Significance
At the amino acid binding and recognition step, some aminoacyl-tRNA synthetases (aaRSs) face the challenge of discrimination between cognate and closely similar noncognate amino acids. To ensure a high fidelity of protein biosynthesis, aaRSs developed an additional editing activity associated with a specific site, where misacylated tRNAs are hydrolyzed. The structure of heterodimeric Thermus thermophilus phenylalanyl-tRNA synthetase (TtPheRS) with the aminonucleoside antibiotic puromycin reveals the detailed architecture of the complex with ligand mimicking the A76 of tRNA misacylated with noncognate Tyr. Based on the crystal structure and using quantum mechanics/molecular mechanics approach, we present a universal hydrolytic mechanism utilizing cyclic 2′,3′-intermediates for TtPheRS, and potentially for other class I and class II aaRSs.
Keywords: biosynthesis, aminoacyl-tRNA synthetases, tRNA, puromycin, editing
Abstract
At the amino acid binding and recognition step, phenylalanyl-tRNA synthetase (PheRS) faces the challenge of discrimination between cognate phenylalanine and closely similar noncognate tyrosine. Resampling of Tyr-tRNAPhe to PheRS increasing the number of correctly charged tRNA molecules has recently been revealed. Thus, the very same editing site of PheRS promotes hydrolysis of misacylated tRNA species, associated both with cis- and trans-editing pathways. Here we report the crystal structure of Thermus thermophilus PheRS (TtPheRS) at 2.6 Å resolution, in complex with phenylalanine and antibiotic puromycin mimicking the A76 of tRNA acylated with tyrosine. Starting from the complex structure and using a hybrid quantum mechanics/molecular mechanics approach, we investigate the pathways of editing reaction catalyzed by TtPheRS. We show that both 2′ and 3′ isomeric esters undergo mutual transformation via the cyclic intermediate orthoester, and the editing site can readily accommodate a model of Tyr-tRNAPhe where deacylation occurs from either the 2′- or 3′-OH. The suggested pathway of the hydrolytic reaction at the editing site of PheRS is of sufficient generality to warrant comparison with other class I and class II aminoacyl-tRNA synthetases.
A key role in genetic code translation play aminoacyl-tRNA synthetases (aaRSs), providing linkage of amino acids to tRNAs. Before activation, at the amino acid recognition step, some aaRSs face a challenge of discrimination among amino acids with closely similar chemical structure. The rate of erroneous aminoacylation products generated in vivo is no more than one error per 104−105 correct reactions (1). To ensure such an extent of accuracy, aaRSs developed a multisieve mechanism of proofreading (2, 3). The existence of a proofreading activity has been demonstrated for both class I and class II aaRSs. Among aaRSs on record, about half of them are capable of selecting between amino acids resembling each other (4).
The class aminoacyl-tRNA synthetases (aaRSs), namely IleRS, ValRS, and LeuRS, are characterized by a conserved connective polypeptide 1 (CP1) editing domain forming insertion into the catalytic core, except in cases of bacterial and mitochondrial LeuRSs, where the CP1 occurs at a different point of insertion (5). MetRS also falls into class I, but its CP1 lacks editing activity (6). The editing domains of class II aaRSs (ThrRS, ProRS, AlaRS, PheRS) are more diverse in amino acid sequence and in the distinguishing features of their folds. Kinetic experiments carried out for SerRS revealed the presence of a tRNA-independent pretransfer editing pathway (7).
Detailed analyses of posttransfer editing were performed for class I LeuRS and class II ThrRS (8, 9). The structures of the LeuRS posttransfer complex imply the existence of water molecules that are specifically coordinated, to play the role of attacking nucleophiles. The alanine-scanning mutagenesis of the editing site has failed to identify key residues directly involved in catalysis (8). Thus, it was proposed that the CP1 domain simply binds the substrates in a configuration that favors attack by a water molecule, which itself is appropriately positioned by the set of key residues. The crystal structure of the editing domain from ThrRS complexed with Ser-A76 reveals two water molecules located on either side of the hydrolyzed bond (9). This study underlines the crucial role played by tRNA in substrate-assisted catalysis, in positioning the catalytic water molecules along with the protein side chains (9, 10).
The 3D structures of Thermus thermophilus phenylalanyl-tRNA synthetase (TtPheRS) and its complexes with functional substrates (11–14) revealed that the catalytic α subunit exerts control over aminoacylation reaction whereas the major role of the β subunit lies in the recognition and binding of cognate tRNAPhe and hydrolysis of misacylated tRNA (Fig. 1A). The early fast kinetic study demonstrated that tyrosine is indeed transferred to tRNAPhe, and the misacylated tRNA is rapidly hydrolyzed (15). Later, it was established that editing activity of the bacterial and archaeal/eukaryotic PheRSs is associated with the active site located at the interface region between B3 and B4 domains in the β subunit (16–18).
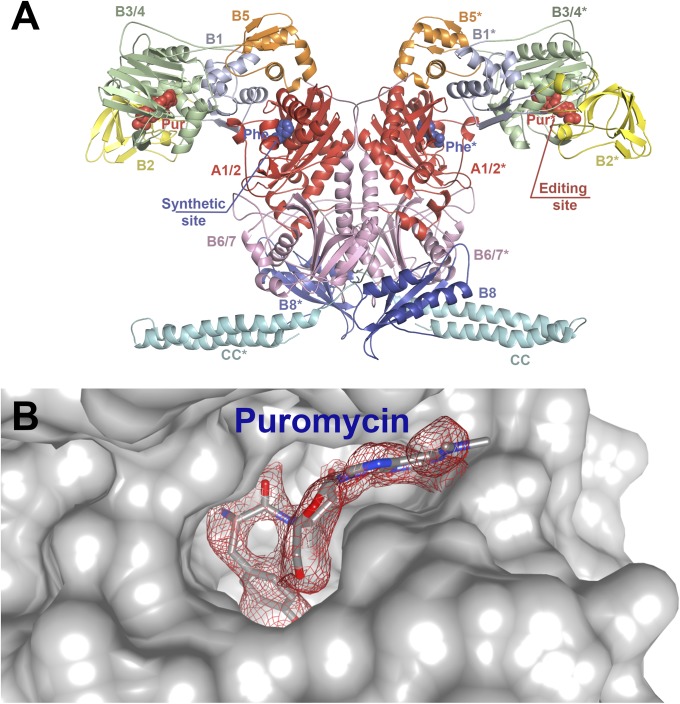
Fig. 1.
