Significance
Methane seeps are natural gas leaks at the seafloor that emit methane to the hydrosphere. The emission rates are controlled by methane-oxidizing microorganisms, which shape the ecosystem by supplying energy sources to other microorganisms and animal symbioses. We provide evidence that methane seeps are island-like habitats, harboring distinct microbial communities that share few organisms with other seafloor ecosystems. The seep communities comprise bacteria and archaea that occur worldwide but are locally selected by the environment. These microorganisms show high relative sequence abundances, suggesting high population densities and global relevance for the control of methane emission from the seafloor. At individual seeps, the cosmopolitan microorganisms are associated with a substantial diversity of rare relatives, turning seeps into hotspots of microbial biodiversity.
Keywords: anaerobic methane oxidation, ANME, deep-sea seafloor ecosystems, microbial community ecology, environmental selection
Abstract
Methane seeps are widespread seafloor ecosystems shaped by the emission of gas from seabed reservoirs. The microorganisms inhabiting methane seeps transform the chemical energy in methane to products that sustain rich benthic communities around the gas leaks. Despite the biogeochemical relevance of microbial methane removal at seeps, the global diversity and dispersion of seep microbiota remain unknown. Here we determined the microbial diversity and community structure of 23 globally distributed methane seeps and compared these to the microbial communities of 54 other seafloor ecosystems, including sulfate–methane transition zones, hydrothermal vents, coastal sediments, and deep-sea surface and subsurface sediments. We found that methane seep communities show moderate levels of microbial richness compared with other seafloor ecosystems and harbor distinct bacterial and archaeal taxa with cosmopolitan distribution and key biogeochemical functions. The high relative sequence abundance of ANME (anaerobic methanotrophic archaea), as well as aerobic Methylococcales, sulfate-reducing Desulfobacterales, and sulfide-oxidizing Thiotrichales, matches the most favorable microbial metabolisms at methane seeps in terms of substrate supply and distinguishes the seep microbiome from other seafloor microbiomes. The key functional taxa varied in relative sequence abundance between different seeps due to the environmental factors, sediment depth and seafloor temperature. The degree of endemism of the methane seep microbiome suggests a high local diversification in these heterogeneous but long-lived ecosystems. Our results indicate that the seep microbiome is structured according to metacommunity processes and that few cosmopolitan microbial taxa mediate the bulk of methane oxidation, with global relevance to methane emission in the ocean.
A microbiome is defined as the microbial community and its genomic diversity associated with a particular ecosystem or habitat, such as soil (1) or the human gut (2). A key question in the study of microbiomes concerns the identification of assembly rules that govern microbial community structure and community function (3). Sampling efforts on local to global scales have been used to determine the key drivers of microbial assembly, which include processes such as dispersal, ecological drift, environmental selection, and diversification (3, 4). Major processes shaping the microbial diversity landscape involve environmental selection of organisms according to their traits, niche preferences, biological interactions, and coevolution with hosts (5, 6). In turn, recent findings suggest that fluctuations of key microbial taxa reflect the dynamics of important biogeochemical processes (7).
Insights into environmental microbiomes have tremendously improved with the use of next-generation sequencing methods and global databases, which have advanced microbial ecology from the identification of rare members of microbial communities (8) to global microbial distribution patterns (9, 10). Benthic (seafloor-hosted) microbial communities of the ocean are very distinct from pelagic (seawater-hosted) communities (9) and are impacted by water depth (11, 12), sediment depth (13, 14), and by energy availability in the form of deposited organic matter (12, 15, 16). Marine sediments are known to host communities as diverse as those found in soils, with pronounced community turnover on small (decimeter to kilometer), intermediate (hundreds of kilometers), and large (thousands of kilometers) spatial scales (17, 18).
Cold seeps are distinct seafloor ecosystems that are defined by the upward advection of methane and other hydrocarbons from the subsurface seabed to the seafloor (19). Gas-emitting methane seeps are found scattered on continental margins worldwide and are separated by large expanses of energy-limited, aerobic deep-sea seafloor where the benthic communities depend on sparse detritus flux. These seeps can be regarded as patches of a certain habitat type, offering niches that differ strongly from the surrounding seafloor (14, 20, 21). Typically, methane seep sediments are highly reduced, and oxygen availability is limited to a few millimeters to centimeters of the sediment surface. Below that thin oxic zone, methane is consumed by microbial consortia of anaerobic methane-oxidizing archaea (ANME) and sulfate-reducing bacteria (SRB) mediating the anaerobic oxidation of methane (AOM) coupled to sulfate reduction (22). This microbial conversion of inorganic energy sources fuels communities of microorganisms and marine invertebrates and thus generates hotspots of biomass and diversity in the deep sea (23). Together, micro- and macroorganisms at methane seeps and sulfate methane transition zones (SMTZ) consume 75% of the methane (0.06 Gt of carbon per year) that reaches the surface seafloor from subsurface zones (19). Hence, they provide a globally relevant ecosystem function by controlling the emission of the potential greenhouse gas methane from the ocean.
