LT/TNF locus was impeded and spatial proximity was reduced (Fig. 3F). Although the frequency of cells that expressed Tnfα was not affected by the treatment with LTA (Fig. 3G), we found that the switch to biallelic gene expression, after 1 h of LPS stimulation, was not observed (Fig. 3H). We next investigated whether the TNFα cytokine, produced by macrophages upon LPS stimulation, was responsible for inducing the homologous spatial proximity of the Tnfα alleles and the subsequent switch from mono- to biallelic gene expression. We treated RAW 264.7 macrophages with mouse recombinant TNFα and found that it failed to induce homologous spatial proximity (Fig. 3I). RNA-DNA FISH experiments on TNFα-treated macrophages revealed that although Tnfα gene transcription was detected (Fig. 3J), biallelic expression was greatly impaired (Fig. 3K). We conclude that the homologous association of the Tnfα alleles is LPS-induced, actin-mediated, and indispensable for the biallelic switch in Tnfα gene expression.
Identification of Two Complementary Long Noncoding RNAs Transcribed from the LT/TNF Locus.
Long noncoding RNAs have been implicated in the regulation of many diverse physiological processes maintaining cell homeostasis (33–36). For example, to compensate for gene dosage differences between sexes, the mechanism of mammalian X inactivation involves the transient pairing of the two X chromosomes, and subsequently the expression of the Xist lncRNA results in the down-regulation of gene expression in the inactive X chromosome (13, 35, 37, 38). The involvement of lncRNAs in mammalian X inactivation, as well as in other processes entailing trans-allelic proximity (39), directed us to examine whether there was evidence of such long transcripts in the LT/TNF locus.
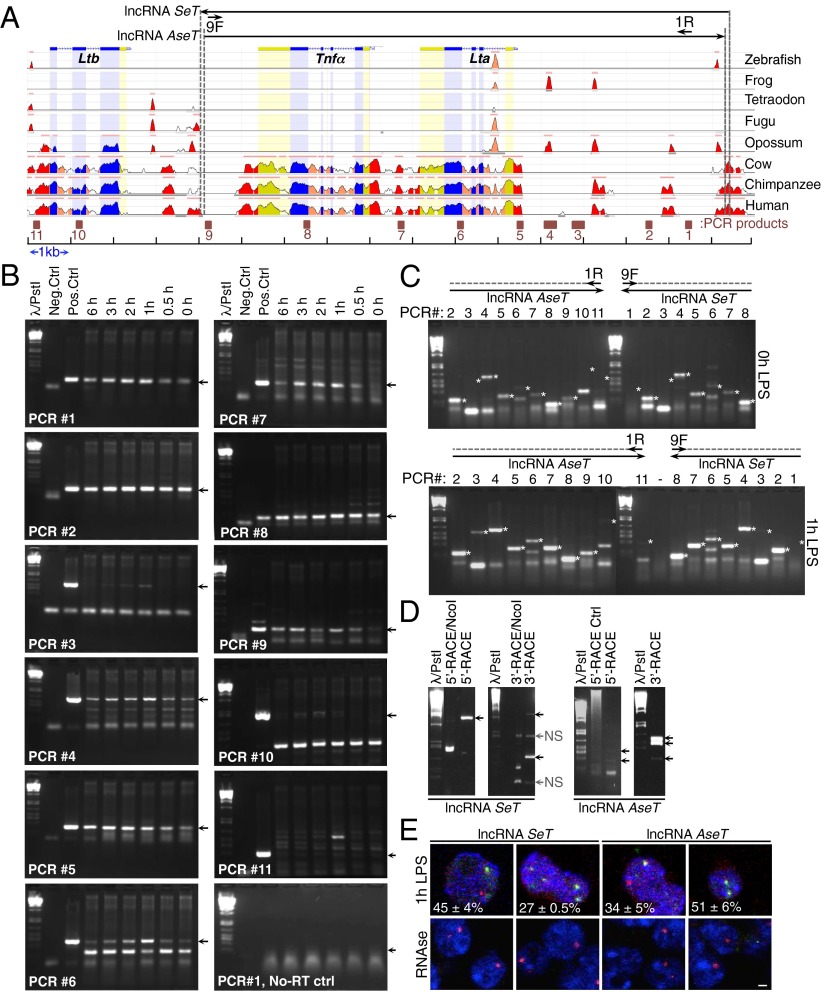
Multiple intergenic regions in the LT/TNF locus are highly conserved among mammals, and the locus itself encompasses a genomic region of 17 kb on murine chromosome 17 (Fig. 4A). To test for the presence of intergenic transcripts in the LT/TNF locus, we isolated total RNA from LPS-stimulated RAW 264.7 cells in a time course of 6 h; RNA samples were treated with DNase I and then subjected to reverse transcription primed by random sequence hexanucleotides. The produced cDNA was used in PCR reactions, using primer pairs that covered the entire LT/TNF locus and mapped to either the coding regions of the genes (such as the primer pairs for the PCR product 6 mapping on the coding region of the Lta gene, PCR product 8 mapping on the coding region of the Tnfα gene, and PCR product 10 mapping on the coding region of the Ltb gene), encompassing the locus, or to intergenic regions (such as PCR products 1–4, 7, 9, and 11) (Fig. 4B). On the basis of this analysis, using random sequence hexanucleotides for the generation of cDNA and the subsequent PCR analysis for the PCR products 1–11, we concluded there was extensive transcription throughout the LT/TNF locus; moreover, these transcripts were up-regulated upon LPS stimulation of the cells. Intergenic transcripts have been detected for PCR reactions 1–9, but not for the PCR reactions 10 and 11, proximal to the Ltb gene.