(A) Structure of the TtPheRS complex with puromycin and phenylalanine. The protein is shown in cartoon representation; the ligands puromycin (red) and phenylalanine (dark blue) are shown in space-filling representation. The domain architecture of one αβ-heterodimer is shown with the N-terminal coiled coil of the α-subunit colored cyan, catalytic domains A1 and A2 colored red, and structural domains of the β-subunit domains from B1 to B8 colored differently. The symmetry-related heterodimer is denoted with asterisks. (B) The editing site cavity of TtPheRS with bound puromycin. The electron density map (colored in red), calculated as described in SI Materials and Methods with coefficients (Fobs − Fcalc) contoured at 2.5σ. Crystal structure of the TtPheRS complex in space-filling representation (colored gray) rendered to show protein surface interacting with puromycin.
Here we present the crystal structure of TtPheRS, in complex with phenylalanine at the “synthetic” (aminoacylation) site and puromycin (mimicking the A76 of tRNA misacylated with Tyr) at the editing site. The natural substrate’s ester moiety represents an isoelectronic analog of the puromycin amide group, wherein the NH group is replaced with an ester oxygen atom. The appearance of puromycin at the editing site is accompanied by changes in the positions of some bound water molecules or even by their loss, compared with TtPheRS complex with Tyr (17). Loss of the water molecules, supposedly underlying the nucleophilic attack on the carbonyl carbon of the ester bond, gives grounds for revisiting the hydrolytic mechanism at work in bacterial PheRSs (17, 19). The suggested pathway of hydrolytic reaction is of sufficient generality to warrant comparison with those of other class I and class II aaRSs.
Results
The Crystal Structure of PheRS Ternary Complex with Puromycin and Phenylalanine.
Before crystal-soaking experiments performed on bacterial PheRS in the presence of puromycin, and X-ray data collection, we assessed the ability of Escherichia coli PheRS (EcPheRS) to bind the ligand, using isothermal titration calorimetry (ITC) and microscale thermophoresis. For the one site-binding model of the αβ heterodimer, a Kd of 0.30 mM was determined by ITC. The thermophoresis of fluorescently labeled EcPheRS differed from that of the complex with puromycin, and yielded Kd of 0.22 mM.
In view of close structural similarity between EcPheRS and TtPheRS (11, 20), and the better resolution limit observed for crystals of the thermophilic homolog, we determined the crystal structure of TtPheRS in complex with puromycin and phenylalanine at 2.6 Å resolution (Table S1). Crystals of TtPheRS were grown as described previously (21). To produce complex with puromycin, crystals were soaked for 48 h in a mixture containing the crystallization buffer (21) and 1 mM puromycin. Notably, although PheRS crystals were not soaked in phenylalanine, the electron density corresponding to the cognate substrate is clearly visible within the synthetic site (Fig. S1). The network of interactions between phenylalanine and the encompassing side chains is in full agreement with those described previously (13, 14).
An electron density that may be unambiguously attributed to the puromycin was identified at the interface between the B3 and B4 domains (Fig. 1B). In puromycin, the OH group in para position is substituted with methoxy group, not favorable for anchoring the analog by Gluβ334 (17). To some extent, this contact is compensated by hydrogen bonding (HB) between the main chain amide of Glyβ315, and oxygen in para position. Furthermore, the binding of puromycin is stabilized by the interaction of its α-amino group with the main chain carbonyl oxygens of Thrβ249 (∼3.2 Å) and Metβ260 (∼2.7 Å). The α-carboxylate of the puromycin forms H bonds with atom ND2 of Asnβ250 (∼2.6 Å) and atom OG1 of Thrβ249 (∼2.9 Å) (Fig. 2A).
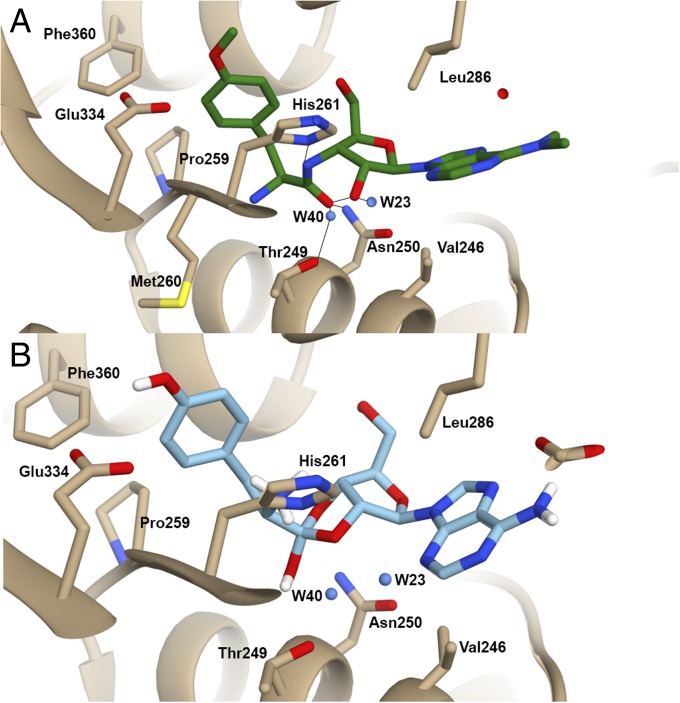
Fig. 2.