At large scales, however, it is unclear which seep microorganisms are important for the removal of methane and which mechanisms govern their community assembly. Defining the methane seep microbiome is needed to identify the microbial key players responsible for these important biogeochemical functions. In this study, we compared the archaeal and bacterial diversity of 23 globally distributed methane seeps to that of 54 globally distributed sites from other seafloor ecosystems, including deep SMTZ, hydrothermal vents, coastal sediments, and deep-sea surface and subsurface sediments. We tested (i) whether methane seeps are island-like habitats that are similar to each other but distinct from their surroundings concerning community richness, evenness, similarity, and composition; (ii) if a core set of associated bacteria and archaea dominates seeps globally; and (iii) whether their assembly patterns follow the metacommunity concept of globally dispersed and locally diversified types (24).
Results and Discussion
Methane Seeps Enhance Seafloor Biodiversity.
Seeps are isolated habitats, as they are highly reduced, sulfidic environments in a vast oxic surrounding, representing biogeochemical barriers to immigration. Methane seep communities hence could reflect island biodiversity patterns with enhanced endemism richness at moderate levels of species richness (25). According to the metacommunity concept (24), methane seeps may select for specific and highly adapted colonists from a global pool of microbes, which disperse across the seafloor with bottom currents or via mobile sediment-feeding fauna. Environmental selection acting over long times could then result in substantial local diversification. This hypothesis was tested by comparing richness, evenness, similarity, and composition of bacterial and archaeal communities of globally distributed methane seeps (Fig. 1).
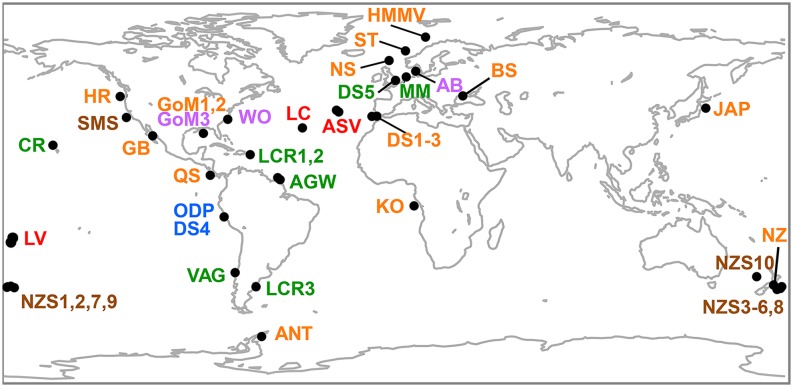
Fig. 1.
Map of seafloor ecosystems. Datasets with spatially distributed sites are numbered (e.g., NZS1-10). Abbreviations of methane seeps (orange) and subsurface sulfate methane transition zones (SMTZ; purple) stand for ANT, Low-activity Antarctic seep; BS, Black Sea microbial reef; DS, Gulf of Cadiz mud volcanoes; GB, Guaymas Basin hot seeps; GoM, Gulf of Mexico seeps; HMMV, Håkon Mosby mud volcano; HR, Hydrate Ridge seeps; JAP, Japanese Trench seep; KO, Congo Basin (REGAB) seep; NS, North Sea seep; NZ, New Zealand seeps; QS, Quepos Slide seep (Costa Rica); and ST, Storegga Slide seep (Norway). Other abbreviations are as follows. Deep-sea surface sediments (brown): SMS, Station M; NZS, New Zealand. Coastal sediments (green): AGW, Amazon–Guiana Waters; CR, Hawaiian Coral Reef; LCR, Latin American Coastal Regions; MM, North Sea Intertidal Microbial Mat; VAG, Chilean Coast sediment. Subsurface sediments (blue): DS4/ODP, Peru Margin Ocean Drilling Core. Hydrothermal Vents (red): ASV, Azores Shallow Vents; LC, Lost City Vent Field; LV, Lau Vent Field.
Microbial community richness and evenness of seafloor ecosystems.
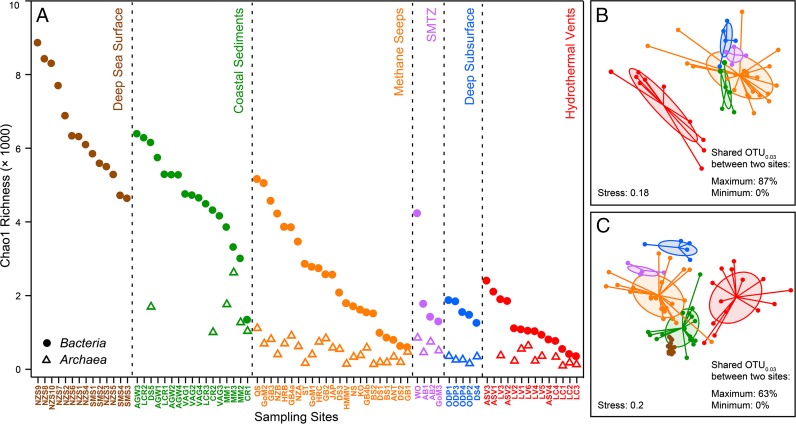
Both observed (S) and estimated (Chao1) richness, as well as Inverse Simpson diversity indices (D) of archaeal and bacterial communities, spanned more than one order of magnitude across seafloor ecosystems based on the 16S ribosomal RNA gene region V6 (Fig. 2A and SI Appendix, Tables S1 and S2). The variation in diversity among methane seeps was large, yet overall their microbial diversity was distinct to that of other seafloor ecosystems, e.g., showing on average lower S, Chao1 (Fig. 2A), and D than the surrounding deep-sea seafloor. Archaeal and bacterial diversity were both extremely low at hydrothermal vents and at some methane seeps and peaked in coastal and deep-sea surface sediments. Across all benthic ecosystems, archaeal diversity was lower than bacterial diversity. However, archaeal and bacterial diversity positively correlated with each other (Spearman’s rank correlation: RS = 0.68, pS < 0.001; RChao1 = 0.62, pChao1 < 0.001; RD = 0.55, pD < 0.001), indicating that diversity patterns of seafloor archaeal and bacterial communities are connected.
