Significance
How organisms adapt to the biotic and abiotic environment is a major question in evolutionary biology that addresses how natural selection shapes biodiversity. Using mass spectrometry, we characterized natural variation in major defense molecules, aliphatic glucosinolates, in hundreds of ecotypes of the model plant Arabidopsis thaliana, spanning the native range of the species. Using extensive genomic resources and field experiments, we provide strong evidence that populations are adapted to local herbivore communities along a striking longitudinal cline. In addition, we show that only a few genes of strong effect govern this natural variation and that alleles at these genes, located on different chromosomes, appear to have coevolved through epistatic selection.
Keywords: Arabidopsis thaliana, glucosinolates, genome-wide association mapping, linkage disequilibrium, adaptation
Abstract
The “mustard oil bomb” is a major defense mechanism in the Brassicaceae, which includes crops such as canola and the model plant Arabidopsis thaliana. These plants produce and store blends of amino acid-derived secondary metabolites called glucosinolates. Upon tissue rupture by natural enemies, the myrosinase enzyme hydrolyses glucosinolates, releasing defense molecules. Brassicaceae display extensive variation in the mixture of glucosinolates that they produce. To investigate the genetics underlying natural variation in glucosinolate profiles, we conducted a large genome-wide association study of 22 methionine-derived glucosinolates using A. thaliana accessions from across Europe. We found that 36% of among accession variation in overall glucosinolate profile was explained by genetic differentiation at only three known loci from the glucosinolate pathway. Glucosinolate-related SNPs were up to 490-fold enriched in the extreme tail of the genome-wide scan, indicating strong selection on loci controlling this pathway. Glucosinolate profiles displayed a striking longitudinal gradient with alkenyl and hydroxyalkenyl glucosinolates enriched in the West. We detected a significant contribution of glucosinolate loci toward general herbivore resistance and lifetime fitness in common garden experiments conducted in France, where accessions are enriched in hydroxyalkenyls. In addition to demonstrating the adaptive value of glucosinolate profile variation, we also detected long-distance linkage disequilibrium at two underlying loci, GS-OH and GS-ELONG. Locally cooccurring alleles at these loci display epistatic effects on herbivore resistance and fitness in ecologically realistic conditions. Together, our results suggest that natural selection has favored a locally adaptive configuration of physically unlinked loci in Western Europe.
Both wild and cultivated plants face a great variety of natural enemies, including herbivores and pathogens, that can negatively impact their growth and yield (1, 2). To afford protection, plants use a wide array of defenses, such as the biosynthesis of toxic secondary metabolites. One such chemical defense is the glucosinolate-myrosinase system that is pervasive in the mustard family Brassicaceae, including crops such as canola and the model plant Arabidopsis thaliana (2–4). Glucosinolates (GSLs) form a diverse class of amino acid-derived thioglycosides. Myrosinases, a corresponding family of glycoside hydrolases, are sequestered from the GSLs in healthy plant tissue. Upon tissue rupture, myrosinases hydrolyze the GSLs to various products including isothiocyanates and nitriles (2), the specific structure and bioactivity of which is dependent upon the GSL side chain and the prevailing chemical conditions during hydrolysis (2). Although GSL hydrolysis products generally inhibit herbivory and the growth of pathogens, many pests have specialized adaptations to avoid toxic effects and use GSLs or their hydrolysis products as feeding or oviposition stimulants (5–7). Biosynthesis of methionine-derived GSLs in the model plant A. thaliana is characterized by three successive multistep phases of side-chain elongation, GSL core construction, and side-chain modification, involving dozens of genes (1, 7). Species in the Brassicaceae display extensive variation for GSL profiles (2, 5, 8–10). Intraspecific variation has been particularly well documented in accessions of A. thaliana, which produces blends of more than 30 GSL molecules (2, 11–13).
A recent study showed that the GS-Elong locus, involved in side-chain elongation of GSLs, is correlated with the abundance of aphids along a longitudinal cline across Europe (14). The role of selection in generating this pattern has been supported with an experimental selection study under controlled conditions (14), although the selective role of particular natural enemies, including aphids, on A. thaliana in Europe is unknown. In addition, although the European cline in GS-Elong suggests that A. thaliana has been selected by herbivores to alter the length of its GSLs, no study has yet examined how well Arabidopsis chemotypes are defended in the field in Europe. Furthermore, little is known about how selection acts on other aspects of glucosinolate diversity. Here, we take a genomic perspective and explore how selection has shaped the loci regulating the biosynthesis and natural variation in a complex suite of GSLs across Europe.
