Abstract
The death-associated protein kinase 1 (DAPK1) is an important regulator of cell death and autophagy. Recently, we have identified that ATF6, an endoplasmic reticulum-resident transcription factor, in association with the transcription factor CEBP-β, regulates the gamma interferon (IFN-γ)-induced expression of Dapk1 (P. Gade et al., Proc. Natl. Acad. Sci. U. S. A. 109:10316–10321, 2012, doi.org/10.1073/pnas.1119273109). IFN-γ-induced proteolytic processing of ATF6 and phosphorylation of C/EBP-β were essential for the formation of a novel transcriptional complex that regulates DAPK1. Here, we report that IFN-γ activates the ASK1-MKK3/MKK6–p38 mitogen-activated protein kinase (MAPK) pathway for controlling the activity of ATF6. The terminal enzyme in this pathway, p38 MAPK, phosphorylates a critical threonine residue in ATF6 upstream of its DNA binding domain. ATF6 mutants defective for p38 MAPK phosphorylation fail to undergo proteolytic processing in the Golgi apparatus and drive IFN-γ-induced gene expression and autophagy. We also show that mice lacking Ask1 are highly susceptible to lethal bacterial infection owing to defective autophagy. Together, these results identify a novel host defense pathway controlled by IFN-γ signaling.
INTRODUCTION
The interferon (IFN) family of cytokines regulates a broad range of biological responses against tumors and pathogens (1) primarily through stimulation of gene expression. Although the JAK-STAT pathway drives a major part of IFN actions, they can stimulate several genes in a STAT-independent manner (2). One example for this is the IFN-γ-induced expression of certain genes through C/EBP-β (3, 4). Additionally, while a great deal has been learned regarding IFN responses that control viral pathogens, relatively less is known about their antitumor mechanisms. DAPK1, a Ca2+/calmodulin-dependent serine/threonine kinase, is known to suppress tumor growth and metastasis by promoting apoptosis and autophagy (5). We recently defined a new motif in the proximal enhancer of DAPK1 through which C/EBP-β regulates transcription in response to IFN-γ (3), in collaboration with activated transcription factor-6 (ATF6), an endoplasmic reticulum (ER) stress regulator. IFN-γ-induced proteolytic processing of ATF6 and the mitogen-activated protein kinase (MAPK) ERK1/2-dependent phosphorylation of Thr189 residue on C/EBP-β were necessary for DAPK1 expression (6). Although proteolysis is critical for the activation of ATF6, it is unclear what other posttranslational modifications control its activity. We hypothesized that posttranslational modifications of ATF6, in addition to modifications on C/EBP-β, by MAP kinases are important for inducing DAPK1 transcription in response to IFN-γ.
A wide variety of intracellular stress conditions disrupt ER homeostasis and cause ATF6 activation (7). At steady state, ATF6 is localized as an ER membrane-anchored transmembrane glycoprotein (8). ER stress causes dissociation of ATF6 from its inhibitor, binding immunoglobulin protein (BiP), exposing the Golgi apparatus localization signals (GLS) of ATF6 and allowing its translocation to the Golgi apparatus (9). In the Golgi apparatus, site-specific endoproteolysis generates transcriptionally active ATF6, permitting its nuclear entry and target gene regulation (10). ER stress is associated with activation of stress-responsive MAPKs. Therefore, we searched for a kinase(s) that mediates this effect and identified the apoptosis-stimulating kinase 1 (ASK1) as a critical regulator of ATF6 in response to IFN-γ.
ASK1 (MAP3K5) is activated by various stress stimuli (11, 12). Although originally identified as a kinase that stimulates apoptosis, ASK1 also contributes to cytokine responses, cell differentiation, and immunity (11). We report, in this study, that IFN-γ activates ASK1-MKK3/MKK6-p38 MAPK for phosphorylating ATF6 at Thr166, which is required for its IFN-γ-induced proteolytic processing. The mutation of the ATF6 Thr166 residue led to retention of the protein in the Golgi apparatus and did not permit its nuclear entry to promote DAPK1 expression and drive autophagy. Consistent with the importance of Ask1 for DAPK1 expression, Ask1−/− mice exhibited defective autophagy and were highly sensitive to lethal bacterial infections. Together, these studies identified a novel pathway that controls the IFN-induced stress response that culminates in autophagy.
MATERIALS AND METHODS
Cell lines, antibodies, plasmids, and other reagents.
Isogenic wild-type (WT), Cebpb−/− Ask1−/− (13), Mkk3−/−, and Atf6−/− mouse embryonic fibroblasts (MEFs) were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum and 1% antibiotic-antimycotic agents as described earlier (6). BEAS-2B, a human bronchial epithelial cell line, was cultured as reported earlier (14). SF9 and SF21 cells were cultured in SF900II medium according to the supplier's instructions (Life Technologies, NY). Human or mouse IFN-γ (PBL Inc., Piscataway, NJ) was used at 500 U/ml, as described in our earlier studies (3, 6). Site-directed mutagenesis of human ATF6 was performed to obtain mutants S16A, S104A, T166A, and T166D using specific primers (Table 1) to express as an N-terminal hemagglutinin (HA)-tagged protein from lentiviral vector pLVX-puro (ATF6 full length) and pTriEx 1.1 Hygro (ATF6-373) (Novagen, Millipore, Billerica, MA). Sequence- and expression-verified mutants and wild-type ATF6 were used in all experiments. Mutants in the context of full-length ATF6 were employed unless mentioned otherwise. Protease-resistant ATF6 mutant lacking the site 1 protease (S1P) cleavage site (HA-ATF6-mut) and C/EBP-β expression vectors were described earlier (3). HA-tagged ASK1 wild-type and kinase-inactive mutant (K709R) constructs were gifts from Hidenori Ichijo (University of Tokyo) (15). Flag-tagged active and inactive mutant constructs of MKK3 were obtained from Addgene (Cambridge, MA) (16). Rabbit antibodies against ATF6 and C/EBP-β (Santa Cruz Biotechnology, Dallas, TX) and rabbit phosphopeptide antibodies against ATF6 T166 were custom developed at GenScript (Piscataway, NJ) using the synthetic peptide PRNKTENGL[pT]PKKK. In these studies, we used rabbit antibodies specific for BiP and actin and mouse monoclonal antibodies specific for DAPK1 (Sigma-Aldrich, St. Louis, MO), fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG, anti-rabbit IgG (Invitrogen, Carlsbad, CA), FLAG epitope, HA epitope, and LC3 (Cell Signaling Technology Danvers, MA). LY294002 (a selective inhibitor of phosphatidylinositol 3 [PI3]-kinase) was from Cell Signaling Technology (Danvers, MA). Chloroquine (an inhibitor of autophagosome and lysosome fusion), SB202190 (potent cell-permeable inhibitor of p38 MAPK), tunicamycin (TM; an inhibitor of protein glycosylation and an inducer of ER stress), and all other reagents were from Sigma-Aldrich, St. Louis, MO. DNA transfection was performed as described in our earlier studies (3, 6). To avoid potential artifacts due to overexpression, all DNA transfection studies were performed with transgene expression levels corresponding to endogenous levels.
TABLE 1.
Oligonucleotides used for site-directed mutagenesis of ATF6
| Construct | Primer name | Primer sequencea (5′→3′) |
|---|---|---|
| S16A | Hs-ATF6-SDM-Fwd1 | AGTCACCTTTTGCCCCGGGACTCTTTCACAGG |
| Hs-ATF6-SDM-Rev1 | GAGTCCCGGGGCAAAAGGTGACTCCATGGTGCC | |
| S104A | Hs-ATF6-SDM-Fwd3 | CAGTCTCAGCTCCTCGGTCAGTGGAC |
| Hs-ATF6-SDM-Rev3 | CCGAGGAGCTGAGACTGAATAACTTGAGG | |
| T166A | Hs-ATF6-SDM-Fwd4 | GAAAATGGACTGGCTCCAAAGAAAAAAATTCAGG |
| Hs-ATF6-SDM-Rev4 | CTTTGGAGCCAGTCCATTTTCAGTCTTGTTCC | |
| T166D | Hs-ATF6-SDM-Fwd5 | GAAAATGGACTG GATCCAAAGAAAAAAATTCAGG |
| Hs-ATF6-SDM-Rev5 | CTTTGGATCCAG TCCATTTTCAGTCTTGTTCC |
Primer sequences were used to generate site-specific ATF6 mutants by PCR. The boldfaced nucleotides designate the specific mutations.
RNA interference (RNAi)-mediated knockdown.
Lentiviral vectors carrying short hairpin RNAs (shRNAs) specific for human p38 MAPK (Table 2) were purchased from Open Biosystems (Huntsville, AL). To deplete all known p38 MAPK isoforms (α, β, γ, and δ), we pooled lentiviral particles carrying target-specific shRNAs to transduce the cells (3). We have employed an empty vector and scrambled shRNA as controls to ascertain the specificity of targeting in all experiments. The depletion of the target gene product was confirmed by performing Western blot analysis of total cell extracts with the indicated antibodies.
TABLE 2.
p38 MAPK isoform-specific shRNAs used in this studya
| p38 MAPK isoform | shRNA sequenceb (5′→3′) |
|---|---|
| α | TGCTGTTGACAGTGAGCGACCGAGCTGTTGACTGGAAGAATAGTGAAGCCACAGA |
| TGTATTCTTCCAGTCAACAGCTCGGCTGCCTACTGCCTCGGA | |
| β | TGCTGTTGACAGTGAGCGCGCCGTTCAATTCCTGGTTTATAGTGAAGCCACAGA |
| TGTATAAACCAGGAATTGAACGTGCTTGCCTACTGCCTCGGA | |
| γ | TGCTGTTGACAGTGAGCGACCTGGATGACTTCACGGACTTTAGTGAAGCCACA |
| GATGTAAAGTCCGTGAAGTCATCCAGGGTGCCTACTGCCTCGGA | |
| δ | TGCTGTTGACAGTGAGCGCCCAGCTGACCCAGATCCTGAATAGTGAAGCCACAGA |
| TGTATTCAGGATCTGGGTCAGCTGGTTGCCTACTGCCTCGGA |
Human p38 MAPK isoform-specific shRNAs are shown.
The boldfaced nucleotides represent the complementary nucleotides of the target.
Reporter gene assays and qPCR.
Reporter gene assays were performed as described earlier (3, 6). After the indicated treatments, luciferase activity was measured and normalized to that of β-galactosidase. Triplicates were used for each sample, and each experiment was repeated thrice. Real-time quantitative PCR (qPCR) was performed by using specific primers as described in our earlier studies (3). The mRNA for ribosomal protein L32 (Rpl32) was used as an internal control. The mean relative abundance values were calculated using the ΔΔCT method (where CT is threshold cycle) as described earlier (3). Triplicate reactions were run for each sample, and each experiment was repeated at least three times with independent preparations of RNA. Statistical significance of the differences was determined using the Student t test, where a P <0.05 was considered significant.
ChIP assays.
Standard chromatin immunoprecipitation (ChIP) assays were performed, using a commercially available kit from Millipore (Billerica, MA), with minor modifications as described in our previous studies (3). The DNA subjected to ChIP was detected by PCR or qPCR (3, 6). DNA from total soluble chromatin was used as a template for PCR for determining input levels in each case. Nonreactive IgG (NR-IgG), no IgG (No-IgG), and specific IgG antibodies were used at 10 μg each per reaction. The former two served as negative controls for ChIP reactions and defined the baseline for all reactions that employed a specific IgG.
Immunoprecipitation and Western blot analysis.
