Significance
Genetic control of the differentiation between Sertoli cells and granulosa cells has been reported previously. However, the relationship between Sertoli cells and Leydig cells in the testis has not yet been definitively determined. In the present study, we demonstrate for the first time, to our knowledge, that these two cell types can be mutually reprogrammed and that Wilms’ Tumor Gene 1 (Wt1) plays a critical role in this process. This study provides a novel concept for cell fate determination in testis development that will improve our understanding of the regulatory mechanisms of gonad development.
Keywords: Wt1, β-catenin, Sertoli cells, Leydig cells, transdifferentiation
Abstract
Sertoli and Leydig cells, the two major somatic cell types in the testis, have different morphologies and functions. Both are essential for gonad development and spermatogenesis. However, whether these cells are derived from the same progenitor cells and the mechanism regulating the differentiation between these two cell types during gonad development remains unclear. A previous study showed that overactivation of Ctnnb1 (cadherin-associated protein, beta 1) in Sertoli cells resulted in Sertoli cell tumors. Surprisingly, in the present study, we found that simultaneous deletion of Wilms’ Tumor Gene 1 (Wt1) and overactivation of Ctnnb1 in Sertoli cells led to Leydig cell-like tumor development. Lineage tracing experiments revealed that the Leydig-like tumor cells were derived from Sertoli cells. Further studies confirmed that Wt1 is required for the maintenance of the Sertoli cell lineage and that deletion of Wt1 resulted in the reprogramming of Sertoli cells to Leydig cells. Consistent with this interpretation, overexpression of Wt1 in Leydig cells led to the up-regulation of Sertoli cell-specific gene expression and the down-regulation of steroidogenic gene expression. These results demonstrate that the distinction between Sertoli cells and Leydig cells is regulated by Wt1, implying that these two cell types most likely originate from the same progenitor cells. This study thus provides a novel concept for somatic cell fate determination in testis development that may also represent an etiology of male infertility in human patients.
In mammals, testes and ovaries arise from the genital ridges, which form as thickening of the epithelial layer on the ventromedial surface of the mesonephros at embryonic day E9.5 in mice. The Sry (sex-determining region Y) gene encodes a high-mobility group (HMG) domain transcription factor that is transiently expressed in the somatic cells of undifferentiated XY gonads from E10–E12. This gene is essential for directing Sertoli cell differentiation in bipotential gonads (1, 2). In XX gonads, which lack Sry expression, the somatic cells instead differentiate into granulosa cells (3).
Sertoli and Leydig cells are two major cell types in the testis, and both play essential roles in spermatogenesis. Sertoli cells are localized within the seminiferous tubules and provide physical and nutritional support for germ cell development. Leydig cells are located in the interstitium between the seminiferous tubules. The testosterone secreted by Leydig cells is necessary for the completion of spermatogenesis and the maintenance of secondary sexual characteristics. Steroidogenic enzymes such as 3β-HSD (3β-hydroxysteroid dehydrogenase), StAR (steroidogenic acute regulatory protein), and P450scc (P450 side-chain cleavage) are specifically expressed in Leydig cells in testes. Cholesterol, a substrate for steroid hormone biosynthesis, accumulates in Leydig cells and can be labeled with Oil Red O (ORO) (4). Sertoli cells are reported to originate from coelomic epithelial cells, whereas cells migrating from the mesonephros represent the putative Leydig cell progenitors (5, 6). However, it is still controversial, and the relationship between Sertoli cells and Leydig cells during testis development remains unclear.
Wt1 encodes a zinc finger nuclear transcription factor that was originally identified as a tumor suppressor gene in WT patients (7–10). During embryonic development, Wt1 is expressed in the coelomic epithelium and the underlying mesenchymal cells of the urogenital ridge (11, 12). Deletion of Wt1 in mouse models results in gonadal agenesis due to the failure of genital ridge development (12).
Our previous study demonstrated that Wt1 plays critical roles in testis development. Inactivation of Wt1 in Sertoli cells after sex determination causes aberrant testis development due to the disruption of testicular cords (13). However, the underlying molecular mechanism is still unclear. Overactivation of Ctnnb1 by deletion of exon3 in Sertoli cells during embryonic development also caused a testicular cord disruption, similar to Wt1 deletion (14), suggesting that Wt1 and Ctnnb1 likely regulate the same signaling pathway in testis development.
To explore the relationship between Wt1 and Ctnnb1 in testis development, Wt1 and Ctnnb1 (exon3) were simultaneously deleted in Sertoli cells using AMH-Cre transgenic mice. Surprisingly, we found that Leydig cell-like tumors, but not Sertoli cell tumors, developed in double knockout (KO) mice. Further studies revealed that Wt1 is required for Sertoli cell lineage maintenance and that inactivation of Wt1 results in Sertoli cell to Leydig cell transdifferentiation. This study thus demonstrates that Sertoli cells and Leydig cells most likely originate from the same progenitor cells and that the differentiation between these two cell types is controlled by Wt1.
Results
Double KO of Wt1 and Ctnnb1 (exon3) in Sertoli Cells Caused Leydig Cell-Like Tumor Development.
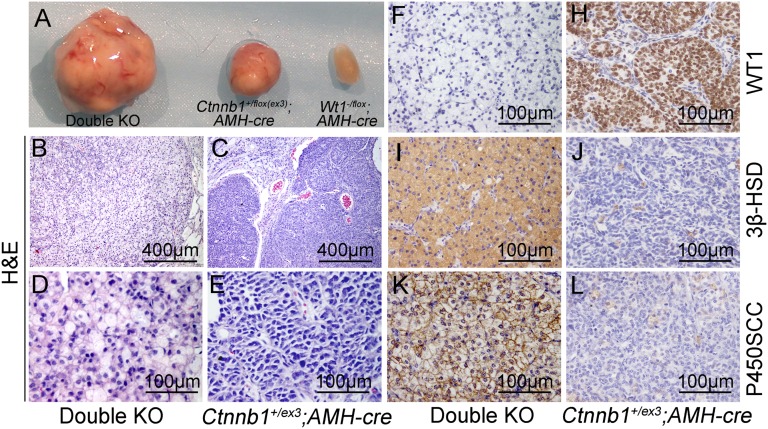
Our previous studies found that both deletion of Wt1 and overactivation of Ctnnb1 induced by deleting exon3 in Sertoli cells using AMH-Cre caused testicular cord disruption (13, 14). However, testicular tumors were observed in Ctnnb1 overactivated mice but not in Wt1 KO mice (14, 15). To test whether these two genes regulate the same signaling pathway in testis development, Wt1 and Ctnnb1 (exon3) were simultaneously deleted in Sertoli cells using AMH-Cre transgenic mice. The male mice were killed at 8 mo of age. We found that ∼80% (13/16) of the Ctnnb1+/flox(ex3)AMH-Cre mice developed testicular tumors, consistent with the previous study (15). Interestingly, 100% (13/13) of the Wt1–/floxCtnnb1+/flox(ex3)AMH-Cre mice (double KO mice) developed testicular tumors, and no tumors were found in Wt1–/floxAMH-Cre mice (Fig. 1A).