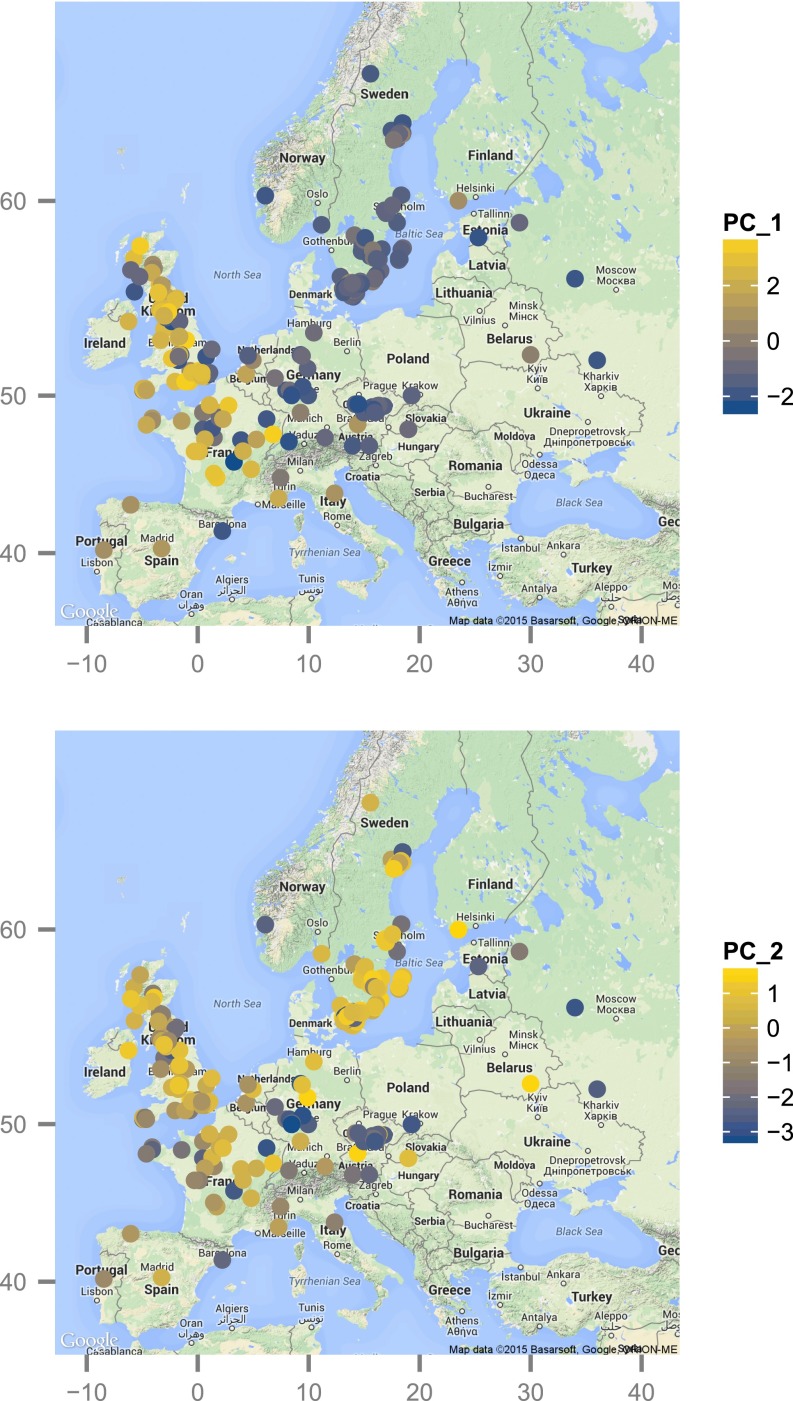
We characterized the relative concentrations of 22 methionine-derived GSLs in the leaves of 595 A. thaliana accessions collected from across its geographic range (15) (Table S1 and Dataset S1). Under controlled greenhouse conditions, natural accessions displayed genetic variation for the blend of molecules they produce. The plant genotype (accession) explained a significant proportion of the phenotypic variation for 16 out of the 22 molecules studied, with broad-sense heritabilities ranging from 5% to 73% (Fig. S1). The first principal component describing this variation explained 58% of the variation among accessions. This component was characterized by high positive loading scores for butenyls, pentenyls, and their respective hydroxylated products. The second principal component explained approximatively 25% of the variation and was mostly positively loaded with short-chain alkenyls, 2-propenyl (sinigrin) and 3-butenyl. Both components (hereafter GSL profile) displayed strong geographical clines (Fig. 1 and Table S2), with accessions from Western Europe containing far more alkenyl and hydroxyalkenyl GSLs than populations across Central Europe, Eastern Europe, and Sweden (Fig. 1 and Fig. S2).
Fig. 1.
Map of methionine-derived GSL variation in Europe. The x and y axes correspond to longitude and latitude, respectively. Dots indicate collection sites, and the color reflects the score of each accession along the first (top map) and second (bottom map) principal component describing GSL profile variation. The gradient from blue to yellow represents an increase in alkenyl and hydroxyalkenyl GSLs (Fig. S2).
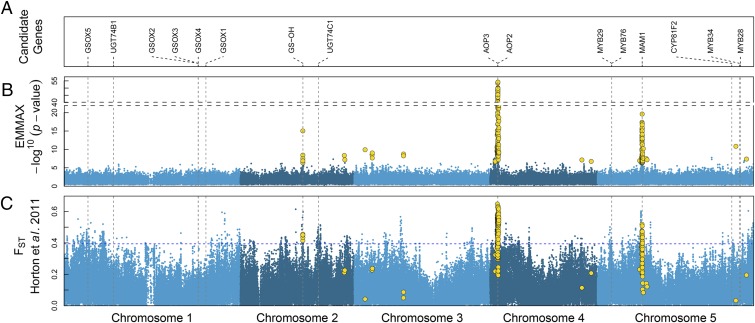
We sought to identify regions of the genome important in shaping GSL profiles by completing a large genome-wide association (GWA) study with our 595 accessions. Each of these accessions has been genotyped for 197,763 single-nucleotide polymorphisms (SNPs) (15, 16). We estimated associations in a mixed-model framework that controlled for confounding due to population structure by including a matrix of kinship among accessions as a random effect (17, 18). Significance was determined by permuting phenotypes 10,000 times for each GSL molecule. A total of 474 significant associations were detected for 12 of the 22 individual GSL molecules used for mapping (Dataset S1). These significant associations represent 227 unique SNPs, all detected for multiple molecules (from 3 to 11). The three main regions corresponded to regions near confirmed GSL biosynthesis genes (Fig. 2 A and B, and Dataset S1), with our strongest association located on chromosome 4 near the GS-AOP/GS-OHP locus, composed of the genes AOP2 (AT4G03060, alkenyl hydroxalkyl producing 2) and AOP3 (AT4G03050) (hereafter “AOP region”) (11). Our second strongest association, located on chromosome 5, was situated near the GS-ELONG locus, containing the genes MAM1 (AT5G23010, methylthioalkylmalate synthase 1) and MAM3 (AT5G23020, methylthioalkylmalate synthase-like) (hereafter “MAM region”). Next, a region of chromosome 2 contained four significantly associated SNPs located within 6 kbp (kb) of the GS-OH locus (AT2G25450), which is known to regulate hydroxylation of alkenyl GSLs (hereafter the “GS-OH” region) (19). Associations near GS-OH were detected when mapping the leaf concentrations of progoitrin (2-hydroxy-3-butenyl), consistent with previous studies (1, 19), as well as napoleiferin (2-hydroxy-4-pentenyl). Although other SNPs distributed throughout the genome were significantly associated with GSL variation, these were primarily individual markers with no obvious candidates located within 20 kbp. The extensive genetic variation captured in our large sample of accessions allowed the identification of the GS-OH locus, which went undetected in a previous GWA study that included fewer accessions, but spanned a similar geographical range (20).