View of the editing site of TtPheRS representing interactions with puromycin and orthoester: (A) The protein residues participating in direct and water-mediated contacts with puromycin and (B) the protein residues participating in direct and water mediated contacts with cyclic orthoester acid 2′,3′-intermediate of the phenylalanine. Dashed lines show direct and water-mediated H bonds. The water molecules are depicted by blue spheres. Residues are numbered as in the PDB entry 1PYS.
In puromycin, the nucleoside and amino acid moieties are linked by an amide bridge, in contrast to the ester bridge occurring in the aminoacyl-tRNA (aa-tRNA). The nitrogen of the amide bridge may be involved in HB with the ND1 atom of Hisβ261, in the event that the latter is protonated. The 2′-OH group interacts with the main chain nitrogen of Alaβ262 via the water molecule W40. The sugar moiety position is further stabilized by linkage between the O5′ atom and the main chain nitrogen of Glyβ318. An electron density associated with adenine is weaker compared with other fragments of puromycin, due to its location at the entrance to the editing site tunnel (Fig. 1B). The anchoring of the adenosine moiety is established by hydrophobic interactions with Ileβ242 and Valβ246 from one side, and Leuβ286 from the other side (Fig. 2A). The plane of the A76 base occupies a position midway between these residues. A puromycin-bound network of interactions is in remarkable agreement with a previously postulated model of CCA translocation and contacts maintaining placement of A76 in the editing site (17). Leuβ286 is localized on the extremity of the B4 domain β-hairpin 282–292, and forms one of the invariant residues located within the editing pockets of both the prokaryotic and archaeal/eukaryal PheRSs. Site-directed mutagenesis of B3/B4 domains from Pyrococcus horikoshii PheRS (PhPheRS), and kinetic studies confirmed the pivotal role of Leu168 (a counterpart of Leuβ286 in TtPheRS) in the editing activity (18).
Comparison of Editing Domains from Different PheRSs.
The B3/B4 module of Homo sapiens PheRS (HsPheRS), which fails to exhibit a significant degree of sequence similarity, resembles the editing module of the TtPheRS: The RMSD for 133 superimposed Cα atoms is 2.28 Å (22) (Fig. S2). The editing domains of PhPheRS and HsPheRS share much higher sequence and structure similarities: a 32% sequence identity and an RMSD that is 1.28 Å over 192 Cα atoms (18, 22).
Previous studies provide evidence that editing modules in both bacterial and archaeal/eukaryal PheRSs preserve the ability to specifically recognize Tyr-tRNAPhe (17, 18). The Gluβ334 in TtPheRS binds tyrosine’s OH group in para position, and discards the phenylalanine. In PhPheRS and HsPheRS, this residue is substituted with Aspβ234 and Gluβ254, respectively. In line with mutation experiments on the PhPheRS, Aspβ234 plays a crucial role in discrimination against Phe (18).
From the above, it might be assumed that the structural resemblance of the editing modules in bacterial and archaeal/eukaryotic PheRSs is indicative of similarity in editing mechanisms as well. The low levels of sequence conservation may imply that specific residues do not participate directly in catalysis but rather coordinate positions of catalytically important water molecules.
In the archaeal enzyme, a water molecule (W461) has been found close to the ester bond coordinated by Gln126 (2.8 Å); similarly, Thrβ249 in TtPheRS creates contact with water molecule W40 (2.9 Å). The water molecule W23 generates an extensive HB network with the amide group of Valβ324 (2.8 Å) and the main chain carbonyl oxygen of Alaβ262 (3.1 Å) as well as with the N3 of the substrate adenine (3.0 Å), and with the O2′ of the ribose (3.3 Å). The W23 molecule is highly conserved in both the apo form of TtPheRS [Protein Data Bank (PDB) ID code 1PYS; W43] and in various complexes with functional ligands [PDB ID codes 1JJC (W138) and 3TEH (F16O)].
Differences in the Location of Puromycin and Other Ligands in the Editing Site of PheRS.
It is evident from comparison of the crystal structures of TtPheRS complexes with puromycin, tyrosine, meta-tyrosine, and l-Dopa that binding sites of the ligands are placed alongside each other (17, 23, 24). The aromatic rings of the ligands are rotated relative to each other, and slightly shifted (Fig. S3). In the complex with puromycin, some of the water molecules previously observed in the vicinity of tyrosine turned out to be displaced from their positions, and are no longer detected. Thus, W20, W51, W80, and W112 water molecules are missing, whereas W12 and W14 preserved their positions in complex with puromycin (PDB ID code 2AMC). Water molecule W99, previously “locked” into position and linking the amino group of Tyr to the hydroxyl of Thrβ354 and the main chain amide of Alaβ356, also disappears, due to the absence of a primary amino group in the puromycin. It is noteworthy that “different methodologies are likely to produce different outcomes for hydrogens for which there is no clear experimental preference” (25). Thus, the concept of a hydrolytic mechanism interpreting water molecule W112 as attacking nucleophile, and W99 as a proton donor stabilizing the leaving group (19), should be amended by the puromycin complex data, and revisited accordingly.
Quantum Mechanics/Molecular Mechanics Simulation and Its Structural Facets.
A vast amount of data implies that an aa-tRNA may exist concurrently in two 2′-O and 3′-O-aminoacylated isomers; i.e., the aminoacyl residue, upon forming an ester bond, migrates between the 2′- and 3′-hydroxyls at the 3′-terminal ribose of the tRNAs (26–29). These findings trace back to results indicative of a solution equilibrium mixture of 2′ and 3′ isomers of ribose esters, including 2′(3′)-O-phenylalanyladenosine and 2′- and 3′-O-anthranoyladenosine (30, 31). The molar ratio of the 2′ and 3′ isomers at equilibrium was reported to be comparable (∼1:2.5) (31). However, migration of the phenylalanine happens four orders of magnitude faster, thus suggesting that the equilibration rate depends on the chemical structure of the acyl group (31). The 2′→3′ transition proceeds via an intramolecular reaction with a formation of cyclic orthoester acid 2′,3′-intermediate of the aminoacyl residue (27). Fast transitions in 2′ ↔ 3′ isomers are associated with proton migration between hydroxyl groups accelerating in the presence of water molecules (30). Formation of the orthoester from the ester is accompanied by proton translocation from the ribose O2′ onto the carbonyl oxygen and subsequent covalent bond formation between the O2′ of the ribose and the carbon atom of the carbonyl group.
