Significance
X-ray crystallography is a powerful approach for understanding the structure and function of biological macromolecules but is largely limited to molecules that form high-quality crystals in the laboratory. Here we present the structure of protein crystals that form naturally in virally infected insects and boost the insecticidal activity of oral pathogens. By proposing a mode of action for these virulence factors based on enzymes degrading chitin by oxidation, our findings may guide their use as synergetic additives to common bioinsecticides. They also reveal that these proteins assemble into ultra-stable crystals stabilized by a 3D network of covalent bonds, a unique strategy for achieving efficient protein crystallization in the complex environment of the cell.
Keywords: microcrystallography, in vivo crystallization, poxvirus, LPMO, pesticide
Abstract
The great benefits that chemical pesticides have brought to agriculture are partly offset by widespread environmental damage to nontarget species and threats to human health. Microbial bioinsecticides are considered safe and highly specific alternatives but generally lack potency. Spindles produced by insect poxviruses are crystals of the fusolin protein that considerably boost not only the virulence of these viruses but also, in cofeeding experiments, the insecticidal activity of unrelated pathogens. However, the mechanisms by which spindles assemble into ultra-stable crystals and enhance virulence are unknown. Here we describe the structure of viral spindles determined by X-ray microcrystallography from in vivo crystals purified from infected insects. We found that a C-terminal molecular arm of fusolin mediates the assembly of a globular domain, which has the hallmarks of lytic polysaccharide monooxygenases of chitinovorous bacteria. Explaining their unique stability, a 3D network of disulfide bonds between fusolin dimers covalently crosslinks the entire crystalline matrix of spindles. However, upon ingestion by a new host, removal of the molecular arm abolishes this stabilizing network leading to the dissolution of spindles. The released monooxygenase domain is then free to disrupt the chitin-rich peritrophic matrix that protects insects against oral infections. The mode of action revealed here may guide the design of potent spindles as synergetic additives to bioinsecticides.
Most entomopoxviruses (EV) produce two types of intracellular crystals. Virus-containing spheroids are the main infectious form of EV (1) and are functionally analogous to polyhedra of cypovirus (2) and baculovirus (3, 4). In contrast, the function of the second type of crystals is less clear. These crystals of the viral fusolin protein, called “spindles” because of their characteristic shape, assemble in the endoplasmic reticulum of infected cells and for some species also occur embedded within the crystalline lattice of spheroids (5). Purified spindles are not infectious but strongly enhance the infectivity of EV by a mechanism that involves disruption of the peritrophic matrix, a physical barrier that protects the midgut epithelium of insects against oral pathogens (6, 7). Remarkably, in larval cofeeding experiments, spindles also enhance the insecticidal activity of unrelated oral pathogens such as baculovirus (8) and the Bacillus thuringiensis (Bt) toxin (9) by up to three orders of magnitude. This effect on virulence prompted their use as synergistic additives to common bioinsecticides, for instance by transgenic expression of spindles in plants to improve the effectiveness of baculovirus insecticides (10).
Fusolin proteins have a signal sequence that targets them to the endoplasmic reticulum, and the mature protein has a mass of 36–44 kDa. Some fusolins are glycosylated, and the glycosylation site of the fusolin produced by Anomala cuprea EV (ACEV) is required for full virulence (11). Sequence analysis shows that the N-terminal region of fusolin is a domain 3 chitin-binding domain, found in more than 500 bacterial and fungal proteins (12). Among viruses, the only recognized homologs are the GP37 proteins produced by baculoviruses, which exhibit 30–40% sequence identity with fusolin (13).
The chitin-binding activity of the N-terminal domain is essential for the virulence enhancement of fusolin. However, it does not explain the mode of action of spindles by itself, because this activity is necessary but not sufficient for virulence (11). No function has been assigned to the variable C-terminal region of fusolin, and this region is not required for the enhancement activity (11).
To investigate how fusolin enhances the virulence of insecticidal agents and assembles into spindles, we used X-ray microcrystallography to determine structures of spindles from three EV that infect major agricultural pests. These structures reveal that the disruption of the chitin-rich peritrophic matrix by fusolin is caused by a globular domain that has the hallmarks of a lytic polysaccharide monooxygenase (LPMO). An extended molecular arm following this domain stabilizes the in vivo crystals of fusolin by interconnecting domain-swapped dimers and forming a unique 3D network of disulfide bonds.
Results
The Structure of Fusolin.
Spindles from three EVs were isolated from their respective insect hosts, the common cockchafer Melolontha melolontha, the cupreous chafer Anomala cuprea, and the porina moth Wiseana spp. These spindles have similar bipyramidal shapes ∼3 μm in length and a smooth surface when viewed by scanning electron microscopy (Fig. S1). Although they generally are described in the literature as paracrystalline, we found that the three types of spindles are single crystals that diffract to high resolution. At a molecular level, they have lattice parameters within 5% of each other (Table S1), and the sequences of the respective fusolin proteins share 55–58% identity. Accordingly, the three derived atomic models for fusolin are similar, with rmsds between corresponding atoms of less than 1 Å (Fig. S1). The following description is based on fusolin from Melolontha melolontha EV (MMEV) unless otherwise specified.