Fig. 2.
Confirmed glucosinolate genes, GWA mapping, and scan. (A) Positions of confirmed genes from the methionine-derived GSL biosynthesis pathway along the five chromosomes of A. thaliana. (B) Manhattan plot of mapping results using EMMAX pooled from all 22 molecules analyzed. SNPs with significant association scores are marked in gold. (C) Wright’s fixation index scan along the Arabidopsis genome, based on 1,080 European accessions, divided into nine populations (15). The blue dotted line represents the 99.5% quantile of the distribution. SNPs significantly associated with GSL variation are marked in gold.
Despite the previously unidentified detection of GS-OH, it was surprising that no other known GSL genes were identified. To explore the possibility that GSL loci were missed due to genetic/allelic heterogeneity or skewed allele frequencies, we performed genome-wide scans in regional mapping populations from France, Sweden, and the United Kingdom (15, 21). These scans identified two additional peaks in Sweden, but no genes related to GSL biosynthesis were located within 20 kbp. We also calculated both GSL ratios for compounds on adjacent steps of the biosynthetic pathway and composite GSLs (1, 22). This strategy also failed to identify GSL loci other than GS-ELONG, GS-AOP/GS-OHP, and GS-OH (see Dataset S1 for all association mapping results). Previous studies found variation among accessions for the conversion of methylthioalkyl to methylsulfinylalkyl and mapped several GS-OX loci using crosses (11, 23–25). We did not find significant associations near known flavin monooxygenases (FMO) genes, now known to constitute the GS-OX loci (25). Genetic heterogeneity at the FMO genes, known to have redundant functions, may explain our failure to detect them (21, 25). Overall, and in agreement with previous studies, our results suggest that only three loci explain most of the natural variation in GSL profiles, even though many genes have been implicated in the biosynthesis of aliphatic GSLs.
The key loci governing variation in GSL profiles are located on three different chromosomes, raising the possibility that a genome-wide signature of selection could be detected. We investigated the degree of overlap between our 227 GSL-related SNPs and three published scans of selection using the RegMap panel (15) from which our accessions derived. Although neither the composite likelihood ratio of the allele frequency spectrum (26) nor the pairwise haplotype sharing test (27) showed significant overlap with GSL-associated SNPs, the 0.5% tail of the (28, 29) distribution displayed 28-fold enrichment for SNPs associated with GSL variation (Fig. 2C) and the 0.1% tail of the distribution displayed 490-fold enrichment. The MAM, GS-OH, and AOP regions all displayed high values, suggesting strong adaptive differentiation among populations. The fact that the genetics of this quantitative trait leaves a clear signature of selection across the genome suggests that variation in the GSL profile is an important adaptive trait.
To confirm the adaptive role of the GSL profile, we conducted field experiments in Lille, France (i.e., in the west of Europe), a region enriched in the hydroxy-alkenyl GSLs that contribute most to natural variation in GSL profiles. First, we scored herbivore damage inflicted on the rosettes of 256 natural accessions in three common garden experiments over 2 successive years. Most of the rosette damage resulted from feeding by insect herbivores, although it is possible that molluscan herbivores also contributed despite efforts to exclude them (Fig. S3). Second, at the end of the field experiments, we estimated the total length of mature fruits produced by each plant, a proxy for lifetime fitness in this mostly selfing, annual species (30). This fitness estimate was highly heritable, with a broad-sense heritability of 51.28% (95% confidence interval, 44.76–58.22) across experiments (Tables S3 and S4).
We explored the adaptive value of the GSL profile by investigating its relationship with herbivore damage and lifetime fitness across the experiments. The level of herbivore damage was negatively correlated with the principal component capturing most of the GSL profile variation among accessions (Spearman ; P = 0.008), despite GSLs being characterized on undamaged plants under greenhouse conditions (31). This indicates that accessions enriched in butenyls, pentenyls, and their hydroxylated derivatives were protected from herbivores in France. In a field study in Ohio, European accessions enriched in alkenyl GSLs were less protected (32). This disparity suggests herbivore communities vary in their GSL preference. The reduced herbivory that we detected translated to significantly higher lifetime fitness, even after including a covariate to control for “home vs. away” effects captured by the distance between Lille and the original collection site of the accessions (Table S5). The geographical distance to the collection site also had a significant, negative impact on plant fitness, consistent with previous studies showing adaptation to climate in natural populations of A. thaliana (33, 34).
