Abstract
The piwi-like 2 (piwil2) gene is widely expressed in tumors and protects cells from apoptosis induced by a variety of stress stimuli. However, the role of Piwil2 in Fas-mediated apoptosis remains unknown. Here, we present evidence that Piwil2 inhibits Fas-mediated apoptosis. By a bacterial two-hybrid screening, we identify a new Piwil2-interacting partner, keratin 8 (K8), a major intermediate filament protein protecting the cell from Fas-mediated apoptosis. Our results show that Piwil2 binds to K8 and p38 through its PIWI domain and forms a Piwil2/K8/P38 triple protein-protein complex. Thus, Piwil2 increases the phosphorylation level of K8 Ser-73 and then inhibits ubiquitin-mediated degradation of K8. As a result, the knockdown of Piwil2 increases the Fas protein level at the membrane. In addition to our previous finding that Piwil2 inhibits the expression of p53 through the Src/STAT3 pathway, here we demonstrate that Piwil2 represses p53 phosphorylation through p38. Our present study indicates that Piwil2 plays a role in Fas-mediated apoptosis for the first time and also can affect p53 phosphorylation in tumor cells, revealing a novel mechanism of Piwil2 in apoptosis, and supports that Piwil2 plays an active role in tumorigenesis.
INTRODUCTION
The PIWI proteins are widely distributed among different animals and have been implicated in functions related to stem-cell self-renewal, gametogenesis, epigenetic modulation, transposon control, and embryogenesis (1–8). The human PIWI family is comprised of four different members, Piwil1/Hiwi, Piwil2/Hili, Piwil3, and Piwil4/Hiwi2 (9). The PIWI proteins are expressed predominantly in testis and embryo (1, 3, 6, 9, 10), and recently, it has been reported that Piwil2 protein is widely detected in tumors and protects cells from apoptosis (11–13). Our previous work showed that human Piwil2 inhibits apoptosis by regulating the transforming growth factor beta (TGF-β) pathway in HEK293 cells and the STAT3/p53 pathway in tumor cells (12, 13). Furthermore, Piwil2 also exhibited resistance in response to other forms of stimuli to apoptosis (14–16).
Apoptosis is the process of programmed cell death that may be initiated by different stimuli, particularly through the stimulation of death receptors (DRs) like FasR, tumor necrosis factor (TNF) receptors (TNFRs), and TNF-related apoptosis-inducing ligand receptors (TRAILRs) or by their respective ligands. The Fas receptor (Fas), also termed Apo-1 or CD95, is a member of the tumor necrosis factor and nerve growth factor (NGF) receptor family (17, 18). Apoptotic cell death induced by the Fas-Fas ligand (FasL) interaction plays a major role in immune modulation (17). The Fas/FasL pathway also plays an important role in tumorigenesis, as many tumor cells exhibit low expression of Fas on the membrane (17, 19).
Keratins are the major intermediate filament proteins and are important for the mechanical stability and integrity of epithelial cells and tissues. Research has shown that keratins participate in intracellular signaling pathways by regulating the cell cycle (20, 21), apoptosis (22–24), and tumorigenesis (25–28). In simple epithelial cells, keratins 8 and 18 (K8/18) typically are coexpressed as the primary keratin pair and play an important cytoprotective role, protecting cells from apoptosis, stress, and injury (23, 24, 29–31). The structure and function of K8/18 probably are regulated through posttranslational modifications, such as phosphorylation, glycosylation, and ubiquitination, in which phosphorylation is considered the major contributing factor (21, 23, 32–35).
Here, we present that human Piwil2 interacts with the p38 pathway in tumor cells, inhibiting Fas-mediated apoptosis by phosphorylating K8 and also suppressing p53 phosphorylation and p53-induced apoptosis.
MATERIALS AND METHODS
Antibodies.
Rabbit monoclonal anti-K8 (2032-1), rabbit monoclonal anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) (5632-1), rabbit monoclonal anti-phospho-K8 (pS73) (1431-s), rabbit monoclonal anti-p38 (3008-1), and rabbit monoclonal anti-phospho-p53 (anti-p-p53) (2190-1) were purchased from Epitomics (Burlingame, CA); rabbit anti-Myc (sc-789), rabbit antihemagglutinin (anti-HA) (sc-805), and rabbit anti-Piwil2 (sc-67303) were from Santa Cruz Biotechnology (Dallas, TX); mouse anti-HA (2367), mouse anti-Myc (2267), rabbit anti-caspase 9 (9502), and rabbit anti-caspase 3 (9662) were from Cell Signaling (Danvers, MA); and mouse anti-p38 (AM065), mouse anti-p-p38 (AM063), mouse anti-caspase 8 (AC056), mouse anti-p53 (AP062), and mouse anti-α-tubulin (AT819) were from Beyotime (Shanghai, China). Mouse anti-Fas (200411) was from Zen BioScience (Chengdu, China). Rabbit antiezrin (E1A6172), rabbit anti-Bax (E1A0120), rabbit anti-p-HSP 27 (E1A6082), and rabbit anti-Na, K ATPase (E1A6109) were from Enogene (Nanjing, China). Goat anti-HA (A00168) was from GenScript (Nanjing, China). EasyBlot anti-rabbit IgG (GTX221666-01), which was used as an immunoprecipitation (IP) secondary antibody, was from GeneTex (Irvine, CA). Western secondary antibodies were from Zhongshan Goldenbridge (Beijing, China). Fluorescent secondary antibodies were from Amyjet (Wuhan, China).
Expression vectors, mutants, and shRNA.
Keratin 8 was cloned into the pcDNA3.1+Myc or pcDNA3.1+HA expression vector. We constructed various K8 mutants by segmented PCR and fusion PCR, taking pcDNA3.1 Myc-K8 as the template. These mutants were cloned into the expression vector pcDNA3.1+Myc. Other expression vectors were constructed in our previous report (13). Short hairpin RNA (shRNA) for PIWIL2 (shPIWIL2) was synthesized and cloned into shRNA expression vector pGPU6/GFP/Neo by GenePharma Inc. (Shanghai, People's Republic of China). The core sequence (sense strand, 5′ CUA UGA GAU UCC UCA ACU ACA GAAG 3′) has been reported in pervious works (12, 13, 36). For rescue experiments, cells were cotransfected with shPiwil2 and WT Piwil2 expression vector.
Cell culture.
HeLa cells and HepG2 cells were maintained in our laboratory as previously described (13). They were cultured in Dulbecco's modified Eagle medium (DMEM)-10% fetal bovine serum (FBS) and transfected by using jetPEI (PolyPlus, Berkeley, CA) according to the manufacturer's protocols. All transfections were performed in 6-well plates. Forty-eight hours after transfection, cells treatments were carried out in reduced-serum medium (0.2% FBS), and FasL (Peprotech, Rocky Hill, NJ) was added as indicated to a final concentration of 100 ng/ml for 8 h. Where specified, cells were treated with the proteasome inhibitor MG132 (Beyotime) at a final concentration of 20 mM. Cells were harvested and analyzed by Western blotting using appropriate antibodies. To inhibit tyrosine kinase activity, HeLa cells were pretreated with 1 μM staurosporine, 5 μM SP600125, 5 μM SB203580, or 5 μM SB202190 (Beyotime) for different periods of time. All of the following experiments were repeated at least three times unless stated otherwise.
Subcellular fractionation.
Cell plasma membrane proteins were isolated using a plasma membrane protein extraction kit (BioVision, Milpitas, CA) as described by the manufacturer. To isolate the cytoplasmic component from the nuclear one, cells were treated with a nuclear protein extraction kit (Beyotime) according to the manufacturer's instructions.
Co-IP and Western blotting.
For coimmunoprecipitation (co-IP) and Western blotting, cells were lysed 48 h after transfected with the designated plasmids in universal protein extraction buffers (Bioteke, Beijing, China). Extracted proteins were immunoprecipitated with special antibody and protein A+G-agarose beads (Beyotime). Bound proteins were separated using SDS-PAGE, transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA), and detected with specific appropriate primary antibodies and horseradish peroxidase-conjugated secondary antibodies. Specific proteins were visualized using an enhanced chemiluminescence (ECL) Western blot detection system (Millipore).