Immunoprecipitation analyses were conducted as described in our earlier studies (3). In brief, 400 μg of total cellular lysate was incubated with 3 μg of the indicated antibody overnight at 4°C and then incubated with protein G-agarose (Santa Cruz) for 2 h at 4°C. Beads were washed, and bound proteins were resolved on 10 to 15% SDS-PAGE and transferred to polyvinylidene difluoride membrane (Millipore, Billerica, MA) for Western blot analyses with the indicated primary antibodies (1:1,000 dilution) overnight at 4°C. They were washed and probed with either anti-mouse or anti-rabbit Alexa Fluor-tagged secondary antibodies (1:2,000 dilution) for 1 h at room temperature. Blots were imaged using the LiCor Odyssey infrared imaging system as described previously (6). Mean intensities of the appropriate bands were quantified using the software provided by the manufacturer.
Expression and purification of ATF6 protein.
N-terminal HA-tagged ATF6-373 WT and mutant constructs were cloned into BamHI and XhoI sites of pTriEx 1.1 Hygro vector. Baculovirus-mediated expression of these constructs in SF9 and SF21 insect cells was performed using the BacVector 3000 transfection kit (Novagen EMD-Millipore, Billerica, MA) per the manufacturer's recommendations. The expressed recombinant proteins were purified by HA-immunoaffinity column chromatography (17). The eluates then were passed through Amicon filters (30-kDa cutoff) to remove any small-molecular-size contaminants. The purity of the protein preparation was confirmed by silver staining per the manufacturer's instructions (Silver Stain Plus reagent; Bio-Rad Laboratories, CA). Aliquots of the purified protein were stored in phosphate-buffered saline (PBS) containing 20% glycerol at −80°C until further use.
Kinase assays.
The purified recombinant ATF6 proteins were incubated with 1 μg of purified active recombinant p38α (Proqinase GmbH, Germany), and the kinase assays were initiated by the addition of 30 μl of kinase buffer containing 1.85 MBq (50 μCi) of [γ-32P]ATP. After incubating for 30 min at 30°C, the reactions were stopped by the addition of SDS sample loading buffer, and the products were separated using SDS-PAGE followed by phosphor imaging (Molecular Dynamics, Inc., Sunnyvale, CA) of the dried gel. In some experiments, p38α was preincubated for 30 min with different concentrations of SB202190 (p38 MAPK-specific inhibitor) before ATF6 was included. Radioactive counts in each band were quantified (ImageQuant software; GE Healthcare Biosciences, Pittsburgh, PA) from 3 separate experiments and plotted. The IFN-γ-induced phosphorylation of native ATF6 by p38 in cultured cells was determined after subjecting the lysates to Western blot analysis with a custom-generated rabbit anti-ATF6 phospho-T166-specific antibody.
Immunofluorescence microscopy.
The immunofluorescence microscopy analyses were conducted as described earlier (6). Anti-ATF6 (1:100), anti-HA (1:100), anti-phospho-p38-MAPK (1:100), anti-GM130 (1:100), or anti-LC3 (1:100) was used as the primary antibody, and Alexa Fluor-conjugated anti-rabbit or anti-mouse IgG (1:50) was used as the secondary antibody. Images were captured using an Olympus fluorescence microscope (BX40–F4) fitted with a QICAM digital camera and were processed with Q-Capture Pro-5.1 (Q Imaging). Mean intensities for ROI (regions of interest) were quantified using the NIH ImageJ software as described in our earlier studies (6).
Cell fractionation.
The rapid, efficient, and practical (REAP) subcellular fractionation method (18) was employed. Briefly, cell pellets were suspended in ice-cold 0.1% NP-40 in PBS and centrifuged for 10 s. The supernatant was removed and collected as the extranuclear fraction. The pellet was resuspended in ice-cold 0.1% NP-40 in PBS, sonicated twice for 5 s each, and labeled as the nuclear fraction. The isolated nuclear and extranuclear fractions were resolved using SDS-PAGE and subjected to Western blot analysis.
Bacterial infection.
Six- to eight-week-old Ask1+/+ and Ask1−/− mice on a C57/BL6 background (13) were used for these studies. Mice were housed at the University of Maryland, Baltimore, animal facility, and all animal experimentation was approved by the Institutional Animal Care and Use Committee. For survival studies, mice (n = 22 per group) were infected intraperitoneally with 1 × 106 Bacillus anthracis Sterne 34F2 spores (this dose is sublethal for wild-type mice) and observed for mortality over the next 9 days (19). In separate experiments, spleen and liver were harvested from Ask1+/+ and Ask1−/− mice (n = 6) at 96 h postinfection for bacterial load analysis. Peritoneal macrophages were collected from these mice and scored for the differences in LC3 puncta formation using immunofluorescence.
Statistical analyses.
Student's t test was performed on the data, and P values are indicated as appropriate.
RESULTS
IFN-γ-stimulated DAPK1 expression requires p38 MAPK.
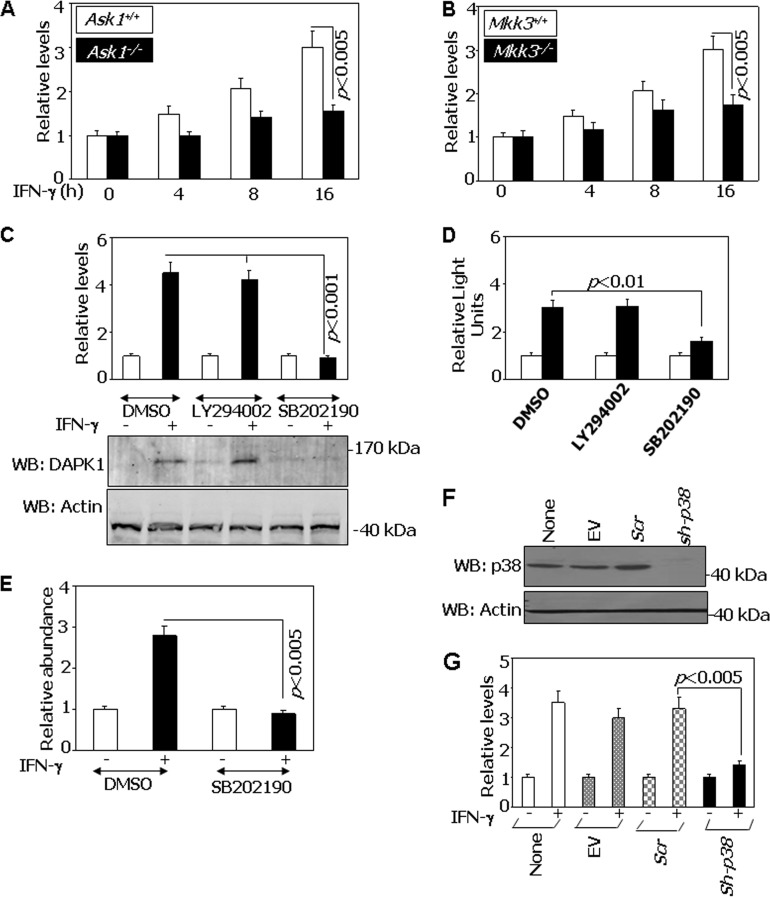
Since p38 is a stress-responsive MAPK and ATF6 participates in ER stress, we first investigated if inhibition of p38 MAPK would have an effect on IFN-γ-stimulated DAPK1 expression. Because p38 activation is dependent on upstream kinases and ASK1 is recruited to one of the ER stress-associated proteins, IRE1, that has been implicated in stimulating another stress-responsive kinase, JNK1/2 (20), we analyzed the expression of Dapk1 mRNA in Ask1+/+ and Ask1−/− mouse embryonic fibroblasts (MEFs). IFN-γ-stimulated Dapk1 mRNA upregulation did not occur in the absence of Ask1 (Fig. 1A). As Mkk3 functions downstream of many MAP3Ks, we also performed a similar experiment in Mkk3−/− MEFs and compared it to control Mkk3+/+ MEFs. Similar to the case for Ask1, the absence of Mkk3 could not upregulate Dapk1 expression upon IFN-γ stimulation (Fig. 1B).
FIG 1.
IFN-γ-induced DAPK1 expression requires p38 MAPK. (A) qPCR analysis of IFN-induced expression of Dapk1 mRNA in Ask1+/+ and Ask1−/− MEFs with gene-specific primers. Cells were treated with IFN-γ (500 U/ml) for the indicated times. Relative levels were calculated with respect to untreated cell values, which were considered 1. (B) qPCR analysis of IFN-induced expression of Dapk1 mRNA in Mkk3+/+ and Mkk3−/− MEFs with gene-specific primers. Relative levels were calculated as described for panel A. (C) Effect of inhibitors on the IFN-γ-induced expression of Dapk1. Wild-type MEFs were treated with IFN-γ in the presence of the indicated inhibitors (SB202190 at 20 μM or LY294002 at 50 μM). qPCR analysis was performed as described for panel A. The bottom panel shows protein levels analyzed after Western blotting (WB) with the indicated antibodies. Open bars, no treatment; filled bars, IFN-γ. (D) Effect of p38 MAPK inhibition on Dapk1 promoter activity. Wild-type MEFs were transfected with the mDapk1.2K-Luc construct (300 ng) along with a β-actin–β-galactosidase reporter (100 ng) and were left untreated (open bars) or were treated with IFN-γ (filled bars). Normalized luciferase activity was plotted as relative light units. (E) qPCR analysis of Dapk1 promoter fragments recovered in ChIP assays with specific primers that can detect the ATF6 binding region. Chromatin was prepared from wild-type MEFs after various treatments and subjected to IP with ATF6, NR-IgG, and No-IgG antibodies. The specific PCR products were normalized to the value for the input across the samples, and the relative levels were calculated with respect to the NR-IgG antibody (n = 6 per sample). (F) p38 MAPK knockdown in BEAS-2B cell lines. Western blot analysis of p38 MAPK was performed using the indicated specific antibodies. EV, empty vector (pLKO1); Scr, pLKO1 vector carrying scrambled shRNA sequences; sh-p38, shRNAs that can target α, β, γ, and δ forms of p38 MAPK expressed from the pLKO1 vector. (G) Effect of p38 MAPK knockdown on the expression of Dapk1 mRNA in the BEAS-2B cell line. After infecting cells with lentiviral vectors carrying the indicated shRNAs, cells were treated with IFN-γ for 16 h. RNA was prepared and a qPCR analysis of Dapk1 transcripts was performed.
The requirement of p38 MAPK for IFN-γ-induced expression of Dapk1 was further ascertained by using chemical inhibitors and RNAi. We first determined the effects of SB202190 (a specific inhibitor of p38 MAPK activity) and an unrelated inhibitor, LY294002 (blocks the PI3-kinase pathways), as a control. Real-time PCR (qPCR) analysis of wild-type MEFs treated with these inhibitors revealed that only SB202190 was capable of blocking Dapk1 mRNA expression (Fig. 1C). Western blot analysis of Dapk1 also mirrored the RNA levels (Fig. 1C, bottom). In the next experiment, wild-type MEFs were transfected with mDapk-1.2K-Luc construct and then stimulated with IFN-γ in the presence of these inhibitors. SB202190, but not LY294002, blocked IFN-γ-induced expression of Dapk1 promoter-driven luciferase (Fig. 1D). Consistent with these results, ChIP analyses showed that IFN-γ-stimulated recruitment of Atf6 to the Dapk1 promoter is ablated only in the presence of SB202190 (Fig. 1E). The control ChIP reactions performed by nonreactive IgG did not yield any significant products.
