Significance
Double-stranded RNA (dsRNA) is a pathogen-associated molecular pattern that triggers the type-I interferon (IFN) response in mammalian cells. The IFN response up-regulates several dsRNA sensors, including closely related oligoadenylate synthetases (OAS). The functional roles of different oligoadenylate synthetases in dsRNA surveillance are not understood. Here, we use X-ray crystallography and biochemistry to demonstrate that human OAS1 and OAS3 recognize dsRNA molecules of different length. We show that domain duplication accompanied by a loss of catalytic activity provides the mechanism for sensing long dsRNA by OAS3. Our studies thus reveal different functions of OAS1 and OAS3 in dsRNA surveillance, identify a key role of domain duplication in the OAS family, and advance the fundamental understanding of the human innate immune system.
Keywords: dsRNA, OAS3, RNase L, interferon, 2-5A
Abstract
The mammalian innate immune system uses several sensors of double-stranded RNA (dsRNA) to develop the interferon response. Among these sensors are dsRNA-activated oligoadenylate synthetases (OAS), which produce signaling 2′,5′-linked RNA molecules (2-5A) that activate regulated RNA decay in mammalian tissues. Different receptors from the OAS family contain one, two, or three copies of the 2-5A synthetase domain, which in several instances evolved into pseudoenzymes. The structures of the pseudoenzymatic domains and their roles in sensing dsRNA are unknown. Here we present the crystal structure of the first catalytically inactive domain of human OAS3 (hOAS3.DI) in complex with a 19-bp dsRNA, determined at 2.0-Å resolution. The conformation of hOAS3.DI is different from the apo- and the dsRNA-bound states of the catalytically active homolog, OAS1, reported previously. The unique conformation of hOAS3.DI disables 2-5A synthesis by placing the active site residues nonproductively, but favors the binding of dsRNA. Biochemical data show that hOAS3.DI is essential for activation of hOAS3 and serves as a dsRNA-binding module, whereas the C-terminal domain DIII carries out catalysis. The location of the dsRNA-binding domain (DI) and the catalytic domain (DIII) at the opposite protein termini makes hOAS3 selective for long dsRNA. This mechanism relies on the catalytic inactivity of domain DI, revealing a surprising role of pseudoenzyme evolution in dsRNA surveillance.
Interferon (IFN)-inducible oligoadenylate synthetases (OASs) are mammalian sensors of double-stranded RNA (dsRNA) that are transcriptionally up-regulated during infections with pathogens, such as Staphylococcus aureus (1) or H1N1 swine flu virus (2). Human cells express four related OAS family members: OAS1, OAS2, OAS3, and OASL. Whereas OASL is catalytically inactive, the remaining family members are dsRNA-activated enzymes synthesizing 2′,5′-linked oligoadenylates (2-5A). The 2-5A serve as chemically unique second messengers that induce regulated RNA decay via RNase L (3, 4) and mediate antiviral and antibacterial innate immunity (5, 6). Here we report the structural and functional mechanism of dsRNA surveillance by the largest 2-5A synthetase, OAS3.
The core building unit of the OAS family is a polymerase beta (pol-β)-like nucleotidyl transferase domain, which shares structural similarity with poly-A polymerase, CCA-adding enzyme, and cytosolic dsDNA sensor cyclic GAMP synthetase (cGAS) (7, 8). Similar to these polymerases, OAS1 and OASL contain a single pol-β domain. However, OAS2 and OAS3 are unusual and contain two and three copies, respectively, acquired apparently by gene duplication (Fig. 1A) (9). The N-terminal pol-β–like domains of OAS2 and OAS3 are thought to have lost their enzymatic activity during evolution (3). In agreement with this hypothesis, mutagenesis of the C-terminal domains in OAS2 and OAS3 has been reported to inactivate these enzymes (10, 11).
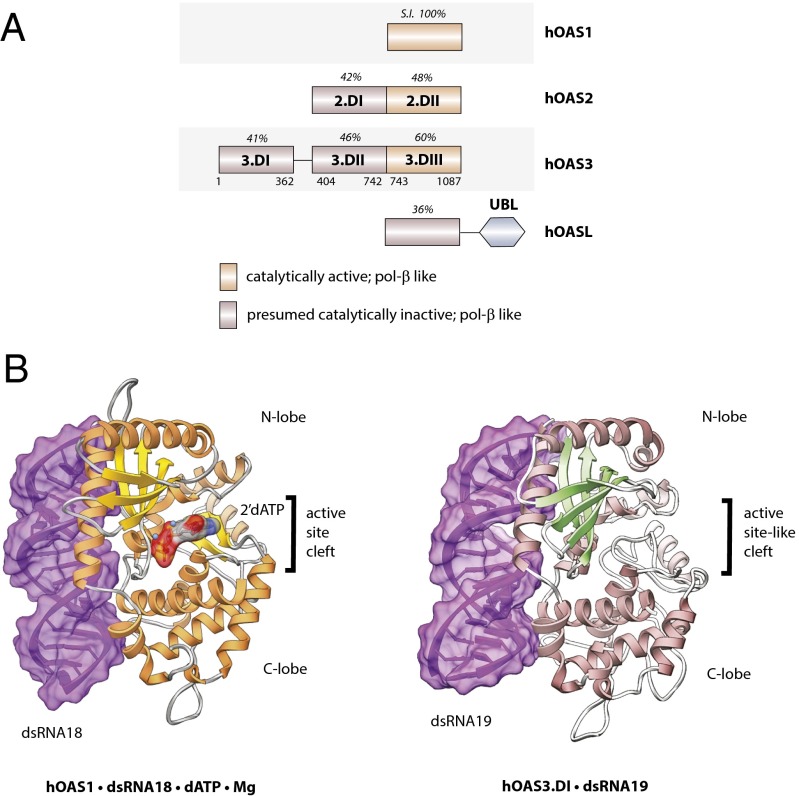
Fig. 1.
Structure of OAS3.DI in complex with dsRNA19. (A) Sequence relationships between human oligoadenylate synthetases. S.I. shows sequence identity (%) calculated from MUSCLE alignments using hOAS1 as a reference (27). Lower row of numbers in hOAS3 indicate amino acid position in the polypeptide chain. (B) Comparison of the previously reported crystal structure of activated OAS1 with dsRNA18, 2'-deoxy-ATP, and magnesium (7) with the crystal structure of hOAS3.DI with dsRNA19.