Fig. 4.
Two complementary long noncoding RNAs are expressed from the LT/TNF locus. (A) Cross-species conservation of the LT/TNF locus. Conservation more than 70% is indicated for gene exons (blue), untranslated regions (yellow), and intergenic regions (red), based on the Evolutionary Conserved Regions browser. Numbered squares: PCR products used to detect transcripts mapped on the locus. (B) Intergenic transcripts are detected on the LT/TNF locus. PCR products spanning the locus in 1–2-kb intervals (sequences 1–11), on random-hexamers primed reverse transcription of total RNA isolated from macrophages stimulated with LPS. Arrows indicate expected PCR products. Additional bands of lower molecular weight for PCR reactions 6 and 10: Lta and Ltb spliced transcripts. Bacterial artificial chromosome DNA was used as a positive control, and no-DNA reactions as a negative control. (C) Long complementary transcripts are detected on the LT/TNF locus. PCR reactions harboring the 11 indicated sequences in A were performed on cDNA templates created with single specific-primer reverse transcription of RNA from macrophages (9F: RT primer for lncRNA SeT; 1R: RT primer for lncRNA AseT). (D) Rapid amplification of cDNA ends for the 5′- and 3′- ends of the lncRNAs SeT and AseT. From left to right: lncRNA SeT 5′-RACE performed with the primer 1F and nested primers (control digestion with NcoI restriction enzyme), lncRNA SeT 3′-RACE performed with primer 9R and nested primers, lncRNA AseT 5′-RACE performed with primer 9R and nested primers, and lncRNA AseT 3′-RACE performed with primer 1F and nested primers. NS = nonspecific. (E) RNA-FISH using strand specific biotinylated riboprobes for the nascent SeT and AseT lncRNAs. (Scale bar, 2 μm.)
We then asked whether the transcripts we have detected were individual short RNA molecules or were part of longer RNA transcripts. To answer this question, we performed reverse transcription experiments on total RNA isolated from either untreated or 1-h LPS-stimulated macrophages, using single primers mapped on the ends of the transcribed region (genomic regions mapped on either PCR product 1 or PCR product 9) of the LT/TNF locus. For the detection of a sense long RNA transcript, we have used primer 9F (Fig. 4A and SI Appendix, Fig. S4) to produce cDNA in a reverse transcription reaction with total RNA as template and then used this single primed cDNA as template for PCR reactions 1–9 (Fig. 4 C and D). To detect an antisense long transcript in the locus, we used primer 1R (Fig. 4A and SI Appendix, Fig. S4) to produce cDNA in a reverse transcription reaction with total RNA as template and then used this single primed cDNA as template for the PCR reactions 1–11 (Fig. 4 C and D). On the basis of the PCR analysis of these single-oligonucleotide-primed cDNAs, we concluded that there are two complementary long RNA molecules, with a length of more than 12.0 kb each, that encompassed the Lta and Tnfα gene loci, and their expression was up-regulated upon LPS stimulation of the cells (Fig. 4C, Upper, for untreated macrophages and Fig. 4C, Lower, for 1-h LPS–treated macrophages). We named these transcripts lncRNA SeT (for LT/TNF locus lncRNA sense transcript) and lncRNA AseT (for LT/TNF locus lncRNA antisense transcript) (Fig. 4 A and C). We then used the rapid amplification of cDNA ends (RACE) approach to characterize the potential 5′- and 3′-ends of each individual transcript and identified both ends of each transcript (Fig. 4D and SI Appendix, Fig. S4). LncRNA SeT had a size of 12,340 bp, and its transcription start site mapped 8,258 bp upstream from the transcription start site of the Tnfα gene, whereas its 3′-end mapped between the Tnfα and Ltb genes. LncRNA AseT had a size of 12,033 bp, its transcription start site mapped 3,835 bp downstream from the transcription start site of the Tnfα gene, and its 3′-end mapped 60 bp downstream from the transcription start site of the lncRNA SeT (Fig. 4A). On the basis of additional RACE reactions performed, we have also identified alternative 5′- and 3′-ends for each transcript, which are presented diagrammatically in SI Appendix, Fig. S4. It is noteworthy that we were able to detect the lncRNA AseT in differentiated human monocytes (THP1 cell line), but not the lncRNA SeT (SI Appendix, Fig. S5). Although informative, a more thorough analysis should be performed for the presence and the transcription start sites of these lncRNAs in human cell types. Moreover, using biotinylated strand-specific riboprobes coupled to extensive signal amplification, we were able to detect each individual SeT and AseT transcript in LPS-activated macrophages (Fig. 4E). On the basis of our results, we conclude there are two long cRNA molecules transcribed from the LT/TNF locus.