The importance of p38 MAPK for IFN-γ-induced expression of DAPK1 was further assessed by the following experiments. BEAS-2B cells (a nononcogenic immortalized human bronchial epithelial cell line) were infected with lentiviral vectors coding for control or specific shRNAs that could target p38 MAPK. Western blot analysis of cellular lysates demonstrated a >95% depletion of p38 MAPK compared to the controls in this experiment (Fig. 1F). DAPK1 mRNA was not induced by IFN-γ when p38 MAPK was depleted (Fig. 1G). As expected, its induction was unaffected in empty vector and scrambled shRNA-transfected cells. Similar results were obtained when p38 MAPK was depleted from MEFs using RNAi (see Fig. S1A and B in the supplemental material). Thus, p38 MAPK upregulates DAPK1 expression upon IFN-γ stimulation.
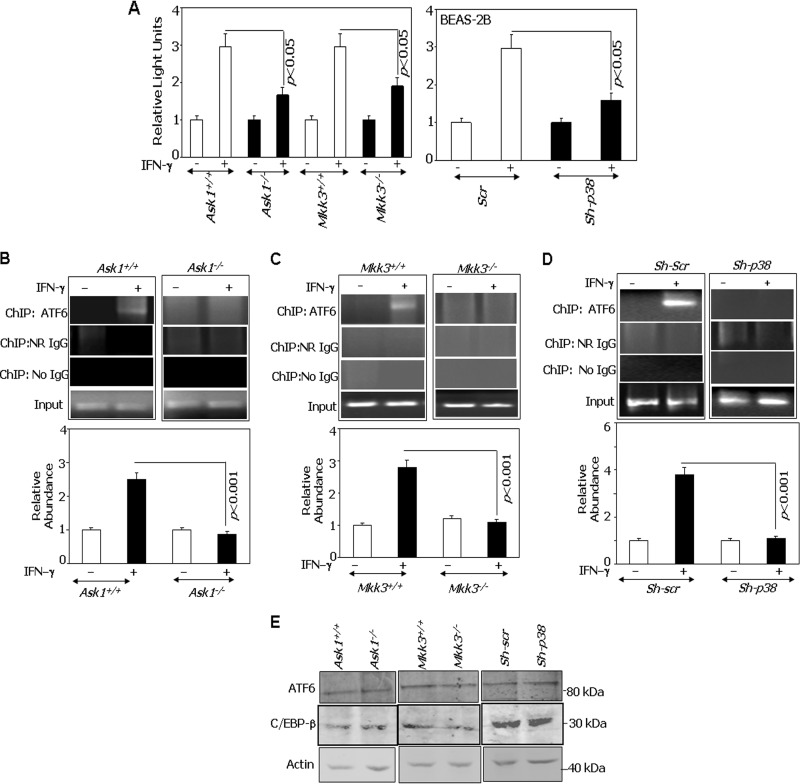
ASK1 is critical for IFN-γ-induced DAPK1 via ATF6 recruitment.
Because ASK1 has been implicated in the ER stress response, we next tested if the loss of this stress-regulated kinase and its downstream components affected DAPK1 expression. Therefore, Ask1+/+, Ask1−/−, Mkk3+/+, Mkk3−/−, wild-type (Scr), and p38 MAPK-depleted (sh-p38) cells were transfected with the Dapk-1.2K-Luc construct, and its response to IFN-γ was measured (Fig. 2A). In each case, the expression of luciferase was induced by IFN-γ only in wild-type cells, while expression was slightly above the baseline in Ask1−/−, Mkk3−/−, and sh-p38 cells. Thus, the Ask1-Mkk3/p38 signaling axis is needed for IFN-γ-induced expression of Dapk1. Since ATF6 is required for Dapk1 expression during stress responses, we next used ChIP assays to determine if ATF6 was recruited to the Dapk1 promoter in response to IFN-γ. In Ask1+/+ MEFs, Atf6 did not bind to the Dapk1 promoter in the unstimulated state. However, IFN-γ treatment induced Atf6 recruitment to the Dapk1 promoter (Fig. 2B). In contrast, Atf6 did not bind to the Dapk1 promoter in Ask1−/− MEFs despite IFN-γ stimulation. A qPCR analysis of the ChIP products showed dramatic differences in ATF6 binding between Ask1+/+ and Ask1−/− MEFs (Fig. 2B, bottom). A similar ChIP experiment was conducted using Mkk3+/+ and Mkk3−/− MEFs. IFN-γ induced the recruitment of ATF6 to the Dapk1 promoter in Mkk3+/+ but not in Mkk3−/− cells (Fig. 2C). As observed with Ask1−/− cells, qPCR analysis of ChIP products indicated an extremely reduced binding of ATF6 to the Dapk1 promoter in Mkk3−/− cells (Fig. 2C, bottom). Similarly, RNAi-mediated depletion of p38 MAPK in BEAS-2B (sh-p38) prevented the IFN-γ-induced ATF6 recruitment to the DAPK1 promoter (Fig. 2D). The No-IgG and NR-IgG controls did not yield any detectable products in either the presence or absence of IFN-γ, confirming the specificity of the binding. Importantly, the difference in the promoter occupancy in these cell lines is not due to lower levels of ATF6 and/or C/EBP-β (Fig. 2E).
FIG 2.
ATF6 is a target of ASK1 in the IFN-induced pathway. (A) The indicated cell lines were transfected with Dapk-1.2K-Luc (300 ng) along with a β-actin–β-galactosidase reporter (100 ng). Cells were treated with IFN-γ, where indicated, and luciferase activity was measured as described in the legend to Fig. 1. Normalized luciferase activity was plotted as relative light units. (B to D) ChIP assays were performed in the indicated cell types as described in the legend to Fig. 1. IFN-γ treatment was for 12 h. Soluble chromatin was used as the input control in each case. The typical PCR patterns after ChIP are shown. (B to D, bottom) qPCR analyses of the ChIP products as described in the legend to Fig. 1. The specific IP products were quantified, and the values were plotted as relative abundance with respect to the NR-IgG control (n = 6 per sample). (E) Western blot analyses of lysates with the indicated antibodies. Specific genotypes of cells used in the experiment are indicated.
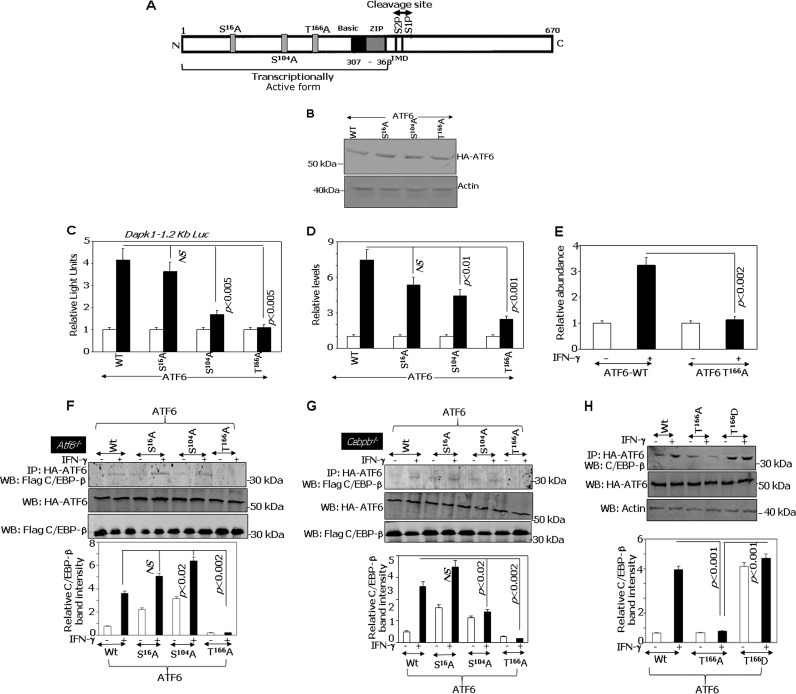
Mutation of putative p38 MAPK phosphorylation sites in ATF6 suppresses its transcriptional induction of DAPK1.
Based on the results described above, we next mapped the targets of ASK1 on ATF6 protein. Sequence analysis suggested 4 putative p38 MAPK phosphorylation sites in ATF6, and we have generated 3 phosphorylation-acceptor site mutants of ATF6, namely, S16A, S104A, and T166A (Fig. 3A). The WT and mutant ATF6 constructs expressed at similar levels, showing that any differences in their activity were not due to their expression levels (Fig. 3B). To understand their functional importance, we performed the following experiments in Atf6−/− MEFs. WT or mutant ATF6 constructs were coexpressed with Dapk-1.2K-Luc, and the impact on IFN-γ-induced Dapk1 expression was examined. Both WT and S16A proteins supported the IFN-γ-induced expression of the Luc reporter equivalently. The T166A mutant completely failed to promote IFN-γ-induced expression of the Dapk1-Luc reporter (Fig. 3C). The S104A mutant significantly lost its capacity compared to that of wild-type ATF6, but it still retained some capacity to drive IFN-γ-induced transcription. These mutants exerted a similar effect on the expression of Dapk1 mRNA (Fig. 3D). Since the T166A mutant completely lost its capacity to induce Dapk1 expression, we used it for subsequent studies.
FIG 3.
Mutation of a p38 MAPK phosphorylation site in ATF6 suppresses transcriptional induction of DAPK1. (A) Cartoon showing ATF6 structure. Putative evolutionarily conserved p38 MAPK phosphorylation sites on ATF6 identified by sequence analysis were indicated along with other important elements. TMD, transmembrane domain; S1P and S2P, proteolytic cleavage sites; N, N terminus; C, C terminus. (B) Western blot analysis of the expression levels of the indicated constructs. (C) Atf6−/− MEFs were transfected with the mDapk1.2K-Luc construct (300 ng) and a β-actin–β-galactosidase reporter (100 ng), along with the indicated ATF6 constructs. Normalized luciferase activity was plotted as relative light units. Each bar represents the means ± standard errors (SE) from triplicates. Open and filled bars correspond to no treatment and IFN-γ treatment, respectively. (D) qPCR analysis of IFN-induced expression of endogenous Dapk1 mRNA in Atf6−/− MEFs in the presence of the indicated ATF6 phosphorylation site mutants. (E) qPCR analyses of the ChIP products was performed in Atf6−/− MEFs in the presence of the indicated ATF6 constructs. The specific IP products were plotted as relative abundance with respect to the NR-IgG (n = 6 per sample). (F and G) Mutation of T166 inhibits IFN-induced interaction of ATF6 with C/EBP-β. For Flag–C/EBP-β interaction, Atf6−/− (F) and Cebpb−/− (G) MEFs transfected with the indicated ATF6 constructs were stimulated with IFN-γ or were left unstimulated. Histograms in the lower part of these panels show the relative intensities of the immunoprecipitated bands after normalization to the expression levels. (H) MEFs transfected with the indicated ATF6 constructs were stimulated with IFN-γ or left unstimulated. Histograms in the lower part of these panels show the relative intensities of the immunoprecipitated bands after normalization to the expression levels.
To further demonstrate the importance of T166 to ATF6 function, WT or T166A constructs were transfected into Atf6−/− MEFs, and we compared their relative capacities to be recruited to the Dapk1 promoter following IFN-γ stimulation using quantitative ChIP PCR. The IFN-γ-induced recruitment of ATF6 to the Dapk1 promoter was completely abolished upon mutating the T166 residue (Fig. 3E).
The T166 residue of ATF6 is required for IFN-γ-induced interaction with C/EBP-β.
