Abstract
Cdt2 is the substrate recognition adaptor of CRL4Cdt2 E3 ubiquitin ligase complex and plays a pivotal role in the cell cycle by mediating the proteasomal degradation of Cdt1 (DNA replication licensing factor), p21 (cyclin-dependent kinase [CDK] inhibitor), and Set8 (histone methyltransferase) in S phase. Cdt2 itself is attenuated by SCFFbxO11-mediated proteasomal degradation. Here, we report that 14-3-3 adaptor proteins interact with Cdt2 phosphorylated at threonine 464 (T464) and shield it from polyubiquitination and consequent proteasomal degradation. Depletion of 14-3-3 proteins promotes the interaction of FbxO11 with Cdt2. Overexpressing 14-3-3 proteins shields Cdt2 that has a phospho-mimicking mutation (T464D [change of T to D at position 464]) but not Cdt2(T464A) from ubiquitination. Furthermore, the delay of the cell cycle in the G2/M phase and decrease in cell proliferation seen upon depletion of 14-3-3γ is partly due to the accumulation of the CRL4Cdt2 substrate, Set8 methyltransferase. Therefore, the stabilization of Cdt2 is an important function of 14-3-3 proteins in cell cycle progression.
INTRODUCTION
The coordinated degradation of proteins in different phases of the cell cycle by the ubiquitin-proteasome system is central to the progression of the cell cycle. The specificity and timely destruction of different proteins are determined by E3 ubiquitin ligase enzymes (1). In eukaryotic cells, two major classes of E3 ligases are HECT and RING domain-containing multiprotein complexes. Among the RING E3 ligases, cullin-Ring E3 ligases (CRLs) represent the largest family, comprised of seven subfamilies based on which cullin protein (Cul1, Cul2, Cul3, Cul4a, Cul4b, Cul5, and Cul7) forms the scaffolding backbone of the enzyme complex (2, 3). CRL4Cdt2 is generally regarded as a master regulator of the cell cycle and of genome stability. The CRL4Cdt2 enzyme complex consists of the cullin 4 (Cul4a or Cul4b) scaffold protein interacting at its C terminus with an E2 enzyme (ubiquitin donor) through Rbx1/2 (Ring domain protein). At its N terminus, Cul4 interaction with Cdt2 is bridged by DDB1 (damaged DNA binding protein 1). Cdt2, belonging to the DCAF group of substrate adaptors, recognizes its substrates usually when they are bound to proliferating cell nuclear antigen (PCNA) on chromatin (4). The CRL4Cdt2 enzyme complex ubiquitinates and degrades p21, Set8, Cdt1, and Cdc6 in a replication-coupled and PCNA-dependent manner during S phase or after DNA damage (5–12). Degradation of these substrates is necessary for cell cycle progression, PCNA-mediated DNA damage repair, chromatin stability, and prevention of abnormal relicensing of origins in the same cell cycle, which would lead to rereplication (4).
Cdt2 is an essential protein important for normal cell proliferation (4, 13). The Cdt2 gene is amplified in Ewing's sarcoma (14), and Cdt2 protein levels are elevated in several human malignancies, such as colon, breast, hepatic, and gastric cancers. The increased Cdt2 in these cancers is correlated with advanced stages of disease progression, metastasis, and poor patient survival (15–18). A homozygous deletion of Cdt2 is lethal at the early embryonic stage in mice (13). Silencing Cdt2 in mammalian cells in culture leads to the accumulation of its substrates, such as Cdt1, Set8, and p21, and is associated with G2 arrest and rereplication (5, 10–12). MLN4924, a small-molecule inhibitor that inhibits the Nedd8 activating enzyme from neddylating and thus activating cullins, phenocopies Cdt2 silencing (19–21). Currently, the drug is in clinical trials for therapy against cancers (4).
Not much is known about the factors regulating the CRL4Cdt2 complex. Cdt2 is phosphorylated in S phase, and a hyperphosphorylated form of Cdt2 associates with chromatin in response to UV irradiation (22). Two recent reports showed that Cdt2 undergoes SCFFbxO11-mediated ubiquitination and degradation, resulting in cell cycle exit (23, 24). Cyclin A/cyclin-dependent kinase 2 (Cdk2) and cyclin B/Cdk1 phosphorylate Cdt2 at threonine 464 (T464) both in vivo and in vitro. This phosphorylation has been implicated in Cdt2 stability, because phosphorylated Cdt2 (at T464) does not interact with SCFFbxO11 ubiquitin ligase complex (24).
14-3-3 proteins are a highly conserved family of low-molecular-mass (28 to 33 kDa) adaptor proteins found in almost all eukaryotes. There are seven different 14-3-3 isoforms in mammals, β, γ, ε, θ, ζ, η, and σ, encoded by seven individual genes. They regulate several cellular pathways and are implicated in cancer development. 14-3-3 proteins usually bind to their ligand proteins at a defined phosphorylation motif, e.g., RSXpSXP or RXYFXpSXP, but there are examples where they bind to their ligands at other phosphorylation sites or even without any phosphorylation (25, 26). 14-3-3 proteins regulate various pathways by different mechanisms, including intra- and intercompartmental sequestration of proteins, activation or inactivation of enzyme activities, and promotion or inhibition of protein interactions (25, 26). In a normal cell cycle, 14-3-3 proteins are effectors of checkpoints that bind to and sequester Cdc25A, Cdc25B, or Cdc25C in the cytoplasm after they have been phosphorylated by the checkpoint kinases. Upon inactivation of the checkpoint, the dephosphorylated Cdc25A or Cdc25B/Cdc25C are released from 14-3-3 proteins and translocate to the nucleus to dephosphorylate and activate Cdk kinases for progression of the cell cycle through G1/S, S, or G2/M (25, 27, 28).
In this study, we identified 14-3-3ε and 14-3-3γ as positive regulators of CRL4Cdt2 E3 ubiquitin ligase. 14-3-3 proteins interact with Cdt2 in a phosphorylation-dependent manner during the S phase of the cell cycle and shield it from ubiquitin-mediated degradation. Depletion of 14-3-3γ destabilizes Cdt2 and causes G2/M arrest in unperturbed cells. Destabilization of Cdt2 by 14-3-3γ depletion is associated with upregulation of the Cdt2 substrates, Set8 methyltransferase, p21, and Cdt1. We found that G2/M arrest and decrease in cell proliferation caused by 14-3-3γ depletion is partly attenuated by preventing the accumulation of Set8. Thus, 14-3-3-mediated stabilization of Cdt2 and the attendant degradation of Set8 are important for normal cell cycle progression.