In addition to testing the influence of the GSL profile on fitness, we tested the effect of GS-OH, MAM, and AOP directly. To capture as much of the complex allelic variation present as possible (35, 36), we characterized the genetic variation at each locus with a pairwise genetic distance matrix (, Fig. S4). We tested the effects of the three genetic distance matrices, as well as their two- and three-way interactions, on a matrix of fitness differences between accessions. Our regression also included a genetic distance matrix calculated for all other SNPs in the genome to guard against confounding due to population structure and/or adaptive genetic variation at other loci. The resulting model succeeded in explaining fitness differences among accessions (; P ≤ 0.00001). In particular, we detected a significant effect of the interaction between allelic differentiation at MAM and GS-OH on fitness, as well as a significant effect of the three-way interaction describing allelic differentiation at MAM, GS-OH, and AOP (Table 1). Thus, fitness variation in the field depends upon alleles at GSL loci and is consistent with the high values detected at those loci in the genome-wide scan. The genome-wide genetic differentiation of accessions also had a significant effect on fitness differences, indicating a residual effect of population structure or adaptive genetic variation unaccounted for in our study (Table 1). Using the same method, we found that, among accessions included in field experiments, the three loci and their interactions explained nearly 36% of the variation in overall GSL profile among accessions. Recent genome-editing techniques may allow formal testing of fitness effects of combinations of GSL alleles in the future (37).
Table 1.
Multiple regression on matrices of fitness variation explained by the three GSL loci and their interactions
| Parameter | Estimate | P value | Significance |
| Intercept | −0.335 | 0.32409 | ns |
| K | 1.941 | 0.00001 | *** |
| MAM | −0.049 | 0.38499 | ns |
| AOP | 0.003 | 0.96037 | ns |
| GS-OH | −0.147 | 0.10116 | ns |
| MAM GS-OH | 0.765 | 0.00008 | *** |
| MAM AOP | −0.063 | 0.66112 | ns |
| GS-OH AOP | 0.301 | 0.11616 | ns |
| MAM GS-OH AOP | −1.326 | 0.00137 | ** |
K stands for the matrix of pairwise distance for all SNPs in the genome not included in the GLS loci. The distance matrices for the GSL loci correspond to MAM, AOP, and GS-OH. Interactions are marked by . (P). Significance: ***P 0.001; **P 0.01; ns, nonsignificant.
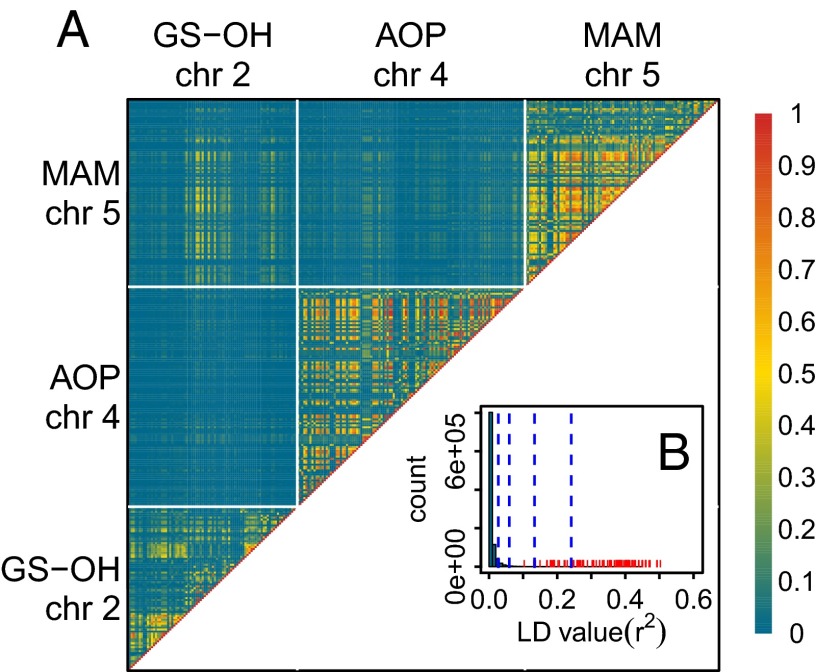
That combinations of GSL alleles impact fitness in the field raises the possibility that epistatic selection played a role in shaping natural variation in GSL profiles (38). A possible signature of such coevolving genes is linkage disequilibrium (LD) (39). Indeed, we found a strong, genome-wide significant correlation between the SNPs in the GS-OH region on chromosome 2 and the SNPs in the MAM region on chromosome 5 (Fig. 3A). The strength of this correlation far exceeds other pairs of SNPs located on different chromosomes in the genome (Fig. 3B), indicating that LD between MAM and GS-OH cannot be explained by demographic history and population structure alone. The clines in GSLs (Fig. 1) were paralleled by clines in the alleles at GS-OH and MAM (Fig. S4). We calculated values comparing Eastern and Western Europe, and found that MAM and GS-OH regions displayed among the strongest differentiation of all loci in the A. thaliana genome (Fig. S5).
Fig. 3.
LD among SNPs located within 20 kb of the three regions associated with GSL variation, MAM, AOP, and GS-OH. (A) Heat map of LD between pairs of SNPs located near the loci associated with GSL variation. GS-OH, AOP, and MAM are located on chromosomes 2, 4, and 5, respectively. Note the high values of LD between SNPs from chromosomes 2 and 5 at the upper left of the heat map. (B) LD between the associated SNPs in the GS-OH (chromosome 2) and MAM (chromosome 5) regions (in red) compared with a null distribution of for 1,000,000 random pairs of SNPs located on different chromosomes (in blue). The vertical dashed lines mark the 0.95, 0.99, 0.999, and 0.9999 quantiles of the null distribution.