Fig. 2.
Richness estimates and NMDS ordinations based on archaeal and bacterial OTU0.03. (A) The symbols represent the mean of 100 Chao1 richness calculations for each of the 77 investigated sampling sites, with each calculation based on rarefaction of 3,000 randomly chosen sequences without replacement. No archaeal data were available for deep-sea surface sediments. (B and C) Similarity of archaeal (B, 48 samples) and bacterial (C, 77 samples) communities visualized by nonmetric multidimensional scaling (NMDS). Each sample (dot) is connected to the weighted averaged mean of the within group distances. Ellipses represent one SD of the weighted averaged mean. All groups were significantly different as tested with a subsampling-based Redundancy Analysis approach (see also SI Appendix). The color code is identical to A.
The wide span of microbial diversity observed across all seep sites is best explained by differences in local heterogeneity of the sampled seep systems, which may determine the number of ecological niches that are available to microbial colonists (for examples, see SI Appendix). Accumulation curves indicate that we captured almost all order-level archaeal and bacterial taxa that are expected at methane-impacted ecosystems (SI Appendix, Fig. S1). However, at OTU0.03 (operational taxonomic units at 97% sequence identity of the 16S rRNA gene region V6) level we retrieved only 55% of the global archaeal and 47% of the bacterial diversity of methane seeps, indicating that a large part of the global phylogenetic diversity remained untapped due to undersampling of seafloor communities.
Microbial community similarity between seafloor ecosystems.
At phylum level, all seafloor ecosystems investigated here shared a high proportion of taxa. The major archaeal phyla Euryarchaeota, Crenarchaeota, and Thaumarchaeota were found at all 77 sites. The sites also shared between 80 and 98% of the 53 bacterial phyla observed in the dataset (SI Appendix, Table S3). However, community similarity based on shared OTU0.03 between most samples was low (Fig. 2 B and C and SI Appendix, Table S4). No single OTU0.03 was found in all seafloor samples. Dissimilarity was even more pronounced between seafloor ecosystems. Pairwise comparisons of seafloor ecosystems resulted in 1–7% shared OTU0.03 for both archaea and bacteria (SI Appendix, Table S3). This result did not change when excluding the rare biosphere (defined here as those OTU0.03 with less than 0.1% total relative sequence abundance) of each seafloor ecosystem. However, communities were more similar to each other within than between ecosystems (Fig. 2 B and C). Hence, we suggest that the microbial community of each of these different seafloor ecosystems can be called a microbiome, analogous to the definition of the soil or human microbiomes.
Community dissimilarity within a seafloor ecosystem was highest for hydrothermal vents and methane seeps, both being associated with a substantial degree of endemism (Fig. 2 B and C and SI Appendix, Tables S1 and S4). Community dissimilarity was lowest in deep-sea surface sediments, which shared a high proportion of bacterial taxa. The maximum percentage of shared archaeal OTU0.03 between any two methane seeps was 66%, but several habitats did not share any OTU0.03 at all. Bacterial OTU0.03 turnover was even more pronounced with a maximum of 36% shared and a minimum of 0% shared OTU0.03. This high degree of endemism at the level of OTU0.03, together with a moderate diversity compared with the surrounding seafloor, confirms our hypothesis that cold seeps are island-like habitats that are shaped by biogeochemical rather than geographical barriers.
Microbial community composition of seafloor ecosystems.