MATERIALS AND METHODS
Cell culture and transfections.
HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS), and 293T cells were grown in DMEM supplemented with 10% donor calf serum (DCS). Small interfering RNA (siRNA) transfection (10 nM) into cells (2.5 × 105 to 3.5 × 105 cells in 10-cm-diameter dishes) was carried out by using RNAiMax reagent (Invitrogen), following the manufacturer's instructions. The sequences of the different double-stranded siRNAs used are given in Table S1 in the supplemental material. Plasmids were transfected into 293T cells by using Lipofectamine 2000 reagent (Invitrogen) as suggested by the manufacturer.
Plasmids.
Flag-Cdt2 and myc-FbxO11 plasmids are described elsewhere (23). Hemagglutinin (HA)-Flag-Cdt2(T464D) and HA-Flag-Cdt2(T464A) plasmids were kindly provided by Michele Pagano (NYU Cancer Institute). myc–14-3-3ε and myc–14-3-3γ plasmids were constructed by amplifying the corresponding cDNAs from a cDNA library with PCR and then cloning them into the pEFM vector backbone with a myc tag.
Yeast two-hybrid assay.
The GAL4 yeast two-hybrid system 3 (Clontech) was used for two-hybrid interactions. Briefly, the human Cdt2 gene was subcloned into the pGBKT7 bait vector in fusion with the Gal4 DNA binding domain and, later, transformed into the AH109 yeast strain. The pGBKT7-Cdt2-transformed AH109 yeasts were mated with Y187 yeast cells that were pretransformed with pGADT7 plasmids carrying a HeLa cDNA library as prey. AH109 cells harboring pGBKT7-p53 were used as a positive control, and cells with pGBKT7-Cdt2 (out of frame) were used for a negative control. The yeast strains used were auxotrophic, and after mating, positive colonies were selected by nutrient dropouts as suggested by the provider (Clontech). The autoactivation of the His gene was prevented by growing cells on plates having 1 mM 3-amino-1,2,4-triazole (3-AT) to eliminate false-positive colonies, though the treatment with this drug slowed the growth of true-positive colonies.
The pGBKT7 plasmid backbone carries a TRP1 gene that allows the transformed cells to grow in medium without tryptophan (Trp− medium). Similarly, pGADT7 has the LEU2 gene and allows transformed cells to grow in medium without leucine (Leu− medium). If the DNA binding domain and activation domain are brought together due to interaction between the fused proteins (Cdt2 and whatever is fused to the bait) in the mated yeast, the HIS3 gene is expressed and allows the yeast cells to grow on medium without histidine (His− medium).
Coimmunoprecipitation.
Cells were lysed on ice for 30 min in a lysis buffer (50 mM Tris-Cl [pH 8.0], 10% glycerol, 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 1 mM dithiothreitol [DTT], 50 mM NaF, 1 mM Na3VO4, 1 mM glycol phosphate, 1× protease inhibitor cocktail), followed by centrifugation at 15,000 rpm for 10 min. The supernatant recovered was immunoprecipitated with the antibodies indicated in the figures. For the coimmunoprecipitations of Cdt2 and FbxO11 and of PCNA and Cdt2, the lysis buffer contained only 100 mM NaCl (29).
Cell synchronization.
For double thymidine block (DTB), HeLa cells were treated with 2 mM thymidine for 17 h, released into fresh medium for 9 h, and then given a second treatment with thymidine solution (2 mM) for the next 18 h. For synchronization with hydroxyurea (HU) or nocodazole (NOC), HeLa cells were treated with 2 mM hydroxyurea (20 or 24 h) or 100 ng/ml nocodazole (18 h), respectively.
Ubiquitination assay.
For the in vivo ubiquitination assay, the plasmid-transfected 293T cells were treated with MG132 (40 μM) for 1 h before harvesting. Cells were harvested in denaturing ubiquitination buffer (50 mM Tris-Cl [pH 8.0], 5 mM DTT, and 1% SDS) and immediately boiled for 10 min at 95°C, followed by cooling on ice for 10 min. The lysate was sonicated, and supernatant recovered after centrifugation at 15,000 rpm for 20 min. The supernatant was diluted with 9 volumes of buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM KCl, 5% glycerol, 0.4% NP-40, and protease inhibitors and subjected to immunoprecipitation followed by Western blotting (30).
Half-life measurements.
Cells were treated with freshly prepared cycloheximide (5 mg/ml in H2O) and harvested at different time points, followed by immunoblotting of the whole-cell lysates.
Western blotting and antibodies.
For Western blotting of cell lysates, cell extracts were prepared by lysing cells in radioimmunoprecipitation assay (RIPA) buffer containing 1× protease inhibitor cocktail (Sigma). The proteins targeted with antibodies and the sources of the antibodies were as follows: Cdt2 (Dutta laboratory); pan-14-3-3, 14-3-3γ, 14-3-3ε, 14-3-3θ, and 14-3-3ζ (Santa Cruz); actin (Santa Cruz); tubulin (Santa Cruz); Set8 (Cell Signaling); FbxO11 (Novus Biologicals); p21 (Santa Cruz); HA (Santa Cruz); Flag (Sigma); myc (Dutta laboratory); PCNA (Santa Cruz); phospho-Cdt2 (p-Cdt2) (Pagano laboratory), Cdt1(Dutta laboratory); cyclin A and B (Santa Cruz); cyclin D (Lab Vision Corp.); monomethylated histone H4-lysine 20 (H4K20me1) (Cell Signaling); and acetylated H4K5 (H4K5ac) (Cell Signaling).
Cell cycle analysis.
Cells were harvested by trypsinization and washed with 1× PBS. The washed cells were fixed with 70% ethanol in 1× PBS at −20°C overnight. Fixed cells were washed with 1× PBS, followed by staining with a solution containing 50 μg/ml propidium iodide plus 50 μg/ml RNase A in PBS. The stained cells were analyzed by using a FACSCalibur flow cytometer (Becton Dickinson).
Real-time q-PCR.
Total RNA was isolated by using TRIzol reagent (Invitrogen), following the manufacturer's instructions, and was used for cDNA synthesis with SuperScript III (Invitrogen). The cDNAs were used as the templates for real-time quantitative PCR (q-PCR) using SYBR green PCR master mix (Applied Biosystems). The sequences of primers used for RT-PCR analysis are given (5′→3′) in Table S2 in the supplemental material.