It is perhaps surprising that AOP did not reveal high or significant LD with other GSL loci because AOP2 and AOP3 are known to be key genes determining the production of alkenyls and hydroxy-alkyls, respectively. Indeed the expression of AOP2 is required to produce the substrate for GS-OH (36), suggesting that AOP should coevolve with the MAM and GS-OH regions (1, 6). An important role of AOP is supported by the significant interaction between the three loci on fitness in our field experiments, and a few SNPs displayed relatively high pairwise LD between AOP and either MAM or GS-OH regions (Fig. 3A) despite the fact that no regional LD could be detected. The fact that SNPs in the AOP region have much lower minor allele frequencies (median frequency, ∼0.11) than those in the MAM and GS-OH regions (for both loci, median frequency, ∼0.32) could explain the generally lower values (Fig. S4). This does not diminish the fact that two, and maybe three, loci responsible for the biosynthesis of aliphatic GSLs show correlations stronger than that produced by population structure and demographic history in the rest of the genome.
Our results provide evidence that GSL profiles and their underlying genetics are under strong selection across Europe and that the signature of selection on this complex defense trait is detectable across the genome. The genes MAM1 and GS-OH appear to be the targets of divergent selection between Eastern and Western Europe, probably mediated by the local herbivore community. The fact that these two genes are part of the same biosynthetic pathway and show significant epistatic effects on fitness estimates suggests that selection played a role in locking the genome into locally favorable combinations of alleles.
Although the extensive population structure observed in Arabidopsis is often assumed to relate to genetic drift and demographic history, a significant proportion may be due to adaptive differentiation along large geographical regions as exemplified here. High-quality genome sequences of a large number of accessions may not only provide a window into the sequence of events shaping the evolution of adaptive traits but also reveal the relative importance of natural selection and random processes in shaping natural variation.
Materials and Methods
Preparation of Plant Material.
A total of 595 accessions of the RegMap panel (15) was grown in three sets between 2008 and 2009. For each set, plants were grown in a randomized design with four replicates per accession. Seeds were cold-stratified for 4 d at 4 °C and sown on a 1:1 mixture of Premier Pro-Mix and MetroMix. Plants were grown under standard greenhouse conditions at the University of Chicago. Seedlings were thinned to one per pot after 1 wk. Rosettes were harvested after 3 wk and flash frozen in liquid nitrogen. Rosettes were then freeze-dried before extraction.
GSL Extraction.
Each sample of dried rosette tissue was weighed (15 ± 5 mg) and transferred to a 1.4-mL Thermo Scientific Matrix storage tube containing two 2.3-mm zirconia/silica beads. Samples were shaken twice for 30 s at 1,750 rpm in a 2010 Geno/Grinder (SPEX SamplePrep) to homogenize the tissues for liquid extraction. A volume of 400 μL of methanol, 10 μL of 0.3 M lead acetate, and 120 μL of deionized water were added sequentially to each of the tubes. Tubes were then shaken twice in the 2010 Geno/Grinder (SPEX SamplePrep) for 30 s at 1,750 rpm. Next, samples were incubated in a rotary shaker for 1 h at 180 rpm at 28 °C, after which they were centrifuged for 10 min at 1,180 × g. The supernatant was filtered through a MultiScreen Solvinert 96-well filter plate in a Millipore MultiScreen HTS vacuum manifold into a deep-well collection plate. The deep-well collection plate was removed from the vacuum manifold and evaporated for 1 h with a 96-pin nitrogen gas drier. The remaining residue was diluted with 200 μL of HPLC-grade water and transferred to an Agilent 96-well sample plate, which was then sealed with a Nalgene preslit silicone 96-well cap mat.
Liquid Chromatography–MS/MS Analysis and Data Extraction.
GSL content was quantified with an Agilent 1200 Series HPLC coupled to an Agilent 6410 triple quadrupole mass spectrometer. Samples were injected in 5-μL aliquots onto a 4.6 × 50-mm Agilent ZORBAX Eclipse Rapid Resolution HT XDB-C18 column with a 1.8-μm particle size. The following HPLC program was used for all samples: 1 min at 5% acetonitrile [aqueous (aq.)], a 5-min gradient from 5% to 100% acetonitrile, 1 min at 100% acetonitrile, a 1-min gradient from 100 to 5% acetonitrile (aq.), and 30 s at 5% acetonitrile for a total of 8.5 min per HPLC run. The mass spectrometer was run in precursor negative-ion electrospray mode, monitoring all parent ions from m/z 350 to 650 with daughter ions of m/z 97, which correspond to the sulfate moiety of the GSL analytes. The fragmentor voltage was optimized using sinigrin (2-propenyl GSL), and maximum detection was achieved with collision energy at 20 and fragmentor voltage at 135. Individual GSLs were identified based on their fragmentation pattern, retention time, and comparison with 2-propenyl injected before and after sets of 40 runs. No internal standards were used and the values recorded for each of the 22 molecules included in this study are parent ion counts per milligram of lyophilized plant tissue. For pairs of molecules with parent ions differing by 1 or 2 mass units, the counts of the parent ion with the highest m/z of the two were corrected by subtracting the counts attributable to the isotopic peaks of the parent ion with lower m/z. After subtracting baselines, the corrected parent ions counts were then denoised by applying a hard threshold at 100 fragments, and summed for the m/z and retention time ranges determined for each molecule (Table S1). The final data were generated by dividing the parent ion counts by the weight of dry leaf tissue used in the extraction.
Genotypes.
The genotypes of our 595 accessions were taken from ref. 15, in which the hybridization of each of 1,307 A. thaliana accessions on a 250,000 SNP chip is described. The 197,763 SNPs with minor allele frequencies above 5% were included in our analyses.
Broad-Sense Heritability of GSLs.