The functions of the nonenzymatic domains in OAS2 and OAS3 are largely unknown. Experimental measurements of dsRNA binding to these domains are unavailable, with the exception of a qualitative study, which reported weak binding of the N-terminal domain of OAS2 to poly I:C-modified Sepharose beads (12). This observation suggests that the noncatalytic domain of OAS2 (and perhaps OAS3) may have lost both the catalytic function and the affinity for dsRNA.
Recent structural and biochemical studies of human OAS1 (hOAS1) revealed the mechanism of dsRNA recognition and activation of this enzyme via a conformational change (7, 13). Here we report, to our knowledge, the first crystal structure of the pseudoenzymatic domain DI of human OAS3 (hOAS3.DI) in complex with dsRNA. This structure and complementary solution studies of hOAS3 and hOAS1 show that the duplicated noncatalytic domains have an essential role in recognition of long dsRNA by hOAS3.
Results and Discussion
Structure of hOAS3.DI Bound to dsRNA.
Human OAS3 is a 121-kDa protein composed of three tandem OAS1-like domains, DI, DII, and DIII (Fig. 1A). To understand how hOAS3 employs these domains for sensing dsRNA, we attempted to crystallize full-length hOAS3 and its domains individually. Only domains DI, DII, and DIII, but not full-length hOAS3, could be purified in sufficient amounts, and only domain DI crystallized. Our crystallization conditions included a nonhydrolyzable ATP analog, AMPCPP, magnesium, and dsRNA with 19 base pairs (dsRNA19; Methods). Electron density for hOAS3.DI and dsRNA19 was clearly observed (Fig. 1B and Fig. S1), and the resulting structure of the hOAS3.DI•dsRNA19 complex was determined at 2.0-Å resolution (Table S1).
As expected, the structure of hOAS3.DI has the nucleotidyl transferase fold and resembles the crystal structures of porcine and human OAS1 published previously (7, 13). The dsRNA19 molecule is bound to both N- and C-lobes of hOAS3.DI and resides on the opposite face from the active site-like pocket (Fig. 1B). In contrast to that of hOAS1 (7), the active site-like pocket of hOAS3.DI is unoccupied with magnesium ions or nucleotide.
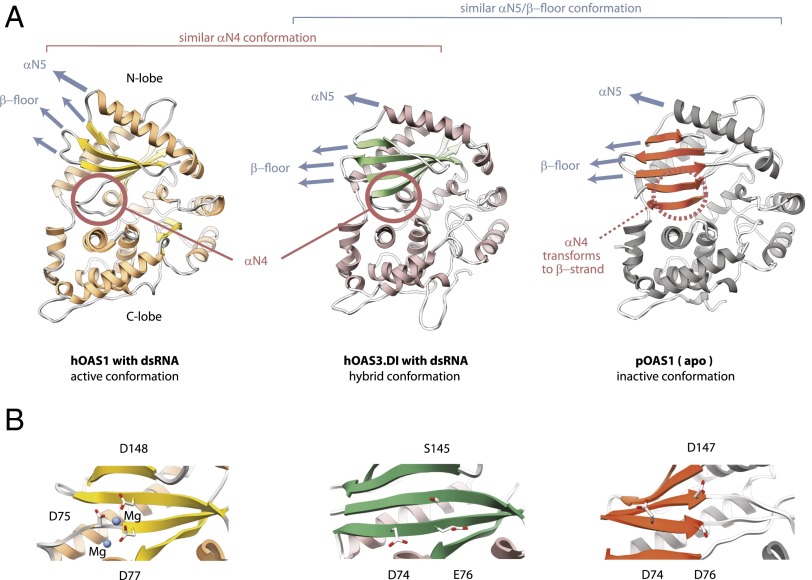
Previous structural analysis of OAS1 (7) revealed that the conformational switch distinguishing the inactive (apo) and active (dsRNA-bound) states of this enzyme involves β-floor sliding, ratchet-like motion of helix αN5, and formation of a new helix αN4 in the active site (Fig. 2A). In the hOAS3.DI•dsRNA19 complex reported here, the helix αN4 is present, but the β-floor/αN5 structure remains in the inactive-like conformation observed with apo-pOAS1. Comparison of hOAS1 with hOAS3.DI reveals that the β-floor/αN5 motion in hOAS3.DI may be limited due to two tryptophan residues, which anchor helix αN5 (Fig. S2A). Therefore, hOAS3.DI has a hybrid conformation, which combines elements of active and inactive states of OAS1.
Fig. 2.
Complex of hOAS3.DI with dsRNA19 has a hybrid conformation. (A) Comparison of three known crystal structures of the OAS proteins. (Left) hOAS1 with dsRNA, 2'-deoxy-ATP, and magnesium (PDB ID code 4IG8). (Center) hOAS3.DI with dsRNA19 (PDB ID code 4S3N). (Right) Porcine OAS1 (pOAS1) without bound ligands (PDB ID code 1PX5). Red circle highlights the αN4 helix. Blue arrows show the orientation of the conformationally mobile secondary structure elements in the N-lobe: β-sheet and helix αN5. (B) Configuration of the catalytic residues in hOAS1/pOAS1, and the equivalent residues in hOAS3.DI. The active configuration places the Mg2+-coordinating active site residues at a close distance, the inactive configuration places these residues apart.
The key to activation of 2-5A synthesis by hOAS1 is the assembly of the active site residues into a compact structure (7), which enables recruitment of Mg2+ ions and ATP (Fig. 2B Left vs. Right). A similar mechanism is responsible for activation of another pol-β–like enzyme, cGAS, which serves as a sensor of cytosolic dsDNA (8). In contrast to these examples, hOAS3.DI does not place the equivalent active site residues compactly even in the presence of dsRNA19 (Fig. 2B). This feature should disable the dsRNA-dependent catalytic switch and block 2-5A synthesis.
Biochemical Dissection of hOAS3 Defines Domain DI as a Pseudoenzyme with Enhanced dsRNA Binding.
Analysis of amino acid conservation revealed that key active site aspartate residues equivalent to D75, D77, and D148 of hOAS1 (Fig. 2B) are not conserved in domains DI and DII of hOAS3, which presumably leads to enzymatic inactivity of these domains (10). To experimentally define how each domain of hOAS3 functions in 2-5A synthesis, we expressed and purified all individual domains and assayed their activity with and without dsRNA. We used poly I:C as a model dsRNA (Fig. 3A).