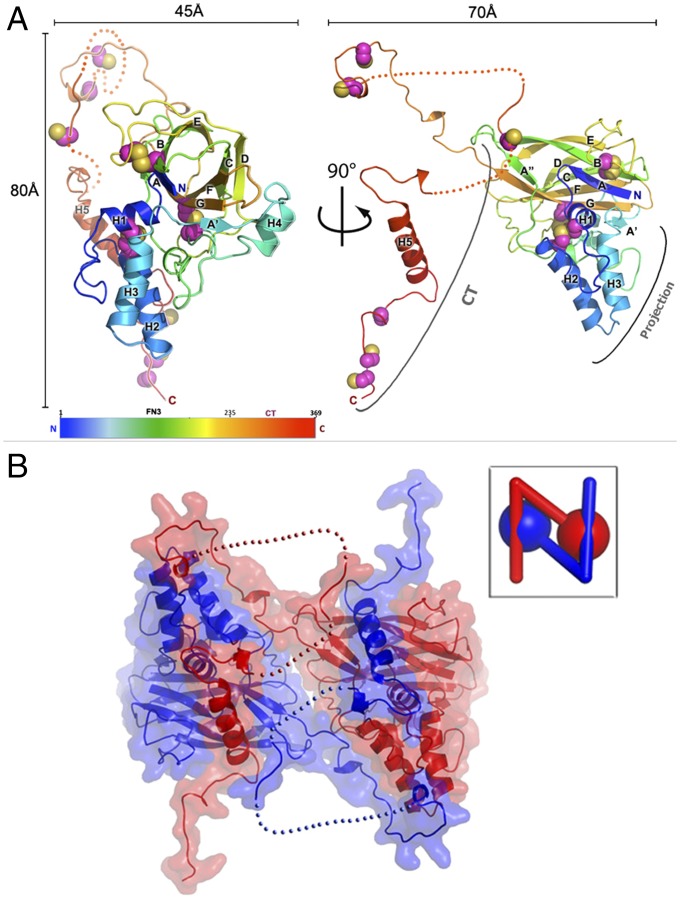
The closest structural homolog of fusolin is the bacterial chitin-binding protein 21 (CBP21) protein of Serratia marcescens (PDB ID code: 2BEM), a member of the AA10 family of copper-dependent LPMOs (14). Like all AA10 proteins, fusolin has a modified fibronectin type III (Fn3) domain with a helical subdomain forming a wedge-shaped projection. This projection subdomain of ∼112 residues is inserted between the first two strands of the Fn3 β-sandwich. In fusolin the AA10 module is followed by a C-terminal (CT) extension composed of an extended linker and a prominent helix located ∼20Å away from the core of the molecule. The CT region varies from 76 to 134 residues, depending on the virus, and contains a central section that appears to be flexible in our three structures (Fig. 1A and Fig. S2). Of the 13 cysteines in MMEV fusolin (see Fig. S2), six form intramolecular disulfide bonds that stabilize the fusolin fold on either side of the β-sandwich (C93–C228 and C139–C189) and within the projection subdomain (C14–C34) (Fig. 1).
Fig. 1.
The structure of fusolin. (A) The structure of fusolin is represented as a ribbon diagram colored in a blue–red gradient from the N to C termini. Cysteine residues are shown as spheres. (B) Fusolin forms a domain-swapped dimer shown as a ribbon diagram within a semitransparent molecular surface. (Inset) A schematic representation of the dimer.
Relationship with Other Viral Fusolin-like Proteins.
The N-terminal domain of EV fusolins is closely related in sequence to baculovirus GP37 proteins, suggesting both homology and a common fold for these virulence factors despite the evolutionary distance between the two viral families (13). For instance, the fusolin-like proteins of EV and baculovirus isolated from the same host, Helicoverpa armigera, share 49% identity over 257 residues. Our results suggest that all viral fusolin-like proteins share two structural characteristics, namely the three disulfide bonds that stabilize the N-terminal domain and a buried salt bridge that stabilizes the interaction between the Fn3 domain and the projection subdomain (residues R10, D230, and W117) (Fig. S2).
An analysis of the sequence conservation at the molecular surface of fusolin reveals an additional patch of conserved residues that do not appear to be involved in the stability of the fusolin dimer or in their assembly into spindles (Fig. 2C). This area overlaps the chitin-binding interface identified in cellular homologs as described in the next section. In contrast, the CT extension generally is much shorter and is poorly conserved in sequence; or it is absent altogether in most baculovirus proteins (Fig. S2).
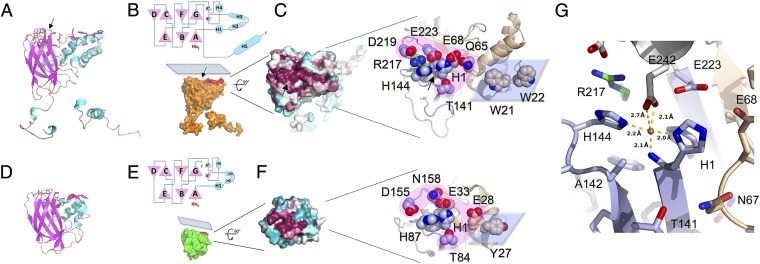
Fig. 2.