Cell viability.
Cell viability was monitored by the trypan blue exclusion method using a 0.4% trypan blue solution (Invitrogen) and a Countess automated cell counter (Invitrogen).
RESULTS
14-3-3 proteins interact with the CRL4Cdt2 E3 ligase adaptor Cdt2.
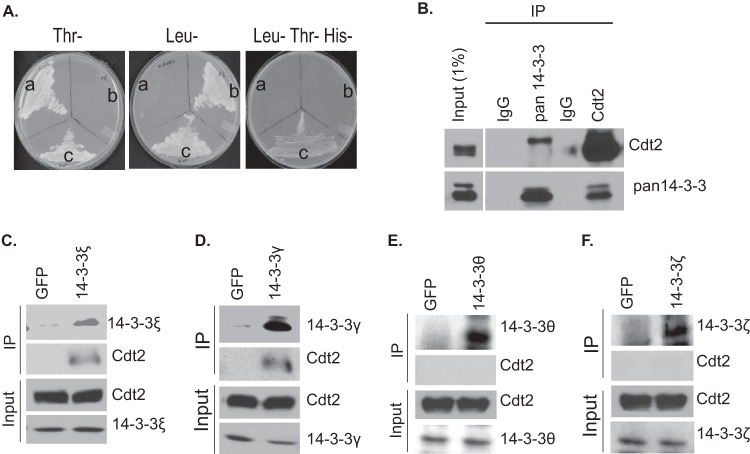
To identify the novel regulators or substrates of CRL4Cdt2 E3 ubiquitin ligase, we performed a yeast two-hybrid screen by using Cdt2 (fused to the DNA binding domain of Gal4 [Gal4-DBD]) as bait against a human cDNA prey library fused to the Gal4 activation domain (Gal4-AD). Full-length 14-3-3ε, along with amino acids contributed by the 5′ untranslated region (5′ UTR) upstream from the initiator methionine, was recovered as an interactor fused in frame with GAL4-AD. To ensure that endogenous 14-3-3ε protein (without the amino acids contributed by the 5′ UTR) interacts with Cdt2, the yeast two-hybrid interaction was confirmed with GAL4-AD fused to the 14-3-3ε open reading frame (Fig. 1A). The interaction activated the HIS3 reporter and allowed the growth of the yeast in histidine-depleted plates.
FIG 1.
14-3-3 proteins interact with Cdt2. (A) Yeast two-hybrid interaction of Cdt2 and 14-3-3ε in auxotrophic yeast cells. (a) AH109 yeast cells transformed with pGBKT7-Cdt2 grow in Trp− medium but not in Leu− medium. (b) Y187 yeast cells transformed with pGADT7-14-3-3ε grow in Leu− medium and not in Trp− medium. (c) Diploid cells resulting from mating the cells grown in sectors a and b grow in Leu− Trp− His− medium. (B) Coimmunoprecipitation of endogenous 14-3-3 and Cdt2. Cell extracts from HeLa cells were immunoprecipitated (IP) with control IgG, pan14-3-3, or Cdt2 antibodies, and the precipitates immunoblotted with the antibodies indicated to the right. (C to F) HeLa cell lysates were immunoprecipitated either with anti-green fluorescent protein (anti-GFP) antibody or antibodies against 14-3-3ε, 14-3-3γ, 14-3-3θ, and 14-3-3ζ, and precipitates were analyzed by Western blotting using antibodies as indicated.
The Cdt2 and 14-3-3 interaction was confirmed biochemically. Immunoprecipitation with pan-14-3-3 antibodies (a blend of antibodies against all seven isoforms of 14-3-3) or anti-Cdt2 antibodies precipitated the cognate antigens but also coimmunoprecipitated Cdt2 or 14-3-3 proteins, respectively (Fig. 1B). Of the seven isoforms of 14-3-3, we used antibodies against 4 individual isoforms (γ, ε, θ, and ζ) to determine whether all isoforms interacted with Cdt2. As expected, immunoprecipitation of 14-3-3ε coprecipitated Cdt2 (Fig. 1C). Interestingly, immunoprecipitation of 14-3-3γ also coprecipitated Cdt2 (Fig. 1D), but immunoprecipitation of 14-3-3θ or 14-3-3ζ did not coprecipitate Cdt2 (Fig. 1E and F). Thus, 14-3-3ε and 14-3-3γ are two new and specific interacting partners of Cdt2 protein.
The interaction of 14-3-3 proteins with Cdt2 is dependent on phosphorylation of Cdt2 at T464.
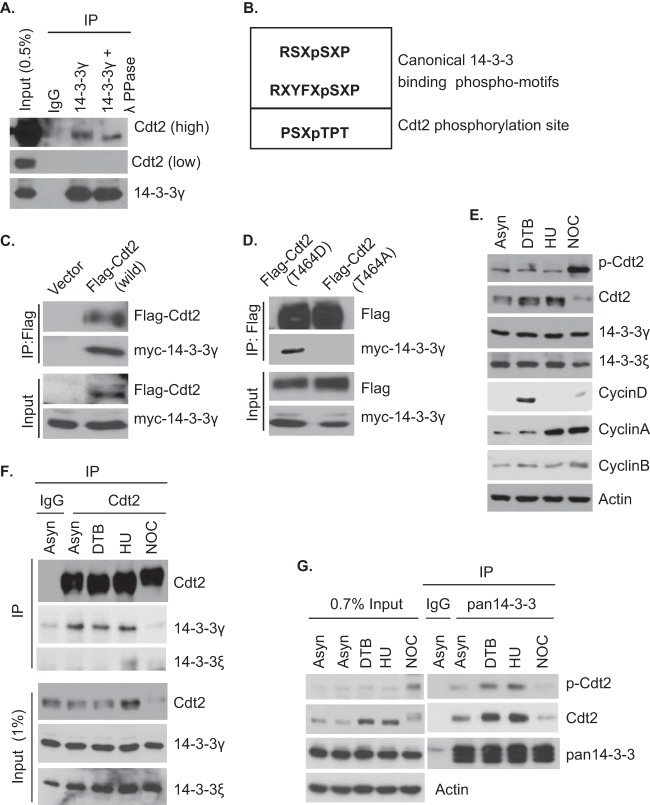
14-3-3 proteins are adaptor proteins that bind to most of their substrate proteins at one of two phosphorylation sites, RSXpSXP and RXYFXpSXP (Fig. 2B), although binding is seen occasionally at noncanonical phosphorylation sites or, even, unphosphorylated sites (27). ATR kinase increases Cdt2 phosphorylation in response to UV irradiation and increases its association with chromatin (22). In addition, cyclin A/Cdk2 and cyclin B/Cdk1 have been implicated as kinases phosphorylating Cdt2 at threonine 464 in vivo and in vitro (24). The slower migration of the Cdt2 that coprecipitated with 14-3-3 relative to the migration of the total pool of cellular Cdt2 (Fig. 1B) led us to test whether the interaction of 14-3-3 with Cdt2 is dependent on phosphorylation of Cdt2. Immunoprecipitates of 14-3-3 from cell lysates were treated with control buffer or λ-protein phosphatase. Phosphatase treatment caused the slower-migrating band of Cdt2 in the 14-3-3 immunoprecipitate to move faster, suggesting that the slower migration was due to phosphorylation (Fig. 2A).