Fig. 1.
Leydig-like cell tumor development in Wt1 and Ctnnb1ex3 double KO mice. (A) The morphology of testes from Wt1–/floxAMH-Cre, Ctnnb1+/flox(ex3)AMH-Cre, and double KO mice (Wt1–/floxCtnnb1+/flox(ex3)AMH-Cre). (B–E) Histological analysis of testicular tumors from Ctnnb1+/flox(ex3)AMH-Cre and double KO mice by H&E staining. Immunohistochemical analysis of testis tumor sections using antibodies to WT1 (F and H), 3β-HSD (I and J), and P450SCC (K and L), showing that WT1 protein was present in tumor cells from Ctnnb1+/flox(ex3)AMH-Cre mice (H) but not in tumor cells from double KO mice (F). 3β-HSD (I and J) and P450SCC (K and L) were expressed in tumor cells from double KO mice (I and K) but not Ctnnb1+/flox(ex3)AMH-Cre mice (J and L).
The histology of tumor cells from Ctnnb1+/flox(ex3)AMH-Cre and double KO mice was examined by hematoxylin and eosin staining. Most of the tumor cells from Ctnnb1+/flox(ex3)AMH-Cre mice were blastema-like with condensed nuclei and reduced eosinophilic cytoplasm (Fig. 1 C and E), consistent with a previous study (15). By contrast, the tumor cells from double KO mice contained increased eosinophilic cytoplasm and fewer condensed round nuclei (Fig. 1 B and D). To further characterize the tumor cells, immunohistochemistry experiments were performed. The tumor cells from Ctnnb1+/flox(ex3)AMH-Cre testes expressed the Sertoli cell marker gene WT1 (Fig. 1H). However, no WT1 signal was detected in the tumor cells from double KO mice (Fig. 1F), despite the fact that it has been demonstrated that the small truncated WT1 protein remaining after the deletion of two exons from the Wt1flox allele is recognized by the antibody used in this study and can be used to trace Wt1 mutant Sertoli cells (13, 16). Surprisingly, the Leydig cell-specific marker genes 3β-HSD and P450SCC were abundantly expressed in double KO tumor cells (Fig. 1 I and K), comparable to normal Leydig cells, but not in the tumor cells from Ctnnb1+/flox(ex3)AMH-Cre testes (Fig. 1 J and L). To further verify the cell type, transmission electron microscopy (TEM) was used to analyze the ultrastructure of the tumor cells from double KO mice. As shown in Fig. S1, numerous large lipid droplet clusters were noted in the Leydig-like tumor cells from the double KO mice (Fig. S1A, circled with black dotted line), similar to normal fetal Leydig cells (Fig. S1D, white arrows) (17, 18). Real-time PCR analysis was also conducted to examine the expression of other Leydig cell marker genes using mRNA from normal Sertoli cells, normal Leydig cells, and tumor cells from double KO mice. The mRNA levels of 3β-HSD, p450scc, Lhr (luteinizing hormone receptor), Star, Sur2 (sulfonylurea receptor 2), and Cyp17a1 were dramatically increased in double KO tumor cells compared with normal Sertoli cells. Some genes, including 3β-HSD, p450scc, and Lhr, were expressed more highly than in normal Leydig cells (Fig. S2A). By contrast, the expression of Sertoli cell-specific genes, such as Sox9 (SRY - box 9), Amh (anti-Mullerian hormone), Dmrt1 (doublesex and mab-3 related transcription factor 1), Gdnf (glial cell line-derived neurotrophic factor), Ptdgs (prostaglandin D2 synthase), Dhh (desert hedgehog), Erbb4 (v-erb-a erythroblastic leukemia viral oncogene homolog 4), Shbg (sex hormone-binding globulin), and Clu (clusterin), was dramatically reduced compared with control Sertoli cells (Fig. S2B). Interestingly, we also found that adult Leydig cell-specific genes [e.g., Est (estrogen sulfotransferase), Vcam1 (vascular cell adhesion molecule 1), PGD, and 17β-HSDIII (17β-hydroxysteroid dehydrogenase III)] were abundantly expressed in the tumor cells from double KO mice, whereas the expression of the fetal Leydig cell marker gene Tsp2 (thrombospondin 2) was not increased compared with control adult Leydig cells (Fig. S2A). These results indicate that the testicular tumor cells in double KO mice are Leydig-like cells. However, based on their histology and gene expression patterns, these cells are not typical fetal or adult Leydig cells.
Leydig-Like Tumor Cells in Double KO Mice Derived from Sertoli Cells.
Given that AMH-Cre is specifically expressed in Sertoli cells (19), Wt1 and Ctnnb1 (exon3) were deleted in only Sertoli cells. Therefore, we surmised that the tumor cells in double KO mice derived either from normal Leydig cells affected by the deletion of Wt1 and Ctnnb1 (exon3) in Sertoli cells or from Sertoli cells that had transdifferentiated into Leydig-like cells.