We have previously reported that IFN-γ induces the interaction of ATF6 with C/EBP-β (6) for stimulating DAPK1 expression. To determine if the disruption of the T166 site affected the IFN-γ-induced interaction of ATF6 with C/EBP-β, we expressed HA-tagged ATF6-373 WT or the S16A, S104A, and T166A mutants along with Flag–C/EBP-β into Atf6−/− MEFs and performed immunoprecipitation with the indicated antibodies (Fig. 3F). IFN-γ-stimulated interaction of C/EBP-β with ATF6 T166A was not evident compared to those of ATF6 WT or S16A and S104A mutants. The middle blots in Fig. 3F show that these differences are not due to the differential expression of either ATF6 WT-373 and mutants or C/EBP-β. The relative intensities of the immunoprecipitated bands are shown in the graph in Fig. 3F. Thus, ATF6 T166 is necessary for its interaction with C/EBP-β in response to IFN-γ. We performed a similar experiment in Cebpb−/− MEFs and observed patterns of interactions similar to those found in Atf6−/− MEFs (Fig. 3G). The upper blot in Fig. 3G shows that IFN-γ-stimulated binding of T166A with C/EBP-β was significantly reduced compared to that of the WT. The middle blot shows that these differences are not due to the differences in the expression levels of either ATF6-373 WT and mutants or C/EBP-β. The relative levels of C/EBP-β immunoprecipitated with ATF6 were plotted (Fig. 3G, lower). Since the IFN-γ-stimulated interaction of T166A with C/EBP-β did not increase, we next tested if the T166D mutant would interact with C/EBP-β better than the wild type. Indeed, the T166D mutant interacted more strongly with the endogenous C/EBP-β even at steady state, and IFN-γ stimulation did not increase it further when immunoprecipitated with HA-ATF6 (Fig. 3H). The middle blot in Fig. 3H shows that these differences are not due to a differential expression of the mutants. The relative levels of C/EBP-β immunoprecipitated with ATF6 were plotted (Fig. 3H, lower).
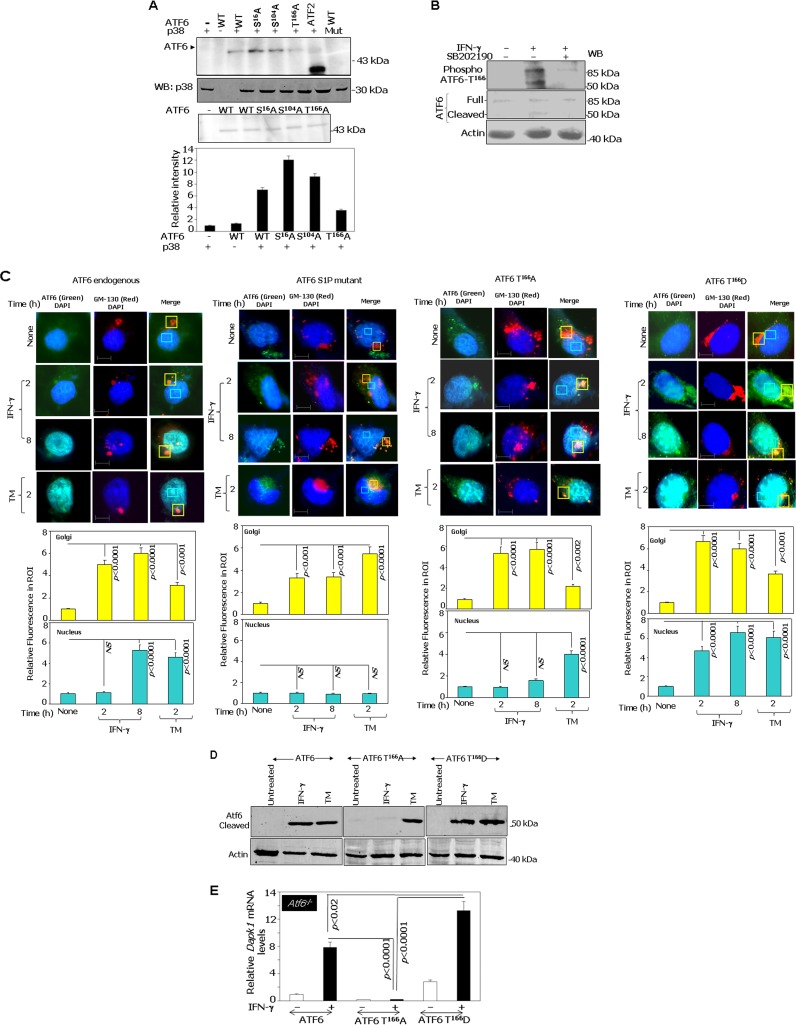
p38 MAPK phosphorylates ATF6.
To directly test if activated p38α (a major form of p38 MAPK) can phosphorylate ATF6, we performed in vitro phosphorylation assays. The WT and S16A, S104A, and T166A mutants of ATF6-373 (active form) were expressed using a baculoviral system and purified using immunoaffinity chromatography with HA tag-specific antibodies as described in Materials and Methods. Glutathione S-transferase (GST)–ATF2, a known substrate of p38α, was used as a positive control in these experiments. Upon incubation with purified active recombinant p38α and [γ-32P]ATP, the WT, S16A, and S104A ATF6 proteins were phosphorylated (Fig. 4A). As expected, 32P incorporation was not observed when WT ATF6 was incubated with an inactive p38α. However, when the T166A mutant was used as a substrate, significant incorporation of 32P comparable to that of WT protein was not observed. Thus, the T166 site appears to be a direct phosphorylation target of p38 MAPK (Fig. 4A). We have ensured the specificity of p38 in phosphorylating ATF6 not only by incubating different concentrations of the purified active p38α with the WT ATF6 but also by performing these assays in the presence of different concentrations of SB202190 (see Fig. S2A and B in the supplemental material). p38α phosphorylated ATF6 in a concentration-dependent manner. While ATF6 phosphorylation was readily detected at 1 μg and 0.5 μg of p38α, it was not detected at 0.1 μg. SB202190 blocked 32P incorporation into ATF6 in a concentration-dependent manner. The lower degree of 32P incorporation into ATF6 versus ATF2 was due to the differences in the concentrations of the substrates. The amount of ATF2 protein used was approximately 10-fold higher than that for ATF6 protein. More importantly, the nature of these two substrates is different. For example, the ATF2 used in this assay was a GST fusion peptide containing the N-terminal amino acids 19 to 96 of the human ATF2 protein containing the p38 target region, whereas the ATF6 protein used was the native 50-kDa version. To provide additional support for T166 as a target of p38 MAPK, we treated MEFs with IFN-γ in the presence or absence of SB202190. Whole-cell lysates were prepared and Western blotted with a custom-generated phosphopeptide-specific antibody (PRNKTENGL[pT]PKKK) that detects only the phosphorylated T166 site of ATF6 (Fig. 4B). We have ensured the phospho-T166 specificity of the antibody (see Fig. S3 in the supplemental material). The preincubation of the antibody with the blocking peptide (phosphopeptide used for raising antibodies) but not an unphosphorylated or a random peptide (generated by the scrambling of the sequence) resulted in the elimination of signal to background levels. Figure 4B, lower portion, shows ATF6 (full-length and processed forms) and actin levels. Interestingly, these analyses revealed that IFN-γ-induced phosphorylation and cleavage of ATF6 is suppressed in the presence of the inhibitor SB202190. This observation not only confirms the specific role for IFN-γ-induced p38 MAPK-dependent phosphorylation of ATF6 at the T166 site but also indicates a connection between phosphorylation and proteolytic activation.
FIG 4.
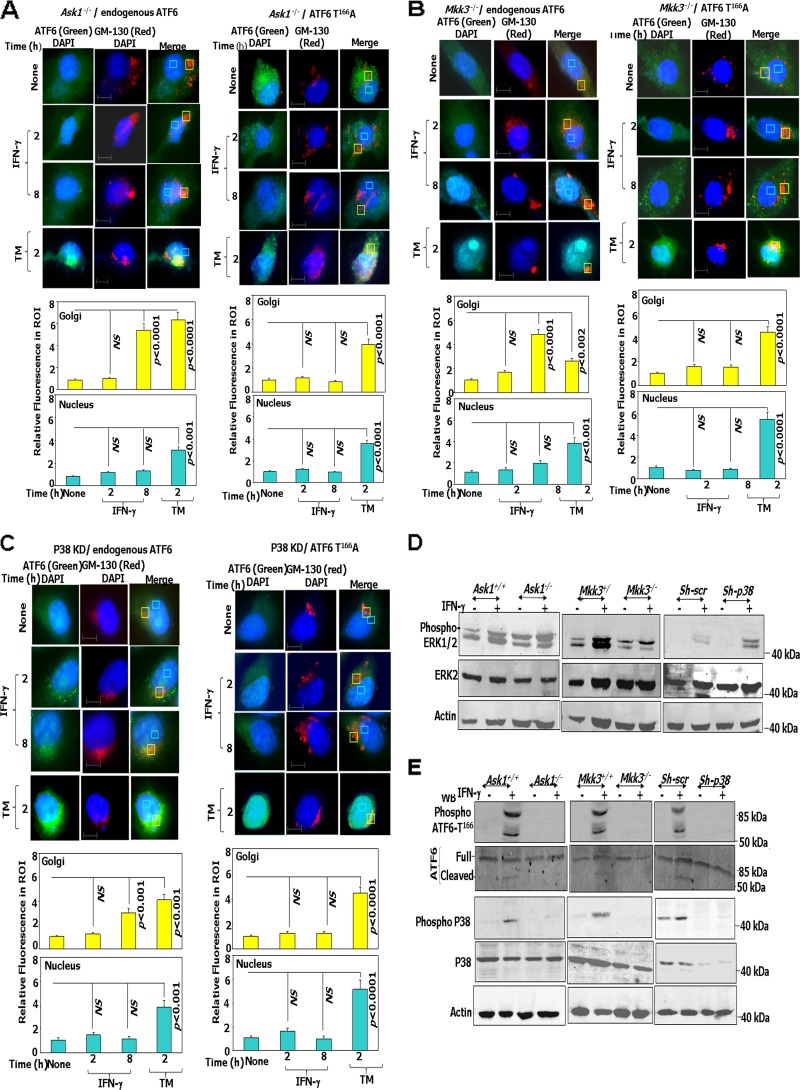
ATF6 is phosphorylated by p38 MAPK in response to IFN-γ and mediates its interaction with C/EBP-β and DAPK1 expression. (A) In vitro kinase assay. Purified recombinant ATF6-373 WT and the indicated phosphorylation site-defective mutants were used as substrates for p38 MAPK. The upper panel shows a representative autoradiograph of the ATF6 mutants along with the positive control, ATF2, which is known to be phosphorylated by p38 MAPK. A kinase-inactive p38 MAPK incubated with wild-type ATF6 served as a negative control. A Western blot for p38 MAPK and a silver-stained gel showing the input levels of recombinant ATF6 proteins are presented in the lower part of this panel. The histogram below shows the relative intensities of the ATF6 bands as measured by phosphorimaging. (B) MEFs were treated in the presence and absence of p38 MAPK inhibitor (SB202190 at 20 μM) and stimulated with IFN-γ, where indicated, and were analyzed by Western blot analysis. Blots were probed with the indicated antibodies. (C, upper) Immunofluorescence images of DAPI (nuclear stain), GM130 (a Golgi apparatus marker), and FITC (ATF6). Scale bar, 10 μm. (Lower) Quantified fluorescence of FITC in regions of interest (ROI) after background correction. Bars represent means ± SE (n = 10 fields). P values are indicated in each case. NS, not significant. TM, tunicamycin A (2 μg/ml for 12 h). (D) Western blot analysis of lysates from Atf6−/− cells transfected with the indicated constructs. Specific treatments are indicated. (E) qPCR analysis of IFN-induced expression of Dapk1 mRNA in ATF6−/− MEFs after reconstituting with the indicated ATF6 constructs.