FIG 2.
Phosphorylation of T464 of Cdt2 mediates interaction of Cdt2 with 14-3-3. (A) HeLa cell extracts were immunoprecipitated using control IgG or anti-14-3-3γ antibodies. The protein G-Sepharose-bound immunoprecipitates were mock treated (buffer) or treated with 400 units of λ-protein phosphatase (λPPase) for 1 h at 30°C. The samples were analyzed by Western blotting using anti-Cdt2 and anti-14-3-3γ antibodies. (B) Comparison of the canonical 14-3-3 binding motifs with the Cdt2 phosphorylation site at threonine 464. (C) 293T cells were transfected with plasmids expressing Flag-Cdt2 and myc–14-3-3γ. The cell lysates were immunoprecipitated with Flag antibodies, followed by Western blotting with anti-Flag and anti-myc antibodies. (D) 293T cells were transfected with plasmids expressing Flag-Cdt2(T464D) or Flag-Cdt2(T464A) in combination with myc–14-3-3γ plasmids. The cell lysates were immunoprecipitated with anti-Flag antibodies, followed by Western blotting with anti-Flag and anti-myc antibodies. (E) HeLa cells were synchronized with double thymidine block (DTB), hydroxyurea (HU), or nocodazole (NOC), and the lysates were probed with the indicated antibodies. (F) Coimmunoprecipitation of Cdt2 and 14-3-3 in asynchronous and synchronized cells. Asynchronous and synchronized HeLa cell lysates were immunoprecipitated with Cdt2 antibody, and the immunoprecipitates probed with anti-Cdt2, anti-14-3-3γ, and anti-14-3-3ε antibodies. (G) Coimmunoprecipitation of 14-3-3 proteins and p-Cdt2/Cdt2 in asynchronous (Asyn) and synchronized cells. Asynchronous and synchronized HeLa cell lysates were immunoprecipitated with pan-14-3-3 antibodies, and the immunoprecipitates probed with anti-p-Cdt2, anti-Cdt2, and pan-14-3-3 antibodies. The signals of p-Cdt2 and Cdt2 were obtained from the same membrane by first probing with anti-p-Cdt2 antibody and then stripping and reprobing with anti-Cdt2 antibody.
To test whether phosphorylation of Cdt2 at threonine 464 has a role in interaction with 14-3-3, we transfected cells with myc–14-3-3 in combination with empty vector or vectors expressing wild-type (WT) Flag-Cdt2, phosphomimetic Flag-Cdt2(T464D), or Flag-Cdt2(T464A), followed by immunoprecipitation with anti-Flag antibodies. WT Flag-Cdt2 and Flag-Cdt2(T464D) coimmunoprecipitated myc–14-3-3, whereas Flag-Cdt2(T464A) did not (Fig. 2C and D).
We further asked when Cdt2 gets phosphorylated at T464 and when 14-3-3 proteins bind Cdt2 during the cell cycle. Analyses of cell extracts prepared from HeLa cells synchronized at G1/S, S, or M phases of the cell cycle using DTB, HU, or NOC blocks indicated that Cdt2 is phosphorylated throughout the cell cycle, with a hyperphosphorylated form seen during M phase (Fig. 2E). To determine the stage of the cell cycle where 14-3-3 proteins bind to Cdt2, asynchronous and synchronized HeLa cell extracts were immunoprecipitated using antibodies against Cdt2. Interestingly, 14-3-3γ coprecipitated with Cdt2 in G1/S phase (DTB-arrested) and early S phase (HU-arrested) extracts, whereas 14-3-3ε coprecipitated with Cdt2 in early S phase (HU-arrested) extracts only. Surprisingly, 14-3-3 proteins did not coprecipitate with Cdt2 in extracts from cells in which Cdt2 was hyperphosphorylated during M phase (Fig. 2F). To further investigate whether the interaction between Cdt2 and 14-3-3 proteins during S phase is phosphorylation dependent, we immunoprecipitated 14-3-3 proteins from asynchronous and synchronized HeLa extracts using pan-14-3-3 antibodies. As indicated by the results shown in Fig. 2G, immunoprecipitates of 14-3-3 proteins coprecipitated phosphorylated Cdt2 in extracts from cells taken during G1/S and early S phase and not in extracts from cells taken during M phase. These results demonstrate that 14-3-3 proteins bind Cdt2 phosphorylated at threonine 464 during S phase even though the sequence of the phosphorylation site is distant from the canonical phosphorylation sites that are normally bound by 14-3-3 (Fig. 2B). In addition, hyperphosphorylation of Cdt2 in M phase dissociates Cdt2 from 14-3-3.
14-3-3 proteins stabilize Cdt2 and increase its half-life.