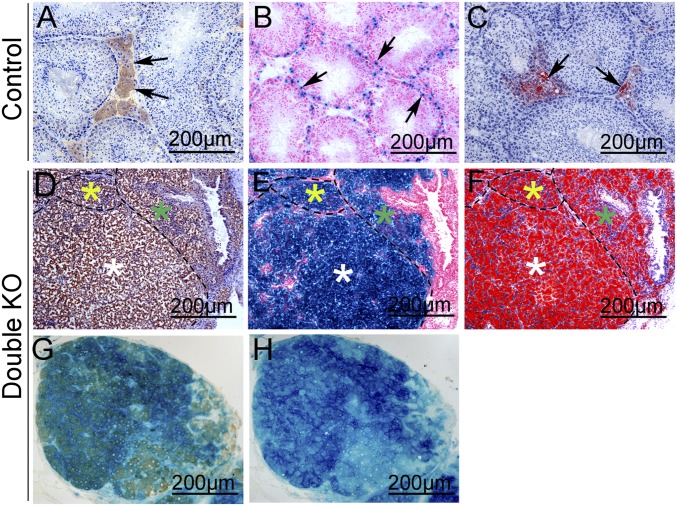
To test these hypotheses, a lineage tracing experiment was performed using Rosa26R mice (20). In control mice (Rosa26R+/floxAMH-Cre), Sertoli cells within seminiferous tubules were stained with X-Gal (Fig. 2B), and no X-Gal signal was detected in Leydig cells, indicating that AMH-Cre is specifically expressed in Sertoli cells. Leydig cells in the interstitium between the seminiferous tubules were labeled with 3β-HSD and ORO (Fig. 2 A and C). To trace the cell lineage of the Leydig-like tumor cells in double KO mice, serial frozen sections from Wt1–/floxCtnnb1+/flox(ex3)Rosa26R+/floxAMH-Cre mice were prepared, and adjacent sections were stained with X-Gal, 3β-HSD, and ORO. As shown in Fig. 2, all of the tumor cells were labeled with X-Gal (Fig. 2E), indicating that these cells originated from Sertoli cells. The X-Gal–positive cells also stained with 3β-HSD (Fig. 2D) and ORO (Fig. 2F) in adjacent sections. These 3β-HSD and X-Gal double staining results further confirm that 3β-HSD and X-Gal were colocalized in the Leydig-like tumor cells in double KO mice (Fig. 2G). These results thus indicate that the Leydig-like tumor cells in double KO mice are derived from Sertoli cells. To further examine the cell fate change of the Sertoli cells in double KO mice, we analyzed tumor development at earlier stages. Because the CTNNB1 protein accumulates in the nucleus after the deletion of exon3, the Ctnnb1 and Wt1 double mutant Sertoli cells were traced by CTNNB1 staining. As shown in Fig. S3, in control testes, CTNNB1 protein was detected at cell junctions between Sertoli and germ cells (Fig. S3 A, C, I, and K), and 3β-HSD protein was expressed in only Leydig cells (Fig. S3 E, G, M, and O, black arrows). In double KO testes, CTNNB1 protein was detected in nuclei, and many CTNNB1-positive tumor-like cell clusters (red dot-line circled) were observed at E15.5 (Fig. S3B), P1 (Fig. S3D), P15 (Fig. S3J), and 1 mo of age (Fig. S3L). However, 3β-HSD protein was not expressed in these CTNNB1-positive tumor-like cell clusters until 1 mo of age (Fig. S3P). These results indicate that the Sertoli cells in double KO mice develop into tumor-like cell clusters first and then transform into 3β-HSD–positive Leydig cells at a later stage.
Fig. 2.
Leydig-like tumor cells in double KO mice derived from Sertoli cells. (A–C) In control testes, Leydig cells were labeled with 3β-HSD (A, arrows) and ORO (C, arrows), and Sertoli cells within seminiferous tubules were labeled with X-Gal (B, arrows). (D–F) In double KO testes, adjacent serial sections were stained with 3β-HSD (D), X-Gal (E), and ORO (F). (G) The tumor cells in double KO testes were double-labeled with X-Gal and 3β-HSD. (H) Adjacent serial section with X-Gal staining but not with 3β-HSD as a negative control.
The Leydig Cell-Specific Gene 3β-HSD Was Expressed in Wt1-Only Inactivated Sertoli Cells.
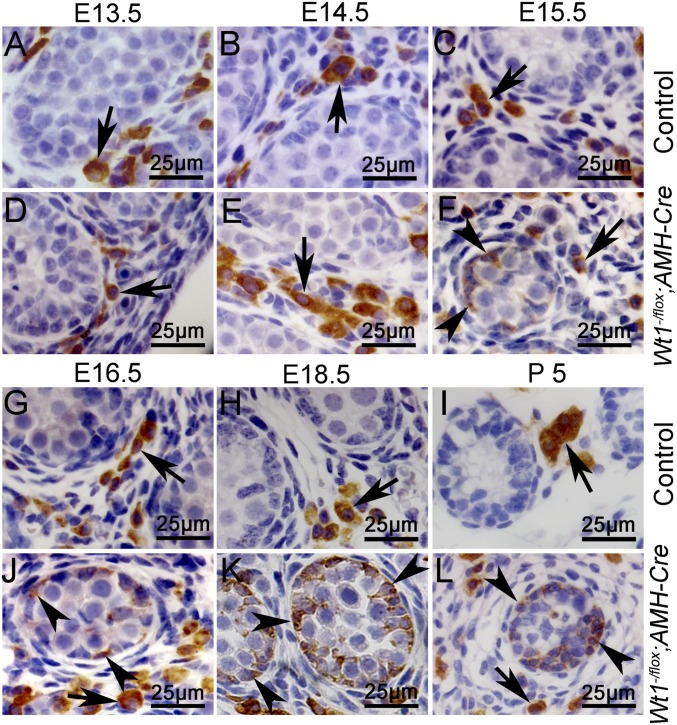
Sertoli cell tumors were observed in Ctnnb1+/flox(ex3)AMH-Cre mice, whereas the Sertoli cells developed into Leydig cell-like tumors in double KO mice, implying that Wt1 is essential for Sertoli cell lineage maintenance. Loss of Wt1 likely results in the Sertoli cell-to-Leydig cell transformation. To further verify these results, Wt1–/floxAMH-Cre mice were generated, and the expression of Sertoli cell-specific genes and 3β-HSD was examined by IHC. As shown in Fig. S4, SOX9 (Fig. S4 A, e, black arrows) and AMH (Fig. S4 B, e, black arrows) proteins were detected in Sertoli cells of Wt1–/floxAMH-Cre mice at E13.5 but were absent from E14.5 to E16.5, consistent with our previous study (13). In control testes, 3β-HSD (Fig. 3 A–C and G–I, arrows) was specifically expressed in Leydig cells from E13.5 to P5. In Wt1–/floxAMH-Cre testes, 3β-HSD was detected in Leydig cells at E13.5 (Fig. 3D, arrows) and E14.5 (Fig. 3E, arrows). Interestingly, 3β-HSD was also detected in the remnant testicular cords (Fig. 3 F and J–L, arrowheads) of Wt1–/floxAMH-Cre testes from E15.5 to P5. The low-magnification images showed that 3β-HSD protein was detected in most of the remnant testicular cords (asterisks) at E16.5 (Fig. S5E, black arrows) and E18.5 (Fig. S5F, black arrows). To further confirm that 3β-HSD was expressed in Wt1-deficient Sertoli cells, a lineage tracing experiment was performed using Wt1–/floxRosa26R+/floxAMH-Cre mice. As shown in Fig. S6, the 3β-HSD–positive cells observed in the remnant testicular cords of Wt1–/floxRosa26R+/floxAMH-Cre mice were also labeled with X-Gal (Fig. S6B, arrowheads). This result indicates that these 3β-HSD–positive cells are Sertoli cells in which Wt1 was inactivated by AMH-Cre.