Fusolin has the hallmarks of a lytic chitin monooxygenase. Comparison of the fusolin and CBP21 proteins reveals similar 3D structures (A and D) and topologies (B, Upper and E, Upper) with a conserved flat platform on the molecular surface (B, Lower and E, Lower). Mapping of sequence variability onto the molecular surface revealed that the flat platform is highly conserved (C, Left and F, Left). The blue–white–red gradient represents low-to-high sequence conservation in 27 viral sequences (C) and 400 CBP21-like proteins (F). The two LPMO active sites are represented (C, Right and F, Right). Side chains of residues mentioned in the text are shown as spheres. The projection and Fn3 domains are colored in brown and light blue, respectively; Arg217 in green; and the polar rim and flat platform in pink and blue, respectively. (G) Representation of the metal-binding site in MMEV spindles with the same color scheme as C. A neighboring molecule capping the site is shown in white. Although this site is only occupied by a water molecule in purified MMEV spindles (Fig. S5), it can be populated by a Cu2+ ion after incubation in CuSO4, shown here as a brown sphere. The metal-binding site also is indicated by black arrows in A, B, and C.
Fusolin Has the Hallmarks of a Chitin Lytic Monooxygenase.
Outside the viral world, fusolin is most closely related to bacterial AA10 LPMOs. Unlike classical chitinases classified as glycosyl hydrolases, AA10 LPMOs are specific for the crystalline form of chitin or cellulose and proceed through an oxidative process involving molecular oxygen activated by copper bound in the active site of the protein (15–18). Despite a low sequence identity between CBP21, a member of the AA10 proteins, and fusolin (14% over 237 residues), the Fn3 domains of the two proteins are structurally similar with an rmsd of 1.55 Å over 145 equivalent residues. The projection subdomain is mostly helical in both proteins but is larger in fusolin by ∼60 residues (Fig. 2). A shallow groove on the β-sandwich is highly conserved in sequence not only between EVs and baculoviruses, two completely unrelated families of insect viruses, but also across AA10 proteins (Fig. 2 C and F). Protruding from this groove, the N-terminal histidine (His1) of the mature fusolin and His144 form a histidine brace (19), a signature of LPMOs consisting of a surface-exposed metal-binding site located between the N-terminal amine of the strictly conserved His1 and the two imidazole side chains (Fig. 2 and Fig. S3).
In material purified from infected insects, only Wiseana EV (WEV) spindles had electron density corresponding to a transition metal within the histidine brace. Because of the limited availability of the material, the identity of this endogenous metal could not be confirmed. However, this site can accommodate Zn2+ or Cu2+ when MMEV spindles are soaked in 2 mM ZnSO4 or CuSO4 solutions (Fig. S4). The structures of these complexes at a resolution of 2.4 Å revealed similar T-shaped coordination spheres consisting of two nitrogen atoms of the imidazole side chains of His1 and His144, and the N-terminal amine of His1 (Fig. 2G and Fig. S5). As in other AA10 and AA11 proteins, an alanine residue, Ala142, partially blocks access to the metal in the axial direction perpendicular to the plane formed by the three nitrogen ligands (Fig. 2G and Fig. S6). This unusual geometry for a transition metal-binding site was first described in CBP21, which also is able to bind a number of divalent metals including Zn2. However, Cu2+ was the only ion able to restore activity in EDTA-inhibited CBP21 (16).
A notable difference between fusolin and other AA10 proteins is the presence in the former of an additional coordination of the metal by residue Glu242 from a symmetry-related molecule that caps the active site in assembled spindles. The carboxylate moiety of the Glu242 side chain coordinates the metal in a bidentate interaction opposite the N-terminal amine compared with the metal. This geometry closely resembles the binding site of the AA10 protein EfaCBM33A with Cu2+, in which two water molecules occupy positions corresponding to the carboxyl oxygen atoms of Glu242 (Fig. S5). These water molecules are absent in the reduced Cu+ structure of EfaCBM33A (20), suggesting that in our structure of fusolin the Cu2+ ion has not undergone complete photoreduction.
In addition, Glu242 fills a small cavity next to the metal site that could accommodate molecular oxygen, as suggested for chitin-specific AA10 and AA11 proteins (Fig. S6). Phylogenetically, fusolin clusters with chitin-specific AA10 proteins (Fig. S3). However, this cavity is shallower than in these proteins and more closely resembles the surface of CelS2, a cellulose-targeting AA10 protein (21). Indeed, the residues delimiting this cavity are conserved between CelS2 and fusolin (Arg217 and Glu223).
Other residues around the metal are appropriately placed to bind the substrate through an outer rim of polar residues and a planar hydrophobic surface on the projection domain. This flat platform is complementary in shape to crystalline chitin (Fig. 2B) and presents two prominent tryptophans, Trp21 and Trp22. The active site copper, Trp21, and Trp22 are approximately aligned and separated by ∼20 Å (metal–Trp21) and ∼30 Å (metal–Trp22). Despite large fold differences in the projection domain where these residues are located, Tryp21 is structurally equivalent to Tyr54 of CBP21 (ring-to-ring distance <4 Å) (Fig. 2 C and F), a residue that is essential for the ability of CBP21 to bind chitin (16, 22).