A combined quantum mechanics/molecular mechanics (QM/MM) approach was exploited to model the editing pathways of the Tyr-tRNAPhe. They were modeled in the presence of water molecules, W40 and W23, and controlled by the stereochemical restraints generated by the side chains (Figs. S4 and S5 and Schemes S1–S6). Formation of the covalent bond between the 2′-oxygen and the carbon atom of the C=O group gives rise to the cyclic (2′,3′-ortho) intermediate, with an architecture complementary to the surface of the misacylated tRNA-binding pocket (Fig. 2B). Notably, the calculated binding energy of the orthoester is higher, than that of native 3′-ester. It is believed that stabilization of the orthoester in the editing site provides a means to control the acyl moiety migration between the vicinal hydroxyls.
Our findings indicate that ribose hydroxyls (2′ or 3′) interact with the juxtaposed ester carbonyl and promote an appearance of three different isomers,
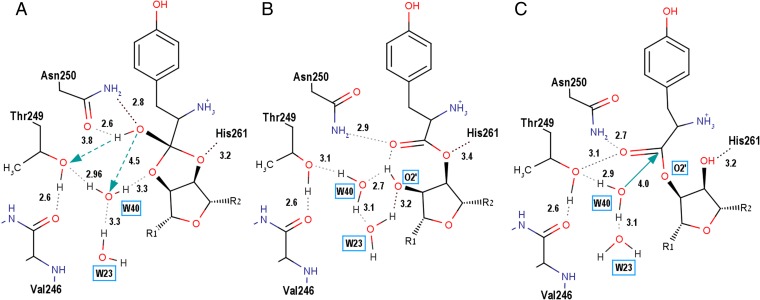
The local energy minimum (S1) corresponds to the 3′-ester analog, with a conformation much like that of puromycin. The 3′-ester/puromycin conformation is stabilized by an intramolecular HB (∼3.2 Å) between the 2′-OH of the ribose and carbonyl oxygen. Polarization of this HB triggers low-barrier proton migration along the sequence of electronegative atoms and converts the 3′-ester to its structural isomers, corresponding to the cyclic 2′,3′-ortho-intermediate, and tyrosyl-2′-ribose ester (S3) (Schemes S1–S6). The 2′,3′-intermediate forms two HBs between the oxygen of the OH group and the amide group of Asnβ250 (Fig. 3A). The water molecule W40 in its original position is located at a distance of ∼4.5 Å from the ortho-OH. The distance between the ortho-OH and the hydroxyl group of Thrβ249 is 3.8 Å. Thus, the S2 structure lacks direct contacts between the labile hydroxyl and a proton acceptor. Two pathways of proton migration immediately follow from QM/MM simulations:
| [1] |
The OG atom of Thrβ249 accepts a proton from ortho-OH (Fig. 3B). The distance between the ortho-OH and Thrβ249 varies from 3.5 Å to 3.1 Å (decreasing from 3.8 Å) and makes possible a proton exchange with Thrβ249 and polarization of the labile hydroxyl ortho-OH (Fig. 3B and Schemes S1–S6). The distance between the hydroxyl of Thrβ249 and the backbone oxygen of Valβ246 for different TtPheRS structures varies within a range of 2.6–2.8 Å. The reaction pathway is accompanied by disruption of the HB between Thrβ249 and Valβ246, and the formation of a new (directed) HB from the donor Thrβ249 to the acceptor W40 (see Scheme S1). Two water molecules, W40 and W23, then form a stable (di)-hydronium ion H5O2+ (Zundel cation) (32) with the proton bridge (Scheme S2). This constitutes an additional factor stabilizing the transition state, and promoting proton transfer toward 2′-OH (Scheme S3). Proton transfer is followed by transition of the cyclic 2′,3′-orthoester to the 3′-ester, and the subsequent hydrolysis of the ester bond formed.
Fig. 3.
Hydrogen bonds stabilizing the 3′-ester (S1), orthoester (S2), and 2′-ester (S3) conformations in the PheRS editing site. (A) Conformation of the ortho-OH group (S2) is stabilized by two HBs: first, between ortho-OH oxygen and the amide group of Asnβ250 and, second, between Hisβ261 and the oxygen atom of the five-membered ring of the orthoester. Proton transfer on the Thrβ249 or W40 (dashed green arrows) can destabilize the system and trigger further transformation into the ester (S3). (B) HB network in the 3′-ester-PheRS complex (S1). The ester oxygen (–O–) forms a weak hydrogen bond with protonated ND1 of Hisβ261. Two key HB water molecules, W40 and W23, are involved in formation of HB network around the ribose O2′. (C) HB network in the 3′-ester-PheRS complex (S3). Carbonyl oxygen forms two hydrogen bonds to Asnβ250 (3.1 Å) and Thrβ249 (2.7 Å); the ribose 3′-oxygen atom can form a hydrogen bond with Hisβ261 (3.2 Å). The reaction coordinate (green arrow) for the nucleophilic attack of W40 is shown.