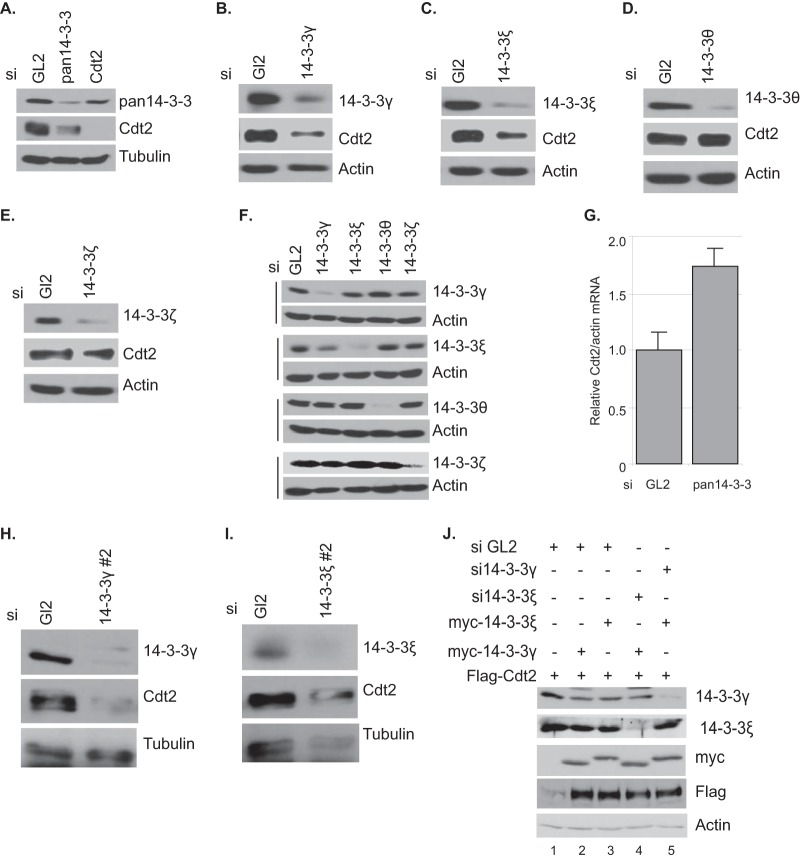
Cdt2 is the substrate adaptor of the CRL4Cdt2 E3 ligase complex and targets substrate proteins, e.g., Set8, p21, and Cdc6, to ubiquitin-mediated degradation (4, 9). We initially hypothesized that 14-3-3 proteins themselves may be targets of CRL4Cdt2 ubiquitination and, thus, knocked down Cdt2 using siRNAs (siCdt2). The depletion of Cdt2 did not increase the 14-3-3 levels (Fig. 3A), suggesting that 14-3-3 proteins are not substrates for Cdt2.
FIG 3.
14-3-3 proteins stabilize Cdt2. (A) 14-3-3 knockdown decreases Cdt2. HeLa cells were transfected with control GL2, pan-14-3-3, and Cdt2 siRNAs and harvested after 60 h. The cell lysates were analyzed by Western blotting with the indicated antibodies. (B to E) HeLa cells were treated with either control GL2 or the indicated siRNAs for 60 h, followed by Western blotting using Cdt2 and the corresponding 14-3-3 antibodies. (F) Lysates from the experiment whose results are shown in panels B to E were analyzed by Western blotting with the indicated antibodies. (G) 14-3-3 knockdown does not affect mRNA of the Cdt2 gene. HeLa cells were treated with control GL2 or pan-14-3-3 siRNAs for 60 h. RNA transcripts were measured by real-time q-PCR. Error bars show standard deviations. (H and I) Western blots of HeLa cell lysates after transfection with indicated siRNAs for 48 h. The antibodies used are indicated. (J) 293T cells were treated with control GL2, 14-3-3γ, or 14-3-3ε siRNAs 24 h before transfection with the indicated plasmids. Cells were again treated with siRNAs 24 h after plasmid transfection and harvested after another 24 h. The cell lysates were analyzed by Western blotting with the indicated antibodies. Note that the blot shows only the slower-migrating overexpressed myc-tagged 14-3-3 proteins.
Surprisingly, depletion of 14-3-3 proteins (with siRNA against pan-14-3-3 [si-pan-14-3-3]) decreases Cdt2 levels (Fig. 3A). To determine which 14-3-3 isoform was important for maintaining Cdt2 levels, we used siRNAs against four individual 14-3-3 isoforms, γ, ε, θ, and ζ, to separately knock down the isoforms. Depletion of the 14-3-3γ or -ε isoform decreased the Cdt2 protein levels, whereas knockdown of the 14-3-3θ or -ζ isoform did not change the level of Cdt2 protein (Fig. 3B to F). Note that the sequences of these siRNAs were different from that of si-pan-14-3-3 (see Table S1 in the supplemental material). In addition, two more siRNAs against different parts of the 14-3-3γ and -ε isoforms also decreased Cdt2 (Fig. 2H and I), diminishing the likelihood that the Cdt2 protein decrease was due to off-target activity of the siRNAs against 14-3-3. The decrease of Cdt2 protein by 14-3-3 depletion was a posttranscriptional effect, as Cdt2 mRNA did not decrease when cells were treated with pan-14-3-3 siRNA (Fig. 3G).
Cdt2 is destabilized by individual knockdown of 14-3-3γ or 14-3-3ε (Fig. 3B and C), so each of the isoforms may independently stabilize the Cdt2. Since 14-3-3γ and 14-3-3ε are known to bind some of their substrates as heterodimers, another possibility is that they work as a heterodimer to stabilize Cdt2. Alternatively, since knockdown of 14-3-3γ decreased 14-3-3ε (Fig. 3F) but knockdown of the ε isoform did not decrease 14-3-3γ, the regulation of Cdt2 could be exclusively by 14-3-3ε. To distinguish between these possibilities, we asked whether the overexpression of 14-3-3 isoforms stabilizes Cdt2 and, if so, whether the stabilization by one isoform requires the presence of the other. When we cotransfected a plasmid expressing Flag-Cdt2 with a vector overexpressing myc–14-3-3γ, the overexpressed myc–14-3-3γ clearly increased the level of the Flag-Cdt2 (Fig. 3J, lanes 1 and 2). The myc–14-3-3γ stabilized Flag-Cdt2 even when the endogenous 14-3-3ε was knocked down by siRNA (Fig. 3J, lane 4). Identical results were obtained in the reciprocal experiment, where cotransfected myc–14-3-3ε stabilized Flag-Cdt2 even in cells lacking endogenous 14-3-3γ (Fig. 3J, lanes 1, 3, and 5). Thus, 14-3-3γ and -ε can stabilize Cdt2 independently, at least when overexpressed.
14-3-3 proteins protect Cdt2 from ubiquitin-mediated degradation.