For trimeric complex coimmunoprecipitation, cells were transfected with HA-Piwil2. Extracted proteins were immunoprecipitated with HA antibody and beads. The complexes were eluted from the beads by 40 μg/ml HA peptide (Zen BioScience) and reimmunoprecipitated with p38 antibody.
Immunofluorescence.
Cells were fixed with 4% paraformaldehyde in PBS for 15 min and permeabilized with 0.5% Triton X-100 for 10 min, blocked with 1% bovine serum albumin (BSA) for 30 min, incubated overnight at 4°C with primary antibody, and finally incubated with DyLight 488-labeled, DyLight 555-labeled, or DyLight 350-labeled secondary antibody for 1 h at room temperature. Each step was followed by two 5-min washes in PBS. To stain Fas on the cell surface, primary and secondary antibodies were treated after 4% paraformaldehyde fixation without permeabilization to stain Fas on the cell surface. For nuclear stain, prepared specimens were counterstained with 5 mg/ml 4′,6-diamidino-2-phenylindole (DAPI) for 2 min. Fluorescent images were obtained with a confocal microscope (Olympus, Japan).
Rabbit anti-K8 (2032-1) and rabbit anti-p-p53 (2190-1) were from Epitomics. Anti-Piwil2 (sc-67303) was from Santa Cruz. Goat anti-HA (A00168) was from GenScript. Mouse anti-HA (2367) was from Cell Signaling. Mouse anti-p38 (AM065) and mouse anti-p-p38 (AM063) were from Beyotime. The antibody staining was validated by shRNA-mediated downregulation of the target protein.
Apoptosis assay.
Apoptotic rates were analyzed with a Coulter Epics XL flow cytometer (Beckman Coulter, CA) by using an annexin V-Alexa Fluor/propidium iodide (PI) apoptosis detection kit (Ebioe, China). Annexin V-Alexa Fluor/PI staining and fluorescence intensity measurements were performed according to the manufacturer's instructions.
Quantitative real-time PCR.
Total RNA was prepared using TRIzol (Invitrogen, Carlsbad, CA) from HeLa cells transfected with Myc-Piwil2 and pcDNA3.1 or shPiwil2 and control mock-transfected shRNA (shNC). Quantitative PCR was performed in an iCycler IQ real-time PCR detection system (Bio-Rad, Hercules, CA) with a denaturation step at 94°C for 10 min, followed by 45 cycles of denaturation at 94°C for 20 s, annealing at 50°C for 30 s, and extension at 72°C for 40 s.
Protein sequence analysis.
The keratin 8 protein sequence was determined by the SMART analysis service at http://smart.embl-heidelberg.de/.
Two-hybrid experiment.
Bacterial two-hybrid experiments were performed as described in the protocol provided with the BacterioMatch II two-hybrid system (Stratagene, Santa Clara, CA). Piwil2 baits were used to screen a HeLa cDNA library. Keratin 8 was obtained and isolated in the screening. For the binding assay, K8 was inserted into pBT plasmid, and p38 and K8 were inserted into pTRG plasmid.
In vitro transcription/translation system.
An in vitro protein binding assay was performed using the TnT quick coupled in vitro transcription/translation system (Promega, Madison, WI) according to the manufacturer's instructions. The reactions were carried out in 25-μl volumes by adding 1 μg of plasmid DNA and 1 μl unlabeled methionine to the TnT mix. We incubated the reaction mix at 30°C for 90 min. Subsequently, 20-μl aliquots of each of the two proteins were mixed together in 200 μl binding buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM dithiothreitol, 0.1% Tween 20) with protease inhibitors (Complete EDTA-free; Roche Molecular Biochemicals) and incubated on a rotating platform at 4°C for 3 h. Interactions were detected by co-IP and Western blotting using a specific antibody.
In vitro kinase assays.
HA-Piwil2 and HA-p38 were expressed using an in vitro transcription/translation system (Promega) and then immunoprecipitated with anti-HA antibody and beads overnight. The protein then was eluted by 40 μg/ml HA peptide from the beads and purified by Amicon Ultra (Millipore). Myc-K8 also was expressed in vitro and immunoprecipitated with anti-Myc antibody and beads. The beads were washed and used for in vitro kinase assays. Beads were mixed with or without purified p38 kinase or Piwil2 and 40 μM ATP (Promega) in kinase buffer (Cell Signaling Technology). Reactions were performed at room temperature for 1 h.
Statistical analysis.
Experiments were repeated three times. Western blot and immunofluorescence results were analyzed using ImageJ software. Differences between experimental groups were determined using Student's t test. Statistical significance was accepted when the P value was <0.05.
RESULTS
Piwil2 knockdown increases cell sensitivity to Fas-mediated apoptosis.
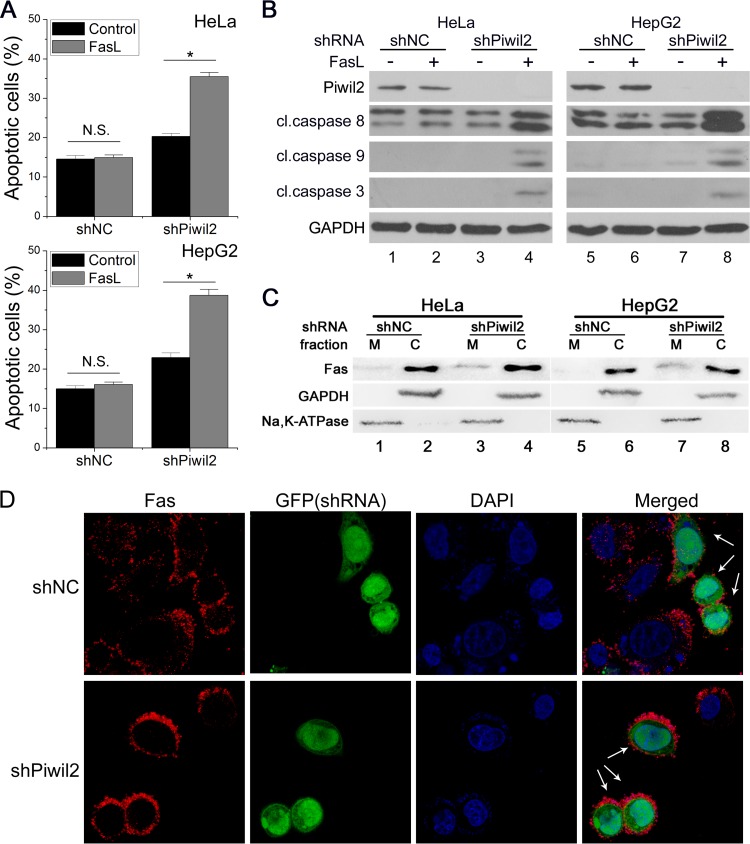
It is known that Piwil2 inhibits apoptosis induced by many different stimuli (12, 13, 16). It has not been reported yet whether Piwil2 can regulate Fas-mediated apoptosis. We examined the effect of Piwil2 knockdown on Fas-mediated apoptosis by fluorescence-activated cell sorting (FACS) analysis. With FasL treatment, an increase in the level of apoptosis was observed in Piwil2 knockdown cells but not in control mock shRNA (shNC)-transfected cells (Fig. 1A). We next detected the activation of caspases, representative members of the Fas signaling pathway. Piwil2 knockdown highly increased the cleavage of proapoptotic caspases 3, 8, and 9 in response to FasL (Fig. 1B, lanes 4 and 8). Meanwhile, a significant increase of Fas at the plasma membrane in Piwil2 knockdown cells was observed after cell surface protein isolation in HeLa (2.4-fold; P < 0.05) (Fig. 1C, lanes 1 and 3) and HepG2 (2.8-fold; P < 0.05) (Fig. 1C, lanes 5 and 7) cells. Immunofluorescence also showed a higher Fas density in the membrane of Piwil2 knockdown cells (74% of transfected cells) than in shNC-transfected cells or untransfected cells without permeabilization for surface stain (Fig. 1D). Given these data, we concluded that Piwil2 knockdown increased cell sensitivity to Fas-mediated apoptosis.
FIG 1.