LncRNAs SeT and AseT Regulate the Homologous Spatial Proximity and Biallelic Expression of Tnfα.
To further characterize the long transcripts SeT and AseT, we examined the genomic regions upstream from the transcription start site of each transcript, using the Evolutionary Conserved Regions browser, and found them to be conserved (>70% homology) between mammalian species (Fig. 5A). To study the chromatin conformation of these conserved genomic regions, we performed DNase I hypersensitivity assays and found them to be hypersensitive in untreated RAW 264.7 murine macrophages, whereas the DNase I hypersensitivity increased upon LPS stimulation of the cells (Fig. 5B). We used mammalian expression vectors and cloned these genomic regions upstream of a luciferase reporter gene (SI Appendix, Fig. S6). Transient transfection of the two individual constructs in RAW 264.7 cells and subsequent LPS stimulation of the cells revealed that the DNase I hypersensitive site (HSS) 1 upstream from the transcription start site of the lncRNA SeT showed increased promoter-reporter activity rapidly (30 min) upon LPS stimulation of the cells, which remained active 3 h after the initial stimulation. In contrast, HSS9, upstream from the transcription start site of the lncRNA AseT, showed rapid and transient promoter-reporter activity after 30 min of LPS stimulation of the cells (Fig. 5C).
Fig. 5.
The complementary lncRNAs SeT and AseT are LPS-regulated and mediate opposite effects on homologous spatial proximity and Tnfα allelic expression. (A) Mapping of the lncRNAs SeT and AseT. Radiolabeled DNA probes were used to hybridize to the PstI (P) fragments spanning the 5′ ends of each long transcript. (B) DNase I hypersensitivity mapping at the transcription start site of each long transcript. Arrows indicate the DNase I HSSs. NS, non specific. (C) Luciferase reporter assays in either untreated or LPS-stimulated macrophages, transiently transfected with promoter-reporter plasmids harboring either the HSS1 or HSS9 region. SDs are from three independent experiments. (D) qRT-PCR results of the relative RNA levels of the lncRNAs SeT and AseT. cDNA was reverse transcribed, using single oligonucleotide primers specific for SeT or AseT. (E) LNA-mediated silencing of the lncRNA SeT has no effect on the LT/TNF homologous spatial proximity, whereas silencing of the lncRNA AseT abolishes homologous spatial proximity. Frequency of cells expressing Tnfα in untreated cells (white) and cells treated with LNA for lncRNA SeT (black) or AseT (gray). (n) = 3,329 cells. (F) LNA-mediated silencing of the lncRNA SeT increases the biallelic Tnfα gene expression, and silencing of AseT abolishes biallelic Tnfα gene expression. (n) = 2,173 cells.
To determine the relative RNA expression levels of the two lncRNAs, we performed qRT-PCR upon specific primer reverse transcription and found that the lncRNAs were both expressed in untreated mouse macrophages and are rapidly up-regulated upon LPS stimulation of the cells (Fig. 5D). Interestingly, the two lncRNAs had distinctly different roles in the Tnfα homologous spatial proximity and allelic expression profile. Silencing of the lncRNA SeT (SI Appendix, Fig. S7A), using locked nucleic acid (LNA) technology, had no effect on the LT/TNF locus homologous spatial proximity (Fig. 5E) and rendered Tnfα transcription mainly biallelic (Fig. 5F) upon LPS stimulation of macrophages, with no apparent effect on Tnfα monoallelic expression (SI Appendix, Fig. S8). In contrast, LNA-mediated silencing of lncRNA AseT (SI Appendix, Fig. S7B) impaired both LT/TNF locus homologous spatial proximity and its biallelic expression (Fig. 5 E and F). Therefore, the specific silencing of each individual of the two complementary lncRNAs had opposite effects on the homologous spatial proximity and allelic Tnfα expression profile.
Identification of the Pyruvate Kinase Muscle Isoform 2 Protein with GA Binding Activity.