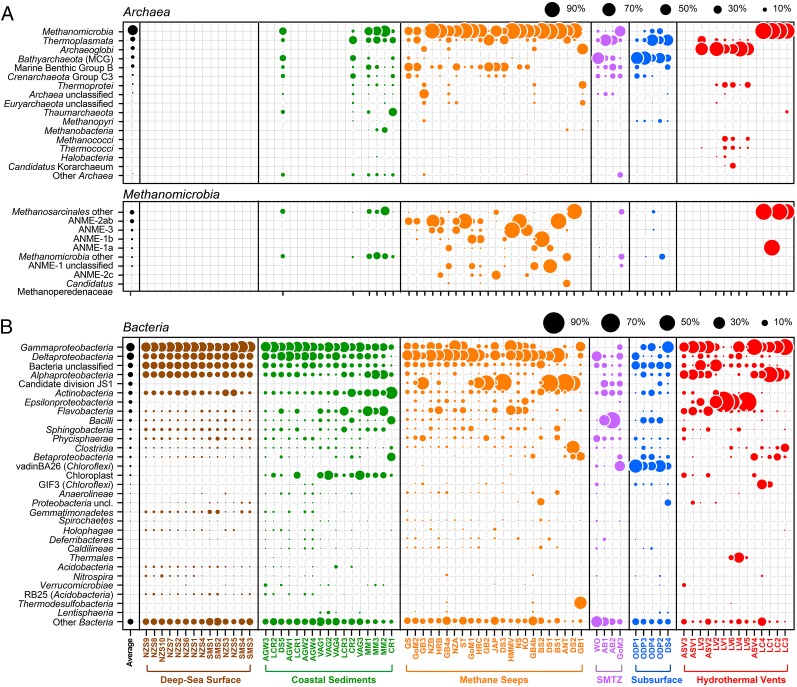
The 10 most sequence-abundant bacterial phyla of the investigated seafloor ecosystems accounted for 84% of all bacterial sequences. Seven of those ten phyla were cosmopolitan (SI Appendix, Fig. S2), matching the classical abundance–range relationship (26). The most sequence-abundant bacterial phyla were Proteobacteria (44% of all bacterial sequences), Bacteroidetes (8%), Firmicutes (6%), and Chloroflexi (6%). At the class (Fig. 3) and order levels (SI Appendix, Fig. S3) we found substantial differences in microbial community composition between the seafloor ecosystems and identified indicator taxa for each ecosystem (SI Appendix, Table S5). Methanomicrobia and Deltaproteobacteria, as well as candidate division Hyd24-12 and candidate division JS1 (Fig. 3), were typical for methane seeps. Gammaproteobacteria, Flavobacteria, Thermoplasmatales, and Marine Benthic Group B were frequently found at seeps, but they were also important in coastal and deep-sea sediments or in the deep subsurface. Anoxic SMTZ shared taxa, such as Bathyarchaeota, Bacilli, and Chloroflexi (27), with deep subsurface communities (ODP1-4) and microbial communities of high-temperature methane seeps (GB sites) overlapped with those of hydrothermal chimneys (LV1-6), sharing clades such as Thermodesulfobacteria, Thermoprotei, and Archaeoglobi (28). This observed overlap between seeps and other ecosystems at the class level may be explained by species sorting according to biogeochemical conditions.
Fig. 3.
Relative sequence abundance of archaeal and bacterial classes. The graph shows the relative sequence abundance of the most abundant archaeal (A) and bacterial (B) classes in different seafloor ecosystems sorted according to average relative abundance across all samples. Sampling sites and ecosystems are ordered from high (Left) to low (Right) Chao1 richness. The archaeal class Methanomicrobia is shown in the lower panel of A, with subdivisions for the major methanotrophic (ANME) as well as methanogenic (Methanosarcinales; other Methanomicrobia) clades.
At order level we identified two archaeal and ten bacterial taxa that were present at all methane seeps and SMTZ (SI Appendix, Table S6) at a sequence abundance of over 1%. The Methanosarcinales found at cold seeps consisted almost entirely of sequences affiliated to ANME clades and just very few sequences belonged to methanogens (Fig. 3A). Desulfobacterales are sulfate reducers and contain all ANME partner SRBs, except the thermophilic HotSeep-1 clade. At methane seeps the great majority of sequences affiliated to Desulfobacterales belonged to the ANME partner SRBs of the clades SEEP-SRB1 and SEEP-SRB2, as well as seep-associated SEEP-SRB4 (SI Appendix, Fig. S4). Aerobic methanotrophic Methylococcales (14, 29) and the conspicuous members of the Thiotrichales often forming thiotrophic bacterial mats at methane seeps (30) were also found at most seep sites with oxygen supplies from the overlying water column. Apart from the methanotrophs the majority of the heterotrophic core clades of methane seeps, such as the Actinobacteridae, Flavobacteriales, Sphingobacteriales, and Caldilineales (Fig. 3B), were also commonly found in detritus-fueled deep-sea sediments. However, these seep heterotrophs showed little overlap with deep-sea sediments at OTU0.03 level, suggesting that the composition and availability of organic matter and electron acceptors differed between seep and nonseep sediments (31). In addition, we found other core microbial clades at methane seeps with unknown function, including Thermoplasmatales, Firmicutes, Spirochaetes, and members of candidate division JS1 (see also SI Appendix). None of the OTU0.03 were ubiquitous, yet the key ecosystem functions, such as methanotrophy and sulfate reduction, were found at all seeps. Hence, functional redundancy is provided by a few core taxa that are likely selected by environmental conditions.
Environmental Selection at Methane Seeps.
To investigate further the role of environmental selection, we determined how much of the variance in community composition between seep sites could be explained by the measured environmental parameters or by distance decay of community similarity (4, 17, 18). Together the available parameters, sediment temperature, sediment depth, water depth, methane concentration, and space, explained 24% [redundancy analysis (RDA), P = 0.001] of the archaeal and 21% (P = 0.003) of the bacterial community variation on OTU0.03 level, with sediment temperature (Archaea: P = 0.02; Bacteria: P = 0.05) and sediment depth (Archaea: P = 0.05) being most important (see also SI Appendix). Neither water depth nor the prevailing ranges of methane concentrations on their own had explanatory power, suggesting that the supply of electron acceptors was more important for shaping community structure than the supply of methane (19). Only a minor fraction of the variance of community structure between seeps was explained by the decay of community similarity with geographic distance (SI Appendix, Fig. S6), which was in the same range as that reported for deep-sea surface sediments (18).