Piwil2 loss sensitizes cells to Fas-mediated apoptosis. (A and B) NC (shNC) and Piwil2 knockdown (shPiwil2) cells were mock treated or treated with FasL (100 ng/ml) and then harvested for apoptosis analysis using FACS assay and Western blot analysis. Error bars indicate SE (N.S. [not significant], P > 0.05; *, P < 0.05). cl., cleaved. (C and D) The impact of Piwil2 depletion on the membrane localization of Fas was determined by membrane (M)/cytosolic (C) fractionation (C) and immunofluorescence staining (D). Na, K ATPase (membrane) and GAPDH (cytosolic) were used as controls. Green fluorescent protein (GFP) is expressed from shRNA vectors and indicates the cell outline. The arrows indicate the expression of the shRNA vectors. Anti-Fas was used for panel D.
Piwil2 interacts with Fas-mediated apoptosis-related protein K8.
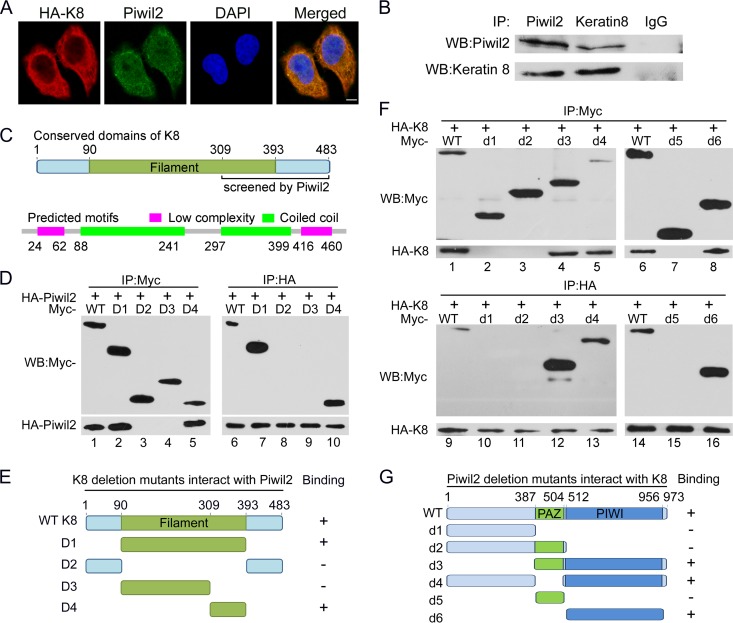
To identify novel Piwil2-interacting proteins and their potential functions in tumor cells, we performed a bacterial two-hybrid screen of a human HeLa cDNA expression library with Piwil2 as the bait. By the sequencing of positive clones, we identified one Piwil2-interacting peptide as C terminal (amino acids [aa] 309 to 483) to K8 which is well known to protect the cell from Fas-mediated apoptosis and injury (23, 24, 31, 37).
We next examined whether Piwil2 could interact with K8 in vivo. Immunofluorescence experiments showed that K8 was located mostly in the cytoplasm and overlapped with Piwil2 (Fig. 2A). To confirm the physical interaction between Piwil2 and K8, we carried out reciprocal immunoprecipitation of endogenous Piwil2 and K8 in HeLa cells. Immunoprecipitation and Western blot data revealed that Piwil2 could be coprecipitated with K8 (Fig. 2B).
FIG 2.
Interaction of Piwil2 and K8. (A) Piwil2 and K8 are colocalized mostly in cytoplasm. Cells were transfected with HA-K8 vector and harvested for immunofluorescence assay with anti-Piwil2 and mouse anti-HA antibodies. Scale bar, 5 μm. (B) Endogenous interactions between Piwil2 and K8 in HeLa cells. Coimmunoprecipitation was performed with anti-Piwil2 or anti-K8, followed by Western blotting. (C) Filament domain of K8 and the predicted domains by SMART analysis. (D) Interaction between HA-tagged Piwil2 and different Myc-tagged K8 mutants. (E) Schematic of different K8 deletion mutants. (F) Interaction between HA-tagged K8 and different Myc-tagged Piwil2 mutants. (G) Schematic of different Piwil2 mutants. All of these assays were performed in HeLa cells. WB, Western blotting; WT, wild type.
The filament domain (pfam00038; aa 90 to 393) is classified as a model that may span more than one domain of K8, as described in GenBank. We performed bioinformatics analysis for K8 protein sequence by SMART analysis and got two low-complexity regions (aa 24 to 62 and aa 416 to 460) and two coiled coils (aa 88 to 241 and aa 297 to 399) (Fig. 2C). Combined with all of the other the information, we constructed four K8 protein deletion mutants (Fig. 2E). To further identify the regions where K8 binds to Piwil2, we examined the ability of Piwil2 to bind with the full-length K8 as well as various Myc-tagged K8 mutants. Our results showed that wild-type K8, the filament domain, and the C terminus of the filament domain interacted with Piwil2 (Fig. 2D). These results suggested that the Piwil2 binding region in K8 was localized in the C terminus of the filament domain (aa 309 to 393).
We next mapped the K8 binding region in Piwil2. There are two functional domains in Piwil2: the PAZ domain and the PIWI domain. Coimmunoprecipitation experiments showed that mutants of Piwil2 lacking the PIWI domain, such as d1, d2, and d5, failed to bind to K8 (Fig. 2F, lanes 2, 3, 7, 10, 11, and 15), while mutants containing the PIWI domain, including d3, d4, and d6, could bind to K8 (Fig. 2F, lanes 4, 5, 8, 12, 13, and 16). Taken together, these findings further revealed that the K8 binding region in Piwil2 was localized in the PIWI functional domain.
The increased sensitivity of Piwil2 knockdown cells to Fas-mediated apoptosis is related to K8 downregulation.
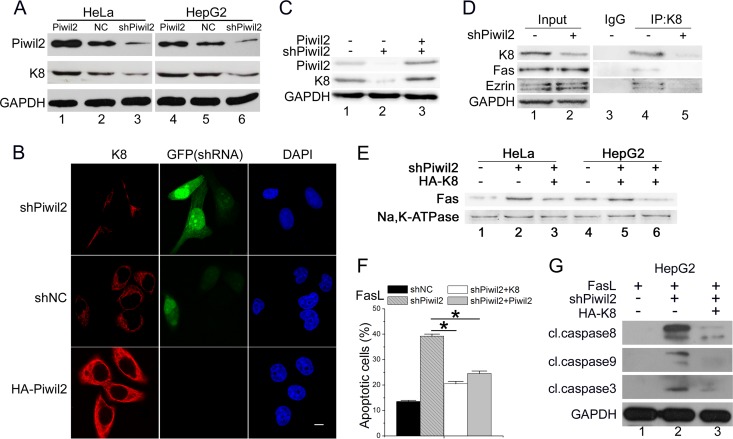
As we have confirmed that Piwil2 can interact with K8, we wanted to know whether K8 was related to the increased Fas-mediated apoptosis in Piwil2 knockdown cells. Western blot analysis showed that the K8 protein level was accumulated by ectopic expression of Piwil2; it was increased 1.79-fold (P < 0.05) in HeLa cells and 1.35-fold (P < 0.05) in HepG2 cells. Piwil2 depletion induced a 0.43-fold decrease (P < 0.05) of K8 in HeLa cells and 0.54-fold decrease (P < 0.05) in HepG2 cells (Fig. 3A). Immunofluorescence results also showed significantly decreased K8 fluorescence intensity in 80% of Piwil2 knockdown cells (Fig. 3B); Piwil2 overexpression, on the other hand, significantly increased K8 fluorescence intensity (65% of total cells). The protein level of K8 could be rescued by the overexpression of Piwil2 in Piwil2-deficient cells (Fig. 3C).
FIG 3.