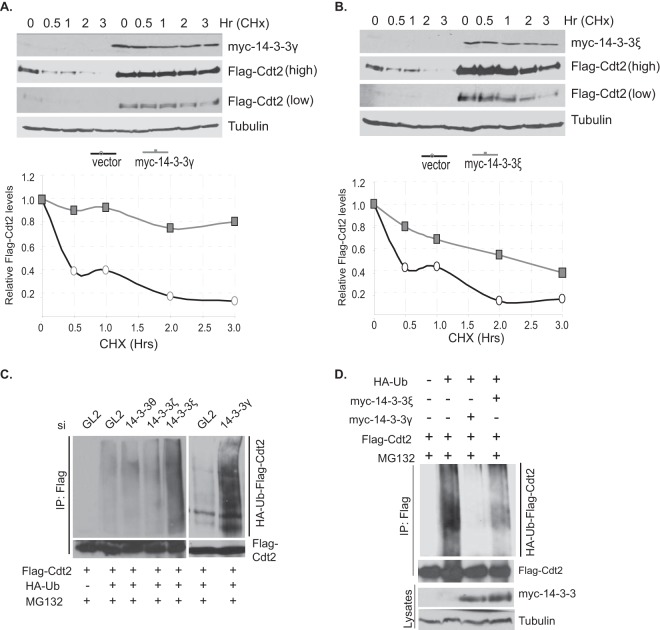
Previous studies have found that Cdt2 is polyubiquitinated in cells and degraded by the 26S proteasomal pathway (23, 24). The posttranscriptional stabilization of Cdt2 by 14-3-3γ and 14-3-3ε suggests that these adaptor proteins protect Cdt2 from degradation. To test this formally, we transiently transfected 293T cells with a plasmid expressing Flag-Cdt2, along with an empty vector or a vector expressing myc–14-3-3γ or myc–14-3-3ε, and measured the half-life of the Flag-Cdt2 after inhibiting new protein translation with cycloheximide (Fig. 4A and B). The half-life of ectopically expressed Flag-Cdt2 was <0.5 h when cotransfected with an empty vector. Coexpression of myc–14-3-3γ or myc–14-3-3ε increased the half-life of Cdt2 significantly, to ≥2 h (Fig. 4A and B).
FIG 4.
14-3-3 proteins increase Cdt2's half-life by protecting it from ubiquitination. (A) 14-3-3γ increases the half-life of Cdt2. 293T cells were transfected with plasmids expressing Flag-Cdt2 in combination with control or myc–14-3-3γ-expressing plasmids. Cells were treated with 80 μg/ml cycloheximide (CHx) for the times indicated. The cell lysates were probed with anti-Flag and anti-myc antibodies. The Flag-Cdt2 bands were quantitated, and the results plotted graphically. (B) 14-3-3ε increases the half-life of Cdt2. 293T cells were transfected with plasmids expressing Flag-Cdt2 in combination with control or myc–14-3-3ε-expressing plasmids. The procedure was otherwise as described for panel A. (C) 14-3-3 depletion increases Cdt2 ubiquitination. 293T cells were treated with control GL2 or 14-3-3 (θ, ζ, ε, and γ) siRNAs 24 h before transfection with the indicated plasmids. Cells were again treated with siRNAs 24 h after plasmid transfection. At 48 h after plasmid transfection, MG132 (40 μM) was added for 1 h before cells were harvested in denaturing ubiquitination buffer. The lysates were immunoprecipitated with anti-Flag antibody and probed first with anti-HA antibody to detect ubiquitinated forms of Cdt2 and then with anti-Flag antibodies to detect the immunoprecipitated Flag-Cdt2. (D) Overexpressing 14-3-3 proteins decreases Cdt2 ubiquitination. 293T cells were transfected with plasmids as indicated. The procedure was otherwise as described for panel C. The bottom bands represent 0.5% of the input lysates.
We next investigated whether 14-3-3 proteins prevent the degradation of Cdt2 by interfering with its polyubiquitination. The 14-3-3θ, -ζ, -ε, and -γ isoforms were individually knocked down using their corresponding siRNAs, and cells transfected with plasmids expressing Flag-Cdt2 and HA-ubiquitin. To measure the polyubiquitination of the Flag-Cdt2 in vivo, the cells were treated with MG132 before harvest, Flag-Cdt2 immunoprecipitated, and the slower moving polyubiquitinated isoforms detected by immunoblotting for HA-ubiquitin (Fig. 4C). Depleting the 14-3-3θ or -ζ isoform from the cells did not change the basal polyubiquitination pattern of Flag-Cdt2, and thus, these cells served as negative controls. In contrast, silencing of the 14-3-3ε or 14-3-3γ isoform drastically increased the polyubiquitinated forms of Flag-Cdt2 (Fig. 4C).
In the converse experiment, we tested whether the overexpression of myc–14-3-3γ or myc–14-3-3ε decreased the polyubiquitination of Flag-Cdt2 in vivo. Cells were cotransfected with plasmids expressing Flag-Cdt2, HA-ubiquitin, and either myc–14-3-3γ or myc–14-3-3ε (Fig. 4D). Overexpression of both myc–14-3-3γ and myc–14-3-3ε decreased the polyubiquitination of Flag-Cdt2. All of these experimental results indicate that 14-3-3γ and -ε protect Cdt2 from polyubiquitination and subsequent proteasomal degradation.
14-3-3 proteins shield Cdt2 from SCFFbxO11 E3 ligase activity.