ANME-2a was the most widespread and sequence-abundant clade and seemed to dominate many cold-temperature seep sites but was less abundant or even absent at hot seeps (Fig. 3A). ANME-2c was predominantly found at sites inhabited by vesicomyid clams [hydrate ridge calyptogena site (HRC), JAP, and KO] (20, 21, 32). ANME-3 was also widespread but dominated only samples from cold-temperature, vigorously emitting methane seeps, such as HMMV in the Barents sea (33) and the REGAB pockmark (KO) in the Congo Basin (21). In addition, the relative abundance of ANME-3 correlated with that of SEEP-SRB4 (Rspearman = 0.79, pspearman < 0.001), indicating that those two organisms may form AOM aggregates in those environments. ANME-1 generally correlated with sediment depth and sediment temperature (SI Appendix, Fig. S5C). The subclade ANME-1b preferentially occurred together with SEEP-SRB2 (Rspearman = 0.68, pspearman < 0.001), that dominated anoxic settings such as the Black Sea microbial mats (BS2) (32) and was abundant in deep sediment layers (DS2/3, GB4b, and GoM1/2) but rare at other seep sites. Interestingly, ANME-1b also cooccurred with a so far unclassified clade of Firmicutes (Rspearman = 0.83, pspearman < 0.001), but this association was not yet confirmed by microscopy. ANME-1a, which cooccurred with HotSeep-1 (Rspearman = 0.70, pspearman < 0.001), and unclassified ANME-1 seemed to inhabit preferably hot seeps (34, 35). The recently described Candidatus Methanoperedenaceae, which uses nitrate as electron acceptor in wastewater sludge (36), was found among others at hot seeps and at a cold, quiescent seep in Antarctica (Fig. 3A), indicating marine relatives. Sequences of Candidatus Methylomirabilis oxyfera, which perform the nitrite-dependent AOM in freshwater habitats (37), occurred at one Guaymas Basin hot seep (GB4a/b). Desulfobacterales dominated shallow, sulfate-rich sediments due to their dependency on sulfate as electron acceptor, and their low relative abundance in hot seeps may indicate sensitivity to high temperature. In turn, hot seep sediments (GB2, 3) and subsurface sediments at methane seeps (ANT, DS) seemed to be dominated by organisms of candidate division JS1 (Fig. 3B and SI Appendix). Methylococcales and Thiotrichales were present at all seeps, except those that were strictly anoxic, which reflects their need for oxygen as an electron acceptor. These differences in the distribution of key functional clades at methane seep sites suggest that the availability of electron acceptors and the habitat temperature are main drivers of environmental filtering.
Global Dispersion and Local Diversification of Key Functional Taxa.
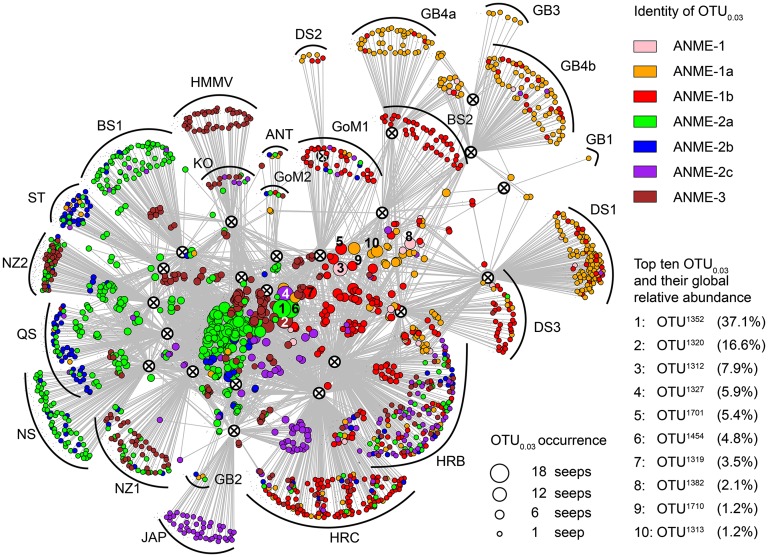
Each of the major functional clades (ANME, Desulfobacterales, Methylococcales, and Thiotrichales) contained one or a few OTU0.03 that largely dominated the sequences affiliated to the respective clade worldwide (SI Appendix, Fig. S7). For instance, OTU1352 (ANME-2a) occurred at 18 of 23 methane seeps and accounted for 37% of all ANME sequences worldwide (Fig. 4). In addition, we found identical ANME-2a sequences in 16S rRNA gene libraries from methane seeps worldwide, including a number of ecosystems that are not included in this study, such as Marmara Sea cold seeps (38).
Fig. 4.