We next investigated whether IFN-γ promoted nuclear translocation of ATF6 T166A via the Golgi apparatus. We transfected HA-tagged ATF6 T166A or ATF6-mut (S1P protease-resistant mutant) into Atf6−/− MEFs and treated them with either IFN-γ or the positive-control agent, tunicamycin A. Immunofluorescence analysis was performed using the HA antibody. As a control, native Atf6 in wild-type MEFs also was subjected to immunofluorescence analysis using Atf6 antibody (Fig. 4C). Anti-GM130, a marker for the Golgi apparatus, and 4′,6-diamidino-2-phenylindole (DAPI), a nuclear stain, were used as internal controls. Steady-state ATF6 localized in the perinuclear and cytoplasmic regions as described earlier (6). Upon IFN-γ treatment for 2 h, ATF6 accumulated in the Golgi apparatus, as indicated by a strong overlap of green (Atf6 or HA tag) and red (GM130) fluorescence. Further, IFN-γ stimulation for 8 h resulted in higher nuclear Atf6 levels (strong overlap of green [ATF6 or HA] and blue [nucleus] fluorescence) (Fig. 4C). ATF6-mut showed higher accumulation in the Golgi apparatus after 8 h of IFN-γ stimulation, as it failed to enter the nucleus (6). Surprisingly, HA-ATF6 T166A, like ATF6-mut, also failed to translocate to the nucleus upon IFN-γ stimulation. In contrast, a higher nuclear level of HA-ATF6 T166A was found with TM treatment. Convincingly, the phosphomimetic mutant ATF6 T166D showed Golgi apparatus and early nuclear accumulation upon IFN-γ stimulation as early as 2 h (Fig. 4C). The histograms at the bottom of Fig. 4C show the quantification of fluorescence intensities in the Golgi apparatus and nucleus from several fields (n = 10) for each mutant. Thus, phosphorylation at T166 appears to be critical for IFN-γ-stimulated nuclear translocation of ATF6. Since the proteolysis-resistant ATF6-mut and the phosphorylation-deficient T166A mutant exhibited similar defects in nuclear entry, we next examined if IFN-γ induced the proteolysis of ATF6 in the absence of T166. Therefore, we expressed the wild type (WT), T166A, or T166D construct into Atf6−/− MEFs and treated them with IFN-γ or TM as indicated. Cell extracts were Western blotted using antibodies specific for the cleaved version of ATF6. WT-ATF6 and T166D were cleaved following IFN-γ and TM treatments, whereas T166A was cleaved only upon TM treatment but not with IFN-γ (Fig. 4D). These results clearly indicate the importance of the T166 site for IFN-γ-induced ATF6 cleavage (i.e., translocation to the nucleus). Consistent with these observations, phospho-p38 MAPK did localize with ATF6 and also with a Golgi marker (GM130) upon IFN treatment, suggesting that phospho-p38 MAPK has an important role in ATF6 translocation to the nucleus upon IFN-γ treatment (see Fig. S4 in the supplemental material). We next determined the effects of T166 on Dapk1 mRNA expression using qPCR. Only the WT and T166D constructs, but not the T166A construct, could induce Dapk1 expression in response to IFN-γ (Fig. 4E). Importantly, the T166D construct induced Dapk1 significantly more strongly than the WT construct in response to IFN-γ, consistent with its phosphomimetic function.
IFN-γ-stimulated ATF6 activation requires ASK1/MKK3/p38 MAPK.
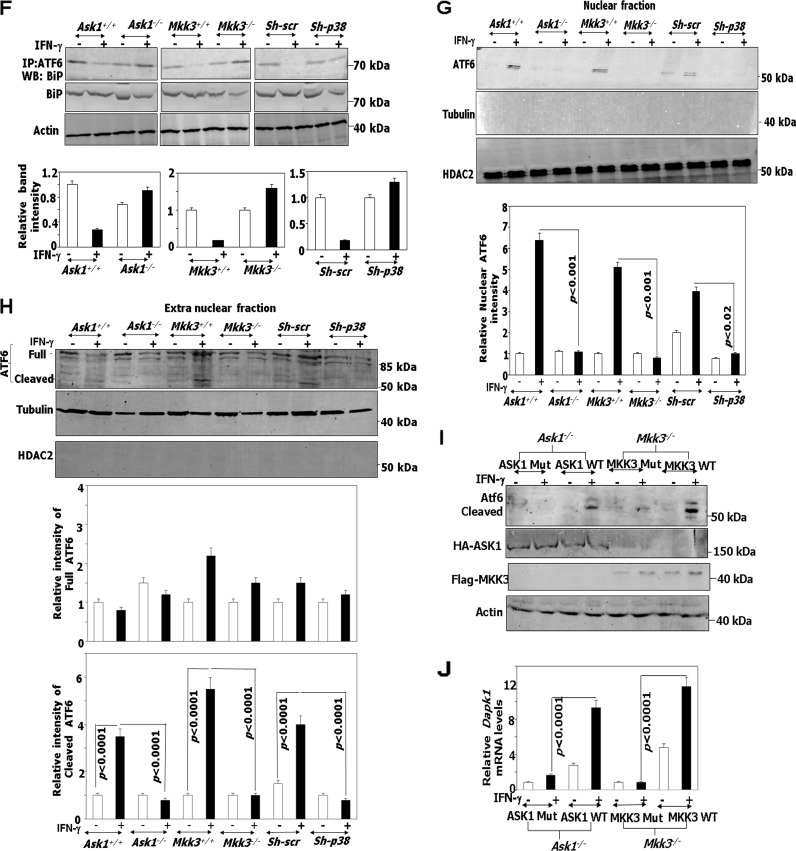
Based on the results described above, we next investigated if ATF6 translocation is dependent on ASK1/MKK3/p38 upon IFN-γ treatment. Using immunofluorescence, we examined the translocation of endogenous ATF6 and overexpressed HA-ATF6 T166A in Ask1−/−, Mkk3−/−, and sh-p38 cells following IFN-γ or TM treatment as a control (Fig. 5A to C). These experiments showed a lag in the translocation of ATF6 and HA-ATF6 T166A in the absence of Ask1, Mkk3, and p38. In contrast to the IFN-γ-stimulated accumulation of ATF6 in the Golgi apparatus by 2 h (strong overlap of green [ATF6] and GM130 [red] fluorescence) (Fig. 4D), a comparable accumulation of it occurred only at 8 h in the Ask1−/− cells (Fig. 5A, left and lower). Strikingly, no significant accumulation of HA-ATF6 T166A was observed in the Golgi apparatus of Ask1−/− cells even after 8 h of IFN-γ treatment (Fig. 5A, right and lower). Interestingly, no significant nuclear accumulation (strong overlap of green [ATF6 or HA] and DAPI [nucleus] fluorescence) was observed following IFN-γ treatment for 8 h with either endogenous ATF6 or HA-ATF6 T166A (Fig. 5A, right and lower). In contrast, TM treatment yielded nuclear accumulation of both ATF6 and HA-ATF6 T166A. Similar patterns of ATF6 and HA-ATF6 T166A translocation were observed in Mkk3−/− and sh-p38 cells (Fig. 5B and C) Thus, the ASK1/MKK3/p38 MAPK axis appears to be critical for the IFN-induced functioning of ATF6. Previously we have shown that IFN-γ-induced phosphorylation of C/EBP-β by ERK1/2 is required for its interaction with ATF6 (6) and the expression of Dapk1. Hence, we examined if there were differences in the ERK1/2 levels in these cells. A comparison of ERK1/2 levels and their phosphorylation in Ask1−/−, Mkk3−/−, and sh-p38 cells did not reveal any significant differences with respect to their wild-type counterparts (Fig. 5D). Consistent with the immunofluorescence observations (Fig. 5A to C), Western blot analysis showed that phosphorylation and cleavage of ATF6 occurred only in IFN-γ-treated wild-type cells but not in Ask1−/−, Mkk3−/−, and sh-p38 cells (Fig. 5E). In Mkk3−/− cells a slightly lower level of total p38 was observed compared to that of Mkk+/+, although no such differences were found between Ask1+/+ and Ask1−/− cells. In contrast to normal p38 activation (as determined by Western blot analysis with anti-phospho-p38 antibodies) in wild-type cells, it was severely diminished in the corresponding knockout/depleted cells. ATF6 activation and translocation requires dissociation from its inhibitory regulator BiP/GRP78 (in the ER), which unmasks its Golgi apparatus localization signals (9). IFN-γ causes the dissociation of ATF6/BiP complexes. The lack of translocation of ATF6 to the Golgi apparatus in the Ask1−/−, Mkk3−/− and sh-p38 cells may be due to an increase in BiP expression, which could retain ATF6 in the ER, or a failure to dissociate the BiP/ATF6 complexes. There was no significant difference in BiP levels between wild-type and kinase-deficient cells. Therefore, we examined if ATF6 is dissociated from BiP in these cell lines when treated with IFN-γ (6). These analyses showed that IFN-γ caused the dissociation of ATF6/BiP complex in wild-type but not in Ask1−/−, Mkk3−/−, and sh-p38 cells (Fig. 5F). Finally, in agreement with the in situ observations, Western blot analysis with the ATF6 antibody in nuclear and extranuclear fractions (cytosol plus membrane) revealed that both proteolytic cleavage and nuclear entry of ATF6 was blocked in Ask1−/−, Mkk3−/−, and sh-p38 cells compared to their corresponding wild-type controls (Fig. 5G and H). These experiments together not only identified the requirement for ASK1/MKK3/p38 MAPK for driving such IFN-γ-induced signals but also established a connection between phosphorylation and proteolytic activation. To determine if these are the direct effects of ASK1/MKK3 on ATF6, we reconstituted the corresponding wild-type and kinase-inactive constructs in Ask1−/− and Mkk3−/− cells to determine if IFN stimulated the cleavage of endogenous ATF6 and Dapk1 induction (Fig. 5I and J). The wild-type constructs, but not the kinase-inactive mutants, restored IFN-induced ATF6 activation and Dapk1 expression.
FIG 5.
IFN-γ-stimulated ATF6 activation requires ASK1/MKK3/p38 MAPK. (A to C, upper) Immunofluorescence images of DAPI, GM130 (Golgi apparatus marker [red]), and FITC-ATF6 in the indicated cell types. Scale bar, 10 μm. (Lower) Quantified fluorescence of FITC in regions of interest (ROI) after background correction. Bars are means ± SE (n = 10 fields). (D and E) Western blot analyses of lysates with the indicated antibodies. Specific genotypes of the cell lines used in the experiment are indicated. (F, upper) Immunoprecipitation and Western blot analysis of lysates prepared from IFN-treated cells (2 h) with the indicated antibodies. Histograms in the lower part of these panels show the relative intensities of the immunoprecipitated bands after normalization to input BiP levels. (G and H, upper) Western blot analysis of lysates from the indicated cell types after fractionation of cells into nuclear and extranuclear fractions with and without IFN treatment. Cell fractionation with 0.1% NP-40 yielded some background noise on these Western blots. The correct-size bands corresponding to ATF6 are indicated. Histograms in the lower part of these panels show the relative intensities of the bands after normalization to total ATF6 levels in the case of cleaved ATF6 and to the tubulin levels in the case of full-length ATF6. (I) Western blot analyses of lysates with the indicated antibodies. Specific genotypes of the cell lines and the transfected constructs are indicated. (J) qPCR analysis with Dapk1 gene-specific primers. Transfection of vector alone did not change Dapk1 expression (not shown).
Importance of p38 MAPK phosphorylation site of ATF6 in driving IFN-γ-induced autophagy.