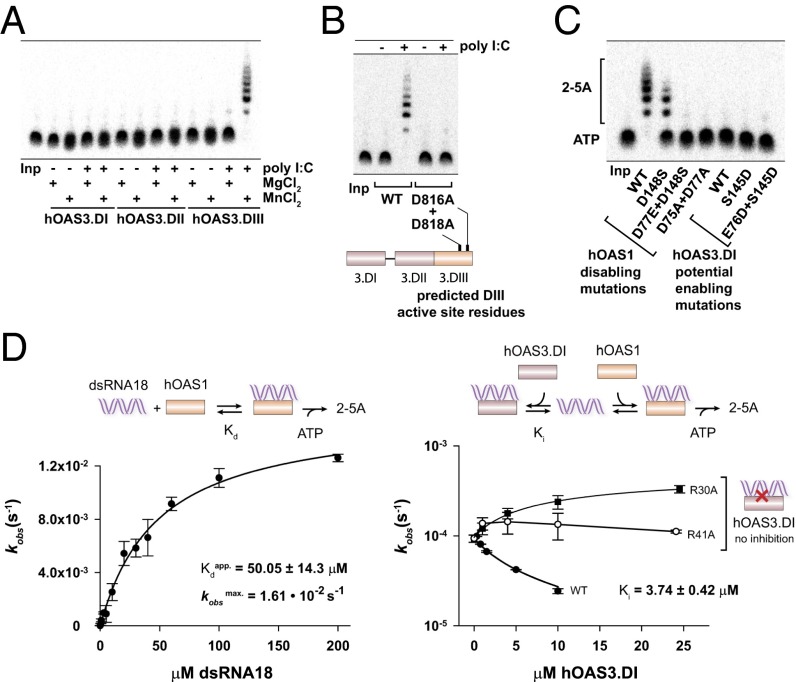
Fig. 3.
Roles of the individual domains of hOAS3 in sensing dsRNA. (A) Catalytic properties of individual domains of hOAS3 in the presence of dsRNA (2.5 OD260 poly I:C) and two divalent metal ions, Mg2+ or Mn2+. (B) Inactivation of hOAS3 by point mutations in the active site of domain DIII. Reactions contained full-length OAS3 (0.1 μM). (C) Active site mutagenesis in hOAS1 and hOAS3. Reactions contained 0.1 μM hOAS1 or hOAS3.DI, 2.5 OD260 poly I:C, 1 mM ATP and trace 32P-α-ATP, and were conducted at 37 °C for 2 h. Samples were resolved by 20% denaturing PAGE and visualized by phosphorimaging. (D, Left) activation of hOAS1 by dsRNA18 (Methods). (Right) Competition of hOAS3.DI and hOAS1 for binding to dsRNA18. WT hOAS3.DI (Ki = 3.74 μM) binds dsRNA stronger than hOAS1 (Kdapp = 50 μM), whereas two hOAS3.DI mutants that disrupt contacts to dsRNA (R30A and R41A) do not exhibit a detectable competition. Reactions contained 0.5 μM hOAS1 (Left), 1 μM hOAS1 (Right), and 100 nM dsRNA18 (Right). Data points show mean ± SD determined from at least two replicate time courses.
None of the purified domains synthesized 2-5A under the conditions that activate hOAS1 (7) (Fig. 3A; poly I:C and MgCl2 data). A similar observation has been reported for hOAS2: neither domain hOAS2.DI nor hOAS2.DII is active when expressed individually (12). Together, these results suggest that 2-5A synthesis by hOAS2 and hOAS3 may require the intact proteins. However, upon further experimentation we found that addition of Mn2+ ions activates rapid, dsRNA-dependent 2-5A synthesis by hOAS3.DIII (Fig. 3A, last lane). The mechanism of hOAS3.DIII activation by Mn2+ ions is presently unknown. However, reactions in the presence of Mn2+ allow, to our knowledge for the first time, studies of 2-5A synthesis by a single domain from the OAS family, other than OAS1. Our data show that the isolated domain hOAS3.DIII is fully sufficient for 2-5A synthesis and is functionally similar to hOAS1. Intact hOAS3, which contains all three domains, is not strictly required for 2-5A synthesis.
Domains DI and DII remain inactive under all conditions tested (Fig. 3A). Complementary analysis shows that domains DI and DII do not carry out 2-5A synthesis in the context of full-length hOAS3 as well. The active site mutations D816A+D818A in domain DIII render full-length hOAS3 catalytically inactive (Fig. 3B), in full agreement with an independently conducted recent study of hOAS3 (11).
The catalytic inactivity of domains DI and DII could be most simply explained by the loss of sequence conservation and mutations of the catalytic aspartate residues (10). To test this hypothesis, we mutated the active site aspartates in hOAS1 to the corresponding residues of hOAS3 (Fig. 2B, Left vs. Center). A single mutation D148S had a minor effect, but the double mutant D148S+D77E, and the control double mutant D75A+D77A inactivated hOAS1, as expected (Fig. 3C, hOAS1). Notably, the reverse experiment that restores the aspartate residues in the active site of hOAS3.DI, does not rescue 2-5A synthesis (Fig. 3C, hOAS3.DI). Therefore, the loss of the aspartate conservation does not fully explain the catalytic inactivity of hOAS3.DI. These results and the crystal structure of hOAS3.DI•dsRNA19 indicate that hOAS3.DI lacks enzymatic activity largely due to the inactive conformation of the catalytic center (Fig. 2B).
Although hOAS3.DI is catalytically inactive, the cocrystal structure with dsRNA19 demonstrates that this protein retains the ability to bind dsRNA (Fig. 1B). It is unknown whether the dsRNA-binding affinity is comparable to that of the catalytically active 2-5A synthetases, such as hOAS1, or compromised as a result of hOAS3.DI becoming a pseudoenzyme. To resolve this uncertainty and understand the function of the pseudoenzymatic domains of OAS3, we evaluated the binding of dsRNA to hOAS3.DI. To this end we used dsRNA with 18 base pairs (dsRNA18; Methods) as a model RNA duplex, and inhibition of hOAS1 activity by hOAS3.DI as a readout. We found that dsRNA18 binds to hOAS3.DI ∼14-fold stronger than to hOAS1 (KdhOAS1 = 50 μM vs. KihOAS3.DI = 3.7 μM, Fig. 3D; a similar ∼10-fold difference is observed using Ki of an inactive mutant hOAS1(D75N, D77N) for the comparison (KihOAS1(D75N, D77N) = 26 μM; Fig. S2 B and C). To confirm that the inhibition by hOAS3.DI results from the binding to dsRNA, we prepared two control mutants, R30A and R41A. Both mutants disrupt the hOAS3.DI/dsRNA interface in the crystal structure (Fig. S2D). As expected, hOAS3.DI with R30A or R41A mutations no longer inhibits hOAS1 (Fig. 3D, Right). The modest (∼3.5-fold) rate enhancement observed with the R30A mutant (Fig. 3D) must arise from modulating the activity of hOAS1, as the mutation per se does not render hOAS3.DI catalytically active (Fig. S2E).