We estimated the broad-sense heritability of each GSL by fitting a model including a single random intercept effect for the identity of the accessions to the log-transformed ion counts per milligram of leaf tissue (R package lme4; Eq. 1):
| [1] |
where is the random effect of the identity of the accessions, . We computed 95% confidence intervals for each molecule by performing 1,000 parametric bootstraps. The best unbiased linear predictors (blups) were generated to be used in the GWA analysis.
Geographical Variation of the GSL Profile.
To investigate the geographical pattern of the overall GSL profiles of the 595 accessions, we ran a principal-component analysis on the blups generated with the mixed model detailed in Eq. 1. The score of each accession along PC1 and PC2 of the GSL profile was used in a linear regression following Eq. 2:
| [2] |
where is the vector scores along PC1 (or PC2) of the GSL profile for each accession and . and stand for latitude and longitude, respectively. This analysis was restricted to Europe, within the native range of A. thaliana (−15 Longitude 50 and 30 Latitude 75; N = 587 accessions). Significance of the effects of longitude and latitude was assessed using a t test (R functions lm and summary).
GWA Mapping.
GWA mapping analyses were performed using a mixed-model (EMMAX) (17, 18), accounting for population stratification by including an identity-by-state kinship matrix among accessions, computed from all SNPs included in the analysis as a random effect. For phenotypes, we used blups generated by the mixed model following Eq. 1. For each molecule, the phenotypes were resampled without replacement and associations were tested using EMMAX. The lowest P value was recorded and the procedure was repeated 10,000 times. The threshold considered was the 95% quantile of the empirical minimum P value distribution (40).
Signals of Selection and Enrichment Ratio.
scores were calculated following ref. 28. Enrichment in associated SNPs in the tail of the distribution was calculated following Eq. 3:
| [3] |
where n is the number of SNPs considered in the tail of the distribution, is the number of SNPs significantly associated with GSLs in the tail of the distribution, N is the total number of SNPs tested genome-wide, and Na is the total number of SNPs significantly associated with GSL variation genome-wide. The scan presented in Fig. 2 was published in ref. 15 and contrasts nine populations. The scan of contrasting Eastern and Western Europe was performed using the RegMap populations (15); the Western populations included in the analysis were France, Iberia, and the British Isles, and the Eastern populations were Austria–Hungary, Fennoscandia, and Northwest and South Central Europe.
LD Calculations.
LD was calculated as values, following Eq. 4 (41):
| [4] |
where , , , and are the observed frequencies of alleles at the two loci tested, and , where is the expected frequency of a given genotype based on the allele frequencies at the two loci. To test for genome-wide significance of LD between SNPs on different chromosomes, we formed 1,000,000 pairs of randomly selected SNPs from different chromosomes to create a null distribution against which the observed LD values could be compared.
Field Experiments.
Three field experiments were established at the University of Lille (Northern France) in August 2010, September 2010, and September 2011. Each experiment was composed of two complete randomized blocks. A total of 398 accessions from the RegMap panel (15) was included, and GSL data were available for 256 out of the 398 accessions. In each experiment, each block included 19 trays of 66 individual wells (TEKU, JP 3050/66) and contained one replicate per genetic line. Seeds were sown on August 30, 2010, September 27, 2010, and September 26, 2011. In each block, at least five seeds were sown per accession on standard culture soil and stratified for 4 d at 4 °C. After stratification, trays were preventively treated against dark-winged fungus gnats (Vectobac; 8 mL per L) and placed in a frost-free greenhouse without additional light or heating. Eighteen days after the stratification treatment, seedlings were thinned to two per well and trays were transported outside to a common garden located at the University of Lille. Vertebrate herbivores were excluded with the use of two successive fences. Molluscicide (PhytorexJ, Bayer Jardin) was added around experimental blocks to reduce slug attacks. Damage on the rosette leaves was scored from photographs taken before the onset of flowering October 26, 2010, November 26, 2010, and November 22, 2011, for the three experiments, respectively. These scores were assigned according to the percentage of rosette area attacked and whether the meristem was damaged (Fig. S3). The lifetime fitness of each plant was estimated by the total length of siliques produced, which strongly correlates with seed count (30). A total silique length value of 0 was assigned to plants that died during the course of the experiment.
Fitness Estimates, Herbivore Damage, and GSL Profile.
Fitness estimates from the field experiments were modeled with a mixed linear model following Eq. 5:
| [5] |
where is the vector of log-transformed fitness estimates for individual plants; is the fixed experiment effect, and are the random intercept and random slopes for each accession, respectively, with , ; is the random effect of the blocks within experiments with ; and is the random error term with . Variance components were estimated by REML, and the proportion of variance explained by the random accession effects (broad-sense heritability, ) was calculated following Eq. 6:
| [6] |
We computed the 95% confidence interval by performing 1,000 parametric bootstraps. Herbivore damage was scored on the same plants used to estimate fitness. Scores were averaged across three experiments for each accession.
Low levels of replication and the zero-inflated nature of the data prevented accurate estimation of heritability of herbivore damage scores.
Investigating the Relationship Between Herbivory, GSL Profile, and Fitness.
For each accession, we calculated the mean fitness and the mean herbivore damage score across all experiments. The relationship between herbivore damage and the score of each accession along the PC1 describing the GSL profile was investigated by calculating Spearman’s rank coefficient. The relationship between fitness and herbivore damage was investigated in a linear model including the distance between the location of the experiment and the original collection site. This last term is included to account for a potential “home vs. away” effect confounding the contribution of herbivore damage to fitness (Eq. 7):
| [7] |
where is the log-transformed fitness mean per accession; is the geographical distance in kilometers between the location of the experiment and the original collection site of each accession; and is the mean herbivore score per accession. Significance of model terms was tested with a t test.