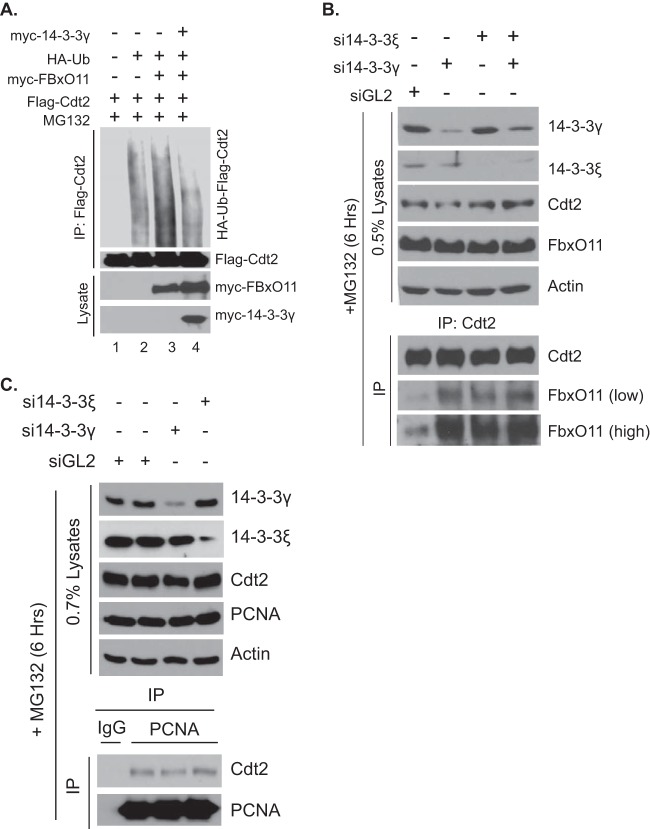
Recent studies have shown that Cdt2 is the target of SCFFbxO11 E3 ligase activity (23, 24) and that the ubiquitination is dependent on a degron that includes residues 456 to 464 of Cdt2. Since we have already shown that phosphorylation of T464 of Cdt2 is important for the interaction of Cdt2 with 14-3-3, we examined whether a 14-3-3 protein can shield Cdt2 from FbxO11-mediated polyubiquitination. Cells were cotransfected with plasmids expressing Flag-Cdt2, myc-FbxO11, and myc–14-3-3γ, as indicated in Fig. 5A, followed by measuring Flag-Cdt2 polyubiquitination in vivo. Overexpression of myc-FbxO11 stimulated the polyubiquitination of Flag-Cdt2 (Fig. 5A, lane 3), and this enhancement in the polyubiquitination of Cdt2 was overcome by co-overexpressing myc–14-3-3γ (Fig. 5A, lane 4). To investigate the molecular mechanism by which 14-3-3 proteins decrease FbxO11-stimulated polyubiquitination of Cdt2, we hypothesized that 14-3-3 proteins act as a barrier to prevent FbxO11 from interacting with Cdt2. As 14-3-3γ and 14-3-3ε interact with and stabilize Cdt2 (Fig. 1C and D, 3B and C, and 4A and B), we tested whether silencing of 14-3-3γ or -ε increased the interaction between Cdt2 and FbxO11 (Fig. 5B). Cells were transfected with control siRNA or siRNA against 14-3-3γ or 14-3-3ε for 48 h and with MG132 for 6 h to prevent the degradation of Cdt2 as a consequence of 14-3-3 silencing. Immunoprecipitation of Cdt2 copurified FbxO11 under control conditions, but the amount of coimmunoprecipitated FbxO11 increased significantly when 14-3-3γ or 14-3-3ε or both were silenced with their corresponding siRNAs (Fig. 5B).
FIG 5.
14-3-3 proteins block FbxO11 from binding and ubiquitinating Cdt2. (A) 293T cells were transfected with plasmids as indicated. The procedure was otherwise as described in the legend to Fig. 4B. The bottom bands represent 0.5% of the input lysates. (B) 293T cells were treated with the indicated control GL2, 14-3-3γ, or 14-3-3ε siRNAs for 48 h. Before harvesting, cells were treated with MG132 (40 μM) for 6 h to allow the accumulation of Cdt2. The lysates were immunoprecipitated with anti-Cdt2 antibodies and probed with anti-Cdt2 and anti-FbxO11 antibodies (bottom). Top, input lysates. (C) Coimmunoprecipitation of PCNA and Cdt2. 293T cells were treated with the siRNAs indicated and MG132 as described for panel B. The extracts were immunoprecipitated with anti-PCNA antibodies and probed with anti-Cdt2 and anti-PCNA antibodies (bottom). Top, input lysates.
PCNA is an important factor for the degradation of many chromatin-bound CRL4Cdt2 substrates, and the depletion of PCNA itself destabilizes Cdt2 protein (4, 23). To investigate whether 14-3-3 depletion interferes with the interaction between Cdt2 and PCNA and, thus, destabilizes Cdt2, cells were transfected with 14-3-3 siRNAs and MG132 as described above. Immunoprecipitation of PCNA copurifies Cdt2 irrespective of the presence or absence of 14-3-3, indicating that 14-3-3 depletion does not destabilize Cdt2 protein through dissociation of PCNA (Fig. 5C). This result also rules out the possibility that 14-3-3 depletion leads to mislocalization of Cdt2 in a nonnuclear cellular compartment. In addition, subcellular fractionation showed that Cdt2 was not mislocalized after overexpression of 14-3-3 (data not shown). Overall, these results demonstrate that 14-3-3 proteins prevent Cdt2 from interacting with FbxO11 and, thus, shield Cdt2 from CRL1FbxO11-mediated polyubiquitination.
Phosphorylation of T464 stabilizes Cdt2 in a 14-3-3-dependent manner.
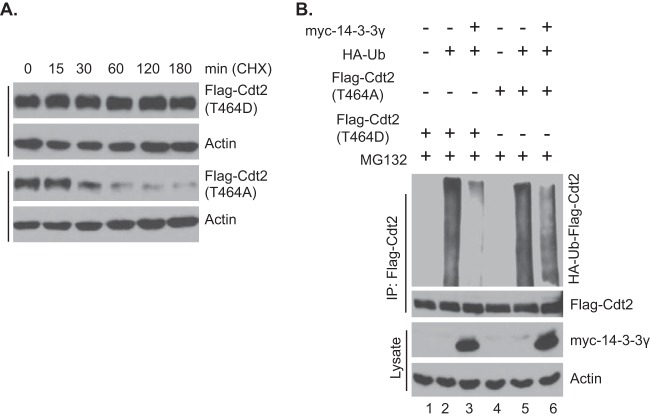
T464 of Cdt2 was believed to be essential for the degradation of Cdt2 because it is required for stable interaction of Cdt2 with FbxO11 (24). Mutating T464 into phosphomimetic D464 or nonphosphorylatable A464 does not allow Cdt2 to associate with FbxO11 in coimmunoprecipitation experiments (24), and so, one would have expected that Cdt2(T464D) and Cdt2(T464A) would both be equally stable in vivo. However, a different result emerged when we compared the half-lives of Flag-Cdt2(T464D) and Flag-Cdt2(T464A) by treating cells with cycloheximide (Fig. 6A): Flag-Cdt2(T464A) had a half-life of <0.5 h, whereas Flag-Cdt2(T464D) was enormously stable, with a half-life of >3 h. These results suggest that, although Cdt2(T464A) does not stably associate with FbxO11 for the latter to appear in coimmunoprecipitation experiments, the Cdt2 is still susceptible to the E3 ligase, perhaps through transient interactions. On the other hand, 14-3-3 proteins interact with Cdt2(T464D) but not with Cdt2(T464A) (Fig. 2D), consistent with the differential stability of the two mutants in vivo. Thus, the phosphorylation of T464 most likely protects Cdt2 from degradation because the phosphorylation promotes interaction with 14-3-3, which shields the Cdt2 from even transient interaction with FbxO11.