Fig. 3.
3β-HSD was expressed in the remnant seminiferous tubules of Wt1–/floxAMH-Cre mice. (A–C and G–I) In control testes, 3β-HSD was detected only in the Leydig cells from E13.5 to P5 (arrows). (D–F and J–L) In Wt1–/floxAMH-Cre testes, in addition to Leydig cells (arrows), 3β-HSD was also detected in the remnant seminiferous tubules (F and J–L, arrowheads).
Our previous studies demonstrated that the Wt1flox allele, after Cre-mediated deletion of two exons, produces a truncated WT1 protein that is still recognized by the antibody used in this study (13, 16). A double staining experiment was thus performed using anti-WT1 and anti–3β-HSD antibodies. We found that the 3β-HSD–positive cells in the remnant testicular cords of Wt1–/floxAMH-Cre mice were also labeled with the anti-WT1 antibody (Fig. S6H, white arrowheads). In control testes, WT1 was detected in Sertoli cells but not in Leydig cells (Fig. S6G). These results further confirm that 3β-HSD is expressed in Wt1-deficient Sertoli cells.
Sertoli Cell-Derived 3β-HSD–Positive Leydig Cells Survived in Adult Testes.
To examine the cell fate of Wt1-deficient Sertoli cells at later developmental stages, the expression of WT1 and 3β-HSD was analyzed in Wt1–/floxAMH-Cre testes at P15 and 1 mo. As shown in Fig. S7, several WT1 and 3β-HSD double-positive cell clusters (Fig. S7 D–F, white arrows) were observed at P15, but the number was reduced compared with embryonic stages (Fig. S5). WT1 and 3β-HSD double-positive cell clusters (Fig. S7 J–L, white arrows) were also observed in Wt1–/floxAMH-Cre testes at 1 mo.
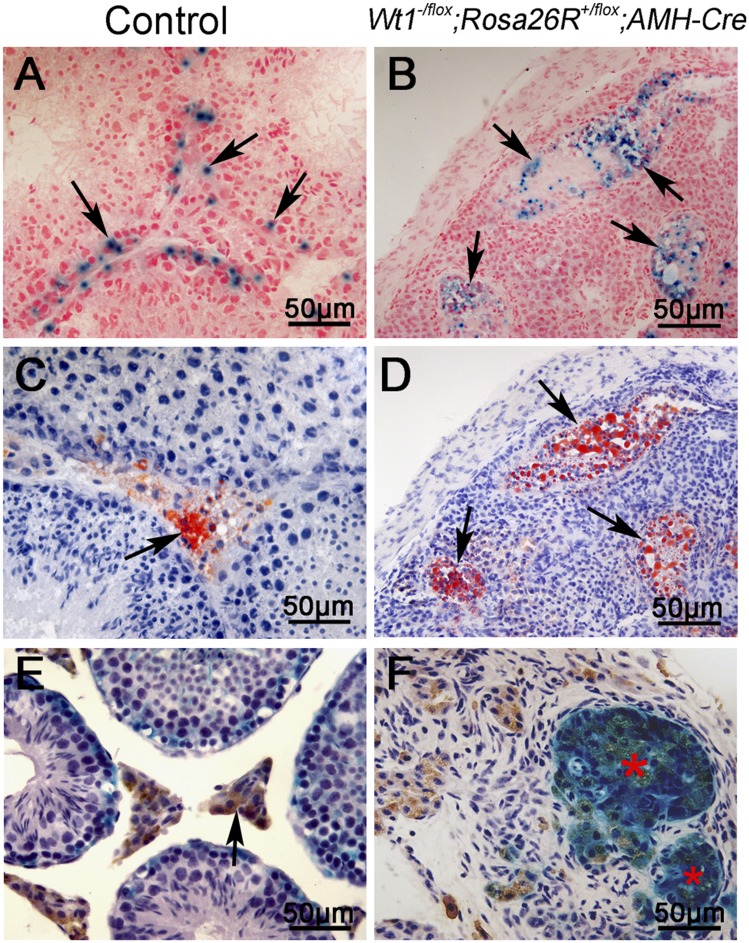
We also examined the development of Sertoli cell-derived 3β-HSD–positive Leydig cells at 4 mo of age using Wt1–/floxRosa26R+/floxAMH-Cre mice. The testis sections from control and Wt1–/floxRosa26R+/floxAMH-Cre mice were stained with X-Gal, 3β-HSD, and ORO. In control testes, X-Gal was observed in only Sertoli cells within seminiferous tubules (Fig. 4A), and Leydig cells were stained with ORO (Fig. 4C) and 3β-HSD (Fig. 4E, arrows). No seminiferous tubules were observed in Wt1–/floxRosa26R+/floxAMH-Cre testes, but several cell clusters were labeled with both X-Gal (Fig. 4B, arrows, and Fig. S8 A and B) and ORO (Fig. 4D, arrows, and Fig. S8 C and D) in adjacent serial sections. 3β-HSD was also expressed in these X-Gal–positive cell clusters (Fig. 4F, red asterisks). These results indicate that the Wt1-deficient Sertoli cells survive in adult testes and continually express Leydig cell-specific marker genes.