To unravel the functional mechanism behind the regulation of homologous spatial proximity of Tnfα alleles and its effect on the regulation of allelic expression profile of the Tnfα gene, we have chosen to purify and characterize protein complexes that mediate such a phenomenon. We thus resorted to the protein factors that play a major role in transvection. GAF, a protein encoded by the Trithorax-like (Trl) gene in Drosophila, is a transcription factor with DNA binding and transactivation properties that binds to GAGAG motifs and locally remodels chromatin to enable enhancer–promoter interactions (40–42). Because there is no biochemically or functionally characterized mammalian homolog of the Drosophila GAF, we performed a series of biochemical assays to identify a protein factor with similar activity.
We selected (GA)n repetitive DNA stretches from the LT/TNF locus or an oligonucleotide harboring a single GAGAG motif mapping on the Tnfα gene promoter and performed electrophoretic mobility shift assays, using nuclear protein extracts prepared from LPS-stimulated mouse macrophages. We identified a specific DNA binding activity, which was dependent on LPS stimulation of the cells, and reached maximal binding on the Tnfα promoter oligonucleotide after 30 min of LPS stimulation (Fig. 6A). In accordance with these experiments, we performed Southwestern blotting experiments and were also able to detect maximal (GA)n oligonucleotide binding of several proteins present in nuclear protein extracts prepared from 30-min LPS-stimulated macrophages (Fig. 6B).
Fig. 6.
Identification of the PKM2 protein from murine macrophages with (GA)n binding activity. (A) electrophoretic mobility shift assay with nuclear extracts from LPS-stimulated macrophages shows specific (GA)n binding activity upon LPS stimulation (GA-repeat: 25-bp oligonucleotide of repeated GA nucleotides, single-GA: specific sequence from the Tnfα promoter with a single GAGAG element; mut.single GA: similar to the latter with mutated GAGAG sequence). (B) Southwestern blot indicating DNA binding activity on a GA 25-mer labeled oligonucleotide from LPS-stimulated macrophage nuclear extracts. (C) SDS polyacrylamide gel electrophoresis of nuclear extracts from 30-min LPS-stimulated murine macrophages fractionated on a P11 phosphocellulose column and eluted with increasing NaCl concentration buffer. (D) Electrophoretic mobility shift assay indicating the presence of the desired DNA binding activity of proteins in specific eluted fractions from C. (E) DNA affinity chromatography. Nuclear protein extracts from mouse macrophages, precleared and fractionated (fractions 3 and 4), were incubated with concatamerized biotinylated oligonucleotides immobilized on streptavidin magnetic beads. Lanes 1/10: molecular weight marker, 2: first flow-through, 3: GA-bound proteins, 4: wash, 5/6: CA-bound, 7: second flow-through, 8: GA-bound, 9: input. Asterisks: bands analyzed. (F) PKM2 has been identified by mass spectrometry. Shown is the amino acid sequence of pyruvate kinase muscle isozyme protein. Green indicates the peptides identified by MS analysis (underlined: PKM2 specific).
To isolate and characterize the protein or proteins with (GA)n binding activity, we have undertaken a series of approaches. Nuclear polyadenylated mRNA from LPS-stimulated macrophages (for 30 min) was used for the construction of a cDNA library in a yeast one-hybrid screening, using a sequence of 25 GA nucleotides as bait (SI Appendix, Fig. S9). In parallel, fractionated nuclear protein extracts from LPS-stimulated RAW 264.7 macrophages (Fig. 6 C and D) were used for DNA affinity chromatography coupled to mass spectrometry. Mouse pyruvate kinase muscle isozyme 2 (PKM2) was identified by both approaches as a protein with the ability to bind specifically on GA-repetitive elements (Fig. 6 E and F). On the basis of our results, we suggest that the PKM2 protein binds GA-containing sequences from the Tnfα locus either directly or indirectly, via its interaction with other protein factors.
Homologous Spatial Proximity Depends on PKM2 and T-Helper–Inducing POZ-Krüppel-Like Factor Proteins.