Additional residues in the polar rim that were shown experimentally to participate in chitin binding in CBP21 (16, 22) are highlighted in Fig. 2F and Fig. S3. In particular, the threonine residue Thr141 preceding one of the active site histidines (His144) and the occluding alanine (Ala142) are strictly conserved across all AA10 and AA11 proteins.
A Crystalline LPMO.
Compared with cellular LPMOs, the mode of action of fusolin is unique, because this virulence factor is naturally produced as ultra-stable crystals (Fig. 3A). Our analysis of native spindles purified from infected insects revealed that they adopt a complex molecular architecture in which fusolin is inactive but that allows its regulated release in an active form upon spindle dissolution.
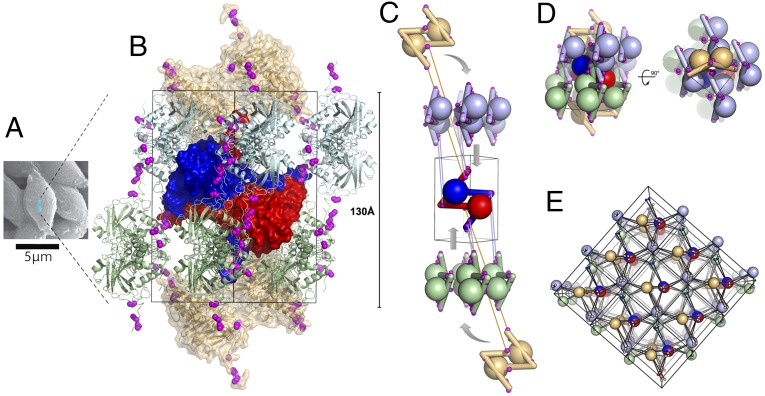
Fig. 3.
Spindles are single-chain crystalline polymers of fusolin dimers crosslinked by a 3D network of disulfide bonds. (A) Scanning electron micrograph of spindles purified from EV-infected cockchafer larvae. The blue box represents a unit cell (not to scale). (B) Representation of a spindle unit cell viewed along a twofold crystallographic axis. Cysteines involved in interdimer crosslinks are shown as magenta spheres. The central dimer is represented as a blue-red molecular surface; two crowns of four dimers are shown in green and cyan (one dimer of each crown is masked because of its location on the opposite side of the unit cell). The two capping dimers are shown in brown as a semitransparent molecular surface. (C) Schematic representation of the unit cell assembly. Interdimer disulfide bonds involving the central dimer are indicated by links between the interacting cysteines. (D) The full unit cell is shown in the same orientation as in C and in an orthogonal view. (E) Representation of 18 unit cells with each dimer shown as a large sphere and cysteines as smaller spheres. Intersphere links highlight the 3D network of covalent bonds crosslinking spindles.
Spindles are formed by an intricate assembly of dimeric building blocks in which the CT extension mediates domain swapping within dimers (Fig. 1B) and crosslinks the entire crystalline matrix (Figs. 3 and 4B). The dimer is formed by interactions of fusolin molecules around a crystallographic twofold axis through the side of the β-sandwich opposite the active site (Fig. 1B). The amphipathic helix H5 of the CT molecular arm is swapped between subunits of the dimer and fits into a hydrophobic groove of the adjacent β-sandwich. The dimeric interface is relatively large with a buried area of 3,530 Å2, representing 36% of the accessible surface of the fusolin protein. However, without the CT extension the dimeric interface only represents 404 Å2, with insufficient interactions to stabilize the assembly in solution as predicted by the Protein Interfaces, Surfaces and Assemblies (PISA) server (Fig. S7).
Fig. 4.