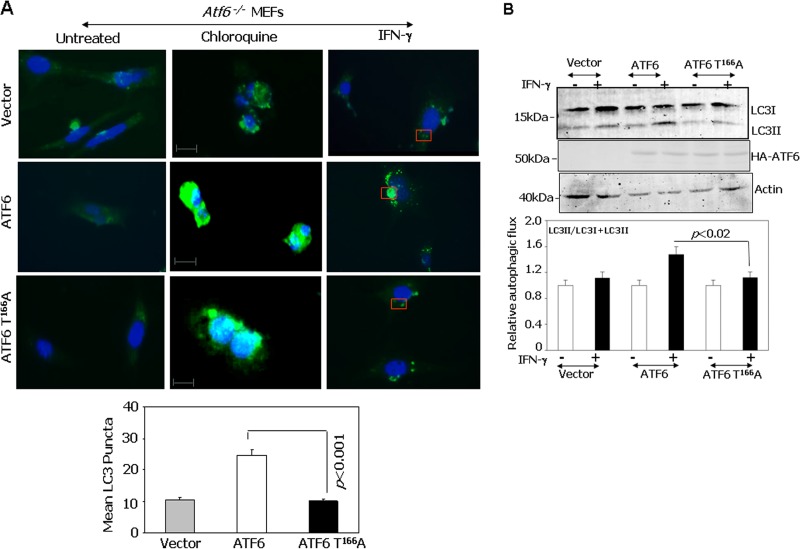
Since functional ATF6 in association with C/EBP-β induces DAPK1, which participates in autophagy, we asked whether the ATF6 T166A mutant can drive IFN-γ-induced autophagy. Autophagy was measured by LC3 puncta formation, which includes redistribution of cytoplasmic LC3. Atf6−/− MEFs were individually transfected with ATF6 WT-373 or the ATF6 T166A-373 mutant and then treated separately with chloroquine (a blocker of autophagosome lysosome fusion) or IFN-γ. Only ATF6 WT-transfected cells yielded maximum puncta upon IFN-γ treatment compared to either the vector control or ATF6 T166A-373 mutant-transfected MEFs (Fig. 6A). Chloroquine-stimulated puncta formation was not affected by the status of ATF6. Thus, there appears to be no global deficiency of puncta formation in these cells. The mean LC3 puncta formed upon IFN-γ treatment showed significant differences in each case (Fig. 6A, bottom). Alternatively, LC3II formation from LC3I, in response to IFN-γ treatment, was analyzed by Western blotting in ATF6 WT-373 and ATF6 T166A-373 mutant-transfected MEFs (Fig. 6B). The fold increase in LC3II levels correlated well with the number of puncta observed in situ. The differences in driving autophagic flux were not due to the differences in expression of ATF6-373 WT or the ATF6-373T166A mutant (Fig. 6B). These results demonstrate the critical role for p38 MAPK phosphorylation of ATF6 (i.e., ATF6 T166) in driving IFN-γ-induced autophagy.
FIG 6.
Role of p38 phosphorylation site of ATF6 (T166) in mediating IFN-γ-induced autophagy. (A) Immunofluorescence images of DAPI and FITC-LC3 (green) after restoring Atf6−/− MEFs with the indicated constructs. Scale bar, 15 μm. Cells were transfected with the indicated ATF6-373 mutants. The lower panel shows mean LC3 puncta formation (n = 10 different fields/sample). (B) Western blot analysis of Atf6−/− MEFs transfected with the indicated ATF6-373 constructs and stimulated with IFN-γ or left unstimulated. LC3 conversion to LC3II was monitored. The graph below shows the quantified autophagic flux on the basis of 3 separate blots.
Increased susceptibility of Ask1−/− mice to Bacillus anthracis infection.
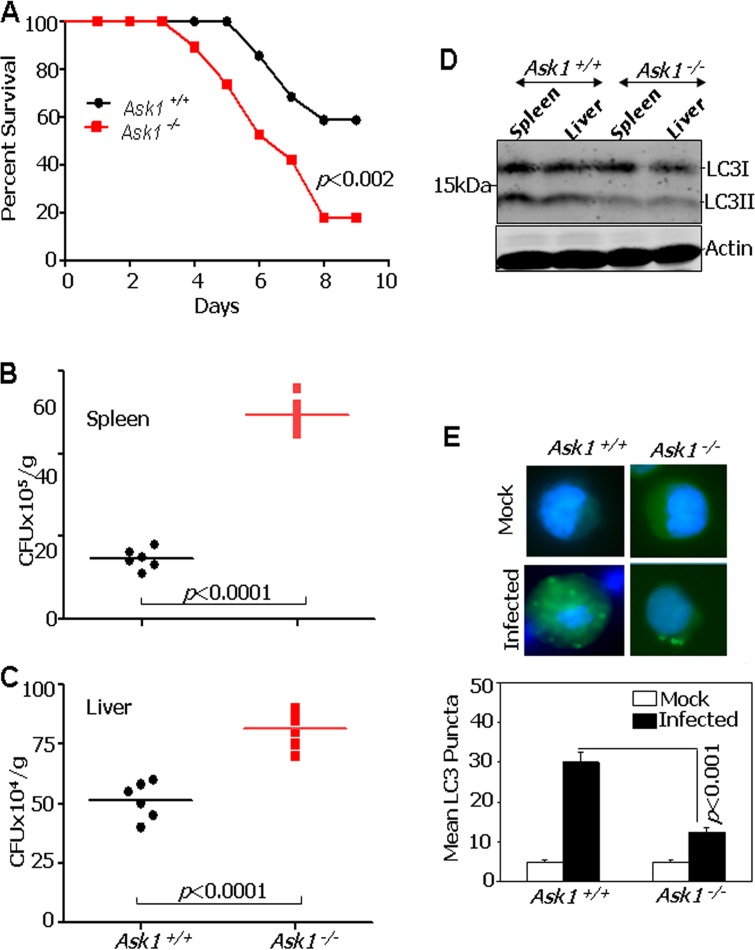
Since IFN-γ plays a critical role in host responses against B. anthracis infection, we next investigated whether Ask1−/− mice are sensitive to B. anthracis infection. Age-matched Ask1+/+ and Ask1−/− mice (n = 22/genotype) were infected with B. anthracis Sterne 34F2 spores (106 spores per mouse; a dose sublethal to wild-type mice) and monitored for signs of infection and survival (Fig. 7A). A significant number of Ask1−/− mice succumbed to infection compared to the number of Ask1+/+ mice. Importantly, mice started to die as early as day 4 after infection in the Ask1−/− group compared to day 6 in the Ask1+/+ group. The differences between Ask1+/+ and Ask1−/− in their susceptibility to infection remained throughout the study period. Analyses of these data with the Mantel-Cox log rank test (P < 0.0138) and Gehan-Breslow-Wilcoxon test (P < 0.0115) also affirmed the significance of these differences. To identify the basis for differences between Ask1−/− and Ask1+/+ mice in their responses to B. anthracis infection, bacterial loads were measured on day 4 after infection in the liver and spleen (Fig. 7B and C). A significant increase in the bacterial burden was observed in spleens and livers of Ask1−/− mice compared to those of Ask1+/+ mice. More importantly, Western blot analyses of the spleen and liver extracts from these mice with LC3-specific antibodies revealed a dramatic difference in LC3II formation between Ask1−/− and Ask1+/+ mice. LC3II formation was high in the Ask1+/+ mice compared to that in Ask1−/− mice (Fig. 7D). Consistent with these observations, peritoneal macrophages collected from infected Ask1−/− mice demonstrated fewer LC3 puncta than those from Ask1+/+ mice (Fig. 7E).
FIG 7.
Increased mortality of Ask1−/− mice upon infection with Bacillus anthracis. (A) Ask1+/+ and Ask1−/− mice were challenged with B. anthracis Sterne 34F2 spores (1 × 106 spores per mouse; a dose sublethal to WT mice), and percent survival (Kaplan-Meier analysis) was plotted. (B and C) Mice were intraperitoneally infected with 1 × 106 spores of B. anthracis Sterne 34F2, and bacterial titers in spleen and liver were determined 96 h postinfection. CFU/g, CFU per gram of tissue. Lines indicate mean values (n = 6 per group). (D) LC3 conversion to LC3II in tissues infected with B. anthracis. Western blot analyses were performed with the indicated antibodies. (E) LC3 puncta formation in the peritoneal macrophages of Ask1+/+ and Ask1−/− at 96 h postinfection. The bottom panel shows mean LC3 puncta formation at 96 h postinfection (n = 3 mice per group).
DISCUSSION
The critical importance of DAPK1 for apoptotic and autophagic forms of cell death induced by various signals, including IFN-γ, is emerging as a major player in cell fate decisions (5). DAPK family members ZIPK and DAPK2 also are known to function in different cell death pathways, including autophagy (21). Since high DAPK1 levels cause toxicity, cellular mechanisms control its protein levels. For example, the KLHL20-Cul3-ROC1 E3 ligase complex targets DAPK1 to ubiquitylation and subsequent proteasome-mediated degradation (22). DAPK1 employs several substrates to exert its biological effects. For example, DAPK1 recently has been reported to phosphorylate Beclin-1, a central player in autophagy and tumor suppression (23). DAPK1 activity also was implicated in the regulation of the pyruvate kinase M2, which is involved in driving the Warburg effect (24). Thus, dysfunction of DAPK1 or its expression may affect multiple cellular processes. Interestingly, in a wide variety of human cancers DAPK1 expression is downregulated via unknown mechanisms (5). Hence, there is a need to understand how DAPK1 is regulated.