PKM2 is a cytoplasmic protein with a role in glycolysis and was also recently shown to translocate into the nucleus and directly regulate transcription as a protein kinase (43–45). Using specific antibodies, we performed immunostaining experiments and found that PKM2 protein was mainly localized in the cytoplasm, but also in the nucleus of macrophages, with a distinct speckled pattern after 30 min of LPS stimulation (Fig. 7A and SI Appendix, Fig. S10A). T-helper–inducing POZ-Krüppel–like factor (ThPOK), in silico predicted to be the GAF mammalian homolog because of its structural and sequence similarity (46), was also tested for its involvement in the Tnfα homologous spatial proximity. ThPOK protein also displayed a distinct speckled pattern in the nucleus of macrophages (Fig. 7B). SiRNA-mediated knock-down of PKM2 (SI Appendix, Fig. S10B) or ThPOK in LPS-stimulated RAW 264.7 murine macrophages resulted in reduced Tnfα mRNA levels (Fig. 7 C and D) and, more importantly, disrupted the homologous spatial proximity of the Tnfα alleles, as depicted by DNA FISH analysis, based on the reduced frequency of cells with a Tnfα allele ND less than 0.1 (Fig. 7E). The reduced Tnfα mRNA levels could be explained by RNA-DNA FISH experiments, which showed that although the frequency of cells depicting Tnfα monoallelic expression was not remarkably affected by the knockdown of PKM2 or ThPOK, the frequency of biallelically expressing cells was greatly reduced (Fig. 7F). Taken together, these data show that PKM2 and ThPOK proteins mediate Tnfα homologous spatial proximity, and silencing either of them disrupts the association and consequently blocks the switch from mono- to biallelic Tnfα expression.
Fig. 7.

PKM2 and ThPOK mediate Tnfα homologous spatial proximity and subsequent biallelic gene expression. (A) PKM2 protein subnuclear localization. (B) ThPOK protein nuclear localization. (Scale bar, 2 μm.) (C) Tnfα mRNA levels in LPS-stimulated macrophages either before or upon siRNA-mediated knockdown of PKM2. (D) Tnfα mRNA levels in LPS-stimulated macrophages either before or upon siRNA-mediated knockdown of ThPOK. (E) Effect of PKM2 or ThPOK siRNA treatment on Tnfα homologous spatial proximity. Frequencies of cells with allele ND < 0.1, for untreated cells (black), cells treated with siRNA for PKM2 (red), and siRNA for ThPOK (blue). (n) = 1,958. *Value not detected. (F) Frequencies of cells with monoallelic Tnfα expression for untreated (black), cells treated with siRNA for PKM2 (red), ThPOK (blue), or scrambled siRNA (gray). (n) = 5,683 cells. *Not detected. (G) Spatial proximity of the Tnfα alleles is LPS-induced, actin-mediated, and necessary for the biallelic switch in Tnfα expression. The complementary lncRNAs SeT and AseT transcribed from the LT/TNF locus have diverse effects on the PKM2- and ThPOK-mediated spatial proximity and biallelic Tnfα expression.
Discussion
Taken together, our data revealed the LPS-induced, transient and rapidly established homologous spatial proximity of Tnfα alleles that did not occur to maintain allelic exclusion, as in X chromosome inactivation or the Ig loci, but regulates mRNA dose control through a mono- to biallelic switch in Tnfα gene expression (Fig. 7G). Used as a way of information exchange in trans, Tnfα homologous spatial proximity ensures the production of maximal Tnfα mRNA levels necessary for macrophage immune responses.
Although evidence for regional pairing of homologous chromosomes has increased over recent years, it still remains unclear how the two alleles find each other and what mediates and/or sustains these associations. It has been speculated to be either a result of the properties of a larger region of the chromosome or a result of the specific settings provided by distinct genomic elements. Homologous pairing has been documented in several studies, the most prominent of which is the establishment of monoallelic silencing of the X chromosome. In X inactivation, the two chromosomes pair and the lncRNA Tsix is transiently down-regulated to allow the monoallelic expression of the lncRNA Xist to reach levels sufficient for the coating and silencing of the inactive X chromosome (12). Pairing was also shown to occur between the Ig loci. In this case, one of the two alleles undergoes recombination-dependent cleavage, and the other is heterochromatinized (23). In imprinted loci, it is easier to distinguish the two loci, as they are differentially premarked through DNA methylation. Homologous pairing in the cases of Prader-Willi/Angelman region in humans or the Kcnq1 cluster has been extensively studied, and in these cases too, the result of the association was monoallelic expression (24, 25). These examples are associated with allelic exclusion, and it has been suggested that homologous pairing is a feature of regions in which one allele is silenced and monoallelic expression is maintained.
Moreover, homologous pairing has also been associated with DNA repair of double-strand breaks. Although nonhomologous end joining is more commonly used in mammals for the repair of such DNA damage, homologous recombination, which is predominantly used in yeast, is also found in mammals in the case of replication-induced breaks (47). It was recently shown that homologous pairing is not dependent on imprinting or allelic exclusion; in fact, loss of imprinting did not change pairing frequency. Instead, it was shown that somatic homologous pairing, although rare, depends on chromosomal position and transcriptional activity (48). Our data are in line with these findings, as we describe allelic spatial proximity, which is transient and rapidly established, and thus independent of imprinting status, and does not occur to maintain allelic exclusion but, instead, activates the expression of the Tnfα gene from the second allele as well.