The stronger binding of dsRNA to hOAS3.DI than to hOAS1 cannot be straightforwardly explained by comparing the respective protein/RNA interfaces. In the crystal structures, hOAS3.DI buries less surface area (2,588 Å2 for hOAS3.DI vs. 3,039 Å2 for hOAS1, PDB ID code 4IG8), and forms noticeably fewer contacts with dsRNA (Fig. S3). The enhanced binding of dsRNA to hOAS3.DI presumably arises from the conformational properties of hOAS3.DI, which binds dsRNA without fully abandoning the inactive-like conformation (Fig. 2A). Because the dsRNA-induced conformational changes in the proteins and the binding affinity of dsRNA are thermodynamically coupled, a less costly conformational rearrangement in hOAS3.DI should lead to a stronger binding of dsRNA. Together, our structural and functional data suggest that the hybrid conformation of hOAS3.DI not only disables 2-5A synthesis, but also favors the binding of dsRNA.
Domain DI Drives Recognition of Long dsRNA by hOAS3.
Domain DI is located at the N terminus, whereas the only catalytically active domain DIII resides at the C terminus of hOAS3. It is possible that hOAS3 wraps around the dsRNA target and uses domain DI to enhance the binding affinity, providing avidity (11). Alternatively, hOAS3 could remain extended, as suggested by SAXS envelope obtained with free OAS3 (11), which would use domain DI for recognition of longer dsRNA (Fig. S4A). To understand the mechanism of dsRNA sensing by hOAS3, we analyzed recognition of long, medium, and short dsRNA targets by this receptor. As a reference, we examined activation of hOAS1, which binds to ∼17 base pairs in the crystal structure (7) and should optimally recognize short dsRNA with fewer than 20 base pairs.
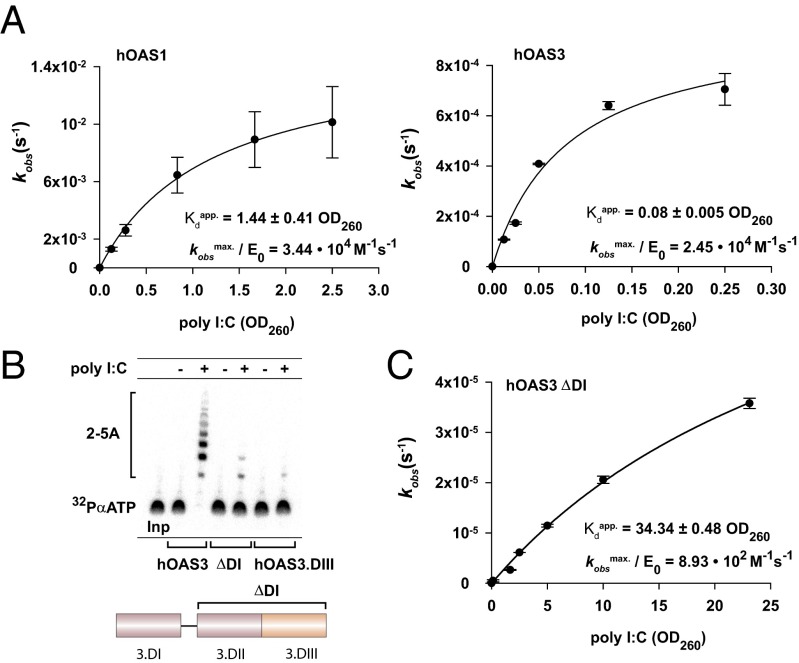
To examine recognition of long dsRNA, we used poly I:C as a model activator. Due to the heterogeneity of poly I:C (Fig. S5A; average length ∼120–130 bp), we expressed the dsRNA concentration in units of OD260. At saturating concentrations of poly I:C, hOAS1 and hOAS3 have similar specific activities of 2-5A synthesis (Fig. 4A). However, poly I:C titration reveals that hOAS3 binds long dsRNA ∼18-fold stronger than does hOAS1, indicating that hOAS3 is considerably more sensitive to long dsRNA. Notably, deletion of domain DI nearly abolished the catalytic activity of hOAS3 (Fig. 4B). The ΔDI mutant (residues 404–1087) exhibits a ∼30-fold smaller specific activity at saturating concentrations of poly I:C, and binds dsRNA ∼430-fold weaker (Fig. 4C). A manganese rescue experiment confirmed that the loss of dsRNA sensing by the ΔDI mutant is not due to nonspecific effects, such as protein misfolding, as ΔDI is intrinsically functional (Fig. S5B). Therefore, domain DI is critical for activation of hOAS3 by poly I:C, and must serve as a dsRNA-binding module. Deletion of the second pseudoenzymatic domain, DII, has a negligible impact on 2-5A synthesis, suggesting that domain DII serves as a spacer between domains DI and DIII (Fig. 4B, ΔDI vs. hOAS3.DIII). This model is further supported by mutagenesis of full-length hOAS3. Mutation K435A in domain DII, which removes a predicted dsRNA-binding basic residue, has a minor effect on activation of hOAS3 (Figs. S3 and S5C).
Fig. 4.
Importance of domain DI for the activation of hOAS3 by dsRNA. (A) Activation of hOAS1 (0.5 μM; Left) and hOAS3 (40 nM; Right) by poly I:C. (B) Comparison of 2-5A synthesis activity of hOAS3 full-length, isolated domains DII+DIII (ΔDI), and isolated domain DIII. Reactions contained 0.1 μM proteins and 2.5 OD260 poly I:C, as indicated, and were conducted at 37 °C for 2 h. (C) Dependence of the rate of 2-5A synthesis by ΔDI hOAS3 on poly I:C concentration. Reactions contained 0.1 μM ΔDI hOAS3. Apparent binding constants are specified in OD260 units. Data points represent mean ± SD determined from at least two replicate time courses. E0 is the total concentration of enzyme used.