In this study, we have demonstrated that ASK1, a member of the MAPK family, regulates the expression of DAPK1 in response to IFN-γ and aids in the execution of autophagy. Accumulating evidence shows that MAPKs have a role in ER stress responses. Some MAPK pathways, such as p38 and JNK, are activated in response to ER stress (11, 25). We have shown recently that IFN-γ induces ER stress, as indicated by the proteolytic cleavage of ATF6 and transcription (6). It was shown earlier that ER stress activates ASK1 through the formation of the IRE1-TRAF2-ASK1 complex (26); its downstream pathway targets are not known. ASK1 is activated by various stimuli, including calcium influx, ER stress, oxidative stress, lipopolysaccharide (LPS), and receptor-mediated signaling through tumor necrosis factor receptor (TNFR) and Toll-like receptor 4 (TLR4), TLR7, and TLR8 (11, 12). Notably, IFN-γ has been reported to cause Ca2+ influx and ER and oxidative stress in a variety of cell lines (27, 28). Activated ASK1 further stimulates the downstream p38 and JNK pathways for driving various cellular responses. In this report, we have demonstrated a direct role for ASK1 in regulating DAPK1 according to the following pieces of evidence: (i) a loss of IFN-γ-induced DAPK1 expression in Ask1−/−, Mkk3−/−, and p38 MAPK-depleted (sh-p38) cells and in the presence of the p38 MAPK-specific inhibitor (Fig. 1); (ii) the failure to recruit IFN-γ-induced ATF6 to the Dapk1 promoter in Ask1−/−, Mkk3−/−, and sh-p38 cells compared to their corresponding WT cells (Fig. 2); (iii) the ability of p38 MAPK to phosphorylate recombinant WT ATF6 protein in vitro but not the p38 MAPK phosphorylation-defective T166A mutant (Fig. 4A); (iv) the phosphorylation of endogenous ATF6 at T166 as detected by specific antibodies and its blockade by a p38 MAPK inhibitor (Fig. 4B); (v) the failure of nuclear entry by the T166A mutant (Fig. 4C and D); (vi) the reexpression of kinase-active mutants induced IFN-stimulated cleavage of ATF6 and DAPK1 induction, showing a direct role for ASK1 and MKK3 in this pathway; (vii) the restoration of autophagy by WT ATF6 but not the T166A mutant in Atf6−/− MEFs (Fig. 6A and B). Interestingly, no detectable cleavage of ATF6 occurred in the presence of a p38 MAPK inhibitor. Thus, it appears that ATF6 phosphorylation occurs prior to its proteolysis (Fig. 4B). This is consistent with the inability of phosphorylation-defective T166A and the enhanced ability of the T166D mutant to enter the nucleus and undergo cleavage in the Golgi apparatus following IFN-γ treatment. This requirement appears to be specific for IFN-γ given the normal activation of T166A by tunicamycin (an ER stress activator) (Fig. 4C and D). Indeed, upon IFN stimulation, the T166D mutant not only enters into the nucleus but also induces Dapk1 (Fig. 4E). The T166D mutant indeed induced DAPK1 expression better than wild-type protein upon reconstitution in Atf6−/− cells. Thus, these studies showed that T166 of ATF6 is essential for its activation and Dapk1 induction. In Ask1−/−, Mkk3−/−, and sh-p38 cells, ATF6 proteolysis was blocked compared to their wild-type counterparts (Fig. 5G and H). The restoration of kinase-active ASK1 or MKK3 in the knockout cell lines supported IFN-stimulated cleavage of ATF6 and Dapk1 induction (Fig. 5I and J). Thus, the phosphorylation of ATF6 appears to guide its proteolytic activation and subsequent transcriptional induction of Dapk1. We noticed a delayed translocation of wild-type ATF6 to the Golgi apparatus in Ask1−/−, Mkk3−/−, and sh-p38 cells compared to their wild-type counterparts. These observations also suggest that an IFN-γ-driven undefined late signal(s) causes ATF6 translocation to the Golgi apparatus in ASK1/MKK3/p38 MAPK-deficient cells (Fig. 5A to C). In the native state, ATF6 exists in a complex with an ER-resident inhibitor, BiP (9). In response to IFN-γ, this complex is dissociated, permitting the translocation of ATF6 to the Golgi apparatus (6). We have found a normal dissociation of ATF6 from BiP in wild-type cells following IFN-γ treatment. However, unlike the wild-type cells, the BiP/ATF6 complex was not dissociated in the Ask1-, Mkk3-, and p38-deficient cells (Fig. 5F). Thus, BiP appears to retain ATF6 in the ER and prevent its translocation to the Golgi apparatus in ASK1/MKK3/p38-deficient cells. The delayed Golgi apparatus translocation of ATF6 in the kinase-deficient cells may be due to unknown IFN-stimulated signals whose nature is unclear at this time. Despite such delayed translocation to the Golgi apparatus, no phosphorylation, proteolysis, and nuclear translocation of ATF6 occurred in the kinase-deficient cells. Thus, the IFN-induced ASK1/MMK3/p38 signaling axis seems to contribute to two critical events: (i) BiP/ATF6 dissociation, which allows ATF6 migration from the ER to the Golgi apparatus, and (ii) phosphorylation of ATF6 at T166, which allows the proteolysis and nuclear translocation of ATF6.
Our studies demonstrate the critical importance of ATF6 T166 for its IFN-γ-induced association with C/EBP-β (Fig. 3F to H) and the induction of DAPK1 (Fig. 3C). We have previously reported that IFN-γ-induced ERK1/2-dependent phosphorylation of C/EBP-β is critical for its interaction with ATF6 and induction of DAPK1 expression (3). Since the levels of ERK2, C/EBP-β, and ATF6 in Ask1−/−, Mkk3−/−, and sh-p38 cells were comparable and IFN-γ-induced ERK1/2 activation was normal in these cells, except for a slightly diminished activation in Mkk3−/− cells, and were comparable to those of the controls, the observed effects in gene expression cannot be due to a defective stimulation of this arm of the pathway (Fig. 2E and 5D). Although ER stress promotes IRE1-dependent activation of ERK1/2, we showed earlier that IFN-γ-dependent gene expression through C/EBP-β depended on MEKK1/MEK1/ERK1 for promoting gene expression (29). Thus, it appears two separate arms of MAPK signaling, viz., ASK1/MKK3/p38 and MEKK1/MEK1/ERK1, activate ATF6 and C/EBP-β, respectively, which converge on the DAPK1 enhancer to drive its expression. The normal cleavage and phosphorylation at T166 of ATF6 in WT but not in Ask1−/−, Mkk3−/−, and sh-p38 cells also suggests the importance of phosphorylation preceding ATF6 activation via proteolysis (Fig. 5E).
Taken together, the findings in the present study show that IFN-γ-induced, ASK1-MKK3-dependent, p38 MAPK-mediated phosphorylation of ATF6 is critical for its cooperation with C/EBP-β and DAPK1 activation and induction of autophagy. Interestingly, CHOP (C/EBP homologous protein), a direct target of ATF6, requires phosphorylation by p38 MAPK for its activity (30). CHOP acts as a dominant-negative inhibitor of C/EBP-β by heterodimerizing with it (31). Thus, CHOP is another well-characterized example which links p38 MAPK, ATF6, and C/EBP-β in ER stress pathways. Although a role for p38 MAPK in ATF6 phosphorylation in regulating the atrial natriuretic factor gene in cardiac myocytes was reported (32), those studies did not critically map precise residues that get activated in ATF6 and/or identify any upstream kinases involved. In contrast to those, where ATF6 was suggested to act as a homodimer for activating gene expression, our studies identified a novel pathway in which two separate transcription factors, C/EBP-β and ATF6, collaborate to induce gene expression and precisely mapped the p38 MAPK phosphorylation site on ATF6.
IFN-γ plays a critical role in mediating protection against infectious pathogens (1, 33). We have recently shown a connection between ER stress and autophagy in mediating ATF6-dependent antibacterial defenses (6). Emerging evidence shows that a number of steps in autophagy are signal dependent (34), and some of the autophagy-associated proteins also participate in driving inflammation (35, 36) and in several nonautophagic activities (37). Importantly, in ER-dependent pathways the Ca2+ release from ER stores could activate DAPK1, while the ER membrane supports the formation of autophagophores (38). The Ca2+ influx resulting from ER stress also permits the association of ASK1 with ER resident proteins like IRE1, which further leads to activation of ASK1 and the downstream JNK and p38 MAPKs (11, 25). ASK1 regulation is more complex than those of most known MAPKs. Multiple stress signals, including proinflammatory cytokines and bacterial LPS, induce ASK1 activity (39). In the steady state, the reduced form of Trx (a redox-responsive protein) binds to the N terminus of ASK1 and inhibits its kinase activity (40). Cytokine/LPS-induced production of reactive oxygen species causes the dissociation of this complex, allowing ASK1 activation. Interestingly, Trx acts as a death promoter in response to IFN-γ (41). Thus, it is likely that Trx limits the proapoptotic effects of ASK1 by acting as a feedback inhibitor after its initial IFN-γ-induced activation. Additionally, ubiquitylation of ASK1 targets it to proteasome-driven degradation to limit its activity (42). This event, coupled with DAPK1 phosphorylation of Thr119 in the BH3 domain of Beclin1, promotes autophagy by freeing Beclin1 from Bcl2 (43), which restricts bacterial infection.
In this study, we have shown that Ask1−/− mice are significantly more sensitive to infection with B. anthracis than Ask1+/+ mice at sublethal doses (Fig. 7A to C). Such sensitivity appears to be due to a poorer execution of autophagy by Ask1−/− macrophages than by Ask1+/+ macrophages (Fig. 7E), which also is consistent with high bacterial loads and low autophagic flux in the liver and spleen of Ask1−/− mice (Fig. 7D). Indeed, the loss of NSY1, an ASK1 orthologue of Caenorhabditis elegans, increased susceptibility to bacterial infections (44), consistent with our observations in Ask1−/− mice. Bacterial LPS-induced activation of TLR4 also has been shown to promote autophagy in RAW264.7 macrophages via the recruitment of Beclin1 to MyD88 and TRIF (45). LPS and tumor necrosis factor alpha have been shown to activate ASK1 (39). Thus, activated ASK1 may induce Beclin1 phosphorylation via DAPK1 to promote autophagy. Notably, ER-associated, but not mitochondrion-associated, Bcl2 blocked Beclin1-driven autophagy (46).
Beyond these findings, Beclin1 also is implicated in tumor suppression (47). Monoallelic loss of Beclin1 is sufficient to promote tumor formation in mice (48, 49). It is likely that ASK1-dependent autophagy via ATF6 contributes to tumor suppression. ASK1 plays contrasting roles in tumorigenesis. In the skin and gastric cancers, ASK1 was involved in tumor promotion (50, 51), whereas in liver it exerted tumor-suppressive functions (52). Lastly, mutations in ASK1 have been documented in certain metastatic melanomas (53). In conclusion, it appears that ASK1 can exert so-called yin-yang effects, like autophagy does, in tumorigenesis. At the least, our studies demonstrate that the IFN-γ-ASK1/MKK3/P38-ATF6-DAPK1 axis may regulate bacterial growth via autophagy.
Supplementary Material
ACKNOWLEDGMENTS
These studies were supported by NIH grant CA78282 to D.V.K.
We thank Hidenori Ichijo for supplying Ask1-deficient mice and ASK1 expression plasmids.
P.G., S.B.M., S.C.N., and U.B.M. performed experiments; P.G., A.S.C., and D.V.K. designed the experiments. P.G. analyzed the data, and P.G. and D.V.K. wrote the manuscript.
Footnotes
Published ahead of print 18 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00397-14.