We have also identified two protein factors mediating the homologous spatial proximity of the two Tnfα alleles: a protein kinase with transactivation potential and a transcription factor considered to be the mammalian homolog of the Drosophila GAGA factor. PKM2, a kinase involved in glycolysis and in cancer metabolism, is surprisingly capable of functioning as a protein kinase in the nucleus. It has been involved in several phosphorylation and transactivation events and has been found to bind DNA either directly or indirectly. It is intriguing, however, how a glycolytic enzyme is able to simultaneously function as a protein kinase. Studies of PKM2 in tumor cells, where it is predominantly found in its dimeric form, unable to convert phosphoenolpyruvic acid to pyruvate (49), give a possible answer to this question. The tetrameric form of PKM2 in the cytoplasm interacts with several glycolytic enzymes or oncoproteins, which are able to promote the conversion of PKM2 to a dimer (50). In addition, the binding of phosphorylated tyrosine peptides to PKM2 decreases its enzymatic activity (43). This was possibly caused by the exposure of a hydrophobic part of the dimeric protein, able to bind a protein substrate, in contrast to the tetrameric form, where this site would be inaccessible (51, 52). Thus, even without a prominent DNA binding domain, it is possible that LPS stimulation induces the switch to the dimeric form of PKM2 via an interaction in the cytoplasm, facilitating PKM2 to bind the LT/TNF locus, maybe through an interaction with ThPOK. Although a phosphorylation event may be occurring during the binding of PKM2 to ThPOK, altering the function of the latter (ThPOK contains tyrosine residues predicted to be phosphorylated by protein kinases), we have shown that both proteins are functionally necessary for the establishment of Tnfα homologous spatial proximity, as well as the switch in allelic expression.
Furthermore, we have implicated two long complementary transcripts, expressed by the LT/TNF locus, in the control of the switch in allelic expression of the Tnfα gene. The two lncRNAs involved seem to counteract one another by means of transcript quantity and localization. In correspondence to the function of the lncRNA Xist in X inactivation, we may suggest that lncRNA SeT blocks Tnfα transcription from the second allele by coating the locus after 30 min of LPS stimulation, supported by the fact that LNA-mediated knock-down of this transcript allowed both alleles to express Tnfα. In parallel, lncRNA AseT functioned oppositely by displacing its complementary lncRNA SeT after 1 h of LPS stimulation (RNA-DNA FISH experiments show lncRNA SeT to be dispersed around the locus after 30 min of LPS stimulation), allowing the switch to biallelic expression. LNA-mediated knockdown of the lncRNA AseT could both disrupt homologous spatial proximity as well as render Tnfα expression monoallelic, possibly by allowing lncRNA SeT to coat the locus.
Further investigation in the mechanisms involved in the induction and maintenance of TNFα maximal levels is of great importance for both basic research and clinical practice. First, because a mechanism controlling somatic homologous pairing and allelic expression may be occurring in a wide range of inducible systems, and second, because the identification of ways to exploit such a mechanism could be used in the future to study and possibly resolve the deregulation of gene expression in disease models.
Materials and Methods
Detailed experimental procedures are available in the SI Appendix.
Cell Treatments.
Murine monocyte-derived macrophage RAW264.7 cells were stimulated with 50 ng/mL LPS (InvivoGen) or 10 ng/mL TNFα (R&D Systems) or pretreated with 10 μM Latrunculin A (Sigma) where stated. Knockdown experiments for PKM2 and ThPOK were performed with the use of 5 nM Silencer Select siRNAs incubated with siPORT NeoFX Transfection Agent (Ambion, Applied Biosystems) in OPTI-MEM media (GIBCO) according to the manufacturer’s instructions. The LT/TNF locus long transcripts were knocked down using Locked Nucleic Acid oligonucleotides (Exiqon). Thioglycollate-elicited (Brewer’s medium, LAB064, Lab M) peritoneal macrophages were harvested from C57BL/6, B6.129P2-Ltb/Tnf/Ltatm1Dvk/J (Delta3−/−), or Myd88−/− mice, plated overnight, and stimulated with LPS as described earlier.
RNA-DNA FISH.
For DNA FISH experiments, cells were fixed in 4% paraformaldehyde/1× PBS and permeabilized in 0.5% Triton X-100/1× PBS. For RNA-DNA FISH, cells were treated with cytoskeletal buffer prior to fixation. Hybridizations of genomic loci and nascent RNA were performed with bacterial artificial chromosome or cDNA probes labeled with spectrum orange/green dUTP. Nuclear DNA was stained with ToPro3 (ToPro3 Iodide 642/661) and pseudocolored blue.
Imaging Analysis and Statistics.
The analyses and measurements of allele distances and nuclear volumes were performed with the use of the Volocity software (Improvision, Perkin-Elmer). Statistical analysis for the randomness of the distance distributions was performed in a pairwise manner, using the Kolmogorov-Smirnov test.