Piwil2 increases K8 protein levels. (A) HeLa and HepG2 cells were transfected with Myc-Piwil2, shNC, or shPiwil2 vector. After 48 h, cell lysates were used for Western blotting with anti-K8. (B) Piwil2 knockdown decreased the K8 fluorescence intensity significantly. HeLa cells were transfected with shNC or shPiwil2 vector and harvested for immunofluorescence assay with anti-K8 antibodies. GFP is expressed from shRNA vectors. Scale bar, 5 μm. (C) Rescue experiments of the knockdown of Piwil2 on the protein level of K8. HepG2 cells were transfected with shPiwil2 or Piwil2 expression vector as indicated. (D) Piwil2 knockdown decreased the interaction between K8 and Fas-ezrin complex. HeLa cells were transfected with shNC or shPiwil2 vector, and a coimmunoprecipitation assay was performed using anti-K8, followed by Western blotting. (E) The impact of K8 on Fas localization in Piwil2 knockdown cells. The level of membrane Fas was determined by membrane protein separation followed by Western blotting. (F) Both Piwil2 and K8 overexpression reduced the sensitivities of Fas-mediated apoptosis enhanced by Piwil2 knockdown. (G) Western blot analysis of caspases activation after FasL induction. Cells were transfected with shNC, shPiwil2, pcDNA3.1, HA-K8, and Myc-Piwil2 plasmids as indicated 48 h before Western blotting. Cells were treated with FasL (100 ng/ml) as indicated. Error bars indicate SE (*, P < 0.05).
Previous research showed that actin, K8/18 intermediate filaments, plectin, and ezrin form a well-organized network on wild-type hepatocyte surfaces. As ezrin interacts with Fas, the network prevents Fas from targeting the membrane. The loss of K8/18 alters the early steps of Fas activation through perturbation of the ezrin/actin (38). Our results showed that Piwil2 knockdown also decreased the binding of K8 to the Fas-ezrin complex (Fig. 3D), while no significant changes were observed in the protein level of Fas and ezrin. To confirm that the increased Fas-mediated apoptosis in Piwil2 knockdown cells was mediated by K8, we induced K8 expression by transfected K8 vector in Piwil2 knockdown cells. Western blot analysis showed that in Piwil2 knockdown cells, the overexpression of K8 reduced the level of Fas on the plasma membrane by 65% (P < 0.05) in HeLa cells and 79% (P < 0.05) in HepG2 cells (Fig. 3E). The ectopic expression of both K8 and Piwil2 rescued the increased apoptosis in Piwil2 knockdown cells in response to FasL (Fig. 3F). The level of cleaved caspases in response to FasL in the Piwil2 knockdown cells also was decreased by overexpression of K8 (Fig. 3G). All of these data confirmed that the loss of Piwil2 sensitized cells to Fas-mediated apoptosis by downregulating K8.
Piwil2 inhibits K8 degradation through facilitating the phosphorylation of K8 by p38 kinase.
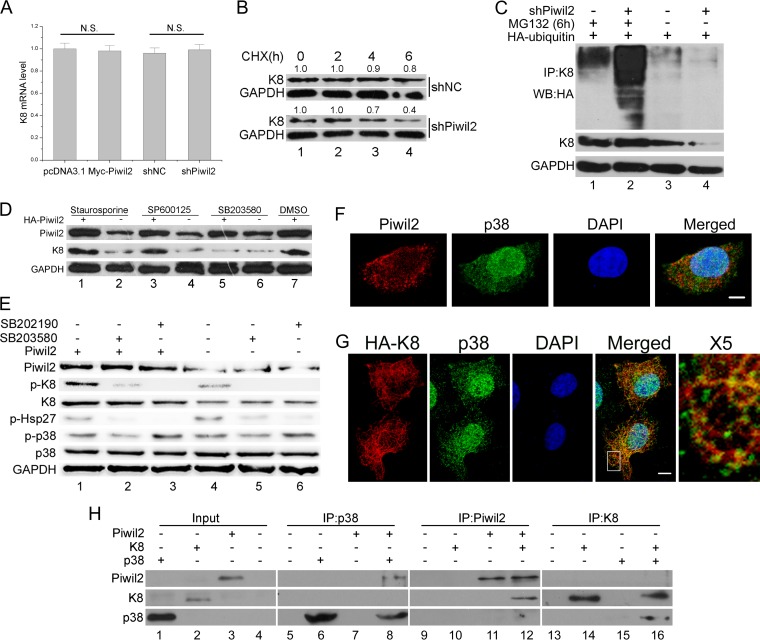
As we have confirmed that Piwil2 could affect the K8 protein level, we next explored the underlying molecular mechanism. In contrast to the protein level, quantitative real-time PCR showed that Piwil2 had no effect on K8 mRNA levels (Fig. 4A). Thus, the decreased level of K8 in Piwil2 knockdown cells was due to protein degradation. To further evaluate the stability of K8, we employed the strategy of suppressing protein translation by utilizing cycloheximide (CHX) to study the degradation of K8. After a 6-h treatment with CHX, less than half of the K8 remained in Piwil2 knockdown cells, in contrast to the higher level of stability of K8 in control cells (Fig. 4B). The addition of MG132 nearly recovered K8 to control levels in the presence of shPiwil2 and facilitated the detection of K8 ubiquitination. Notably, knockdown of Piwil2 promoted K8 ubiquitination (Fig. 4C, lanes 1 and 2). Transfection with shPiwil2 alone promoted the dramatic degradation of K8, so less ubiquitination was detected compared to that with controls (Fig. 4C, lanes 3 and 4). These results imply that Piwil2 decreased K8 ubiquitination and then prevented its ubiquitin-controlled degradation.
FIG 4.
Piwil2 inhibits degradation of K8 in a phosphorylation-modulated fashion. (A) Real-time PCR confirmed that the K8 mRNA level remains unchanged after Piwil2 overexpression or Piwil2 knockdown. N.S., P > 0.05 by the Student's t test. (B) Stability analysis of K8 protein. The cells transfected with shNC or shPiwil2 were treated with cycloheximide (CHX) at 5 mg/ml for 0 to 6 h. (C) Piwil2 knockdown improved K8 ubiquitination. HeLa cells were cotransfected with shPiwil2 and HA-ubiquitin plasmids. After 48 h, they were treated with MG132 for 6 h as indicated. K8 was precipitated with anti-K8 antibody, and the ubiquitination and degradation of K8 were determined by anti-HA-ubiquitin and anti-K8 immunoblotting. (D) P38 inhibitor blocks Piwil2 from upregulating K8. Cells were transfected with Myc-Piwil2 or pcDNA3.1 and treated with p38, PKC, and JNK inhibitors for 6 h. DMSO, dimethylsulfoxide. (E) P38 is involved in Piwil2-induced K8 phosphorylation. The inhibition of p38 using SB203580 or SB202190 (1 h of treatment) reduces the Ser-73 phosphorylation of K8, as shown by Western blotting. The level of p-HSP27 showed that inhibitors are working. (F) Colocalization between endogenous Piwil2 and p38. Cells were stained by anti-Piwil2 and anti-p38. (G) Colocalization between K8 and p38. Cells were transfected with HA-K8 vector and harvested for immunofluorescence assay with anti-HA and anti-p38 antibodies. Scale bars, 5 μm. The indicated region in the merged panel was shown at a higher resolution (500%) in the X5 panel. (H) Piwil2, p38, and K8 directly interact in vitro. Cell-free Piwil2, p38, and K8 protein expression was carried out using a TnT system separately, mixed together in binding buffer with protease inhibitors, and incubated on a rotating platform at 4°C for 3 h. Error bars indicate SE (N.S., P > 0.05).