Network graph of 23 methane seeps based on occurrence of ANME OTU0.03. The colored nodes (circles) represent all 1,765 ANME OTU0.03 that were found in this study. The colors indicate the taxonomic assignment of each ANME OTU0.03. Nodes marked as circles with black crosses represent the 23 investigated methane seeps to which the gray lines connect their respective ANME OTU0.03. The distance between methane seep nodes reflects their OTU0.03 connectivity;, e.g., methane seeps that share many ANME OTU0.03 are plotted close to each other, whereas those that share less are more distant. ANME OTU0.03 that occurred at many seeps are shown as larger nodes found in the middle of the plot, whereas OTU0.03 that occurred only at one seep are small nodes on the periphery. There are 10 nodes with numbers from 1 to 10, which represent the 10 most sequence-abundant ANME OTU0.03. These 10 OTU0.03 were responsible for around 85% of all sequenced ANME reads and had a cosmopolitan distribution. The majority of the ANME diversity, however, was rare and locally restricted.
Despite the pronounced environmental differences between seep habitats, the 10 most sequence-abundant ANME OTU0.03 were responsible for 86% of all ANME sequences but represented only 0.6% of their global diversity (total: 1,765 ANME OTU0.03). Interestingly, five of these ANME OTU0.03 occurred as only one sequence in at least one seep but largely dominated other seeps. We observed a similar trend for the 10 most frequent OTU0.03 of the Desulfobacterales, Methylococcales, and Thiotrichales that represented 0.3–3% of each clade’s total diversity but accounted for 38–96% of all its sequences. Also for these clades, environmental filtering was an important force because on average eight of the ten OTU0.03 were conditionally rare taxa, being singletons at some seeps but abundant at others.
The existence of cosmopolitan ANME populations (Fig. 4) raises the question as to their dispersal mechanisms. ANME are sensitive to oxygen and are only known to have a pelagic lifestyle in fully anoxic water bodies such as the Black Sea (39). However, ANME may disperse via passive transport in the guts of benthic fauna or they may have globally distributed during oceanic anoxic events (40). Interestingly, the endemic ANME OTU0.03, which occurred only at one methane seep, were low in sequence abundance and were mostly related to the seep-dominating clade (Fig. 4). This suggests a strong pressure for local diversification on ANME organisms caused by isolation due to the prevailing environmental conditions, as proposed by the island biodiversity theory (25). Altogether, these patterns of microbial community structure at cold seeps suggest that methane seeps are patchy habitats with some connectivity of key functional clades that disperse globally and diversify locally.
Conclusion
Seafloor microbiomes overlap at phylum level but show increasing dissimilarity at increasing phylogenetic resolution below class level, with clear distinctions between surface and subsurface, vent, and seep communities. Methane seeps harbor microbial communities with moderate species richness but high levels of endemism and thus resemble islands in terms of diversity and diversification patterns. Despite this endemism our results indicate that all methane seeps share a core community of few species-level taxa that are globally distributed but locally selected and diversified. Environmental controls, foremost seafloor temperature and electron acceptor availability, were key factors associated with the diversification of the global seep microbiome. The few cosmopolitan taxa were also the most sequence-abundant organisms and seem to mediate the bulk of methanotrophy, thiotrophy, and sulfate reduction. This suggests that only few microbial taxa at seeps potentially impact the global carbon budget today. However, the redundancy of those taxa, being dominant at most seep sites, yet rare at others, indicates that local diversity may be relevant to provide resilience to environmental dynamics.
Materials and Methods
Dataset Specification.
We analyzed archaeal and bacterial sequence data from a sample set of 23 globally distributed methane seeps and four SMTZ combined with data from the International Census of Marine Microbes (ICoMM) project database (14 hydrothermal vent samples, 17 coastal sediment samples, and 14 deep-sea surface and 5 deep subsurface sediment samples (Fig. 1). The full 16S rRNA gene V6 pyrotag dataset is available on VAMPS (https://vamps.mbl.edu); for contextual data, see SI Appendix, Table S7.
Generation of Pyrotags, Quality Control, and Taxonomic Annotation.
DNA extraction from all samples was carried out by a standardized protocol, as described on the MICROBIS project pages (icomm.mbl.edu/microbis), using commercial extraction kits. The variable V6 region of the 16S rRNA gene was PCR-amplified, using archaeal (Arch958F/Arch1048Rmix) and bacterial (Bac967Fmix/Bac1064Rmix) primers (see SI Appendix, Supplementing Materials and Methods). Massively parallel tag sequencing of the PCR products was carried out on a 454 Life Sciences GS FLX sequencer at Marine Biological Laboratory, Woods Hole, MA. The sequences were submitted to a rigorous quality control procedure (see also SI Appendix) based on mothur v24 (41), which included denoising of the flow grams, single-linkage preclustering, and the removal of chimeras. Archaeal and bacterial sequences were clustered at 97% sequence identity (OTU0.03) and taxonomically assigned based on SILVA [release 102, 02–2010 (42)]. To minimize biases, all steps were performed with the same protocols and infrastructure.
Data Analyses.