Fig. 4.
Sertoli cell-derived 3β-HSD–positive cells were detected in the testes of adult Wt1–/floxRosa26R+/floxAMH-Cre mice. In control testes, Sertoli cells were labeled with X-Gal (A, arrows), whereas Leydig cells were labeled with ORO (C, arrows) and 3β-HSD (E, arrows). In Wt1–/floxRosa26R+/floxAMH-Cre testes, serial sections were prepared and stained with X-Gal (B, arrows) and ORO (D, arrows). (F) X-Gal and 3β-HSD double-positive cells (red asterisks) were observed in Wt1–/floxRosa26R+/floxAMH-Cre testes.
Differential Expression of Leydig Cell- and Sertoli Cell-Specific Genes in Wt1-Deficient Sertoli Cells.
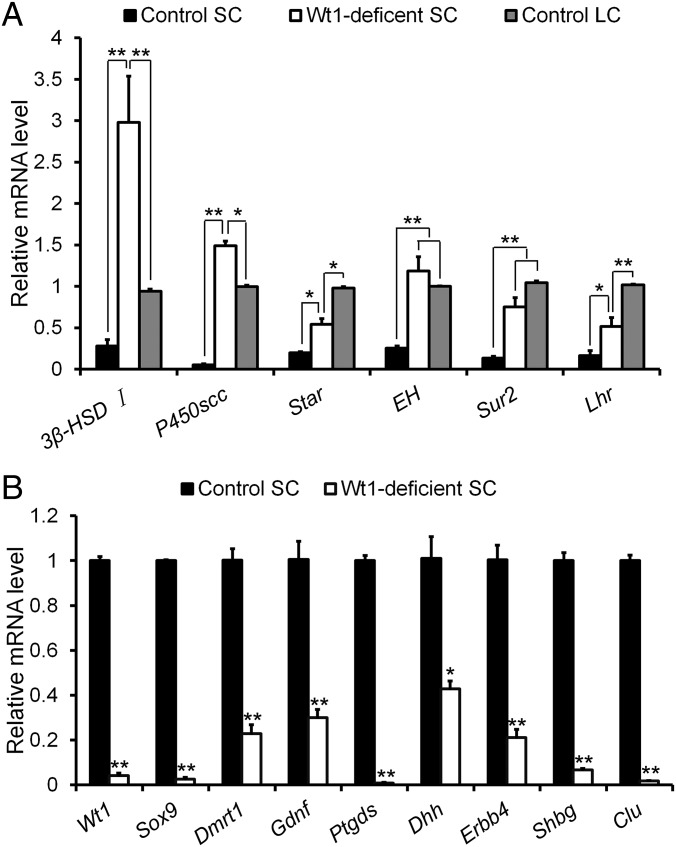
The above experiments showed that Wt1-deficient Sertoli cells transdifferentiate into Leydig-like cells and start to express 3β-HSD. To examine the expression of other Leydig cell-specific genes, Leydig cells, Sertoli cells, and Wt1-deficient Sertoli cells were isolated from control and Wt1–/floxAMH-Cre testes at P1 by flow cytometry. The relative mRNA levels of Leydig cell- and Sertoli cell-specific genes were analyzed by real-time PCR. As shown in Fig. 5, the expression of Sertoli cell-specific genes (e.g., Sox9, Dmrt1, Gdnf, Dhh, Ptgds, Erbb4, Shbg, and Clu) was dramatically reduced in Wt1-deficient Sertoli cells compared with control Sertoli cells (Fig. 5B). By contrast, the expression of Leydig cell-specific genes [e.g., 3β-HSD, p450scc, Lhr, Star, EH (epoxide hydrolase), and Sur2] was dramatically increased relative to control Sertoli cells, comparable to control Leydig cells (Fig. 5A).
Fig. 5.
The expression of Leydig cell and Sertoli cell-specific genes in Wt1-deficient Sertoli cells. (A) Leydig cell-specific genes were abundantly expressed in Wt1-deficient Sertoli cells at levels that were comparable to those of control Leydig cells. (B) The expression of Sertoli cell-specific genes was dramatically reduced in Wt1-deficient Sertoli cells. Data are presented as the mean ± SEM. *P < 0.05; **P < 0.01.
Overexpression of Wt1 in Leydig Cells Caused a Leydig Cell to Sertoli Cell Transformation.
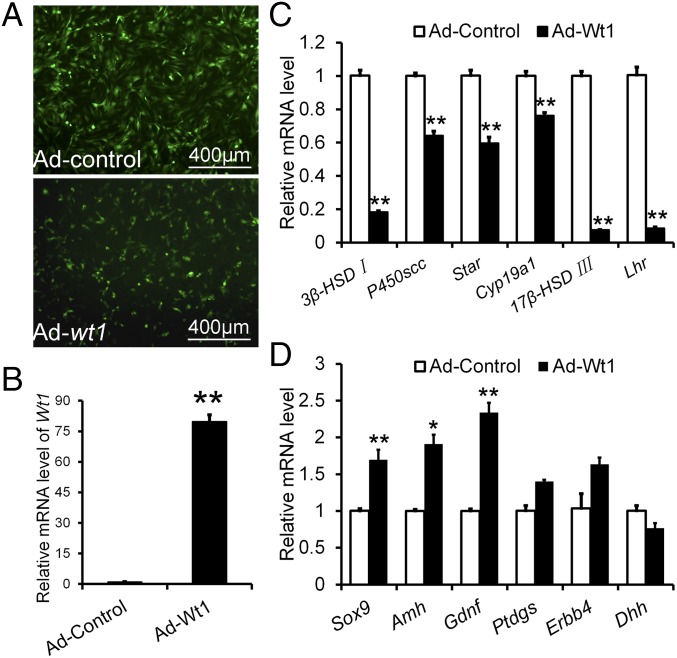
To test whether overexpression of Wt1 could reprogram Leydig cells into Sertoli cells, Leydig cells isolated from 21-d-old testes were cultured in vitro and transfected with a Wt1-expressing adenovirus (Fig. 6 A and B). The cells were collected 48 h after transfection, and gene expression was analyzed by real-time PCR. As shown in Fig. 6C, the expression of 3β-HSD, p450scc, Star, Cyp19a1, 17β-HSD III, and Lhr was significantly reduced in Wt1-overexpressing Leydig cells. However, Sertoli cell-specific genes, including Sox9, Amh, Gdnf, and Ptdgs, were markedly up-regulated (Fig. 6D), indicating that the Leydig cells were transformed into Sertoli-like cells upon Wt1 overexpression. These results further confirmed that the differentiation between Sertoli cells and Leydig cells is controlled by Wt1.