The second pathway of proton migration results from dynamic fluctuation of the distance between ortho-OH and W40 (from 4.5 Å to 3.5 Å):
| [2] |
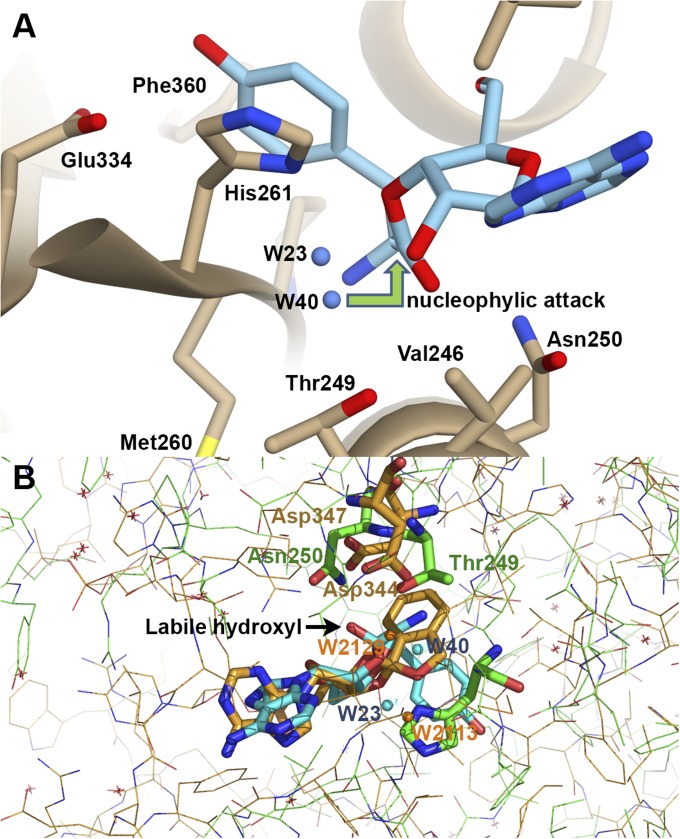
The reaction pathway demonstrates a very low, if any, energy barrier for migration of the W40 molecule. Reversible transformations between the orthoester, S1 ester and S3 ester are accompanied by a reduction in distances to the nearby proton acceptors, Thr249 and W40. Deprotonation of the ortho-OH group and formation of the transient tetrahedral oxyanion (Schemes S2–S5) lead to uncoupling of the chemical bond between the cyclic/central carbon atom and either O2′ or O3′ atoms. These rearrangements transfer the system to the tyrosyl-3′-ribose ester (S1) (Scheme S6) or, alternatively, to the tyrosyl-2′-ribose ester (S3). Thus, proton migration triggers rearrangement of water molecule W40 playing a central role in catalysis, being always available for nucleophilic attack on the carbonyl group of the 2′/3′-ester (see Figs. 3C and 4A), or for proton transfer on the O2′ atom of the orthoester (Scheme S5).
Fig. 4.
(A) The editing site of TtPheRS with modeled tyrosyl-3′-ribose ester (S1). The 3′-ester analog resembles the experimentally observed puromycin. Two water molecules, W23 and W40, are located on one side from the cleaved ester bond. (B) Superposition of the spiroborate/ribose structure of adduct (golden sticks) with two water molecules, W2113 and W2129, from editing site of LeuRS onto the orthoester moiety (blue sticks) with water molecules, W23 and W40, from TtPheRS editing site. Key residues Thrβ249 and Asnβ250 in TtPheRS are colored green, while the partner residues in LeuRS Asp344 and Asp347 are colored golden. Distances between W40-W2129 and W23-W2113 are 1.44 Å and 2.69 Å, respectively. Positions of the water molecule W2113 (LeuRS) and the ND1 atom of Hisβ261 (TtPheRS) are juxtaposed.
Interpretation of the Site-Directed Mutagenesis Data in the Light of TtPheRS Structure Complexed with Puromycin.
When the –NH– group of puromycin is substituted with –O– in the amide, one of the HBs disappears, and repulsion between lone electron pairs (localized at the ester oxygen and the ND1 nitrogen of Hisβ261) occurs at a distance of 3.2 Å. Nevertheless, two stable low-energy structures with neutralized repulsion turn out to be feasible; in either instance, the ND1 atom is in the protonated state. Hisβ261 makes direct contacts with the ester and with the cyclic oxygen of the orthoester in both the protonated and unprotonated states. However, the interaction energy of Hisβ261 with orthoester turned out ∼3 kcal/mol stronger than that seen with the isomeric ester. Molecular modeling/dynamics studies demonstrate that substitution of Hisβ261 with alanine results in the emergence of a water molecule in this area that appears as a compensatory effect of the mutation. These findings correlate well with the mutational experiments on EcPheRS: replacement Hisβ261 for Ala did not affect the editing activity of the mutant (19).
The distance between the hydroxyl of Thrβ249 and the labile hydroxyl of the orthoester is at most 3.7–3.8 Å. Substitution of Thrβ249 with Ala has little effect on the substrate binding: Lack of the Thrβ249 hydroxyl may easily be counterbalanced by rearrangement of the W40 position. The water molecule moves toward the orthoester’s labile hydroxyl, and a new distance of 3.3–3.5 Å enables formation of an HB with the orthoester hydroxyl. Notably, upon rearrangement, the distance between W40 and W23 changes insignificantly. In view of direct contacts of Asnβ250 with the ester and orthoester, it is conceivable that it may exert influence on hydrolytic activity. However, its replacement with alanine in EcPheRS demonstrated only a 1.6-fold reduction in editing activity (19). The QM/MM simulation with alanine in place of Asnβ250 reveals a decrease in distance between the Thrβ249 hydroxyl and the carbonyl oxygen of the ester (or the labile hydroxyl in the orthoester). In these conditions, attacking nucleophile W40 could not be moved out of its original position (Fig. 4A). Thus, Thrβ249 and Asnβ250 seek to maintain the network of HB interactions controlling the deacylation reaction.
Combined QM/MM was applied to evaluate the contribution of 2′/3′-OH groups into editing reaction (ER). Because puromycin offers the experimental root for the 3′-OH ester isomer, we replaced the 2′-OH group with fluorine and hydrogen atoms in turn, and carried out simulations. When the 2′-OH group is substituted with F, the ester conformation turns out to be unstable in view of electrostatic repulsion between the F and the carbonyl oxygen atoms. In contrast, substitution of the 2′-OH with hydrogen leads to relatively stable “editing” conformation that may be the integral part of the ER pathway. However, at a given local energy minimum, the position of the attacking nucleophile W40 undergoes significant displacement, and, as such, can’t be properly arranged for subsequent nucleophilic attack. On the other side, experiments testify that replacements of the 3′-OH with hydrogen or fluorine significantly decrease the posttransfer editing activity of PheRS (19). Thus, from the experimental and theoretical points of view, we can conclude that deacylation in PheRS, which is associated with 2′ or 3′ isomers only, leads to considerable/total lack of the hydrolytic activity.