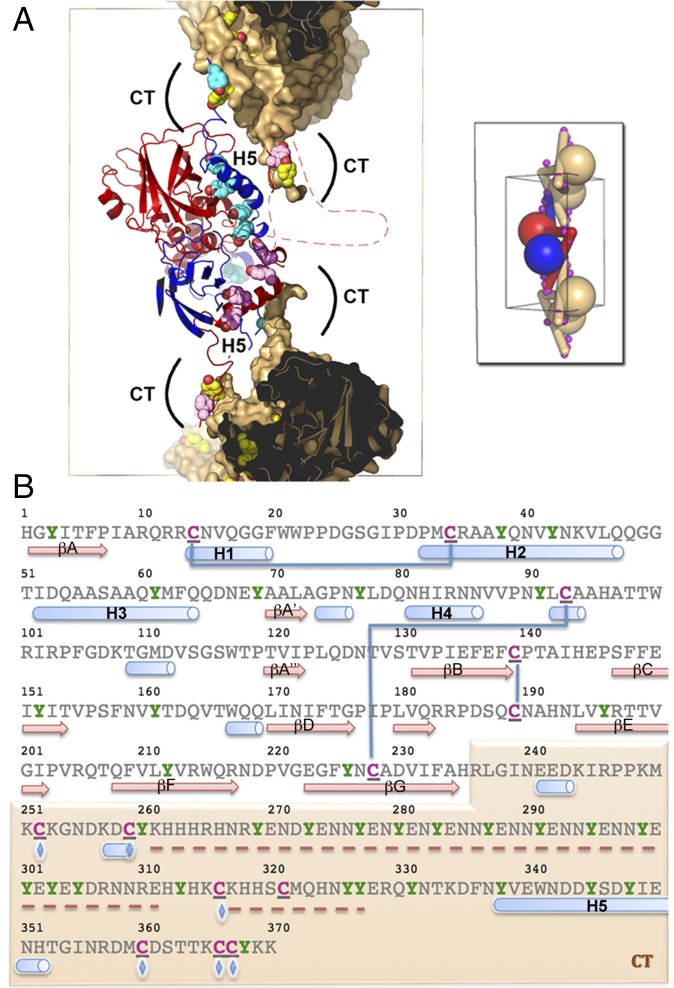
The CT extension controls the stability and dissociation of spindles through intermolecular disulfide bonds and multiple tyrosine residues, respectively. (A) Deprotonated tyrosines located at dimeric interfaces and crystal contacts may participate in the dissociation of spindles in alkaline conditions. The central dimer is shown as ribbons colored in blue and red. The molecular surface of the capping dimers is shown in brown but is omitted for CT tyrosine residues. The side chains of tyrosine residues in the CT region (including the H5 helix) are represented as spheres colored according to the rest of the molecule. The front part of the molecules has been clipped to reveal the interface. Regions clipped and absent from the model are indicated by a dashed line. (Inset) A schematic representation of connectivity. Magenta spheres indicate intermolecular disulfide bonds that crosslink spindles. (B) Sequence of the MMEV fusolin highlighting cysteine and tyrosine residues in magenta and green, respectively. These residues are particularly abundant in the CT arm shaded in brown. Blue lines represent intramolecular disulfide bonds, and blue diamonds indicate residues involved in intermolecular disulfide bonds (C252–C366; C259–C360; C316–C367). Secondary structure elements are shown under the sequence with missing regions indicated by a red dashed line.
In spindles, fusolin dimers assemble according to the P41212 symmetry of the crystal to form two crowns of four dimers that encase a central dimer (Fig. 3). Each crown is capped by a dimer from neighboring cells that projects its CT extensions through the central opening of the crown toward the central dimer. The central dimer is connected to both capping dimers and two dimers of each crown by disulfide bonds between the respective CT extensions (Fig. 3 B and C).
The CT Molecular Arm Stabilizes Spindles Through a 3D Network of Covalent Bonds.
Remarkably, the 12 interdimer disulfide bonds form a 3D network that covalently crosslinks the entire crystal into a single polymer (Fig. 3 C and E). The network relies exclusively on the CT extension where the linker preceding helix H5 is connected to three cysteines present in the last 11 residues of a fusolin molecule from a neighboring dimer (Fig. 4B). This network also may be present in ACEV, although disulfide bonds are not visualized in our structure because of missing electron density. The cysteines involved in interdimer crosslinking in MMEV and ACEV spindles are not present in WEV spindles, but an alternate interdimer disulfide bond between Cys80 and Cys255 plays a similar role.
In the absence of the CT sequence, no interdimer disulfide bonds would be formed; the size and nature of the remaining interdimer buried surfaces would be typical of crystal contacts with no biological relevance; and no higher assemblies of such truncated fusolin would be predicted by interface analysis (SI Methods, and Fig. S7).
Glycosylation Site.
Most fusolin and GP37 proteins contain at least one N-glycosylation site within the N-terminal conserved regions, although not all fusolins are glycoproteins. Well-defined electron density for the (N-acetyl-glucosamine)2-mannose base of the carbohydrate structure is visible at Asn173 in the WEV fusolin and at Asn175 in the ACEV fusolin electron density maps (Fig. S1). In ACEV spindles, the carbohydrate chain plays a role in stability in the host digestive tract, perhaps explaining the decreased virulence when glycosylation of the ACEV fusolin is prevented (11). In the ACEV spindle structure the carbohydrate chain points into a solvent channel, away from the dimeric interface or crystal contacts. The first three moieties attached to Asn175 cover a small hydrophobic patch on the surface of fusolin, which may increase the folding efficiency and solubility. No electron density corresponding to carbohydrate or protein is visible at the equivalent position in the MMEV structure. In WEV spindles, the carbohydrate chain fits between two dimers and interacts with Glu263, hydrogen-bonded to O6 and O7 of NAG1, and Phe259, forming a stacking interaction with NAG1 (Fig. S1). This bridging interaction forms additional crystal contacts that may compensate for the lower amount of covalent crosslinking in WEV spindles, which lack Cys252, Cys259, and Cys316 involved in intermolecular disulfide bonds in the MMEV fusolin.
Fusolin Release from Spindles.