Previous studies have shown that phosphorylation protects K8 against ubiquitination and subsequent degradation, but this protection is not site specific and involves K8 residues S23, S73, and/or S431, which undergo phosphorylation (34). For this reason, we next examined whether Piwil2 protects K8 from ubiquitination via phosphorylation. Previous research showed that K8 could be phosphorylated by different kinases, such as protein kinase C (PKC), p38, and Jun N-terminal protein kinase (JNK) (34, 39–41). To search for potential factors in the accumulation of K8 induced by Piwil2, HeLa cells were treated with different kinase inhibitors after transfection of the Piwil2 expression plasmid. Western blot analysis showed that after treatment with p38 inhibitor, K8 accumulation was blocked in Piwil2-overexpressed cells (Fig. 4D, lanes 5 and 6). However, the overexpression of Piwil2 still increased the K8 protein level in cells treated with PKC and JNK inhibitors. This indicated that p38, rather than PKC and JNK, plays a role in Piwil2-induced accumulation of K8, which was further confirmed by Western blotting using phosphorylated K8 antibody. The phosphorylation level of K8 was significantly increased in Piwil2-overexpressing cells (2.79-fold; P < 0.05) (Fig. 4E, lanes 1 and 4) and could be reduced by two p38 inhibitors. However, phosphorylated p38 (p-p38) was not increased in ectopically expressed Piwil2 cells (Fig. 4E, lanes 1 and 4). p38 localized both in the cytoplasm and nucleus, while K8 was colocalized with p38 mostly in the cytoplasm (Fig. 4F). Endogenous Piwil2 also was observed to colocalize with p38 mostly in the cytoplasm (Fig. 4G). Furthermore, we asked whether Piwil2 is required for the interaction of K8 and p38. Therefore, an in vitro protein binding assay was conducted to detect direct interaction between Piwil2, K8, and p38. We translated Piwil2, K8, and p38 in vitro and performed coimmunoprecipitations with each of them. Our results showed that Piwil2 and K8, p38 and K8, as well as Piwil2 and p38 can be coimmunoprecipitated in vitro (Fig. 4H). In addition, interaction between p38, K8, and Piwil2 also was validated by the two-hybrid experiment (not shown). These findings suggested that Piwil2 protected K8 from ubiquitination-mediated degradation by enhancing p38-induced K8 phosphorylation.
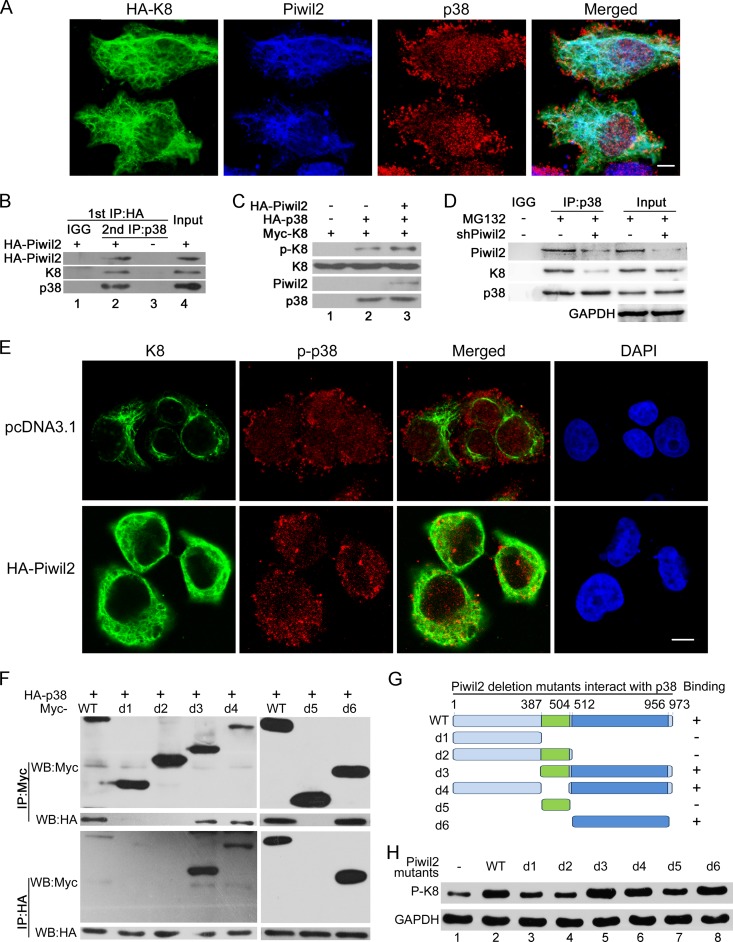
Piwil2 enhances the formation of K8-p38 complexes.
We next investigated whether Piwil2 was able to enhance the endogenous interaction between K8 and p38 to increase K8 phosphorylation. An obvious colocalization among Piwil2, K8, and p38 was observed mostly in the cytoplasm (Fig. 5A). Coimmunoprecipitation experiments also revealed that K8 protein was physically associated with Piwil2-p38 complex (Fig. 5B). These pieces of evidence suggested that p38, Piwil2, and K8 formed a trimeric complex. An in vitro experiment showed that Piwil2 facilitated the phosphorylation of K8 induced by p38 (Fig. 5C). Meanwhile, the absence of Piwil2 decreased p38 binding to K8 (Fig. 5D). This result showed that the knockdown of Piwil2 suppressed the formation of K8-p38 complexes. To further confirm that Piwil2 promotes p38 binding to K8, we performed immunofluorescence assays. The overexpression of Piwil2 significantly increased p-p38/K8 colocalization in the cytoplasm and decreased p-p38 nuclear translocation (63% of total cells) (Fig. 5E). Together, these results suggested that Piwil2 participated in and enhanced the formation of K8-p38 complex.
FIG 5.
Piwil2 facilitates the interaction of K8 and p38. (A) Colocalization of K8, Piwil2, and p38. Cells were transfected with HA-K8 vector and harvested for immunofluorescence assay with goat anti-HA, rabbit anti-Piwil2, and mouse anti-p38 antibodies. (B) Co-IP assay of the Piwil2-p38-K8 complex. Cells were transfected with HA-Piwil2, the lysates were subjected to anti-HA antibody immunoprecipitation and eluted by HA peptide, and then they were reimmunoprecipitated by anti-p38 antibody followed by Western blotting. (C) In vitro phosphorylation of K8. HA-Piwil2 and HA-p38 were translated and purified in vitro. Myc-K8 was translated in vitro and immunoprecipitated by Myc antibody with beads. The beads with Myc-K8 were subjected to in vitro phosphorylation in kinase buffer for 1 h with p38 or Piwil2 as indicated, followed by Western bolt analysis. (D) After 48 h, cells were transfected with shPiwil2 or shNC vector and treated with MG132 for 6 h. The lysates were subjected to anti-p38 immunoprecipitation followed by WB. (E) Piwil2 enhanced the colocalization of p-p38 and K8 in cytoplasm. HeLa cells were transfected with pc-DNA3.1 or HA-Piwil2 vector and harvested for immunofluorescence assay with anti-K8 and anti-p-p38 antibodies. Scale bars, 5 μm. (F) Interaction between different HA-p38 and different Piwil2 mutants with Myc tag. (G) Schematic of different Piwil2 mutants binding to p38. (H) The effect of different Piwil2 mutants on phosphorylation of K8 by Western blotting.
We next mapped the p38 binding region in Piwil2. As with K8, coimmunoprecipitation experiments showed that Piwil2 and its mutants containing the PIWI domain, including d3, d4, and d6, remained capable of binding p38 (Fig. 5F). These results suggested that the p38 binding region also was localized in the PIWI domain. Furthermore, the PIWI-deleted mutants failed to increase K8 phosphorylation (Fig. 5H, lanes 3, 4, and 7), confirming the potential role of the PIWI domain of Piwil2.
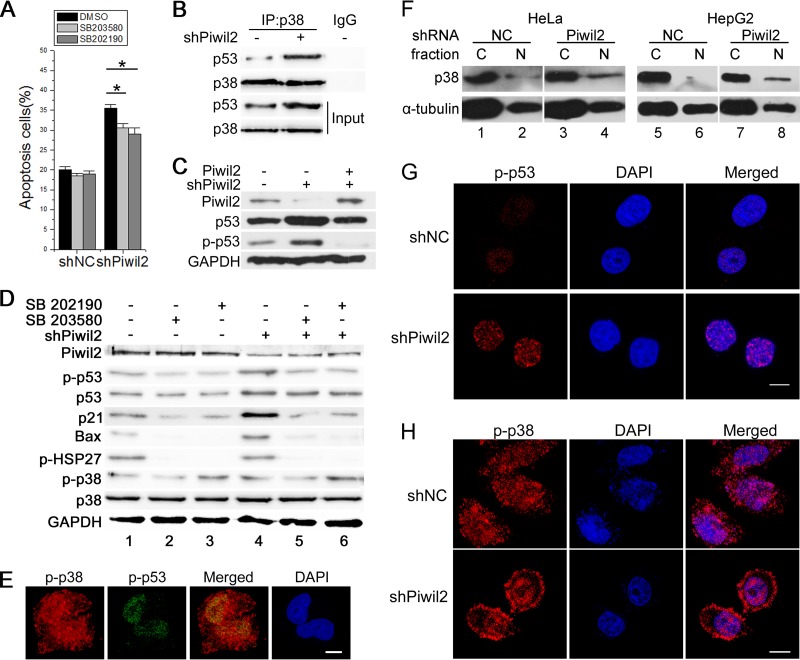
Piwil2 knockdown increases the phosphorylation of p53.