Discussion
The fraction of mischarged 2′-OH aminoacylated tRNAPhe dissociates from the PheRS before translocation to the editing site, and may be delivered to the ribosome in a ternary complex of aa-tRNA•GTP•EF-Tu (33). However, PheRS proved to be a competitor with EF-Tu for Tyr-tRNAPhe, resulting in the rebinding of the mischarged species at the editing site. Notice that EF-Tu also acts as an isomerase, using a mixture of the 2′(3′)-aminoacyl tRNA isomers as a substrate, and converting them to uniform 3′ complexes (28, 29, 34). Therefore, the editing site of PheRS has to be in a position to hydrolyze either the 2′- or 3′-OH misacylated tRNA species. Occurrence of an equilibrium mixture of two isomers displays the robustness of the editing mechanism: Either of the two misacylated aa-tRNA isomers can bind at the editing site to undergo hydrolysis. The two six-membered HB networks (Fig. 3B) provide a stable configuration well adapted to the intermediate states generated by misacylated tRNA.
The orthoester is known to be unstable in solution, due to labile hydroxyls formed from the ester’s carbonyl of 2′ or 3′ isomers. Nevertheless, NMR studies of EF-Tu complex with tRNA reported that the protein can stabilize an orthoester intermediate between the two isomeric forms (27). QM/MM simulations of deacylation pathway for the TtPheRS complex with puromycin also testify that the labile hydroxyl will be locked into position by interactions with side chains and water molecules. Thereby, the substrate moiety reveals the ability to control the cyclic 2′,3′-orthoester state. A stabilizing role of protein in formation of the high-energy intermediates was also detected in QM/MM dynamic simulation of LeuRS (35).
Of particular interest is the comparison of the modeled cyclic 2′,3′-orthoester intermediate in the editing site of PheRS with the experimentally observed benzoxaborole compounds, forming adducts with terminal A76 ribose of tRNALeu in LeuRS (36). The 2′/3′-hydroxyl groups on the ribose covalently bind to the boron atom from the oxaborole ring, forming tetrahedral “spiroborate” (Fig. S6A). Superimposition of fused spiroborate moiety and orthoester’s ribose with the tetrahedral carbon reveals their close resemblance (Fig. S6 B and C). The only difference is the orientation of the labile hydroxyl at the chiral carbon center. One can hypothesize that “chiral discrimination” is conditioned by the structure of the editing site. The stereochemical resemblance of two systems comes into particular prominence when comparing the arrangement of the water molecules and key residues immediately adjacent to benzoxaborole adducts in TtLeuRS with those in the vicinity of orthoester at the editing site of TtPheRS (Fig. 4B). When superimposed, positions of the key residues Thrβ249 and Asnβ250 in TtPheRS are closely approximating in space the conserved residues Asp344 and Asp347 in TtLeuRS (36–38). The W2113 of LeuRS is found at the position of protonated ND1 atom of Hisβ261. Near the cleavage site, the benzoxaborole from one side is clamped by Thr247, Thr248, and Thr252. The opposite site of the ligand is also clamped by Asp344, Asp347, and Arg346. Residues in both triads form HB with environmental water molecules. Single mutations of either of these conserved threonine residues had minimal effect on editing in EcLeuRS, whereas double mutations of neighboring threonines abolished editing activity (38). Notably, QM/MM metadynamic simulation of TtLeuRS demonstrates involvement of Asp344 in water-mediated ER (39). In LeuRS complexes (PDB ID codes 4ARI, 4AS1, and 2WFG), one water molecule is always located near the O2′ ribose atom (∼2.6 Å), while the other is HB (∼2.8 Å) to the oxygen, analog of the labile hydroxyl. In TtPheRS, the water molecule W23 interacts with the O2′ ribose and the W40 accepts proton from the labile hydroxyl of the orthoester.
Single atom substitutions at the 2′- and 3′-hydroxyls of a variety of mischarged RNAs have been carried out for two close homologs: IleRS and ValRS (40). These experiments revealed that, although acylation is at the 2′-OH for both enzymes, IleRS catalyzes deacylation specifically from the 3′-OH. Note that in normal conditions, transacylation from the 2′- to the 3′-hydroxyl appears to be required for deacylation of Val-tRNAIle by IleRS (41). As opposed to IleRS, ValRS can deacylate noncognate amino acids from the 2′-OH. For LeuRS, no electron density was observed in the editing site upon soaking with the mischarged 3′-OH tRNA analog. This suggests that in the course of editing, LeuRS preserves for Leu its initial point of attachment: the 2′-OH of the ribose (8). Moreover, LeuRS binds pretransfer and posttransfer editing substrates for the case of noncognate norvaline, using a single amino acid discriminatory pocket, thus suggesting a similar mechanism of hydrolysis for both of them (37). All these findings suggest that the editing sites of the above-cited class I aaRSs have a remarkable degree of editing site (CP1) plasticity upon substrate recognition (40).
The enzyme-controlled hydrolysis is nearly 5 × 102- to 103-fold slower, compared with the rate of spontaneous transacylation of 2′(3′)-O-phenylalanyladenosine in a water solution (26). Elongation of the polypeptide chain on the ribosome occurs at a faster rate (by 15–20 s−1) than spontaneous acyl migration (29). Not considered as a rate-limiting step in chain elongation, the joint activities of transacylation and hydrolysis should keep the supply of charged tRNA ahead of its transportation to the ribosomal peptidyl transferase center with EF-Tu. The catalytic mechanism using the orthoester, presented herein and applicable to the hydrolysis of any given 2′- or 3′-misacylated tRNA, provides a higher-yield process, as against that involving scanning of the necessary configuration at the binding site for subsequent hydrolysis.