The sequence abundance tables were used to calculate diversity indices, using mothur v24. Dissimilarities between all samples were calculated, and the resulting beta-diversity matrices were used for two-dimensional nonmetric multidimensional scaling (NMDS) ordinations. RDA (Redundancy Analyses) based on Hellinger-transformed OTU0.03 datasets were carried out to evaluate the combined effects of sediment depth, sediment temperature, water depth, and ranges of methane and sulfate concentrations on the microbial community composition in methane seep habitats. The significance of combined and pure effects was assessed by analysis of variance (ANOVA). Indicator taxa of the different seafloor microbiomes were calculated based on relative abundance and frequency of occurrence. Distance decay was based on pairwise community dissimilarities, using the Sørensen index, and assessed in a logarithmic transformed space to enhance the linear fitting. The OTU0.03 network was based on presence–absence, frequency of occurrence, and identity of OTU0.03. A detailed description of data analyses is provided in the SI Appendix.
Supplementary Material
Acknowledgments
We gratefully acknowledge Mitchell Sogin, Hilary Morrison, and Susan Huse of Marine Biological Laboratory for their support with sequencing according to the ICoMM pipeline. We thank Angélique Gobet, Lucie Zinger, and Pier L. Buttigieg for help with statistical analyses and Jillian M. Petersen, Matthias Winkel, and Dimitri V. Meier for helpful discussions and comments on the manuscript. This study is a contribution to the ICoMM and to the Deep Carbon Observatory project funded by the Alfred P. Sloan Foundation. Further support for this study was supplied by the Max Planck Society, the Leibniz program of the Deutsche Forschungsgemeinschaft (to A.B.), and the European Research Council Adv G ABYSS (294757) (to A.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Sequence raw data has been deposited in the National Center for Biotechnology Information Sequence Read Archive; for the list of accession numbers, see Dataset S1. Contextual data are archived in VAMPS (https://vamps.mbl.edu).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1421865112/-/DCSupplemental.
References
- 1.Fierer N, et al. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci USA. 2012;109(52):21390–21395. doi: 10.1073/pnas.1215210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levy R, Borenstein E. Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proc Natl Acad Sci USA. 2013;110(31):12804–12809. doi: 10.1073/pnas.1300926110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nemergut DR, et al. Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev. 2013;77(3):342–356. doi: 10.1128/MMBR.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JBH. Beyond biogeographic patterns: Processes shaping the microbial landscape. Nat Rev Microbiol. 2012;10(7):497–506. doi: 10.1038/nrmicro2795. [DOI] [PubMed] [Google Scholar]
- 5.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJM, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336(6086):1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lax S, et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. 2014;345(6200):1048–1052. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCalley CK, et al. Methane dynamics regulated by microbial community response to permafrost thaw. Nature. 2014;514(7523):478–481. doi: 10.1038/nature13798. [DOI] [PubMed] [Google Scholar]
- 8.Hugoni M, et al. Structure of the rare archaeal biosphere and seasonal dynamics of active ecotypes in surface coastal waters. Proc Natl Acad Sci USA. 2013;110(15):6004–6009. doi: 10.1073/pnas.1216863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zinger L, et al. Global patterns of bacterial beta-diversity in seafloor and seawater ecosystems. PLoS ONE. 2011;6(9):e24570. doi: 10.1371/journal.pone.0024570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sul WJ, Oliver TA, Ducklow HW, Amaral-Zettler LA, Sogin ML. Marine bacteria exhibit a bipolar distribution. Proc Natl Acad Sci USA. 2013;110(6):2342–2347. doi: 10.1073/pnas.1212424110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hewson I, Jacobson Meyers ME, Fuhrman JA. Diversity and biogeography of bacterial assemblages in surface sediments across the San Pedro Basin, Southern California Borderlands. Environ Microbiol. 2007;9(4):923–933. doi: 10.1111/j.1462-2920.2006.01214.x. [DOI] [PubMed] [Google Scholar]
- 12.Bienhold C, Boetius A, Ramette A. The energy-diversity relationship of complex bacterial communities in Arctic deep-sea sediments. ISME J. 2012;6(4):724–732. doi: 10.1038/ismej.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Böer SI, et al. Time- and sediment depth-related variations in bacterial diversity and community structure in subtidal sands. ISME J. 2009;3(7):780–791. doi: 10.1038/ismej.2009.29. [DOI] [PubMed] [Google Scholar]
- 14.Ruff SE, et al. Microbial communities of deep-sea methane seeps at Hikurangi continental margin (New Zealand) PLoS ONE. 2013;8(9):e72627. doi: 10.1371/journal.pone.0072627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruff SE, et al. Indications for algae-degrading benthic microbial communities in deep-sea sediments along the Antarctic Polar Front. Deep Sea Res Part II Top Stud Oceanogr. 2014;108:6–16. [Google Scholar]