The tumor suppressor p53 is a well-known regulator of apoptosis. Our previous work showed that Piwil2 could suppress apoptosis by inhibiting p53 expression through the Src-STAT3 pathway (12). We then examined whether Piwil2 also could affect p53 and p53-induced apoptosis through binding to p38, since p53 is also an important substrate of p38 (42). The apoptosis ratio induced by Piwil2 knockdown was partly decreased by p38 inhibitor in Piwil2 knockdown cells (Fig. 6A). The decrease of Piwil2 increased the p53 protein level detected in p38 immunoprecipitates (Fig. 6B). Overexpression of Piwil2 rescued the p53 level in Piwil2 knockdown cells (Fig. 6C). However, when treated with p38 inhibitors, the p53 level was slightly decreased after knockdown of Piwil2 (0.80-fold and 0.73-fold; P < 0.05) (Fig. 6D). We also examined the phosphorylation of p53 and several other proteins that may have roles in p53-induced apoptosis. The loss of Piwil2 increased the levels of p21, Bax, and phosphorylated p53 proteins. When cells were treated with p38 inhibitor, the levels of p21, Bax, and phosphorylated p53 were significantly reduced in Piwil2 knockdown cells (Fig. 6D). Once activated, p38 mitogen-activated protein kinase (MAPK) either phosphorylates cytoplasmic targets or translocates into the nucleus, leading to the regulation of transcription factors involved in cellular responses (43). For example, after activation by UV irradiation, p38 moves to the nucleus and phosphorylates p53 (42). In HeLa cells, p-p53 is colocalized with p-p38 in the nucleus (Fig. 6E). Although Piwil2 did not affect the total protein and phosphorylation level of p38 (Fig. 6D), we found that there was an increase in the level of p38 protein in the nucleus when Piwil2 was knocked down (Fig. 6F). Immunofluorescence results also showed that Piwil2 knockdown increased p-p53 fluorescence intensity in the nucleus (73% of transfected cells) (Fig. 6G) and the level of p-p38 nuclear translocation (76% of transfected cells) (Fig. 6H). These results demonstrated that phosphorylation of p53 protein increased after Piwil2 knockdown and suggested that p38 also plays a role as an upstream factor of p53 in the loss of Piwil2-induced apoptosis.
FIG 6.
Piwil2 inhibits apoptosis by impairing p53 phosphorylation. (A) Increased apoptosis in Piwil2 knockdown cells rescued by p38 inhibitor. NC (shNC) and Piwil2 knockdown (shPiwil2) cells were mock treated or treated with 5 μM SB203580 or 5 μM SB202190 for 6 h and then collected for flow cytometry using annexin double staining. Error bars indicate SE (*, P < 0.05). (B) Piwil2 suppresses interaction between p38 and p53. Cell lysates were prepared and subjected to IP assays. (C) Piwil2 overexpression rescued the increase of p53 in Piwil2 knockdown cells. (D) P38 is involved in Piwil2 knockdown-induced p53 phosphorylation. For p38 inhibition, HeLa cells were transfected with shNC or shPiwil2 vector and treated with 5 μM SB203580 or 5 μM SB202190 for 1 h as indicated. (E) Colocalization between p-p38 and p-p53. (F) The impact of Piwil2 knockdown on p38 nuclear localization was determined by Western blotting by cytosolic (C)/nuclear (N) fractionation followed by Western blotting. (G and H) Piwil2 knockdown increased p-p53 fluorescence intensity and p-p38 nuclear localization. HeLa cells were transfected with shNC or shPiwil2 vector and harvested for immunofluorescence assay with anti-p-p53 or anti-p-p38 antibodies. Scale bars, 5 μm.
DISCUSSION
Recently, more and more pieces of evidence have confirmed that Piwil2 is expressed in various tumors or cancer cell lines, playing important roles in tumorigenesis and protecting cells from apoptosis (11, 14, 36). Our previous results also indicate that Piwil2 inhibits apoptosis through TGF-β signaling in HEK293 cells and suppresses p53 in tumor cells (12, 13). Our current study shows that Piwil2 knockdown significantly increases epithelial cancer cell sensitivity to Fas-mediated apoptosis. The protection against Fas-mediated apoptosis of Piwil2 is partly due to K8. Keratins are the intermediate filament proteins of epithelial cells. K8/18 are found mainly in simple-type epithelia and protect cells from injury and apoptosis (23, 31, 44). K8 or K8/18 loss increases the membrane targets of Fas and the sensitivity to FasL and cisplatin (24, 37, 38). Although Piwil2 and keratin 8 both provide resistance to apoptosis, by now there is no definite evidence on their relationship. Our results indicate that Piwil2 promotes the accumulation of K8, reducing the localization of Fas at the membrane (Fig. 1C and D and 3E). The overexpression of K8 partly rescues the Fas-mediated apoptosis ratio of Piwil2 knockdown cells (Fig. 3F and G). We conclude that Piwil2 provides resistance to Fas-mediated apoptosis through the accumulation of K8.
Our research shows that Piwil2 promotes K8 stability by attenuating the ubiquitination and degradation of K8 (Fig. 4C). Although the E3 ligase associated with keratin intermediate filament protein degradation has not been identified, previous studies have shown that the phosphorylation of K8 protects it from ubiquitination and regulates its reorganization (34, 35). Many in vivo K8/18 phosphorylation kinases have been identified, such as p38 MAPK, JNK, and PKC δ (35, 39, 41). Our present study uncovers that Piwil2 enhances the phosphorylation of K8 to protect K8 from ubiquitination (Fig. 4E). Immunoprecipitation assays demonstrated that Piwil2 knockdown decreased the association of K8 and p38 (Fig. 5D). Previous work showed that p38 kinase interacted with K8 in vitro (35), and we confirmed this by immunoprecipitation in vitro and the two-hybrid experiment. We also find that Piwil2 directly binds to K8 and p38 (Fig. 4H). All these results suggest that Piwil2 is not necessary for the phosphorylation of K8 by p38, but it does promote it.
Many studies have shown that p38 activation is involved in apoptosis (45, 46). However, we find that Piwil2 does not activate p38 but regulates its combining with the substrate. Our previous research showed that Piwil2 repressed p53 expression by the c-src/STAT pathway, and in the present work we reveal that Piwil2 also inhibits p53 phosphorylation by p38. These kinases that phosphorylate K8 also activate membrane receptors, transcription factors, and other downstream protein kinases which regulate gene expression under various stimuli. For example, MAPK signaling pathways activate transcription factors (e.g., p53 and c-jun) and protein kinases (e.g., MK2 and MK3) directly or indirectly (47–49). The highly abundant K8 can protect tissue from injury by absorbing stress-activated kinases from untoward substrates (50). We also confirmed that overexpressed K8 could absorb cytoplasmic p38 to the K8 filaments (Fig. 4G, X5). Our results show that the overexpression of Piwil2 recruits p-p38 to K8 in the cytoplasm (Fig. 5E), while Piwil2 knockdown increases the formation of p38-p53 complexes and the nuclear translocation of p-p38 (Fig. 6B and H). Thus, Piwil2 prevents the phosphorylation of p53 induced by p38 and then inhibits p53, which is involved in apoptosis.