RACE.
The FirstChoice RLM-RACE kit (Invitrogen, AM1700M) was used to identify the 5′- and 3′- ends of specific capped mRNA molecules from RAW 264.7 macrophages treated with 50 ng/mL LPS for 1 h.
Protein Identification and Mass Spectrometry.
For the isolation of DNA binding proteins, the Yaneva and Tempst protocol was followed (53). Coomassie-stained polyacrylamide gel bands were destained, reduced, alkylated, and digested with trypsin (proteomics grade, Sigma, T6567). The subsequent mass spectrometric analysis involved nano-liquid chromatography–MS/MS analysis (54).
Supplementary Material
Acknowledgments
We thank N. Tavernarakis, I. Talianidis, and C. Mamalaki for discussion and their comments on the manuscript. This project is implemented under the “ARISTEIA” Action of the “OPERATIONAL PROGRAMME EDUCATION AND LIFELONG LEARNING” and is cofunded by the European Social Fund (ESF) and National Resources. This work was also supported by a Cancer Research Institute Investigator Award (to C.G.S.), a Marie Curie International Reintegration Grant within the 7th European Community Framework Programme (FP7/2007-2013) under Grant Agreement 239339 (to C.G.S.), and the European Commission through Seventh Framework Programme Agreement No. 229823, Capacities-FP7-REGPOT-2008-1/project “ProFI.”
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1502182112/-/DCSupplemental.
References
- 1.Lelli KM, Slattery M, Mann RS. Disentangling the many layers of eukaryotic transcriptional regulation. Annu Rev Genet. 2012;46:43–68. doi: 10.1146/annurev-genet-110711-155437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spector DL. The dynamics of chromosome organization and gene regulation. Annu Rev Biochem. 2003;72:573–608. doi: 10.1146/annurev.biochem.72.121801.161724. [DOI] [PubMed] [Google Scholar]
- 3.Hübner MR, Spector DL. Chromatin dynamics. Annu Rev Biophys. 2010;39:471–489. doi: 10.1146/annurev.biophys.093008.131348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumaran RI, Thakar R, Spector DL. Chromatin dynamics and gene positioning. Cell. 2008;132(6):929–934. doi: 10.1016/j.cell.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013;49(5):773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krijger PH, de Laat W. Identical cells with different 3D genomes; cause and consequences? Curr Opin Genet Dev. 2013;23(2):191–196. doi: 10.1016/j.gde.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 7.Henikoff S, Comai L. Trans-sensing effects: The ups and downs of being together. Cell. 1998;93(3):329–332. doi: 10.1016/s0092-8674(00)81161-7. [DOI] [PubMed] [Google Scholar]
- 8.Higgs DR, Wood WG. Long-range regulation of alpha globin gene expression during erythropoiesis. Curr Opin Hematol. 2008;15(3):176–183. doi: 10.1097/MOH.0b013e3282f734c4. [DOI] [PubMed] [Google Scholar]
- 9.Palstra RJ, de Laat W, Grosveld F. Beta-globin regulation and long-range interactions. Adv Genet. 2008;61:107–142. doi: 10.1016/S0065-2660(07)00004-1. [DOI] [PubMed] [Google Scholar]
- 10.Krueger C, Osborne CS. Raising the curtains on interchromosomal interactions. Trends Genet. 2006;22(12):637–639. doi: 10.1016/j.tig.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Ling JQ, et al. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312(5771):269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 12.Anguera MC, Sun BK, Xu N, Lee JT. X-chromosome kiss and tell: How the Xs go their separate ways. Cold Spring Harb Symp Quant Biol. 2006;71:429–437. doi: 10.1101/sqb.2006.71.012. [DOI] [PubMed] [Google Scholar]
- 13.Xu N, Tsai CL, Lee JT. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311(5764):1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- 14.Augui S, et al. Sensing X chromosome pairs before X inactivation via a novel X-pairing region of the Xic. Science. 2007;318(5856):1632–1636. doi: 10.1126/science.1149420. [DOI] [PubMed] [Google Scholar]
- 15.Phillips JE, Corces VG. CTCF: Master weaver of the genome. Cell. 2009;137(7):1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lomvardas S, et al. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126(2):403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 17.Fuss SH, Omura M, Mombaerts P. Local and cis effects of the H element on expression of odorant receptor genes in mouse. Cell. 2007;130(2):373–384. doi: 10.1016/j.cell.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 18.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435(7042):637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 19.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489(7414):91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489(7414):109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489(7414):75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hewitt SL, et al. RAG-1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci. Nat Immunol. 2009;10(6):655–664. doi: 10.1038/ni.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LaSalle JM, Lalande M. Homologous association of oppositely imprinted chromosomal domains. Science. 1996;272(5262):725–728. doi: 10.1126/science.272.5262.725. [DOI] [PubMed] [Google Scholar]