Fig. 6.
Differentially expressed genes in Wt1-overexpressing Leydig cells. (A) The efficiency of adenovirus virus transfection of Leydig cells. (B) Relative mRNA level of Wt1 after adenovirus transfection. (C) The mRNA levels of steroidogenic enzymes were significantly reduced in Leydig cells after Wt1 transfection. (D) The expression of Sertoli cell-specific genes was significantly increased in Wt1-overexpressing Leydig cells. Data are presented as the mean ± SEM. *P < 0.05; **P < 0.01.
Discussion
Sex determination in mammals depends on the fate decision of supporting cells in the gonads. These cells can either become Sertoli cells in the testes or granulosa cells in the ovaries. Sertoli cells and granulosa cells are derived from the coelomic epithelium (21), and the differentiation between them is genetically controlled. Inactivation of Foxl2 in granulosa cells results in granulosa cell-to-Sertoli cell transdifferentiation (22). By contrast, deletion of Dmrt1 results in Sertoli cell-to-granulosa cell transdifferentiation (23). Leydig cells, another major somatic cell type in the testis, were reported to migrate into the fetal testis from the mesonephros and the coelomic epithelium (6, 17, 24). However, Leydig cell origins are still controversial, and the regulation of Leydig cell differentiation has not been determined. In this study, we found that Leydig cell-specific steroidogenic genes were abundantly expressed in tumor cells from double KO mice, at levels comparable to those of control Leydig cells, whereas the expression of Sertoli cell marker genes was virtually absent. Based on these results, we conclude that the tumor cells in double KO mice are “Leydig-like” cells.
Rosa26R mouse strains are frequently crossed to Cre transgenic mice for lineage tracing experiments (20). In the present study, only Sertoli cells were labeled with X-Gal in control testes (Rosa26R+/floxAMH-Cre), indicating that AMH-Cre was specifically expressed in Sertoli cells in testes (19). In double KO mice, all of the Leydig-like tumor cells were labeled with X-Gal, indicating that these cells were derived from Sertoli cells in which AMH-Cre had been activated. Because of the complexity of the double KO mice, these results only suggest that loss of Wt1 results in a Sertoli-to-Leydig cell transformation. The function of Wt1 in Sertoli cell lineage maintenance was further confirmed using a Wt1–/floxAMH-Cre mouse model. As expected, Leydig cell marker genes were dramatically increased in Wt1-deficient Sertoli cells, whereas the Sertoli cell-specific genes were virtually absent. In this study, Sertoli cell-specific genes were down-regulated at E14.5, whereas 3β-HSD was first detected in Wt1-deficient Sertoli cells at E15.5, indicating that the transition from Sertoli cells to Leydig cells occurs between E14.5 and E15.5. Most Sertoli cells were 3β-HSD–positive in the remnant testicular cords of Wt1–/floxAMH-Cre mice during embryonic stages. However, the number of X-Gal and 3β-HSD double-positive Leydig-like cell clusters was reduced in the adult mutant testes, suggesting that, although most Sertoli cells were transdifferentiated into Leydig-like cells after Wt1 inactivation, not all of them survive in adult testes.
Sertoli cells were transformed into Leydig cells in both double KO and Wt1 single KO mice. However, in double KO mice, Sertoli cells developed into tumor-like cell clusters during embryonic stages, then transformed into 3β-HSD–positive Leydig cells at 1 mo of age, whereas in Wt1 single KO mice, the Sertoli cells were transformed into Leydig cells at E15.5. These results suggest that Ctnnb1 and Wt1 probably participate in testis development by regulating different signaling pathways. The functions of overactivated CTNNB1 are dominant at the early stage, which promote the proliferation of Sertoli cells, and the transformation from Sertoli to Leydig cells is delayed in double KO mice. The interaction of Wt1 and Ctnnb1 in testes development is complicated and needs further investigation.
In the present study, we found that Wt1 plays an essential role in Sertoli cell lineage maintenance and that inactivation of Wt1 in Sertoli cells during embryonic stages resulted in Sertoli cell-to-Leydig cell transdifferentiation, whereas overexpression of Wt1 in Leydig cells significantly inhibited the expression of steroidogenic enzymes and induced the expression of Sertoli cell-specific genes. These results further confirm that the differentiation between Sertoli cells and Leydig cells is regulated by Wt1, as shown by the model (Fig. S9). However, the underlying mechanism is still an open question. The expression of Sertoli cell-specific genes such as Sox9, Amh, Dmrt1, Dhh, Ptgds, Erbb4, Shbg, and Clusterin was dramatically reduced in Wt1-deficient Sertoli cells. However, inactivation of these genes does not result in a Sertoli-to-Leydig cell transdifferentiation (23, 25–28). The reduction of these genes’ expression is most likely a consequence of cell fate alteration after Wt1 inactivation. A recent study found that overexpression of Wt1 in the adrenal gland inhibited the development of steroidogenic cells (29), implying that during testis development, Wt1 probably functions in Sertoli lineage maintenance by antagonizing steroidogenesis-related enzyme expression. However, the expression of Sertoli cell-specific genes was induced in cultured Leydig cells upon overexpression of Wt1, suggesting that the down-regulation of steroidogenesis-related enzyme expression is probably also a consequence of cell fate alteration.
Wt1 has been reported to be involved in the development of multiple organs, including the adrenal glands (30), retina (31), liver (32), spleen (33), heart (12, 34), and kidney (35). Most, if not all, of these phenotypes are linked to a role for Wt1 in the control of EMT or MET. Our previous study also demonstrated that inactivation of Wt1 in mature Sertoli cells resulted in down-regulation of E-cadherin and loss of polarity (16). Therefore, Wt1 probably participates in Sertoli cell lineage maintenance by regulating the balance between mesenchyme and epithelium. However, the downstream target genes through which the Wt1 transcription factor acts to regulate the lineage transition between Sertoli and Leydig cells are still unclear and require further investigation.