Our structure of spindles shows that fusolin must be released from the crystal to function because the metal-binding site is capped by residue Glu242 in assembled spindles, and neighboring molecules occlude the planar platform containing the active site (Fig. S6). This controlled release is indeed observed in the midgut of larvae and has been attributed to the combined action of the alkaline environment and proteolytic degradation (11). In addition to the intermolecular disulfide bonds, the CT region contains an unusually high proportion of tyrosines, which are located strategically at the dimer interface within helix H5 and around interdimer crystal contacts next to stabilizing disulfide bonds (Fig. 4). Thus, dissolution may be facilitated by the concerted deprotonation of the hydroxyl group of tyrosines (pKaTyr = 10.1), introducing destabilizing buried charges, and the weakening of disulfide bonds (pKaCys = 8.0) in the alkaline midgut. A similar mechanism was proposed for both classes of viral polyhedra, where tyrosine clusters are located in close proximity to molecular arms analogous to the CT extension (23).
In vitro, alkaline dissolution of spindles does not dissociate fusolin oligomers completely, and some noncovalent, SDS-resistant oligomers are observed even after incubation at pH 13 (Fig. S8 A and B). Limited proteolysis of dissolved spindles by chymotrypsin produces a stable core that corresponds in size to the AA10 domain and no longer forms SDS-resistant oligomers (Fig. S8C). In vivo, the released fusolin is processed by host serine proteases into an N-terminal fragment with a cleavage site situated between residues 250 and 289 of the mature ACEV fusolin (11). In our structure, this site is located at the beginning of the CT sequence within a disordered region that follows conserved proline Pro247. The resulting fragment corresponds to the globular LPMO module identified in this study (Fig. 5).
Fig. 5.
Mode of action of spindles. (A) Spindles crystallize in the endoplasmic reticulum of infected cells. (B) A schematic view of the unit cell highlights the crosslinking role of the CT molecular arm and the approximate region of proteolytic cleavage (arrowheads). (C) Upon ingestion by the new host, the CT extension is proteolytically removed, abolishing the 3D network of intermolecular disulfide bonds and disrupting most crystal contacts. (D and E) Dissolution of spindles releases active fusolin in the insect midgut (D) leading to disruption of the chitin-rich peritrophic matrix, facilitating invasion of the host by spheroid-derived virions (E).
Discussion
From an evolutionary standpoint, the close similarity of fold and topology between fusolin and AA10 proteins establishes homology between these viral and bacterial proteins. Functionally, our structural analysis indicates that fusolin harbors all the hallmarks of functional AA10 proteins, including a characteristic N-terminal histidine residue, a surface-exposed metal-binding site, and a flat protrusion suitable for substrate binding. Overall, the active site of fusolin more closely resembles AA10 proteins that target chitin, such as CBP21 (22) and AA11 proteins (24), than AA9 proteins that degrade cellulose (25). In particular, the fusolin protein sequence clusters phylogenetically with chitin-specific AA10 proteins, and its surface presents a characteristic cavity next to the histidine brace that is thought to accommodate dioxygen. Accordingly fusolin and GP37 proteins bind purified chitin (11, 26), and ingestion of fusolin results in the disruption of the chitin-rich peritrophic matrix (6, 7). Importantly, the LPMO activity itself appears to be essential, because an IHE145-AAA145 fusolin mutant with a disrupted metal-binding site remains able to bind chitin but fails to enhance the insecticidal activity of baculovirus in cofeeding experiments (11). Similarly, fusolin loses its ability to enhance virulence when it is expressed recombinantly with a heterologous N-terminal extension, consistent with the proposed essential role of His1, as in all other LPMOs (11, 27). Taken together, these results indicate that a chitin LPMO activity of fusolin underpins the ability of poxvirus spindles to disrupt the peritrophic matrix of their host, providing a molecular basis for their broad-spectrum enhancement of virulence.
This role of an LPMO module in viral pathogenicity differs from the primary role of many bacterial LPMOs, which is the degradation of recalcitrant polysaccharides as a source of nutrients. However, several proteins containing LPMO modules also have been linked to virulence in bacteria (28), and it will be interesting to see whether bacterial and viral LPMO factors affect virulence through similar mechanisms in insect pathogens.
Fusolin is unique among AA10 proteins—and in fact among all characterized LPMOs—in that it is produced in an inactive, crystalline form. Our structure reveals that this ability to crystallize in vivo results at least in part from the addition of a CT extension to the LPMO domain. The role of the CT arm in the assembly and stability of spindles is evidenced further by the fact that most structural homologs of fusolin lack CT extensions, are monomeric, and do not form in vivo crystals. Four baculovirus GP37 proteins also are known to form spindle-like crystals, and all have cysteine-rich C-terminal extensions that are comparable in length to the CT arm of EV fusolin (Fig. S2). The amino acid sequences of these CT-like extensions are highly divergent, suggesting that in vivo crystallization may have evolved independently in these viruses. Intriguingly, the role of the CT region in assembly is analogous to the function of H1 of the cypovirus polyhedrin and the CT loop of baculovirus polyhedrin that stabilizes polyhedra, virus-containing crystals unrelated to spindles (23). Crystals of cathepsin B grown in vivo for X-ray free-electron laser analysis using a recombinant baculovirus expression system present a similar extension in which the last three residues interlock neighboring molecules in the crystal packing (29). These diverse examples suggest that crosslinking using molecular arms may represent a general feature of stable in vivo crystals.