- 25.Thatcher KN, Peddada S, Yasui DH, Lasalle JM. Homologous pairing of 15q11-13 imprinted domains in brain is developmentally regulated but deficient in Rett and autism samples. Hum Mol Genet. 2005;14(6):785–797. doi: 10.1093/hmg/ddi073. [DOI] [PubMed] [Google Scholar]
- 26.Duncan IW. Transvection effects in Drosophila. Annu Rev Genet. 2002;36:521–556. doi: 10.1146/annurev.genet.36.060402.100441. [DOI] [PubMed] [Google Scholar]
- 27.Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9(10):692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 28.O’Shea JJ, Ma A, Lipsky P. Cytokines and autoimmunity. Nat Rev Immunol. 2002;2(1):37–45. doi: 10.1038/nri702. [DOI] [PubMed] [Google Scholar]
- 29.Croft M. The role of TNF superfamily members in T-cell function and diseases. Nat Rev Immunol. 2009;9(4):271–285. doi: 10.1038/nri2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waters JP, Pober JS, Bradley JR. Tumour necrosis factor and cancer. J Pathol. 2013;230(3):241–248. doi: 10.1002/path.4188. [DOI] [PubMed] [Google Scholar]
- 31.Falvo JV, Tsytsykova AV, Goldfeld AE. TNF Pathophysiology. Molecular and Cellular Mechanisms. In: Kollias G, Sfikakis PP, editors. Current directions in autoimmunity. Karger; Basel: 2010. pp. 27–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsytsykova AV, et al. Activation-dependent intrachromosomal interactions formed by the TNF gene promoter and two distal enhancers. Proc Natl Acad Sci USA. 2007;104(43):16850–16855. doi: 10.1073/pnas.0708210104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 34.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 35.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338(6113):1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 36.Batista PJ, Chang HY. Long noncoding RNAs: Cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masui O, Heard E. RNA and protein actors in X-chromosome inactivation. Cold Spring Harb Symp Quant Biol. 2006;71:419–428. doi: 10.1101/sqb.2006.71.058. [DOI] [PubMed] [Google Scholar]
- 38.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152(6):1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 39.Alleman M, et al. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442(7100):295–298. doi: 10.1038/nature04884. [DOI] [PubMed] [Google Scholar]
- 40.Gilmour DS, Thomas GH, Elgin SC. Drosophila nuclear proteins bind to regions of alternating C and T residues in gene promoters. Science. 1989;245(4925):1487–1490. doi: 10.1126/science.2781290. [DOI] [PubMed] [Google Scholar]
- 41.Strutt H, Cavalli G, Paro R. Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J. 1997;16(12):3621–3632. doi: 10.1093/emboj/16.12.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Omichinski JG, Pedone PV, Felsenfeld G, Gronenborn AM, Clore GM. The solution structure of a specific GAGA factor-DNA complex reveals a modular binding mode. Nat Struct Biol. 1997;4(2):122–132. doi: 10.1038/nsb0297-122. [DOI] [PubMed] [Google Scholar]
- 43.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452(7184):181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 44.Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell. 2012;45(5):598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang W, et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150(4):685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matharu NK, Hussain T, Sankaranarayanan R, Mishra RK. Vertebrate homologue of Drosophila GAGA factor. J Mol Biol. 2010;400(3):434–447. doi: 10.1016/j.jmb.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 47.Dion V, Gasser SM. Chromatin movement in the maintenance of genome stability. Cell. 2013;152(6):1355–1364. doi: 10.1016/j.cell.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 48.Krueger C, et al. Pairing of homologous regions in the mouse genome is associated with transcription but not imprinting status. PLoS ONE. 2012;7(7):e38983. doi: 10.1371/journal.pone.0038983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol. 2005;15(4):300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 50.Zwerschke W, et al. Modulation of type M2 pyruvate kinase activity by the human papillomavirus type 16 E7 oncoprotein. Proc Natl Acad Sci USA. 1999;96(4):1291–1296. doi: 10.1073/pnas.96.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muirhead H, et al. The structure of cat muscle pyruvate kinase. EMBO J. 1986;5(3):475–481. doi: 10.1002/j.1460-2075.1986.tb04236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44(27):9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- 53.Yaneva M, Tempst P. Isolation and mass spectrometry of specific DNA binding proteins. Methods Mol Biol. 2006;338:291–303. doi: 10.1385/1-59745-097-9:291. [DOI] [PubMed] [Google Scholar]
- 54.Papanastasiou M, et al. The Escherichia coli peripheral inner membrane proteome. Mol Cell Proteomics. 2013;12(3):599–610. doi: 10.1074/mcp.M112.024711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.