Testing the Effect of Allelic Variation at Genes Associated with GSL Variation on Fitness.
We investigated the effect of major GSL loci detected in both the GWA study and the scan (MAM, AOP, and GS-OH regions) on the estimate of fitness measured in the field. We extracted all of the SNPs located within a 10-kb window around each locus (for the AOP region, the window started 10 kb before AOP3 and ended 10 kb after AOP2, and for the MAM region the window was centered on MAM1). We then computed matrices of the pairwise distance between accessions (1 − kinship) for each locus (emma.kinship in the R package emma) (17). We also computed a genome-wide distance matrix for all of the SNPs in the genome, leaving out the SNPs located near the three GSL loci. We performed multidimensional scaling and k-means clustering to visualize the allelic variation captured by the SNPs at each locus (R function mdscale and kmeans). To investigate relationships between the fitness estimates measured in the field and the GSL loci, we performed a multiple regression on matrices following Eq. 8:
| [8] |
where is the pairwise euclidian distance matrix between accessions for the log-transformed fitness mean; , , are the pairwise distance matrices between accessions included in the common garden experiments, for each of the three GSL loci; indicates interactions, and , the genome-wide pairwise distance matrix. Interaction matrices were computed by multiplying the corresponding matrices with each other. Significance of the terms of the regression and of the model were obtained by permuting the rows and columns of the fitness distance matrix 10,000 times (R function MRM in the package Ecodist). To estimate the variance of GSL profile differentiation explained by the three loci and their interactions, we also fitted a model following Eq. 8, but this time was the pairwise euclidian distance between accessions for the first two principal components describing GSL variation.
Supplementary Material
Acknowledgments
Many thanks to Timothée Flutre, Andy Gloss, Matt Horton, Richard Hudson, Leah R. Johnson, Talia Karasov, Marcus Kronforst, Thomas Mitchell-Olds, Jonathan Pritchard, Molly Przeworski, Matthew Stephens, Noah Whiteman, and three anonymous reviewers for helpful discussions and comments on earlier versions of the manuscript. This work was funded by grants from the National Science Foundation (MCB 0603515) and NIH (GM 083068) (to J.B.), by a Dropkin Foundation Fellowship (to B.B.), a Graduate Assistance in Areas of National Need Scholarship in Evolutionary Genomics (to C.G.M.), by a PhD fellowship from the University of Lille 1 (to R.V.), and by Laboratoire d’Excellence TULIP (ANR-10-LABX-41; ANR-11-IDEX-0002-02) (to F.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1421416112/-/DCSupplemental.
References
- 1.Sønderby IE, Geu-Flores F, Halkier BA. Biosynthesis of glucosinolates—gene discovery and beyond. Trends Plant Sci. 2010;15(5):283–290. doi: 10.1016/j.tplants.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Fahey JW, Zalcmann AT, Talalay P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry. 2001;56(1):5–51. doi: 10.1016/s0031-9422(00)00316-2. [DOI] [PubMed] [Google Scholar]
- 3.Kliebenstein DJ. A quantitative genetics and ecological model system: Understanding the aliphatic glucosinolate biosynthetic network via QTLs. Phytochem Rev. 2008;8:243–254. [Google Scholar]
- 4.Textor S, et al. Biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana: Recombinant expression and characterization of methylthioalkylmalate synthase, the condensing enzyme of the chain-elongation cycle. Planta. 2004;218(6):1026–1035. doi: 10.1007/s00425-003-1184-3. [DOI] [PubMed] [Google Scholar]
- 5.Lankau RA. Specialist and generalist herbivores exert opposing selection on a chemical defense. New Phytol. 2007;175(1):176–184. doi: 10.1111/j.1469-8137.2007.02090.x. [DOI] [PubMed] [Google Scholar]
- 6.Burow M, Halkier BA, Kliebenstein DJ. Regulatory networks of glucosinolates shape Arabidopsis thaliana fitness. Curr Opin Plant Biol. 2010;13(3):348–353. doi: 10.1016/j.pbi.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Halkier BA, Gershenzon J. Biology and biochemistry of glucosinolates. Annu Rev Plant Biol. 2006;57:303–333. doi: 10.1146/annurev.arplant.57.032905.105228. [DOI] [PubMed] [Google Scholar]
- 8.Rodman JE, Kruckeberg AR, Al-Shehbaz IA. Chemotaxonomic diversity and complexity in seed glucosinolates of Caulanthus and Streptanthus (Cruciferae) Syst Bot. 1981;6(3):197–222. [Google Scholar]
- 9.Rodman JE. Population variation and hybridization in sea-rockets (Cakile, Cruciferae): Seed glucosinolate characters. Am J Bot. 1980;67(8):1145–1159. [Google Scholar]
- 10.Windsor AJ, et al. Geographic and evolutionary diversification of glucosinolates among near relatives of Arabidopsis thaliana (Brassicaceae) Phytochemistry. 2005;66(11):1321–1333. doi: 10.1016/j.phytochem.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 11.Kliebenstein DJ, Gershenzon J, Mitchell-Olds T. Comparative quantitative trait loci mapping of aliphatic, indolic and benzylic glucosinolate production in Arabidopsis thaliana leaves and seeds. Genetics. 2001;159(1):359–370. doi: 10.1093/genetics/159.1.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reichelt M, et al. Benzoic acid glucosinolate esters and other glucosinolates from Arabidopsis thaliana. Phytochemistry. 2002;59(6):663–671. doi: 10.1016/s0031-9422(02)00014-6. [DOI] [PubMed] [Google Scholar]
- 13.Brown PD, Tokuhisa JG, Reichelt M, Gershenzon J. Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry. 2003;62(3):471–481. doi: 10.1016/s0031-9422(02)00549-6. [DOI] [PubMed] [Google Scholar]