- 16.Jamieson RE, Heywood JL, Rogers AD, Billett DSM, Pearce DA. Bacterial biodiversity in deep-sea sediments from two regions of contrasting surface water productivity near the Crozet Islands, Southern Ocean. Deep Sea Res Part I Oceanogr Res Pap. 2013;75:67–77. [Google Scholar]
- 17.Martiny JBH, Eisen JA, Penn K, Allison SD, Horner-Devine MC. Drivers of bacterial β-diversity depend on spatial scale. Proc Natl Acad Sci USA. 2011;108(19):7850–7854. doi: 10.1073/pnas.1016308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zinger L, Boetius A, Ramette A. Bacterial taxa-area and distance-decay relationships in marine environments. Mol Ecol. 2014;23(4):954–964. doi: 10.1111/mec.12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boetius A, Wenzhöfer F. Seafloor oxygen consumption fuelled by methane from cold seeps. Nat Geosci. 2013;6(9):725–734. [Google Scholar]
- 20.Felden J, et al. Anaerobic methanotrophic community of a 5346-m-deep vesicomyid clam colony in the Japan Trench. Geobiology. 2014;12(3):183–199. doi: 10.1111/gbi.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pop Ristova P, et al. Bacterial diversity and biogeochemistry of different chemosynthetic habitats of the REGAB cold seep (West African margin, 3160 m water depth) Biogeosciences. 2012;9(12):5031–5048. [Google Scholar]
- 22.Knittel K, Boetius A. Anaerobic oxidation of methane: Progress with an unknown process. Annu Rev Microbiol. 2009;63(1):311–334. doi: 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- 23.Levin LA. Ecology of cold seep sediments: Interactions of fauna with flow, chemistry and microbes. In: Gibson RN, Atkinson RJA, Gordon JDM, editors. Oceanography and Marine Biology: An Annual Review. Vol 43. Taylor & Francis; Boca Raton, FL: 2005. pp. 1–46. [Google Scholar]
- 24.Leibold MA, et al. The metacommunity concept: A framework for multi-scale community ecology. Ecol Lett. 2004;7(7):601–613. [Google Scholar]
- 25.Kier G, et al. A global assessment of endemism and species richness across island and mainland regions. Proc Natl Acad Sci USA. 2009;106(23):9322–9327. doi: 10.1073/pnas.0810306106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amend AS, et al. Macroecological patterns of marine bacteria on a global scale. J Biogeogr. 2013;40(4):800–811. [Google Scholar]
- 27.Biddle JF, et al. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc Natl Acad Sci USA. 2006;103(10):3846–3851. doi: 10.1073/pnas.0600035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flores GE, et al. Inter-field variability in the microbial communities of hydrothermal vent deposits from a back-arc basin. Geobiology. 2012;10(4):333–346. doi: 10.1111/j.1472-4669.2012.00325.x. [DOI] [PubMed] [Google Scholar]
- 29.Tavormina PL, Ussler W, 3rd, Orphan VJ. Planktonic and sediment-associated aerobic methanotrophs in two seep systems along the North American margin. Appl Environ Microbiol. 2008;74(13):3985–3995. doi: 10.1128/AEM.00069-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grünke S, et al. Mats of psychrophilic thiotrophic bacteria associated with cold seeps of the Barents Sea. Biogeosciences. 2012;9(8):2947–2960. [Google Scholar]
- 31.Pohlman JW, Bauer JE, Waite WF, Osburn CL, Chapman NR. Methane hydrate-bearing seeps as a source of aged dissolved organic carbon to the oceans. Nat Geosci. 2010;4(1):37–41. [Google Scholar]
- 32.Knittel K, Lösekann T, Boetius A, Kort R, Amann R. Diversity and distribution of methanotrophic archaea at cold seeps. Appl Environ Microbiol. 2005;71(1):467–479. doi: 10.1128/AEM.71.1.467-479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Felden J, Wenzhöfer F, Feseker T, Boetius A. Transport and consumption of oxygen and methane in different habitats of the Håkon Mosby Mud Volcano (HMMV) Limnol Oceanogr. 2010;55(6):2366–2380. [Google Scholar]
- 34.Holler T, et al. Thermophilic anaerobic oxidation of methane by marine microbial consortia. ISME J. 2011;5(12):1946–1956. doi: 10.1038/ismej.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biddle JF, et al. Anaerobic oxidation of methane at different temperature regimes in Guaymas Basin hydrothermal sediments. ISME J. 2012;6(5):1018–1031. doi: 10.1038/ismej.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haroon MF, et al. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature. 2013;500(7464):567–570. doi: 10.1038/nature12375. [DOI] [PubMed] [Google Scholar]
- 37.Ettwig KF, et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464(7288):543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 38.Chevalier N, Bouloubassi I, Birgel D, Taphanel MH, López-García P. Microbial methane turnover at Marmara Sea cold seeps: A combined 16S rRNA and lipid biomarker investigation. Geobiology. 2013;11(1):55–71. doi: 10.1111/gbi.12014. [DOI] [PubMed] [Google Scholar]
- 39.Schubert CJ, et al. Aerobic and anaerobic methanotrophs in the Black Sea water column. Environ Microbiol. 2006;8(10):1844–1856. doi: 10.1111/j.1462-2920.2006.01079.x. [DOI] [PubMed] [Google Scholar]
- 40.Hesselbo SP, et al. Morgans Bell HS Massive dissociation of gas hydrate during a Jurassic oceanic anoxic event. Nature. 2000;406(6794):392–395. doi: 10.1038/35019044. [DOI] [PubMed] [Google Scholar]
- 41.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quast C, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41(D1):D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.