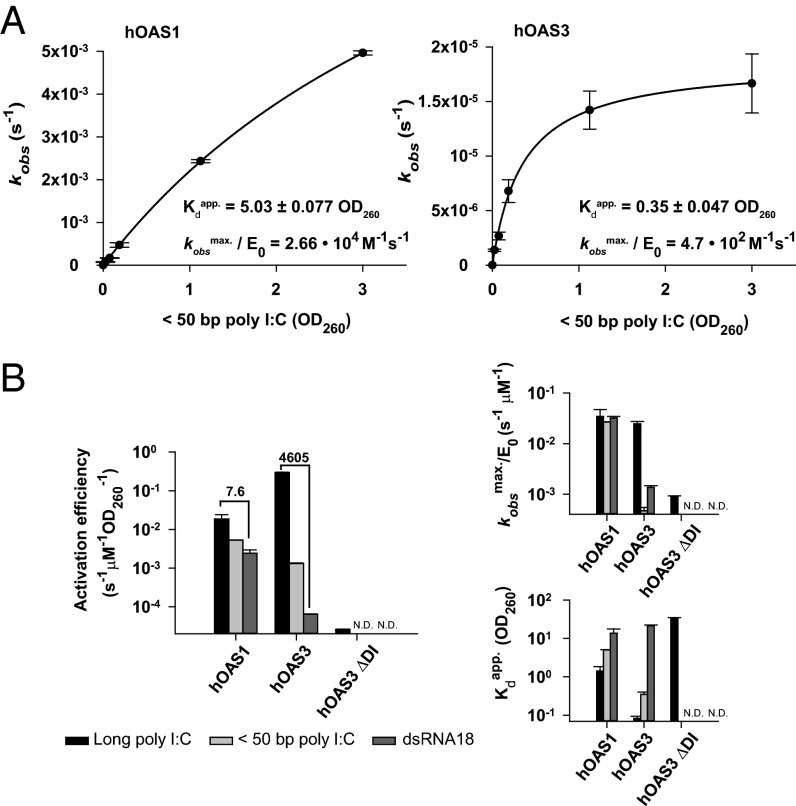
To probe activation of hOAS3 by medium-size dsRNA, we partially digested poly I:C with RNase III and purified products that contain 50 base pairs or less (Fig. S5A). The medium-size poly I:C had nearly the same binding preference for hOAS3 versus hOAS1 (∼14-fold; Fig. 5A) as observed above with long poly I:C (∼18-fold; Fig. 4A). The maximum specific activity of hOAS1 bound to medium and long poly I:C was also similar (Fig. 5B). However, the maximum specific activity measured with hOAS3 was ∼50-fold lower with medium-size poly I:C compared with long poly I:C (Fig. 5B). These results show that hOAS1 does not discriminate between long and medium-size poly I:C, whereas hOAS3 has a strong preference for dsRNA with more than 50 base pairs.
Fig. 5.
Differential sensing of long and short dsRNA by hOAS1 and hOAS3. (A) Activation of 2-5A synthesis by hOAS1 (0.5 μM; Left) and hOAS3 (40 nM; Right) in response to size-fractionated poly I:C with less than 50 base pairs. (B) Comparison of the activation parameters of hOAS1 and hOAS3 with dsRNA of different length. Activation efficiency (Left) is defined as the ratio of specific activity (kobsmax/E0) to the binding affinity (Kdapp), plotted individually on the smaller graphs (right). N.D., not determined. Brackets indicate the fold-difference between reactions with poly I:C and dsRNA18. Data represent mean ± SD determined from at least two independent time courses. E0 is the total concentration of enzyme used.
The preference for long poly I:C suggests that hOAS3 should have a linear arrangement of domains upon binding to dsRNA (Fig. S4A). Considering that a single domain of a 2-5A synthetase can accommodate ∼17–20 base pairs (Fig. 1B), <50-bp poly I:C should dock to two hOAS3 domains at a time and bind either to domains DI+DII or to domains DII+DIII (Fig. S5D). We have shown that the tandem DII+DIII binds dsRNA poorly (Fig. 4 B and C). Therefore, poly I:C with fewer than 50 base pairs should dock predominantly to domains DI+DII. This mechanism leaves the catalytic domain DIII mostly unoccupied and explains the low specific activity of hOAS3 observed in Fig. 5. Of note, the sensitivity of the OAS enzymes to dsRNA length may additionally arise, at least in part, from mechanisms that include assembly of hOAS3 oligomers or filaments on long dsRNA, as observed with MDA-5 (14). We do not expect that hOAS3 oligomerization has a major role, as we do not observe a cooperative activation of this protein (Fig. S4B).
Finally, we compared activation of hOAS3 and hOAS1 by transcribed dsRNA of defined sequence, using dsRNA18 and dsRNA54 as model substrates. Human OAS3 again exhibited a stronger preference for longer dsRNA compared with hOAS1 (Fig. 5B and Fig. S6 A and B). Together, our data show that hOAS3 is functionally distinct from hOAS1 and serves as a sensor of dsRNA with ∼50 base pairs or more. The different dsRNA length requirements of hOAS3 and hOAS1 suggest nonoverlapping biological functions of these receptors, which could explain the diversity of 2-5A synthetases in genomes of higher vertebrates (Fig. 1A).
Probing with hOAS3 and hOAS1 Reveals Relatively Long Endogenous dsRNA in HeLa Cells.
Pathogens are the most commonly described sources of intracellular dsRNA, however, mammalian cells also contain endogenous dsRNA molecules. It has been shown that nuclear RNA purified from leukemia cells can activate mixed 2-5A synthetases (15), whereas total RNA from HeLa cells can activate purified hOAS1 in vitro (16). The size of the endogenous dsRNA structures detected in these experiments is unknown and difficult to evaluate experimentally. The selectivity of hOAS3 for long dsRNA suggests a simple and potentially broadly applicable approach for detection of long dsRNA elements in heterogeneous RNA samples. We used this approach to examine endogenous dsRNA purified from human epithelial (HeLa) cells.
To evaluate the dsRNA size, we carried out a study with both hOAS1 and hOAS3. Our experiments confirmed the presence of hOAS1 activators in total HeLa RNA (Fig. S7A, lanes 11–13). We selected the concentration of poly I:C and dsRNA18 to match the specific activity observed with HeLa RNA (Fig. S7A, lanes 6–13). Next, we experimentally determined the concentration of hOAS3 that matches the rate of hOAS1 in the presence of poly I:C (Fig. S7A, lanes 20–22 vs. 8–10), and tested the effect of dsRNA18 and total HeLa RNA at this hOAS3 concentration. As expected, dsRNA18 barely activated hOAS3 (Fig. S7A, lanes 17–19 vs. 5–7). However, total HeLa RNA had a noticeably stronger effect (Fig. S7A, lanes 23–25 vs. 18–20), although the activation was not as strong as with poly I:C. These data suggest that the endogenous dsRNA species present in HeLa cells should be shorter than the effective size of poly I:C (<120–130 bp), but longer than dsRNA18 (>20 bp).