Materials and Methods
Puromycin was purchased from Sigma-Aldrich. The E. coli PheRS was cloned, expressed, and purified as described previously (20). TtPheRS was purified and crystallized as described (25). Detailed procedures of structure determination and protocols of QM/MM simulations can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We are very grateful to A. Rabinkov and I. Shin for expert assistance in carrying out ITC and MST experiments and D. Safro for help in preparation of the manuscript. This work was partially supported by the Kimmelman Center for Biomolecular Structure and Assembly. M.G.S. holds the Lee & William Abramowitz Professorial Chair of Molecular Biophysics.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4TVA).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1414852112/-/DCSupplemental.
References
- 1.Perona JJ, Gruic-Sovulj I. Synthetic and editing mechanisms of aminoacyl-tRNA synthetases. Top Curr Chem. 2014;344:1–41. doi: 10.1007/128_2013_456. [DOI] [PubMed] [Google Scholar]
- 2.Eldred EW, Schimmel PR. Rapid deacylation by isoleucyl transfer ribonucleic acid synthetase of isoleucine-specific transfer ribonucleic acid aminoacylated with valine. J Biol Chem. 1972;247(9):2961–2964. [PubMed] [Google Scholar]
- 3.Fersht AR. Editing mechanisms in protein synthesis. Rejection of valine by the isoleucyl-tRNA synthetase. Biochemistry. 1977;16(5):1025–1030. doi: 10.1021/bi00624a034. [DOI] [PubMed] [Google Scholar]
- 4.Yadavalli SS, Ibba M. Quality control in aminoacyl-tRNA synthesis its role in translational fidelity. Adv Protein Chem Struct Biol. 2012;86:1–43. doi: 10.1016/B978-0-12-386497-0.00001-3. [DOI] [PubMed] [Google Scholar]
- 5.Cusack S, Yaremchuk A, Tukalo M. The 2 Å crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J. 2000;19(10):2351–2361. doi: 10.1093/emboj/19.10.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crepin T, Schmitt E, Blanquet S, Mechulam Y. Three-dimensional structure of methionyl-tRNA synthetase from Pyrococcus abyssi. Biochemistry. 2004;43(9):2635–2644. doi: 10.1021/bi0356247. [DOI] [PubMed] [Google Scholar]
- 7.Gruic-Sovulj I, Rokov-Plavec J, Weygand-Durasevic I. Hydrolysis of non-cognate aminoacyl-adenylates by a class II aminoacyl-tRNA synthetase lacking an editing domain. FEBS Lett. 2007;581(26):5110–5114. doi: 10.1016/j.febslet.2007.09.058. [DOI] [PubMed] [Google Scholar]
- 8.Palencia A, et al. Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase. Nat Struct Mol Biol. 2012;19(7):677–684. doi: 10.1038/nsmb.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dock-Bregeon AC, et al. Achieving error-free translation: The mechanism of proofreading of threonyl-tRNA synthetase at atomic resolution. Mol Cell. 2004;16(3):375–386. doi: 10.1016/j.molcel.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Hussain T, Kamarthapu V, Kruparani SP, Deshmukh MV, Sankaranarayanan R. Mechanistic insights into cognate substrate discrimination during proofreading in translation. Proc Natl Acad Sci USA. 2010;107(51):22117–22121. doi: 10.1073/pnas.1014299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mosyak L, Reshetnikova L, Goldgur Y, Delarue M, Safro MG. Structure of phenylalanyl-tRNA synthetase from Thermus thermophilus. Nat Struct Biol. 1995;2(7):537–547. doi: 10.1038/nsb0795-537. [DOI] [PubMed] [Google Scholar]
- 12.Goldgur Y, et al. The crystal structure of phenylalanyl-tRNA synthetase from Thermus thermophilus complexed with cognate tRNAPhe. Structure. 1997;5(1):59–68. doi: 10.1016/s0969-2126(97)00166-4. [DOI] [PubMed] [Google Scholar]
- 13.Reshetnikova L, Moor N, Lavrik O, Vassylyev DG. Crystal structures of phenylalanyl-tRNA synthetase complexed with phenylalanine and a phenylalanyl-adenylate analogue. J Mol Biol. 1999;287(3):555–568. doi: 10.1006/jmbi.1999.2617. [DOI] [PubMed] [Google Scholar]
- 14.Fishman R, Ankilova V, Moor N, Safro M. Structure at 2.6 Å resolution of phenylalanyl-tRNA synthetase complexed with phenylalanyl-adenylate in the presence of manganese. Acta Crystallogr D Biol Crystallogr. 2001;57(Pt 11):1534–1544. doi: 10.1107/s090744490101321x. [DOI] [PubMed] [Google Scholar]
- 15.Lin SX, Baltzinger M, Remy P. Fast kinetic study of yeast phenylalanyl-tRNA synthetase: Role of tRNAPhe in the discrimination between tyrosine and phenylalanine. Biochemistry. 1984;23(18):4109–4116. doi: 10.1021/bi00313a015. [DOI] [PubMed] [Google Scholar]
- 16.Roy H, Ling J, Irnov M, Ibba M. Post-transfer editing in vitro and in vivo by the beta subunit of phenylalanyl-tRNA synthetase. EMBO J. 2004;23(23):4639–4648. doi: 10.1038/sj.emboj.7600474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotik-Kogan O, Moor N, Tworowski D, Safro M. Structural basis for discrimination of L-phenylalanine from L-tyrosine by phenylalanyl-tRNA synthetase. Structure. 2005;13(12):1799–1807. doi: 10.1016/j.str.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Sasaki HM, et al. Structural and mutational studies of the amino acid-editing domain from archaeal/eukaryal phenylalanyl-tRNA synthetase. Proc Natl Acad Sci USA. 2006;103(40):14744–14749. doi: 10.1073/pnas.0603182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ling J, Roy H, Ibba M. Mechanism of tRNA-dependent editing in translational quality control. Proc Natl Acad Sci USA. 2007;104(1):72–77. doi: 10.1073/pnas.0606272104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mermershtain I, et al. Idiosyncrasy and identity in the prokaryotic Phe-system: Crystal structure of E. coli phenylalanyl-tRNA synthetase complexed with phenylalanine and AMP. Protein Sci. 2011;20(1):160–167. doi: 10.1002/pro.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chernaya MM, Korolev SV, Reshetnikova LS, Safro MG. Preliminary crystallographic study of the phenylalanyl-tRNA synthetase from Thermus thermophilus HB8. J Mol Biol. 1987;198(3):555–556. doi: 10.1016/0022-2836(87)90301-9. [DOI] [PubMed] [Google Scholar]