From a biochemical point of view, the specificity of the fusolin crystals is their stabilization by a 3D network of disulfide bonds that results in a fully crosslinked matrix. To our knowledge, this is the first visualization of such a stabilization strategy in protein crystals, which has also been postulated to exist in other in vivo crystals such as Bt parasporal δ-endotoxins and poxvirus spheroids. This covalent crosslinking may explain why spindles are even more stable than polyhedra (30) despite forming a crystalline matrix that is much less compact (solvent contents of 33–40% for spindles vs. ∼20% for polyhedra) (23). In keeping with their primarily structural role, a large proportion of the surface of both classes of polyhedrin proteins has evolved to make strong crystal contacts. In contrast, the ultimate role of fusolin is to enhance virulence. The modular crystallization strategy based on the CT extension offers an elegant solution to allow an efficient transition from an ultra-stable crystalline form to a soluble, active LPMO domain.
Methods
Spindles were isolated from infected insects that were collected from the field (Melolontha melolontha (13) and Wiseana spp. larvae) (SI Methods) or from insects reared as previously described (Anomala cuprea) (6). Spindles were purified by centrifugation through a stepwise sucrose gradient and were resuspended in water. X-ray diffraction data were collected on multiple crystals, typically ∼3 µm in diameter, and the structure of MMEV fusolin was determined using the Rosetta ab initio molecular replacement implementation in PHENIX (rosetta_mr) with the CBP21 protein structure (PDB ID code: 2BEM) as template (31). The three models of fusolin were built in COOT (32) and refined with BUSTER 2.10 (33) at resolutions of 1.9–2.0 Å (Table S1). The structures of MMEV fusolin in complex with Cu2+ and Zn2+ were refined at a resolution of 2.4 Å (Table S2).
Supplementary Material
Acknowledgments
Diffraction data were collected on the MX2 beamline of the Australian Synchrotron and the X06SA beamline of the Swiss Light Source. F.C. is supported by Future Fellowship FT1210893 and Discovery Projects Grants DP1214039 and DP1314885 of the Australian Research Council. E.C., V.Y., V.K.W., and P.M. were supported by the Royal Society of New Zealand Marsden Fund. Travel funding was provided by the International Synchrotron Access Program managed by the Australian Synchrotron and the New Zealand Synchrotron group.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 4OW5 (Melolontha melolontha entomopoxvirus, MMEV), 4YN1 (Anomala cuprea entomopoxvirus), 4YN2 (Wiseana spp. entomopoxvirus), 4X27 (MMEV spindles soaked in CuSO4), and 4X29 (MMEV spindles soaked in ZnSO4)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1418798112/-/DCSupplemental.
References
- 1.Perera S, Li Z, Pavlik L, Arif B. In: Insect Virology. Asgari S, Johnson K, editors. Caister Academic; Norwich, UK: 2010. pp. 83–102. [Google Scholar]
- 2.Coulibaly F, et al. The molecular organization of cypovirus polyhedra. Nature. 2007;446(7131):97–101. doi: 10.1038/nature05628. [DOI] [PubMed] [Google Scholar]
- 3.Coulibaly F, et al. The atomic structure of baculovirus polyhedra reveals the independent emergence of infectious crystals in DNA and RNA viruses. Proc Natl Acad Sci USA. 2009;106(52):22205–22210. doi: 10.1073/pnas.0910686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ji X, et al. How baculovirus polyhedra fit square pegs into round holes to robustly package viruses. EMBO J. 2010;29(2):505–514. doi: 10.1038/emboj.2009.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergoin M, Devauchelle G, Vago C. [Fusiform inclusions associated with the entomopoxvirus of the beetle Melolontha melolontha] J Ultrastruct Res. 1976;55(1):17–30. doi: 10.1016/s0022-5320(76)80078-0. [DOI] [PubMed] [Google Scholar]
- 6.Mitsuhashi W, Miyamoto K. Disintegration of the peritrophic membrane of silkworm larvae due to spindles of an entomopoxvirus. J Invertebr Pathol. 2003;82(1):34–40. doi: 10.1016/s0022-2011(02)00203-3. [DOI] [PubMed] [Google Scholar]
- 7.Mitsuhashi W, et al. Spindles of an entomopoxvirus facilitate its infection of the host insect by disrupting the peritrophic membrane. J Virol. 2007;81(8):4235–4243. doi: 10.1128/JVI.02300-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu J, Hukuhara T. Enhanced infection of a nuclear polyhedrosis virus in larvae of the armyworm, Pseudaletia separata, by a factor in the spheroids of an entomopoxvirus. J Invertebr Pathol. 1992;60(3):259–264. [Google Scholar]
- 9.Mitsuhashi W, Asano S, Miyamoto K, Wada S. Further research on the biological function of inclusion bodies of Anomala cuprea entomopoxvirus, with special reference to the effect on the insecticidal activity of a Bacillus thuringiensis formulation. Pest Manag Sci. 2014;70(1):46–54. doi: 10.1002/ps.3521. [DOI] [PubMed] [Google Scholar]
- 10.Hukuhara T, Hayakawa T, Wijonarko A. Increased baculovirus susceptibility of armyworm larvae feeding on transgenic rice plants expressing an entomopoxvirus gene. Nat Biotechnol. 1999;17(11):1122–1124. doi: 10.1038/15110. [DOI] [PubMed] [Google Scholar]