We note that our analysis of dsRNA presence is accurate only if hOAS1 and hOAS3 are not activated by single-stranded RNA (ssRNA) present in HeLa samples. However, OAS1 has been reported to synthesize 2-5A in the presence of both dsRNA and ssRNA (17). On close inspection, we noticed that ssRNA activators of OAS1 were obtained using transcription with T7 RNA polymerase, which can produce contaminating dsRNA by reading the primary transcript as a template (18, 19). To verify whether hOAS1 or hOAS3 can indeed be activated by ssRNA, we examined two ssRNA samples. One was transcribed with T7 RNA polymerase (ssT7) and the other obtained by chemical synthesis (ssD). Only ssT7 could activate hOAS1 and hOAS3, whereas ssD had no detectable activity (Fig. S7B). Therefore, hOAS1 and hOAS3 are exquisitely selective for dsRNA and can be used for dsRNA detection even in the presence of ssRNA.
Double-stranded RNA elements with fewer than 20 base pairs are relatively abundant in coding RNA (7); however, the dsRNA molecules detected in total HeLa RNA are longer and could arise from other sources, such as transcription of centromeric satellite DNA, antisense transcription, or additional mechanisms that can produce complementary RNA (20). If these dsRNA elements can become accessible to the OAS receptors in vivo, they could have homeostatic functions, which may explain increased basal OAS expression in human organs, such as thymus and bone marrow (21).
Conclusions
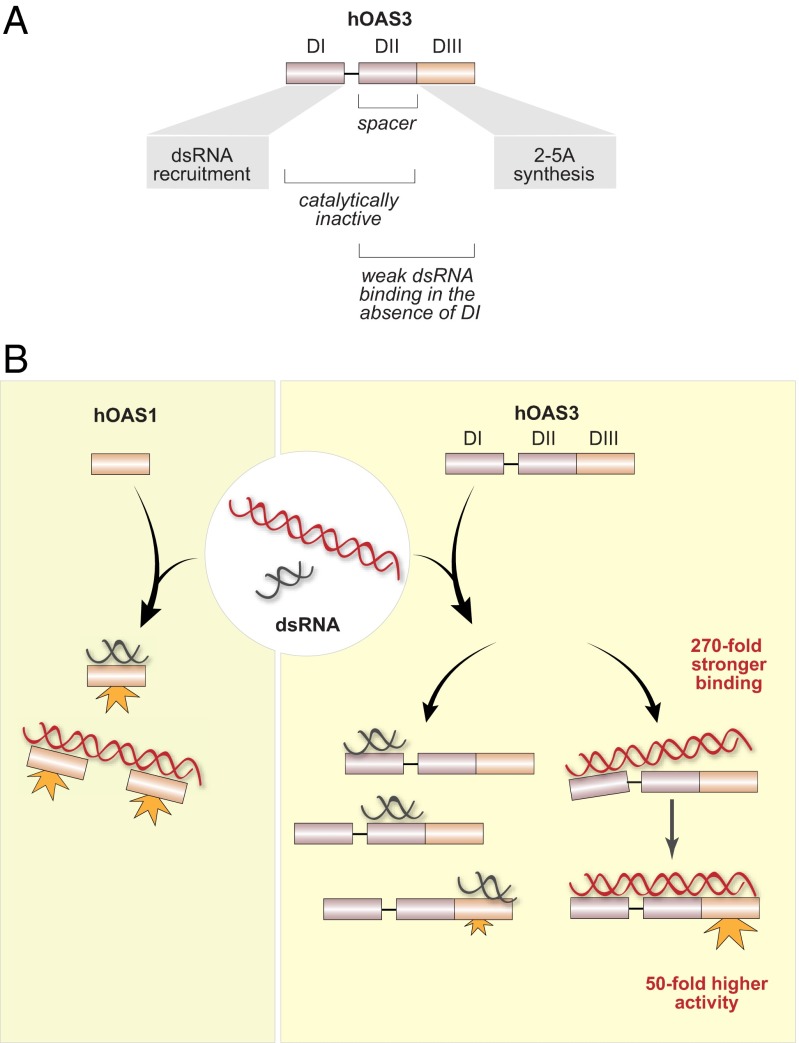
We show that hOAS3 is potently activated by dsRNA with more than 50 base pairs, which suggests that the specific role of hOAS3 is to sense long dsRNA. The discrimination between short and long dsRNA requires all three domains of hOAS3. The C-terminal domain DIII is the only catalytically active domain that carries out 2-5A synthesis in response to dsRNA (Fig. 6A). Domain DII is a pseudoenzyme, which not only lacks the enzymatic activity, but also makes no major contribution to activation of 2-5A synthesis in the absence of domain DI. Domain DI is also a pseudoenzyme, but with a high affinity for dsRNA. This arrangement makes hOAS3 sensitive only to sufficiently long dsRNA molecules, which span from domain DI to domain DIII. Domain duplication and the loss of catalysis in the duplicated domains thus provide a surprising mechanism for bypassing short and medium-size dsRNA, and activation of OAS3 only by long dsRNA.
Fig. 6.
Model of dsRNA surveillance by hOAS1 and hOAS3. (A) Roles of the individual domains of hOAS3 in sensing dsRNA. (B) Human OAS1 recognizes long and short dsRNA indiscriminately. Multiple hOAS1 copies are allowed to bind to long dsRNA, however they are unlikely to functionally interact as hOAS1 exhibits similar specific activity with long dsRNA (>50 bp) and short dsRNA (<20 bp). In contrast, hOAS3 exhibits a strong (∼104-fold) preference for long dsRNA. Short dsRNA binds to the high-affinity domain DI, but not to the low-affinity domains DII/DIII, which is insufficient to activate hOAS3. Long dsRNA binds to the high-affinity domain DI and docks intramolecularly to the catalytically active domain DIII, activating hOAS3.
The crystal structure of the hOAS3.DI•dsRNA19 complex provides, to our knowledge, the first atomic view of a pseudoenzyme from the OAS family. The unusual conformation of hOAS3.DI prevents catalysis even in the presence of dsRNA, but favors the binding of dsRNA. This observation suggests an evolutionary tradeoff between regulated 2-5A synthesis and strong dsRNA binding, both of which depend on the conformational properties of hOAS3.DI. This tradeoff is consistent with the escape from adaptive conflict model, wherein a protein with more than one function cannot improve one function (dsRNA binding) without sacrificing the other (2-5A synthesis), thereby necessitating gene or domain duplication (22).