Transdifferentiation between Sertoli cells and Leydig cells has never been reported previously. Our study demonstrates that the differentiation between Sertoli cells and Leydig cells is controlled by Wt1. Inactivation of Wt1 resulted in Sertoli-to-Leydig cell transdifferentiation, and overexpression of Wt1 caused a Leydig-to-Sertoli cell transformation. This study provides a new concept for cell fate determination in testis development, which should shed light on the regulation of somatic cell differentiation in testis development and etiological causes of male infertility in human patients.
Methods
Tissue Collection and Histological Analysis.
All animal studies were carried out in accordance with the protocols approved by the Institutional Animal Care and Use Committee at the Institute of Zoology, Chinese Academy of Sciences (CAS). Testes were dissected from mutant and control mice immediately after euthanasia, fixed in 4% (mass/vol) paraformaldehyde (PFA) for up to 24 h, stored in 70% (vol/vol) ethanol, and embedded in paraffin. We prepared 5-μm -thick sections were prepared and mounted them on glass slides. After deparaffinization, slides were stained with H&E for histological analysis.
Immunohistochemical Analysis and Immunofluorescence.
Immunohistochemistry (IHC) and immunofluorescence (IF) were performed on tissues from at least three mice for each genotype using the Vectastain ABC (avidin–biotin–peroxidase) kit (Vector Laboratories) as recommended. After staining, the slides were examined with a Nikon Microscope, and images were captured with a Nikon DS-Ri1 CCD camera.
ORO Staining.
We prepared 10-mm-thick frozen sections. After air drying, the sections were fixed in 4% PFA for 15 min and rinsed with 60% (vol/vol) isopropanol. The sections were stained with ORO (Sigma) solution (ORO saturated solution in isopropanol:water, 3:2) for 15 min as previously described (36). The sections were then washed with 70% alcohol for 5 s to remove background staining. After rinsing with tap water, the sections were counterstained with Harris hematoxylin and mounted in glycerol-PBS (9:1) for further analysis.
X-Gal Staining.
For frozen sections, slides were fixed in ice-cold fixation buffer [2% (vol/vol) formaldehyde, 0.2% glutaraldehyde, 0.02% Nonidet P-40 in PBS] for 15 min and washed in permeabilization solution (0.01% sodium deoxycholate, 0.02% Nonidet P-40 in PBS) three times before staining with X-Gal solution (1 mg/mL X-Gal, 5 mM potassium ferricyanide, and 5 mM potassium ferrocyanide in PBS) overnight at 37 °C. For whole-mount staining, testes were collected and fixed in ice-cold fixation buffer for 2 h at 4 °C on a shaking platform and stained with X-Gal solution overnight at 37 °C. Tissues were then fixed in 4% PFA and embedded in paraffin.
RNA Isolation and Real-Time PCR Analysis.
Total RNA was extracted from testes using a TRIzol kit following the manufacturer’s instructions. To quantify gene expression, the Real-Time SYBR Green assay was performed. Gapdh was used as an endogenous control. The relative levels of candidate gene expression were calculated using the formula 2–ΔΔ CT as described in the SYBR Green user manual. Primers used for RT-PCR are listed in Table S1.
Primary Leydig Cell Isolation and Culture.
Leydig cells collected from 21-d-old testes were cultured in vitro and transfected with Wt1 adenovirus after 24 h of culture. The cells were collected at 48 h after transfection and prepared for gene expression assays.
Statistical Analysis.
Experiments were repeated at least three times. Three to five control or mutant testes were used for immunostaining at each time point. The quantitative results are presented as the mean ± SEM. The data were evaluated for significant differences using Student’s t test and one-way ANOVA. P values < 0.05 were considered to be significant.
Supplementary Material
Acknowledgments
We thank Dr. Peter Koopman for his constructive comments on the manuscript. We also thank Shiwen Li and Tong Zhao for their assistance in fluorescence image capture and fluorescence-activated cell sorting. This work was supported by the Major Research Plan (2013CB945001 and 2011CB944303), the Scientific Innovation Project of the Chinese Academy of Science (XDA01040106), the National Natural Science Foundation of China (31471348, 31171373, and 31071271), and the Collaborative Innovation Center for Cardiovascular Disorders.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1422371112/-/DCSupplemental.