- 11.Takemoto Y, Mitsuhashi W, Murakami R, Konishi H, Miyamoto K. The N-terminal region of an entomopoxvirus fusolin is essential for the enhancement of peroral infection, whereas the C-terminal region is eliminated in digestive juice. J Virol. 2008;82(24):12406–12415. doi: 10.1128/JVI.01605-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dall D, Luque T, O’Reilly D. Insect-virus relationships: Sifting by informatics. BioEssays. 2001;23(2):184–193. doi: 10.1002/1521-1878(200102)23:2<184::AID-BIES1026>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 13.Gauthier L, Cousserans F, Veyrunes JC, Bergoin M. The Melolontha melolontha entomopoxvirus (MmEPV) fusolin is related to the fusolins of lepidopteran EPVs and to the 37K baculovirus glycoprotein. Virology. 1995;208(2):427–436. doi: 10.1006/viro.1995.1173. [DOI] [PubMed] [Google Scholar]
- 14.Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels. 2013;6(1):41. doi: 10.1186/1754-6834-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaaje-Kolstad G, et al. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science. 2010;330(6001):219–222. doi: 10.1126/science.1192231. [DOI] [PubMed] [Google Scholar]
- 16.Aachmann FL, Sørlie M, Skjåk-Bræk G, Eijsink VGH, Vaaje-Kolstad G. NMR structure of a lytic polysaccharide monooxygenase provides insight into copper binding, protein dynamics, and substrate interactions. Proc Natl Acad Sci USA. 2012;109(46):18779–18784. doi: 10.1073/pnas.1208822109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hemsworth GR, et al. The copper active site of CBM33 polysaccharide oxygenases. J Am Chem Soc. 2013;135(16):6069–6077. doi: 10.1021/ja402106e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Beeson WT, 4th, Phillips CM, Marletta MA, Cate JH. Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases. Structure. 2012;20(6):1051–1061. doi: 10.1016/j.str.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quinlan RJ, et al. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc Natl Acad Sci USA. 2011;108(37):15079–15084. doi: 10.1073/pnas.1105776108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gudmundsson M, et al. Structural and electronic snapshots during the transition from a Cu(II) to Cu(I) metal center of a lytic polysaccharide monooxygenase by X-ray photoreduction. J Biol Chem. 2014;289(27):18782–18792. doi: 10.1074/jbc.M114.563494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forsberg Z, et al. Structural and functional characterization of a conserved pair of bacterial cellulose-oxidizing lytic polysaccharide monooxygenases. Proc Natl Acad Sci USA. 2014;111(23):8446–8451. doi: 10.1073/pnas.1402771111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaaje-Kolstad G, Horn SJ, van Aalten DMF, Synstad B, Eijsink VGH. The non-catalytic chitin-binding protein CBP21 from Serratia marcescens is essential for chitin degradation. J Biol Chem. 2005;280(31):28492–28497. doi: 10.1074/jbc.M504468200. [DOI] [PubMed] [Google Scholar]
- 23.Chiu E, Coulibaly F, Metcalf P. Insect virus polyhedra, infectious protein crystals that contain virus particles. Curr Opin Struct Biol. 2012;22(2):234–240. doi: 10.1016/j.sbi.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Hemsworth GR, Henrissat B, Davies GJ, Walton PH. Discovery and characterization of a new family of lytic polysaccharide monooxygenases. Nat Chem Biol. 2014;10(2):122–126. doi: 10.1038/nchembio.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forsberg Z, et al. Cleavage of cellulose by a CBM33 protein. Protein Sci. 2011;20(9):1479–1483. doi: 10.1002/pro.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z, et al. Characterization of a chitin-binding protein GP37 of Spodoptera litura multicapsid nucleopolyhedrovirus. Virus Res. 2003;96(1-2):113–122. doi: 10.1016/s0168-1702(03)00179-5. [DOI] [PubMed] [Google Scholar]
- 27.Hukuhara T, Hayakawa T, Wijonarko A. A bacterially produced virus enhancing factor from an entomopoxvirus enhances nucleopolyhedrovirus infection in armyworm larvae. J Invertebr Pathol. 2001;78(1):25–30. doi: 10.1006/jipa.2001.5036. [DOI] [PubMed] [Google Scholar]
- 28.Frederiksen RF, et al. Bacterial chitinases and chitin-binding proteins as virulence factors. Microbiology. 2013;159(Pt 5):833–847. doi: 10.1099/mic.0.051839-0. [DOI] [PubMed] [Google Scholar]
- 29.Redecke L, et al. Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser. Science. 2013;339(6116):227–230. doi: 10.1126/science.1229663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitsuhashi W, Murakami R, Takemoto Y, Miyamoto K, Wada S. Stability of the viral-enhancing ability of entomopoxvirus spindles exposed to various abiotic factors. Appl Entomol Zool (Jpn) 2008;43(4):483–489. [Google Scholar]
- 31.Terwilliger TC, et al. phenix.mr_rosetta: Molecular replacement and model rebuilding with Phenix and Rosetta. J Struct Funct Genomics. 2012;13(2):81–90. doi: 10.1007/s10969-012-9129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blanc E, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.