- 14.Züst T, et al. Natural enemies drive geographic variation in plant defenses. Science. 2012;338(6103):116–119. doi: 10.1126/science.1226397. [DOI] [PubMed] [Google Scholar]
- 15.Horton MW, et al. Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat Genet. 2012;44(2):212–216. doi: 10.1038/ng.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atwell S, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465(7298):627–631. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang HM, et al. Efficient control of population structure in model organism association mapping. Genetics. 2008;178(3):1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang HM, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42(4):348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen BG, et al. A novel 2-oxoacid-dependent dioxygenase involved in the formation of the goiterogenic 2-hydroxybut-3-enyl glucosinolate and generalist insect resistance in Arabidopsis. Plant Physiol. 2008;148(4):2096–2108. doi: 10.1104/pp.108.129981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan EK, Rowe HC, Corwin JA, Joseph B, Kliebenstein DJ. Combining genome-wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana. PLoS Biol. 2011;9(8):e1001125. doi: 10.1371/journal.pbio.1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bergelson J, Roux F. Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. Nat Rev Genet. 2010;11(12):867–879. doi: 10.1038/nrg2896. [DOI] [PubMed] [Google Scholar]
- 22.Jensen LM, Halkier BA, Burow M. How to discover a metabolic pathway? An update on gene identification in aliphatic glucosinolate biosynthesis, regulation and transport. Biol Chem. 2014;395(5):529–543. doi: 10.1515/hsz-2013-0286. [DOI] [PubMed] [Google Scholar]
- 23.Kliebenstein DJ, et al. Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol. 2001;126(2):811–825. doi: 10.1104/pp.126.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kliebenstein D, Pedersen D, Barker B, Mitchell-Olds T. Comparative analysis of quantitative trait loci controlling glucosinolates, myrosinase and insect resistance in Arabidopsis thaliana. Genetics. 2002;161(1):325–332. doi: 10.1093/genetics/161.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Hansen BG, Ober JA, Kliebenstein DJ, Halkier BA. Subclade of flavin-monooxygenases involved in aliphatic glucosinolate biosynthesis. Plant Physiol. 2008;148(3):1721–1733. doi: 10.1104/pp.108.125757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen R, et al. Genomic scans for selective sweeps using SNP data. Genome Res. 2005;15(11):1566–1575. doi: 10.1101/gr.4252305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toomajian C, et al. A nonparametric test reveals selection for rapid flowering in the Arabidopsis genome. PLoS Biol. 2006;4(5):e137. doi: 10.1371/journal.pbio.0040137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38(6):1358–2728. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 29.Wright S. The genetical structure of populations. Ann Eugen. 1951;15(4):323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 30.Roux F, Gasquez J, Reboud X. The dominance of the herbicide resistance cost in several Arabidopsis thaliana mutant lines. Genetics. 2004;166(1):449–460. doi: 10.1534/genetics.166.1.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hopkins RJ, van Dam NM, van Loon JJA. Role of glucosinolates in insect-plant relationships and multitrophic interactions. Annu Rev Entomol. 2009;54:57–83. doi: 10.1146/annurev.ento.54.110807.090623. [DOI] [PubMed] [Google Scholar]
- 32.Bidart-Bouzat MG, Kliebenstein DJ. Differential levels of insect herbivory in the field associated with genotypic variation in glucosinolates in Arabidopsis thaliana. J Chem Ecol. 2008;34(8):1026–1037. doi: 10.1007/s10886-008-9498-z. [DOI] [PubMed] [Google Scholar]
- 33.Hancock AM, et al. Adaptation to climate across the Arabidopsis thaliana genome. Science. 2011;334(6052):83–86. doi: 10.1126/science.1209244. [DOI] [PubMed] [Google Scholar]
- 34.Fournier-Level A, et al. A map of local adaptation in Arabidopsis thaliana. Science. 2011;334(6052):86–89. doi: 10.1126/science.1209271. [DOI] [PubMed] [Google Scholar]
- 35.Kroymann J, Donnerhacke S, Schnabelrauch D, Mitchell-Olds T. Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proc Natl Acad Sci USA. 2003;100(Suppl 2):14587–14592. doi: 10.1073/pnas.1734046100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kliebenstein DJ, Lambrix VM, Reichelt M, Gershenzon J, Mitchell-Olds T. Gene duplication in the diversification of secondary metabolism: Tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell. 2001;13(3):681–693. doi: 10.1105/tpc.13.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schiml S, Fauser F, Puchta H. The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J. 2014;80(6):1139–1150. doi: 10.1111/tpj.12704. [DOI] [PubMed] [Google Scholar]
- 38.Olson-Manning CF, Wagner MR, Mitchell-Olds T. Adaptive evolution: Evaluating empirical support for theoretical predictions. Nat Rev Genet. 2012;13(12):867–877. doi: 10.1038/nrg3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clark NL, et al. Coevolution of interacting fertilization proteins. PLoS Genet. 2009;5(7):e1000570. doi: 10.1371/journal.pgen.1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sham PC, Purcell SM. Statistical power and significance testing in large-scale genetic studies. Nat Rev Genet. 2014;15(5):335–346. doi: 10.1038/nrg3706. [DOI] [PubMed] [Google Scholar]
- 41.Flint-Garcia SA, Thornsberry JM, Buckler ES., 4th Structure of linkage disequilibrium in plants. Annu Rev Plant Biol. 2003;54:357–374. doi: 10.1146/annurev.arplant.54.031902.134907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.