REFERENCES
- 1.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227–264. 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 2.Li H, Gade P, Xiao W, Kalvakolanu DV. 2007. The interferon signaling network and transcription factor C/EBP-beta. Cell. Mol. Immunol. 4:407–418 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2782719/. [PMC free article] [PubMed] [Google Scholar]
- 3.Gade P, Roy SK, Li H, Nallar SC, Kalvakolanu DV. 2008. Critical role for transcription factor C/EBP-beta in regulating the expression of death-associated protein kinase 1. Mol. Cell. Biol. 28:2528–2548. 10.1128/MCB.00784-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roy SK, Wachira SJ, Weihua X, Hu J, Kalvakolanu DV. 2000. CCAAT/enhancer-binding protein-beta regulates interferon-induced transcription through a novel element. J. Biol. Chem. 275:12626–12632. 10.1074/jbc.275.17.12626. [DOI] [PubMed] [Google Scholar]
- 5.Bialik S, Kimchi A. 2006. The death-associated protein kinases: structure, function, and beyond. Annu. Rev. Biochem. 75:189–210. 10.1146/annurev.biochem.75.103004.142615. [DOI] [PubMed] [Google Scholar]
- 6.Gade P, Ramachandran G, Maachani UB, Rizzo MA, Okada T, Prywes R, Cross AS, Mori K, Kalvakolanu DV. 2012. An IFN-gamma-stimulated ATF6-C/EBP-beta-signaling pathway critical for the expression of death associated protein kinase 1 and induction of autophagy. Proc. Natl. Acad. Sci. U. S. A. 109:10316–10321. 10.1073/pnas.1119273109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rutkowski DT, Kaufman RJ. 2004. A trip to the ER: coping with stress. Trends Cell. Biol. 14:20–28. 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. 1999. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10:3787–3799. 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen J, Chen X, Hendershot L, Prywes R. 2002. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 3:99–111. 10.1016/S1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 10.Shen J, Prywes R. 2005. ER stress signaling by regulated proteolysis of ATF6. Methods 35:382–389. 10.1016/j.ymeth.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 11.Matsukawa J, Matsuzawa A, Takeda K, Ichijo H. 2004. The ASK1-MAP kinase cascades in mammalian stress response. J. Biochem. 136:261–265. 10.1093/jb/mvh134. [DOI] [PubMed] [Google Scholar]
- 12.Nagai H, Noguchi T, Takeda K, Ichijo H. 2007. Pathophysiological roles of ASK1-MAP kinase signaling pathways. J. Biochem. Mol. Biol. 40:1–6. 10.5483/BMBRep.2007.40.1.001. [DOI] [PubMed] [Google Scholar]
- 13.Tobiume K, Matsuzawa A, Takahashi T, Nishitoh H, Morita K, Takeda K, Minowa O, Miyazono K, Noda T, Ichijo H. 2001. ASK1 is required for sustained activations of JNK/p38 MAP kinases and apoptosis. EMBO Rep. 2:222–228. 10.1093/embo-reports/kve046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gade P, Singh AK, Roy SK, Reddy SP, Kalvakolanu DV. 2009. Down-regulation of the transcriptional mediator subunit Med1 contributes to the loss of expression of metastasis-associated dapk1 in human cancers and cancer cells. Int. J. Cancer 125:1566–1574. 10.1002/ijc.24493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saitoh M, Nishitoh H, Fujii M, Takeda K, Tobiume K, Sawada Y, Kawabata M, Miyazono K, Ichijo H. 1998. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 17:2596–2606. 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raingeaud J, Whitmarsh AJ, Barrett T, Derijard B, Davis RJ. 1996. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol. Cell. Biol. 16:1247–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakamoto S, Taura F, Tsuchihashi R, Putalun W, Kinjo J, Tanaka H, Morimoto S. 2010. Expression, purification, and characterization of anti-plumbagin single-chain variable fragment antibody in Sf9 insect cell. Hybridoma 29:481–488. 10.1089/hyb.2010.0052. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki K, Bose P, Leong-Quong RY, Fujita DJ, Riabowol K. 2010. REAP: a two minute cell fractionation method. BMC Res. Notes 3:294. 10.1186/1756-0500-3-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang TJ, Basu S, Zhang L, Thomas KE, Vogel SN, Baillie L, Cross AS. 2008. Bacillus anthracis spores and lethal toxin induce IL-1beta via functionally distinct signaling pathways. Eur. J. Immunol. 38:1574–1584. 10.1002/eji.200838141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darling NJ, Cook SJ. 2014. The role of MAPK signalling pathways in the response to endoplasmic reticulum stress. Biochim. Biophys. Acta 1843:2150–2163. 10.1016/j.bbamcr.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 21.Gozuacik D, Kimchi A. 2006. DAPk protein family and cancer. Autophagy 2:74–79. [DOI] [PubMed] [Google Scholar]
- 22.Lee YR, Yuan WC, Ho HC, Chen CH, Shih HM, Chen RH. 2010. The Cullin 3 substrate adaptor KLHL20 mediates DAPK ubiquitination to control interferon responses. EMBO J. 29:1748–1761. 10.1038/emboj.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zalckvar E, Berissi H, Eisenstein M, Kimchi A. 2009. Phosphorylation of Beclin 1 by DAP-kinase promotes autophagy by weakening its interactions with Bcl-2 and Bcl-XL. Autophagy 5:720–722. 10.4161/auto.5.5.8625. [DOI] [PubMed] [Google Scholar]
- 24.Mor I, Carlessi R, Ast T, Feinstein E, Kimchi A. 2012. Death-associated protein kinase increases glycolytic rate through binding and activation of pyruvate kinase. Oncogene 31:683–693. 10.1038/onc.2011.264. [DOI] [PubMed] [Google Scholar]
- 25.Matsuzawa A, Nishitoh H, Tobiume K, Takeda K, Ichijo H. 2002. Physiological roles of ASK1-mediated signal transduction in oxidative stress- and endoplasmic reticulum stress-induced apoptosis: advanced findings from ASK1 knockout mice. Antioxid. Redox Signal. 4:415–425. 10.1089/15230860260196218. [DOI] [PubMed] [Google Scholar]
- 26.Nishitoh H, Matsuzawa A, Tobiume K, Saegusa K, Takeda K, Inoue K, Hori S, Kakizuka A, Ichijo H. 2002. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 16:1345–1355. 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuda T, Nagano T, Takemura M, Baba A. 2006. Topics on the Na+/Ca2+ exchanger: responses of Na+/Ca2+ exchanger to interferon-gamma and nitric oxide in cultured microglia. J. Pharmacol. Sci. 102:22–26. 10.1254/jphs.FMJ06002X4. [DOI] [PubMed] [Google Scholar]
- 28.Watanabe Y, Suzuki O, Haruyama T, Akaike T. 2003. Interferon-gamma induces reactive oxygen species and endoplasmic reticulum stress at the hepatic apoptosis. J. Cell. Biochem. 89:244–253. 10.1002/jcb.10501. [DOI] [PubMed] [Google Scholar]
- 29.Roy SK, Hu J, Meng Q, Xia Y, Shapiro PS, Reddy SP, Platanias LC, Lindner DJ, Johnson PF, Pritchard C, Pages G, Pouyssegur J, Kalvakolanu DV. 2002. MEKK1 plays a critical role in activating the transcription factor C/EBP-beta-dependent gene expression in response to IFN-gamma. Proc. Natl. Acad. Sci. U. S. A. 99:7945–7950. 10.1073/pnas.122075799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma Y, Brewer JW, Diehl JA, Hendershot LM. 2002. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J. Mol. Biol. 318:1351–1365. 10.1016/S0022-2836(02)00234-6. [DOI] [PubMed] [Google Scholar]
- 31.Ron D, Habener JF. 1992. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 6:439–453. 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- 32.Thuerauf DJ, Arnold ND, Zechner D, Hanford DS, DeMartin KM, McDonough PM, Prywes R, Glembotski CC. 1998. p38 mitogen-activated protein kinase mediates the transcriptional induction of the atrial natriuretic factor gene through a serum response element. A potential role for the transcription factor ATF6. J. Biol. Chem. 273:20636–20643. [DOI] [PubMed] [Google Scholar]
- 33.Sen GC, Sarkar SN. 2007. The interferon-stimulated genes: targets of direct signaling by interferons, double-stranded RNA, and viruses. Curr. Top. Microbiol. Immunol. 316:233–250. [DOI] [PubMed] [Google Scholar]
- 34.Boya P, Reggiori F, Codogno P. 2013. Emerging regulation and functions of autophagy. Nat. Cell Biol. 15:713–720. 10.1038/ncb2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levine B, Mizushima N, Virgin HW. 2011. Autophagy in immunity and inflammation. Nature 469:323–335. 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castillo EF, Dekonenko A, Arko-Mensah J, Mandell MA, Dupont N, Jiang S, Delgado-Vargas M, Timmins GS, Bhattacharya D, Yang H, Hutt J, Lyons CR, Dobos KM, Deretic V. 2012. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc. Natl. Acad. Sci. U. S. A. 109:E3168–E3176. 10.1073/pnas.1210500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramani S, Malhotra V. 2013. Non-autophagic roles of autophagy-related proteins. EMBO Rep. 14:143–151. 10.1038/embor.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrison B, Kraus M, Burch L, Stevens C, Craig A, Gordon-Weeks P, Hupp TR. 2008. DAPK-1 binding to a linear peptide motif in MAP1B stimulates autophagy and membrane blebbing. J. Biol. Chem. 283:9999–10014. 10.1074/jbc.M706040200. [DOI] [PubMed] [Google Scholar]
- 39.Shiizaki S, Naguro I, Ichijo H. 2013. Activation mechanisms of ASK1 in response to various stresses and its significance in intracellular signaling. Adv. Biol. Regul. 53:135–144. 10.1016/j.jbior.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 40.Fujino G, Noguchi T, Matsuzawa A, Yamauchi S, Saitoh M, Takeda K, Ichijo H. 2007. Thioredoxin and TRAF family proteins regulate reactive oxygen species-dependent activation of ASK1 through reciprocal modulation of the N-terminal homophilic interaction of ASK1. Mol. Cell. Biol. 27:8152–8163. 10.1128/MCB.00227-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deiss LP, Kimchi A. 1991. A genetic tool used to identify thioredoxin as a mediator of a growth inhibitory signal. Science 252:117–120. 10.1126/science.1901424. [DOI] [PubMed] [Google Scholar]
- 42.Nagai H, Noguchi T, Homma K, Katagiri K, Takeda K, Matsuzawa A, Ichijo H. 2009. Ubiquitin-like sequence in ASK1 plays critical roles in the recognition and stabilization by USP9X and oxidative stress-induced cell death. Mol. Cell 36:805–818. 10.1016/j.molcel.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 43.Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A. 2009. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-X(L) and induction of autophagy. EMBO Rep. 10:285–292. 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, Tanaka-Hino M, Hisamoto N, Matsumoto K, Tan MW, Ausubel FM. 2002. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297:623–626. 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- 45.Shi CS, Kehrl JH. 2008. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J. Biol. Chem. 283:33175–33182. 10.1074/jbc.M804478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. 2005. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122:927–939. 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 47.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. 1999. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402:672–676. 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 48.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, Cattoretti G, Levine B. 2003. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Investig. 112:1809–1820. 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. 2003. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. U. S. A. 100:15077–15082. 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iriyama T, Takeda K, Nakamura H, Morimoto Y, Kuroiwa T, Mizukami J, Umeda T, Noguchi T, Naguro I, Nishitoh H, Saegusa K, Tobiume K, Homma T, Shimada Y, Tsuda H, Aiko S, Imoto I, Inazawa J, Chida K, Kamei Y, Kozuma S, Taketani Y, Matsuzawa A, Ichijo H. 2009. ASK1 and ASK2 differentially regulate the counteracting roles of apoptosis and inflammation in tumorigenesis. EMBO J. 28:843–853. 10.1038/emboj.2009.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hayakawa Y, Hirata Y, Nakagawa H, Sakamoto K, Hikiba Y, Kinoshita H, Nakata W, Takahashi R, Tateishi K, Tada M, Akanuma M, Yoshida H, Takeda K, Ichijo H, Omata M, Maeda S, Koike K. 2011. Apoptosis signal-regulating kinase 1 and cyclin D1 compose a positive feedback loop contributing to tumor growth in gastric cancer. Proc. Natl. Acad. Sci. U. S. A. 108:780–785. 10.1073/pnas.1011418108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakagawa H, Hirata Y, Takeda K, Hayakawa Y, Sato T, Kinoshita H, Sakamoto K, Nakata W, Hikiba Y, Omata M, Yoshida H, Koike K, Ichijo H, Maeda S. 2011. Apoptosis signal-regulating kinase 1 inhibits hepatocarcinogenesis by controlling the tumor-suppressing function of stress-activated mitogen-activated protein kinase. Hepatology 54:185–195. 10.1002/hep.24357. [DOI] [PubMed] [Google Scholar]
- 53.Stark MS, Woods SL, Gartside MG, Bonazzi VF, Dutton-Regester K, Aoude LG, Chow D, Sereduk C, Niemi NM, Tang N, Ellis JJ, Reid J, Zismann V, Tyagi S, Muzny D, Newsham I, Wu Y, Palmer JM, Pollak T, Youngkin D, Brooks BR, Lanagan C, Schmidt CW, Kobe B, MacKeigan JP, Yin H, Brown KM, Gibbs R, Trent J, Hayward NK. 2012. Frequent somatic mutations in MAP3K5 and MAP3K9 in metastatic melanoma identified by exome sequencing. Nat. Genet. 44:165–169. 10.1038/ng.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.