- 22.Finarov I, Moor N, Kessler N, Klipcan L, Safro MG. Structure of human cytosolic phenylalanyl-tRNA synthetase: Evidence for kingdom-specific design of the active sites and tRNA binding patterns. Structure. 2010;18(3):343–353. doi: 10.1016/j.str.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 23.Klipcan L, Moor N, Kessler N, Safro MG. Eukaryotic cytosolic and mitochondrial phenylalanyl-tRNA synthetases catalyze the charging of tRNA with the meta-tyrosine. Proc Natl Acad Sci USA. 2009;106(27):11045–11048. doi: 10.1073/pnas.0905212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moor N, Klipcan L, Safro MG. Bacterial and eukaryotic phenylalanyl-tRNA synthetases catalyze misaminoacylation of tRNA(Phe) with 3,4-dihydroxy-L-phenylalanine. Chem Biol. 2011;18(10):1221–1229. doi: 10.1016/j.chembiol.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 25.Forrest LR, Honig B. An assessment of the accuracy of methods for predicting hydrogen positions in protein structures. Proteins. 2005;61(2):296–309. doi: 10.1002/prot.20601. [DOI] [PubMed] [Google Scholar]
- 26.Taiji M, Yokoyama S, Miyazawa T. Transacylation rates of (aminoacyl)adenosine moiety at the 3′-terminus of aminoacyl transfer ribonucleic acid. Biochemistry. 1983;22(13):3220–3225. doi: 10.1021/bi00282a028. [DOI] [PubMed] [Google Scholar]
- 27.Förster C, Limmer S, Zeidler W, Sprinzl M. Effector region of the translation elongation factor EF-Tu.GTP complex stabilizes an orthoester acid intermediate structure of aminoacyl-tRNA in a ternary complex. Proc Natl Acad Sci USA. 1994;91(10):4254–4257. doi: 10.1073/pnas.91.10.4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sprinzl M. Chemistry of aminoacylation and peptide bond formation on the 3′terminus of tRNA. J Biosci. 2006;31(4):489–496. doi: 10.1007/BF02705188. [DOI] [PubMed] [Google Scholar]
- 29.Weinger JS, Strobel SA. Participation of the tRNA A76 hydroxyl groups throughout translation. Biochemistry. 2006;45(19):5939–5948. doi: 10.1021/bi060183n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reese CB, Trentham DR. Acyl migration in ribonucleoside derivatives. Tetrahedron Lett. 1965;6(29):2467–2472. doi: 10.1016/s0040-4039(01)84008-9. [DOI] [PubMed] [Google Scholar]
- 31.Nawrot B, Milius W, Ejchart A, Limmer S, Sprinzl M. The structure of 3′-O-anthraniloyladenosine, an analogue of the 3′-end of aminoacyl-tRNA. Nucleic Acids Res. 1997;25(5):948–954. doi: 10.1093/nar/25.5.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marx D, Tuckerman ME, Hutter J, Parrinello M. The nature of the hydrated excess proton in water. Nature. 1999;397(6720):601–604. [Google Scholar]
- 33.Ling J, et al. Resampling and editing of mischarged tRNA prior to translation elongation. Mol Cell. 2009;33(5):654–660. doi: 10.1016/j.molcel.2009.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nissen P, et al. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270(5241):1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 35.Hagiwara Y, Field MJ, Nureki O, Tateno M. Editing mechanism of aminoacyl-tRNA synthetases operates by a hybrid ribozyme/protein catalyst. J Am Chem Soc. 2010;132(8):2751–2758. doi: 10.1021/ja9095208. [DOI] [PubMed] [Google Scholar]
- 36.Rock FL, et al. An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science. 2007;316(5832):1759–1761. doi: 10.1126/science.1142189. [DOI] [PubMed] [Google Scholar]
- 37.Lincecum TL, Jr, et al. Structural and mechanistic basis of pre- and posttransfer editing by leucyl-tRNA synthetase. Mol Cell. 2003;11(4):951–963. doi: 10.1016/s1097-2765(03)00098-4. [DOI] [PubMed] [Google Scholar]
- 38.Zhai Y, Martinis SA. Two conserved threonines collaborate in the Escherichia coli leucyl-tRNA synthetase amino acid editing mechanism. Biochemistry. 2005;44(47):15437–15443. doi: 10.1021/bi0514461. [DOI] [PubMed] [Google Scholar]
- 39.Boero M. LeuRS synthetase: A first-principles investigation of the water-mediated editing reaction. J Phys Chem B. 2011;115(42):12276–12286. doi: 10.1021/jp2070024. [DOI] [PubMed] [Google Scholar]
- 40.Nordin BE, Schimmel P. Plasticity of recognition of the 3′-end of mischarged tRNA by class I aminoacyl-tRNA synthetases. J Biol Chem. 2002;277(23):20510–20517. doi: 10.1074/jbc.M202023200. [DOI] [PubMed] [Google Scholar]
- 41.Mascarenhas APAS, Rosen AE, Martinis SA, Musier-Forsyth K. 2008. Fidelity mechanisms of the aminoacyl-tRNA synthetases. Protein Engineering, ed Koehrer C, RajBhandary UL (Springer, New York), pp 153−200.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.