The functions of the catalytically inactive domains in OAS2 and OASL await future characterization, but analogy with hOAS3.DI suggests that these domains could serve as potent dsRNA recruitment modules. Recently, it has been established that OASL forms a complex with RIG-I, a sensor of dsRNA with a helicase domain, and enhances its signaling (23). The role of OASL in the OASL•RIG-I complex may parallel that of domain DI in hOAS3, and render RIG-I sensitive to longer dsRNA.
Previous studies of mammalian oligoadenylate synthetases focused largely on the enzymatically active domains. Here we show that the noncatalytic domains of hOAS3 have an essential function and drive recognition of long dsRNA. Among four human oligoadenylate synthetases, three have noncatalytic domains, arranged uniquely in each sensor. Our results suggest that these domains should give distinct specificities to oligoadenylate synthetases and determine their roles in the IFN response.
Methods
DNA Plasmids.
The coding region of human OAS3 was amplified by PCR from an in-house cDNA library using poly I:C transfected HeLa cells and cloned into pGEX-6P vector. Sequencing revealed the clone to have a naturally occurring Arg18Lys polymorphism (GenBank BP285234.1, BP288205.1, BP286010.1). The construct for OAS3 DI was made using site-directed mutagenesis that introduced the Leu372Stop mutation. The DII construct was generated by deleting the coding region for OAS3 residues 1–403 and introducing the Tyr743Stop mutation. The DIII construct was created by deleting the coding region for OAS3 residues 1–742. Plasmids for full length (1–1087) and ΔDI OAS3 (residues 404–1087) encoded a C-terminal 6×-His tag added by site-directed mutagenesis. The human OAS1 construct in pGEX-6P encoding residues 1–346 has been described (7). All constructs used in this study were verified by DNA sequencing.
Protein Purification.
Human OAS1 was purified as described (7). Individual domains of OAS3 were purified as described for human OAS1. Escherichia coli BL21 Codon Plus RIPL (Applied Biosystems) transformed with plasmids encoding C-terminally 6×-His-tagged full-length or ΔDI OAS3 were grown in LB at 22 °C until OD600 = 1 and induced with 1 mL of 60 mg/mL IPTG per L of culture. Bacteria were harvested and the cell pellets were lysed in buffer A (20 mM Hepes pH 7.5, 300 mM NaCl, 10% (vol/vol) glycerol, 0.1 mM EDTA, 2 mM DTT, 30 mM imidazole, 1% Triton X-100, 1× Roche complete protease inhibitors) using Avestin EmulsiFlex C3. Cell lysates were cleared by centrifugation at 35,000 × g for 30 min at 4 °C. Proteins were purified on Nickel Superflow Resin (Clontech) followed by size-exclusion FPLC. Full-length and ΔDI OAS3 were further purified by MonoS ion-exchange chromatography and then exchanged into buffer A without Triton X-100 and imidazole. Protein concentrations were quantified by UV spectrophotometry. ΔDI construct was quantified by band densitometry of SDS/PAGE gels, using BSA as a standard.
Double-Stranded RNA.
Short synthetic dsRNAs used for kinetics assays and crystallography were purchased as individual strands from Dharmacon. Duplexes were generated by annealing the ssRNAs. Poly I:C was purchased from Sigma. All RNAs were dissolved in RNA buffer (20 mM Hepes pH 7.5, 100 mM NaCl). Sequences of dsRNA used: 5′-GGAUUUUGACCUUUAUGC-3′ (top strand of dsRNA18); 5′-GGCUUUUGACCUUUAUGAA-3′ (top strand of dsRNA19). In vitro transcribed ssT7 (Fig. S7B), its reverse complement, and both strands of dsRNA54 were synthesized with the MegashortScript kit (Invitrogen) using purified PCR-DNA template. Transcripts were phenol-chloroform extracted and PAGE-purified before the use in OAS assays.
RNase III Digest.
Poly I:C (200 μg) was incubated with 0.08 U/μL ShortCut RNase III (New England Biolabs) for 1 h at 37 °C. Reactions were stopped with EDTA according to the manufacturer’s protocol, phenol:chloroform (1:1) extracted, and ethanol-precipitated in the presence of 10 μg glycogen. Digested poly I:C was separated by electrophoresis using a 4% agarose gel, and the RNA was excised. Poly I:C was recovered by the freeze and squeeze method. Briefly, gel slices in 1.5-mL microfuge tubes were covered with TE buffer pH 7.5, melted at 95 °C, and frozen at −80 °C for 10 min. After freezing the samples were centrifuged at 21,130 × g for 10 min at room temperature and the supernatants saved. The melt/freeze/centrifuge process was repeated three times. The pooled supernatant was extracted with isobutanol (1:1) to remove SybrSafe dye and the RNA was recovered from the aqueous phase by ethanol precipitation in the presence of 10 μg of glycogen. Recovered poly I:C was resuspended in RNA buffer (20 mM Hepes pH 7.5, 100 mM NaCl) and quantified by UV spectrophotometry.
OAS Activity Assays.
Kinetics analyses were carried out at 37 °C using the concentrations of dsRNA and OAS proteins indicated for each experiment. Reactions contained 20 mM Hepes pH 7.5, 70 mM NaCl, 5 mM MgCl2 (or 5 mM MnCl2 when indicated), 10% (vol/vol) glycerol, 4 mM DTT, and 0.05% Triton X-100. Reactions were done and analyzed as described (7). Observed rate constants (kobs) were determined by single-exponential decay fitting in SigmaPlot. Binding constants were obtained from hyperbolic fit to the equation kobs = (kobsmax•[dsRNA])/(Kdapp + [dsRNA]). Activation efficiency was calculated as (kobsmax/E0)/Kdapp, and represents the ratio of specific activity to dsRNA binding affinity. Of note, although the titrations kobs vs. [dsRNA] result in Michaelis–Menten-like profiles, these titrations measure Kd rather than Km because dsRNA is not a substrate or an enzyme, but an activator in the reaction.
Cell Culture.
HeLa cells were cultured in MEM + 10% FBS at 37 °C with a humidified 5% CO2 atmosphere. Total RNA was extracted using TRIzol from cultures at ∼90% confluence.
OAS3.DI•dsRNA19 Crystallization.