References
- 1.Gubbay J, et al. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346(6281):245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- 2.Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature. 1991;351(6322):117–121. doi: 10.1038/351117a0. [DOI] [PubMed] [Google Scholar]
- 3.Albrecht KH, Eicher EM. Evidence that Sry is expressed in pre-Sertoli cells and Sertoli and granulosa cells have a common precursor. Dev Biol. 2001;240(1):92–107. doi: 10.1006/dbio.2001.0438. [DOI] [PubMed] [Google Scholar]
- 4.Kim SK, Kim JH, Lee HJ, Yoon YD. Octylphenol reduces the expressions of steroidogenic enzymes and testosterone production in mouse testis. Environ Toxicol. 2007;22(5):449–458. doi: 10.1002/tox.20280. [DOI] [PubMed] [Google Scholar]
- 5.Barsoum IB, Yao HH. Fetal Leydig cells: Progenitor cell maintenance and differentiation. J Androl. 2010;31(1):11–15. doi: 10.2164/jandrol.109.008318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeFalco T, Takahashi S, Capel B. Two distinct origins for Leydig cell progenitors in the fetal testis. Dev Biol. 2011;352(1):14–26. doi: 10.1016/j.ydbio.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coppes MJ, et al. Inherited WT1 mutation in Denys-Drash syndrome. Cancer Res. 1992;52(21):6125–6128. [PubMed] [Google Scholar]
- 8.Gessler M, et al. Homozygous deletion in Wilms tumours of a zinc-finger gene identified by chromosome jumping. Nature. 1990;343(6260):774–778. doi: 10.1038/343774a0. [DOI] [PubMed] [Google Scholar]
- 9.Huff V, et al. Evidence for WT1 as a Wilms tumor (WT) gene: Intragenic germinal deletion in bilateral WT. Am J Hum Genet. 1991;48(5):997–1003. [PMC free article] [PubMed] [Google Scholar]
- 10.Pelletier J, et al. Germline mutations in the Wilms’ tumor suppressor gene are associated with abnormal urogenital development in Denys-Drash syndrome. Cell. 1991;67(2):437–447. doi: 10.1016/0092-8674(91)90194-4. [DOI] [PubMed] [Google Scholar]
- 11.Pelletier J, et al. Expression of the Wilms’ tumor gene WT1 in the murine urogenital system. Genes Dev. 1991;5(8):1345–1356. doi: 10.1101/gad.5.8.1345. [DOI] [PubMed] [Google Scholar]
- 12.Kreidberg JA, et al. WT-1 is required for early kidney development. Cell. 1993;74(4):679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- 13.Gao F, et al. The Wilms tumor gene, Wt1, is required for Sox9 expression and maintenance of tubular architecture in the developing testis. Proc Natl Acad Sci USA. 2006;103(32):11987–11992. doi: 10.1073/pnas.0600994103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang H, et al. Wt1 negatively regulates beta-catenin signaling during testis development. Development. 2008;135(10):1875–1885. doi: 10.1242/dev.018572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang H, Guillou F, Taketo MM, Behringer RR. Overactive beta-catenin signaling causes testicular Sertoli cell tumor development in the mouse. Biol Reprod. 2009;81(5):842–849. doi: 10.1095/biolreprod.109.077446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang XN, et al. The Wilms tumor gene, Wt1, is critical for mouse spermatogenesis via regulation of sertoli cell polarity and is associated with non-obstructive azoospermia in humans. PLoS Genet. 2013;9(8):e1003645. doi: 10.1371/journal.pgen.1003645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merchant-Larios H, Moreno-Mendoza N. Mesonephric stromal cells differentiate into Leydig cells in the mouse fetal testis. Exp Cell Res. 1998;244(1):230–238. doi: 10.1006/excr.1998.4215. [DOI] [PubMed] [Google Scholar]
- 18.Haider SG. Cell biology of Leydig cells in the testis. Int Rev Cytol. 2004;233:181–241. doi: 10.1016/S0074-7696(04)33005-6. [DOI] [PubMed] [Google Scholar]
- 19.Lécureuil C, Fontaine I, Crepieux P, Guillou F. Sertoli and granulosa cell-specific Cre recombinase activity in transgenic mice. Genesis. 2002;33(3):114–118. doi: 10.1002/gene.10100. [DOI] [PubMed] [Google Scholar]
- 20.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 21.Karl J, Capel B. Sertoli cells of the mouse testis originate from the coelomic epithelium. Dev Biol. 1998;203(2):323–333. doi: 10.1006/dbio.1998.9068. [DOI] [PubMed] [Google Scholar]
- 22.Uhlenhaut NH, et al. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell. 2009;139(6):1130–1142. doi: 10.1016/j.cell.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 23.Matson CK, et al. DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature. 2011;476(7358):101–104. doi: 10.1038/nature10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishino K, Yamanouchi K, Naito K, Tojo H. Characterization of mesonephric cells that migrate into the XY gonad during testis differentiation. Exp Cell Res. 2001;267(2):225–232. doi: 10.1006/excr.2001.5238. [DOI] [PubMed] [Google Scholar]
- 25.Barrionuevo F, et al. Testis cord differentiation after the sex determination stage is independent of Sox9 but fails in the combined absence of Sox9 and Sox8. Dev Biol. 2009;327(2):301–312. doi: 10.1016/j.ydbio.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 26.Arango NA, Lovell-Badge R, Behringer RR. Targeted mutagenesis of the endogenous mouse Mis gene promoter: In vivo definition of genetic pathways of vertebrate sexual development. Cell. 1999;99(4):409–419. doi: 10.1016/s0092-8674(00)81527-5. [DOI] [PubMed] [Google Scholar]
- 27.Bitgood MJ, Shen L, McMahon AP. Sertoli cell signaling by Desert hedgehog regulates the male germline. Curr Biol. 1996;6(3):298–304. doi: 10.1016/s0960-9822(02)00480-3. [DOI] [PubMed] [Google Scholar]
- 28.Malki S, et al. Prostaglandin D2 induces nuclear import of the sex-determining factor SOX9 via its cAMP-PKA phosphorylation. EMBO J. 2005;24(10):1798–1809. doi: 10.1038/sj.emboj.7600660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bandiera R, et al. WT1 maintains adrenal-gonadal primordium identity and marks a population of AGP-like progenitors within the adrenal gland. Dev Cell. 2013;27(1):5–18. doi: 10.1016/j.devcel.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore AW, McInnes L, Kreidberg J, Hastie ND, Schedl A. YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development. 1999;126(9):1845–1857. doi: 10.1242/dev.126.9.1845. [DOI] [PubMed] [Google Scholar]
- 31.Wagner KD, et al. The Wilms’ tumor gene Wt1 is required for normal development of the retina. EMBO J. 2002;21(6):1398–1405. doi: 10.1093/emboj/21.6.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ijpenberg A, et al. Wt1 and retinoic acid signaling are essential for stellate cell development and liver morphogenesis. Dev Biol. 2007;312(1):157–170. doi: 10.1016/j.ydbio.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 33.Herzer U, Crocoll A, Barton D, Howells N, Englert C. The Wilms tumor suppressor gene wt1 is required for development of the spleen. Curr Biol. 1999;9(15):837–840. doi: 10.1016/s0960-9822(99)80369-8. [DOI] [PubMed] [Google Scholar]
- 34.Martínez-Estrada OM, et al. Wt1 is required for cardiovascular progenitor cell formation through transcriptional control of Snail and E-cadherin. Nat Genet. 2010;42(1):89–93. doi: 10.1038/ng.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davies JA, et al. Development of an siRNA-based method for repressing specific genes in renal organ culture and its use to show that the Wt1 tumour suppressor is required for nephron differentiation. Hum Mol Genet. 2004;13(2):235–246. doi: 10.1093/hmg/ddh015. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, et al. The role of Tyro 3 subfamily receptors in the regulation of hemostasis and megakaryocytopoiesis. Haematologica. 2007;92(5):643–650. doi: 10.3324/haematol.10939. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.