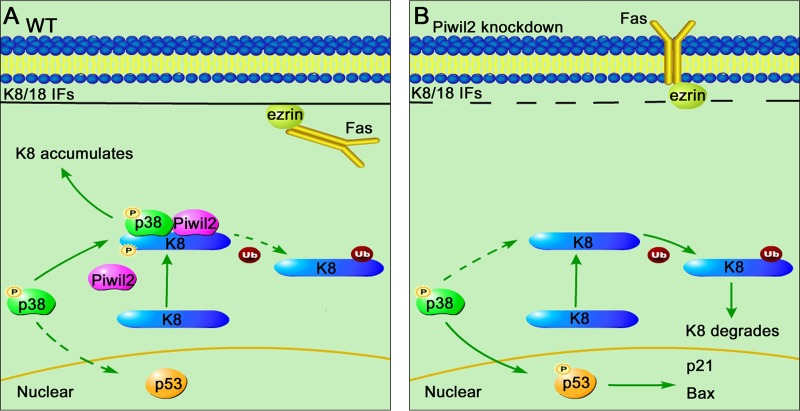
In summary, Piwil2 is a novel regulator of the Fas/FasL signaling pathway. Our research shows that Piwil2 recruits p38 to K8 and increases the phosphorylation of K8 induced by p38. Piwil2 loss decreases K8, increasing the membrane targeting of Fas and Fas-mediated apoptosis. Meanwhile, our results have demonstrated that Piwil2 inhibits p53 phosphorylation and p53-induced apoptosis through binding to p38 (Fig. 7). Considering that p38 controls a variety of signaling pathways, our research provides a new perspective on the study of the participation of PIWI proteins in regulating diverse types of signal transduction.
FIG 7.
Model depicting different patterns of p38 localization depending on the presence or absence of Piwil2. (A) In WT epithelial cancer cells (at least HeLa and HepG2), Piwil2 recruits p38 to K8, increases the phosphorylation of K8, and protects K8 from degradation. As a result, K8 accumulation protects cells from Fas-mediated apoptosis. Another effect of the K8/Piwil2/p38 complex is to decrease the interaction of p38 with p53. (B) While in Piwil2 knockdown cells, p38 phosphorylates p53 rather than K8, increasing FasL sensitivity and p53-induced apoptosis.
ACKNOWLEDGMENTS
We thank Yongqiu Mao for technical support in FACS analysis.
This work was supported by the National Basic Research Program of China (973 Program; 2012CB947600) and the National Natural Science Foundation of China (31070676, 90919006, and 31300961).
Y.M. and S.J. conceived and designed the experiments. S.J., L.Z., and Y.C. performed the experiments. S.J., Y. Lu, H.S., and M.W. analyzed the data. D.T., Y. Liu, and S.Z. contributed reagents/materials/analysis tools. S.J. wrote the paper. Y.M. and Y. Lu revised the manuscript.
Footnotes
Published ahead of print 11 August 2014
REFERENCES
- 1.Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. 1998. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12:3715–3727. 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farazi TA, Juranek SA, Tuschl T. 2008. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development 135:1201–1214. 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- 3.Houwing S, Berezikov E, Ketting RF. 2008. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 27:2702–2711. 10.1038/emboj.2008.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seto AG, Kingston RE, Lau NC. 2007. The coming of age for Piwi proteins. Mol. Cell 26:603–609. 10.1016/j.molcel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 5.Lee JH, Engel W, Nayernia K. 2006. Stem cell protein Piwil2 modulates expression of murine spermatogonial stem cell expressed genes. Mol. Reprod. Dev. 73:173–179. 10.1002/mrd.20391. [DOI] [PubMed] [Google Scholar]
- 6.Sun H, Li D, Chen S, Liu Y, Liao X, Deng W, Li N, Zeng M, Tao D, Ma Y. 2010. Zili inhibits transforming growth factor-beta signaling by interacting with Smad4. J. Biol. Chem. 285:4243–4250. 10.1074/jbc.M109.079533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smulders-Srinivasan TK, Szakmary A, Lin H. 2010. A Drosophila chromatin factor interacts with the Piwi-interacting RNA mechanism in niche cells to regulate germline stem cell self-renewal. Genetics 186:573–583. 10.1534/genetics.110.119081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szakmary A, Cox DN, Wang Z, Lin H. 2005. Regulatory relationship among piwi, pumilio, and bag-of-marbles in Drosophila germline stem cell self-renewal and differentiation. Curr. Biol. 15:171–178. 10.1016/j.cub.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Sasaki T, Shiohama A, Minoshima S, Shimizu N. 2003. Identification of eight members of the Argonaute family in the human genome small star, filled. Genomics 82:323–330. 10.1016/S0888-7543(03)00129-0. [DOI] [PubMed] [Google Scholar]
- 10.Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RH, Hannon GJ, Draper BW, Ketting RF. 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129:69–82. 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 11.Lee JH, Schutte D, Wulf G, Fuzesi L, Radzun HJ, Schweyer S, Engel W, Nayernia K. 2006. Stem-cell protein Piwil2 is widely expressed in tumors and inhibits apoptosis through activation of Stat3/Bcl-XL pathway. Hum. Mol. Genet. 15:201–211. 10.1093/hmg/ddi430. [DOI] [PubMed] [Google Scholar]
- 12.Lu Y, Zhang K, Li C, Yao Y, Tao D, Liu Y, Zhang S, Ma Y. 2012. Piwil2 suppresses p53 by inducing phosphorylation of signal transducer and activator of transcription 3 in tumor cells. PLoS One 7:e30999. 10.1371/journal.pone.0030999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang K, Lu Y, Yang P, Li C, Sun H, Tao D, Liu Y, Zhang S, Ma Y. 2012. HILI inhibits TGF-beta signaling by interacting with Hsp90 and promoting TbetaR degradation. PLoS One 7:e41973. 10.1371/journal.pone.0041973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JH, Jung C, Javadian-Elyaderani P, Schweyer S, Schutte D, Shoukier M, Karimi-Busheri F, Weinfeld M, Rasouli-Nia A, Hengstler JG, Mantilla A, Soleimanpour-Lichaei HR, Engel W, Robson CN, Nayernia K. 2010. Pathways of proliferation and antiapoptosis driven in breast cancer stem cells by stem cell protein piwil2. Cancer Res. 70:4569–4579. 10.1158/0008-5472.CAN-09-2670. [DOI] [PubMed] [Google Scholar]
- 15.Yin DT, Wang Q, Chen L, Liu MY, Han C, Yan Q, Shen R, He G, Duan W, Li JJ, Wani A, Gao JX. 2011. Germline stem cell gene PIWIL2 mediates DNA repair through relaxation of chromatin. PLoS One 6:e27154. 10.1371/journal.pone.0027154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang QE, Han C, Milum K, Wani AA. 2011. Stem cell protein Piwil2 modulates chromatin modifications upon cisplatin treatment. Mutat. Res. 708:59–68. 10.1016/j.mrfmmm.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagata S, Golstein P. 1995. The Fas death factor. Science 267:1449–1456. 10.1126/science.7533326. [DOI] [PubMed] [Google Scholar]
- 18.Muzio M. 1998. Signalling by proteolysis: death receptors induce apoptosis. Int. J. Clin. Lab. Res. 28:141–147. 10.1007/s005990050035. [DOI] [PubMed] [Google Scholar]
- 19.Reichmann E. 2002. The biological role of the Fas/FasL system during tumor formation and progression. Semin. Cancer Biol. 12:309–315. 10.1016/S1044-579X(02)00017-2. [DOI] [PubMed] [Google Scholar]
- 20.Kim S, Wong P, Coulombe PA. 2006. A keratin cytoskeletal protein regulates protein synthesis and epithelial cell growth. Nature 441:362–365. 10.1038/nature04659. [DOI] [PubMed] [Google Scholar]
- 21.Ku NO, Michie S, Resurreccion EZ, Broome RL, Omary MB. 2002. Keratin binding to 14-3-3 proteins modulates keratin filaments and hepatocyte mitotic progression. Proc. Natl. Acad. Sci. U. S. A. 99:4373–4378. 10.1073/pnas.072624299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inada H, Izawa I, Nishizawa M, Fujita E, Kiyono T, Takahashi T, Momoi T, Inagaki M. 2001. Keratin attenuates tumor necrosis factor-induced cytotoxicity through association with TRADD. J. Cell Biol. 155:415–426. 10.1083/jcb.200103078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ku NO, Toivola DM, Strnad P, Omary MB. 2010. Cytoskeletal keratin glycosylation protects epithelial tissue from injury. Nat. Cell Biol. 12:876–885. 10.1038/ncb2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert S, Loranger A, Daigle N, Marceau N. 2001. Simple epithelium keratins 8 and 18 provide resistance to Fas-mediated apoptosis. The protection occurs through a receptor-targeting modulation. J. Cell Biol. 154:763–773. 10.1083/jcb.200102130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long HA, Boczonadi V, McInroy L, Goldberg M, Maatta A. 2006. Periplakin-dependent re-organisation of keratin cytoskeleton and loss of collective migration in keratin-8-downregulated epithelial sheets. J. Cell Sci. 119:5147–5159. 10.1242/jcs.03304. [DOI] [PubMed] [Google Scholar]