Crystallization complexes contained 20 mg/mL DI, 582 μM dsRNA19, 5 mM AMPCPP, 10 mM MgCl2, 142 mM NaCl, and 13.4 mM Hepes pH 7.5. Crystals were grown using the hanging drop vapor diffusion method by mixing the crystallization complex 1:1 with reservoir solution (200 mM sodium citrate, 150 mM NaCl, 12% PEG 3350). Crystals were transferred in cryo-protectant (200 mM sodium citrate, 12% PEG 3350, 25% ethylene glycol) and frozen in liquid nitrogen.
X-Ray Data Collection and Structure Determination.
X-ray diffraction data were collected using beamline X29 at the National Synchrotron Light Source (Brookhaven National Laboratory). Data were collected at wavelength 1.075 Å. Data were processed with the XDS package (24). Crystals contain one DI-dsRNA binary complex in the asymmetric unit and belong to the monoclinic P21 space group (Table S1). The structure was solved by molecular replacement in PHASER using hOAS1 (PDB ID code 4IG8) as the search model. The structure was modeled in COOT (25) and refined by simulated annealing using PHENIX (26). The orientation of dsRNA was determined by refining the data against a model in which the dsRNA was built in the opposite direction (Fig. S1). The final model has good stereochemical parameters and no Ramachandran outliers (Table S1).
Supplementary Material
Acknowledgments
We thank the staff at the National Synchrotron Light Source, beam line X29, for on-site help and members of the A.K. laboratory for insightful discussions and feedback during writing. We also thank Professor Andrei Korostelev for reading and commenting on the finished manuscript and the reviewers for making valuable suggestions. This study was funded by Princeton University, NIH Grant 1R01GM110161-01 (to A.K.), Sidney Kimmel Foundation (to A.K.), and Burroughs Wellcome Foundation (to A.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4S3N).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419409112/-/DCSupplemental.
References
- 1.Koziel J, et al. Phagocytosis of Staphylococcus aureus by macrophages exerts cytoprotective effects manifested by the upregulation of antiapoptotic factors. PLoS ONE. 2009;4(4):e5210. doi: 10.1371/journal.pone.0005210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shapira SD, et al. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139(7):1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8(7):559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han Y, et al. Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response. Science. 2014;343(6176):1244–1248. doi: 10.1126/science.1249845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin RJ, et al. Distinct antiviral roles for human 2′,5′-oligoadenylate synthetase family members against dengue virus infection. J Immunol. 2009;183(12):8035–8043. doi: 10.4049/jimmunol.0902728. [DOI] [PubMed] [Google Scholar]
- 6.Li XL, et al. An essential role for the antiviral endoribonuclease, RNase-L, in antibacterial immunity. Proc Natl Acad Sci USA. 2008;105(52):20816–20821. doi: 10.1073/pnas.0807265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donovan J, Dufner M, Korennykh A. Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. Proc Natl Acad Sci USA. 2013;110(5):1652–1657. doi: 10.1073/pnas.1218528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Civril F, et al. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498(7454):332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Justesen J, Hartmann R, Kjeldgaard NO. Gene structure and function of the 2′-5′-oligoadenylate synthetase family. Cell Mol Life Sci. 2000;57(11):1593–1612. doi: 10.1007/PL00000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarkar SN, Ghosh A, Wang HW, Sung SS, Sen GC. The nature of the catalytic domain of 2′-5′-oligoadenylate synthetases. J Biol Chem. 1999;274(36):25535–25542. doi: 10.1074/jbc.274.36.25535. [DOI] [PubMed] [Google Scholar]
- 11.Ibsen MS, et al. The 2′-5′-oligoadenylate synthetase 3 enzyme potently synthesizes the 2′-5′-oligoadenylates required for RNase L activation. J Virol. 2014;88(24):14222–14231. doi: 10.1128/JVI.01763-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marié I, Rebouillat D, Hovanessian AG. The expression of both domains of the 69/71 kDa 2′,5′ oligoadenylate synthetase generates a catalytically active enzyme and mediates an anti-viral response. Eur J Biochem. 1999;262(1):155–165. doi: 10.1046/j.1432-1327.1999.00361.x. [DOI] [PubMed] [Google Scholar]
- 13.Hartmann R, Justesen J, Sarkar SN, Sen GC, Yee VC. Crystal structure of the 2′-specific and double-stranded RNA-activated interferon-induced antiviral protein 2′-5′-oligoadenylate synthetase. Mol Cell. 2003;12(5):1173–1185. doi: 10.1016/s1097-2765(03)00433-7. [DOI] [PubMed] [Google Scholar]
- 14.Wu B, et al. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152(1-2):276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 15.Hubbell HR, et al. Heterogeneous nuclear RNA from hairy cell leukemia patients activates 2′,5′-oligoadenylate synthetase. Anticancer Res. 1991;11(5):1927–1932. [PubMed] [Google Scholar]
- 16.Dan M, Zheng D, Field LL, Bonnevie-Nielsen V. Induction and activation of antiviral enzyme 2′,5′-oligoadenylate synthetase by in vitro transcribed insulin mRNA and other cellular RNAs. Mol Biol Rep. 2012;39(7):7813–7822. doi: 10.1007/s11033-012-1624-x. [DOI] [PubMed] [Google Scholar]
- 17.Hartmann R, et al. Activation of 2′-5′ oligoadenylate synthetase by single-stranded and double-stranded RNA aptamers. J Biol Chem. 1998;273(6):3236–3246. doi: 10.1074/jbc.273.6.3236. [DOI] [PubMed] [Google Scholar]
- 18.Triana-Alonso FJ, Dabrowski M, Wadzack J, Nierhaus KH. Self-coded 3′-extension of run-off transcripts produces aberrant products during in vitro transcription with T7 RNA polymerase. J Biol Chem. 1995;270(11):6298–6307. doi: 10.1074/jbc.270.11.6298. [DOI] [PubMed] [Google Scholar]
- 19.Schlee M, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31(1):25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leonova KI, et al. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc Natl Acad Sci USA. 2013;110(1):E89–E98. doi: 10.1073/pnas.1216922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dezso Z, et al. A comprehensive functional analysis of tissue specificity of human gene expression. BMC Biol. 2008;6:49. doi: 10.1186/1741-7007-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hahn MW. Distinguishing among evolutionary models for the maintenance of gene duplicates. J Hered. 2009;100(5):605–617. doi: 10.1093/jhered/esp047. [DOI] [PubMed] [Google Scholar]
- 23.Zhu J, et al. Antiviral activity of human OASL protein is mediated by enhancing signaling of the RIG-I RNA sensor. Immunity. 2014;40(6):936–948. doi: 10.1016/j.immuni.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst. 1993;26(6):795–800. [Google Scholar]
- 25.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 27.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.