- 26.Buhler H, Schaller G. 2005. Transfection of keratin 18 gene in human breast cancer cells causes induction of adhesion proteins and dramatic regression of malignancy in vitro and in vivo. Mol. Cancer Res. 3:365–371. 10.1158/1541-7786.MCR-04-0117. [DOI] [PubMed] [Google Scholar]
- 27.Schaller G, Fuchs I, Pritze W, Ebert A, Herbst H, Pantel K, Weitzel H, Lengyel E. 1996. Elevated keratin 18 protein expression indicates a favorable prognosis in patients with breast cancer. Clin. Cancer Res. 2:1879–1885. [PubMed] [Google Scholar]
- 28.Bianchi N, Depianto D, McGowan K, Gu C, Coulombe PA. 2005. Exploiting the keratin 17 gene promoter to visualize live cells in epithelial appendages of mice. Mol. Cell. Biol. 25:7249–7259. 10.1128/MCB.25.16.7249-7259.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oshima RG. 2002. Apoptosis and keratin intermediate filaments. Cell Death Differ. 9:486–492. 10.1038/sj.cdd.4400988. [DOI] [PubMed] [Google Scholar]
- 30.Alam CM, Silvander JS, Daniel EN, Tao GZ, Kvarnstrom SM, Alam P, Omary MB, Hanninen A, Toivola DM. 2013. Keratin 8 modulates beta-cell stress responses and normoglycaemia. J. Cell Sci. 126:5635–5644. 10.1242/jcs.132795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilbert S, Loranger A, Marceau N. 2004. Keratins modulate c-Flip/extracellular signal-regulated kinase 1 and 2 antiapoptotic signaling in simple epithelial cells. Mol. Cell. Biol. 24:7072–7081. 10.1128/MCB.24.16.7072-7081.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaitovich A, Mehta S, Na N, Ciechanover A, Goldman RD, Ridge KM. 2008. Ubiquitin-proteasome-mediated degradation of keratin intermediate filaments in mechanically stimulated A549 cells. J. Biol. Chem. 283:25348–25355. 10.1074/jbc.M801635200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ku NO, Omary MB. 1997. Phosphorylation of human keratin 8 in vivo at conserved head domain serine 23 and at epidermal growth factor-stimulated tail domain serine 431. J. Biol. Chem. 272:7556–7564. 10.1074/jbc.272.11.7556. [DOI] [PubMed] [Google Scholar]
- 34.Ku NO, Omary MB. 2000. Keratins turn over by ubiquitination in a phosphorylation-modulated fashion. J. Cell Biol. 149:547–552. 10.1083/jcb.149.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ku NO, Azhar S, Omary MB. 2002. Keratin 8 phosphorylation by p38 kinase regulates cellular keratin filament reorganization: modulation by a keratin 1-like disease causing mutation. J. Biol. Chem. 277:10775–10782. 10.1074/jbc.M107623200. [DOI] [PubMed] [Google Scholar]
- 36.Ye Y, Yin DT, Chen L, Zhou Q, Shen R, He G, Yan Q, Tong Z, Issekutz AC, Shapiro CL, Barsky SH, Lin H, Li JJ, Gao JX. 2010. Identification of Piwil2-like (PL2L) proteins that promote tumorigenesis. PLoS One 5:e13406. 10.1371/journal.pone.0013406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fortier AM, Asselin E, Cadrin M. 2013. Keratin 8 and 18 loss in epithelial cancer cells increases collective cell migration and cisplatin sensitivity through claudin1 up-regulation. J. Biol. Chem. 288:11555–11571. 10.1074/jbc.M112.428920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilbert S, Loranger A, Lavoie JN, Marceau N. 2012. Cytoskeleton keratin regulation of FasR signaling through modulation of actin/ezrin interplay at lipid rafts in hepatocytes. Apoptosis 17:880–894. 10.1007/s10495-012-0733-2. [DOI] [PubMed] [Google Scholar]
- 39.Ridge KM, Linz L, Flitney FW, Kuczmarski ER, Chou YH, Omary MB, Sznajder JI, Goldman RD. 2005. Keratin 8 phosphorylation by protein kinase C delta regulates shear stress-mediated disassembly of keratin intermediate filaments in alveolar epithelial cells. J. Biol. Chem. 280:30400–30405. 10.1074/jbc.M504239200. [DOI] [PubMed] [Google Scholar]
- 40.Ku NO, Omary MB. 1997. Phosphorylation of human keratin 8 in vivo at conserved head domain serine 23 and at epidermal growth factor-stimulated tail domain serine 431. J. Biol. Chem. 272:7556–7564. 10.1074/jbc.272.11.7556. [DOI] [PubMed] [Google Scholar]
- 41.Park MK, Lee HJ, Shin J, Noh M, Kim SY, Lee CH. 2011. Novel participation of transglutaminase-2 through c-Jun N-terminal kinase activation in sphingosylphosphorylcholine-induced keratin reorganization of PANC-1 cells. Biochim. Biophys. Acta 1811:1021–1029. 10.1016/j.bbalip.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 42.Bulavin DV, Saito S, Hollander MC, Sakaguchi K, Anderson CW, Appella E, Fornace AJ., Jr 1999. Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. EMBO J. 18:6845–6854. 10.1093/emboj/18.23.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koul HK, Pal M, Koul S. 2013. Role of p38 MAP kinase signal transduction in solid tumors. Genes Cancer 4:342–359. 10.1177/1947601913507951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tao GZ, Looi KS, Toivola DM, Strnad P, Zhou Q, Liao J, Wei Y, Habtezion A, Omary MB. 2009. Keratins modulate the shape and function of hepatocyte mitochondria: a mechanism for protection from apoptosis. J. Cell Sci. 122:3851–3855. 10.1242/jcs.051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farley N, Pedraza-Alva G, Serrano-Gomez D, Nagaleekar V, Aronshtam A, Krahl T, Thornton T, Rincon M. 2006. p38 mitogen-activated protein kinase mediates the Fas-induced mitochondrial death pathway in CD8+ T cells. Mol. Cell. Biol. 26:2118–2129. 10.1128/MCB.26.6.2118-2129.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merritt C, Enslen H, Diehl N, Conze D, Davis RJ, Rincon M. 2000. Activation of p38 mitogen-activated protein kinase in vivo selectively induces apoptosis of CD8(+) but not CD4(+) T cells. Mol. Cell. Biol. 20:936–946. 10.1128/MCB.20.3.936-946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ronkina N, Kotlyarov A, Dittrich-Breiholz O, Kracht M, Hitti E, Milarski K, Askew R, Marusic S, Lin LL, Gaestel M, Telliez JB. 2007. The mitogen-activated protein kinase (MAPK)-activated protein kinases MK2 and MK3 cooperate in stimulation of tumor necrosis factor biosynthesis and stabilization of p38 MAPK. Mol. Cell. Biol. 27:170–181. 10.1128/MCB.01456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwong J, Chen M, Lv D, Luo N, Su W, Xiang R, Sun P. 2013. Induction of p38delta expression plays an essential role in oncogenic ras-induced senescence. Mol. Cell. Biol. 33:3780–3794. 10.1128/MCB.00784-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tournier C, Whitmarsh AJ, Cavanagh J, Barrett T, Davis RJ. 1999. The MKK7 gene encodes a group of c-Jun NH2-terminal kinase kinases. Mol. Cell. Biol. 19:1569–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ku NO, Omary MB. 2006. A disease- and phosphorylation-related nonmechanical function for keratin 8. J. Cell Biol. 174:115–125. 10.1083/jcb.200602146. [DOI] [PMC free article] [PubMed] [Google Scholar]