FIG 6.
14-3-3 proteins protect Cdt2 phosphorylated at T464 from degradation. (A) 293T cells were transfected with Flag-Cdt2 containing phosphomimetic or nonphosphorylatable residues at position 464 as indicated and treated with cycloheximide (80 μg/ml) for different times. The cell lysates were probed with anti-Flag antibodies. (B) 293T cells were transfected with plasmids as indicated. The procedure was otherwise as described in the legend to Fig. 4D. Bottom, 0.5% of the input lysates.
This hypothesis predicts that FbxO11 should be able to polyubiquitinate Cdt2(T464A) and Cdt2(T464D) but that polyubiquitination of the latter and not the former will be inhibited by coexpression of 14-3-3 protein. Indeed, transfection of Flag-Cdt2(T464D) and Flag-Cdt2(T464A) with HA-ubiquitin showed that both isoforms were polyubiquitinated in vivo (Fig. 6B, lanes 2 and 5). However, coexpression of myc–14-3-3γ protected Flag-Cdt2(T464D) from polyubiquitination much more than it protected Flag-Cdt2(T464A) (Fig. 6B, lanes 3 and 6). Therefore, 14-3-3 binding to phosphorylated Cdt2(T464) promotes binding to 14-3-3 proteins in vivo, which shields Cdt2 from the E3 ligase.
Depletion of 14-3-3γ arrests the cell cycle at G2/M partly by increasing the CRL4Cdt2 substrate, Set8.
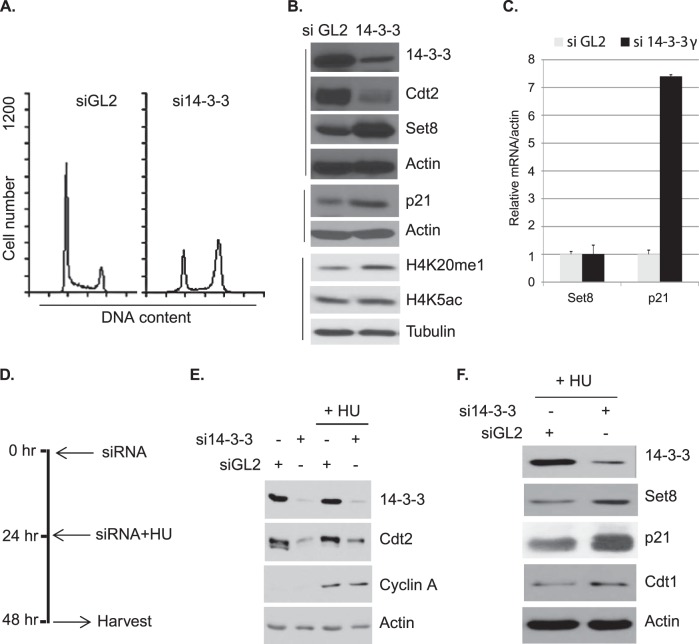
14-3-3 proteins, particularly 14-3-3ε, regulate the cell cycle by binding to various cyclin-dependent kinases (CDKs) and Cdc25 phosphatases (Cdc25A, -B, and -C) at different phases of the cell cycle (27). Likewise, CRL4Cdt2 E3 ligase regulates the cell cycle by degrading various substrate proteins, such as p21, Set8, and Cdt1, at G1/S phase (4). Having discovered that 14-3-3γ is required for Cdt2 stability, we asked whether 14-3-3γ also regulates the cell cycle by protecting the activity of CRL4Cdt2. siRNA-mediated knockdown of 14-3-3γ arrested cells in G2/M phase (Fig. 7A). Analysis of cell lysates showed that 14-3-3γ silencing leads to a decrease in Cdt2 and a concomitant increase in Set8 and its downstream histone H4-lysine 20 monomethylation (H4K20me1) mark (Fig. 7B). There is also an increase in p21 protein (Fig. 7B). Next, we measured the mRNA levels of Set8 and p21 in control and 14-3-3γ-depleted cells. As shown by the results in Fig. 7C, 14-3-3γ knockdown causes a marginal increment in Set8 mRNA, whereas p21 mRNA is increased very significantly. Thus, the increase in Set8 protein upon 14-3-3γ depletion is via destabilization of Cdt2, which leads to stabilization of Set8 protein, whereas the p21 increase could be due to the transcriptional activation of p21, perhaps due to an increased expression of p53.
FIG 7.
Silencing 14-3-3γ decreases Cdt2 in S phase, stabilizes Cdt2 substrates, and arrests the cell cycle at G2/M. (A) Fluorescence-activated cell sorting (FACS) profile of HeLa cells treated with control GL2 or 14-3-3γ siRNAs for 60 h. (B) HeLa cells were treated with control GL2 or 14-3-3γ siRNAs for 60 h, and the lysates probed with the indicated antibodies. (C) Relative q-PCR analysis of Set8 and p21 transcripts in HeLa cells treated with the indicated siRNAs. Error bars show standard deviations. (D) The timeline of siRNA and HU treatment of HeLa cells in the experimental scheme is shown. (E and F) The lysates prepared from HeLa cells treated with or without HU were probed with the indicated antibodies.
The CRL4Cdt2 E3 ligase complex is active during the S phase of the cell cycle and degrades p21, Set8, and Cdt1 for smooth progression of the cell cycle. Since depletion of 14-3-3γ destabilizes Cdt2, with a concomitant increase in Cdt2 substrates (Fig. 7B), we asked whether 14-3-3γ is responsible for the stabilization of Cdt2 during S phase. Silencing of 14-3-3γ in HU-treated HeLa cells destabilizes Cdt2, with a consequent increase in the Cdt2 substrates Set8, p21, and Cdt1 (Fig. 7D to F). The marginal Cdt1 stabilization seen after 14-3-3γ knockdown in S phase cells is due to the fact that another E3 ligase, CRL1Skp2, degrades Cdt1 during S phase (31, 32). These results (Fig. 7E and F) and the phosphorylation-dependent binding of 14-3-3 proteins with Cdt2 in S phase (Fig. 2F and G) suggest that 14-3-3 proteins are required for maintaining Cdt2 protein stability during the S phase of the cell cycle.
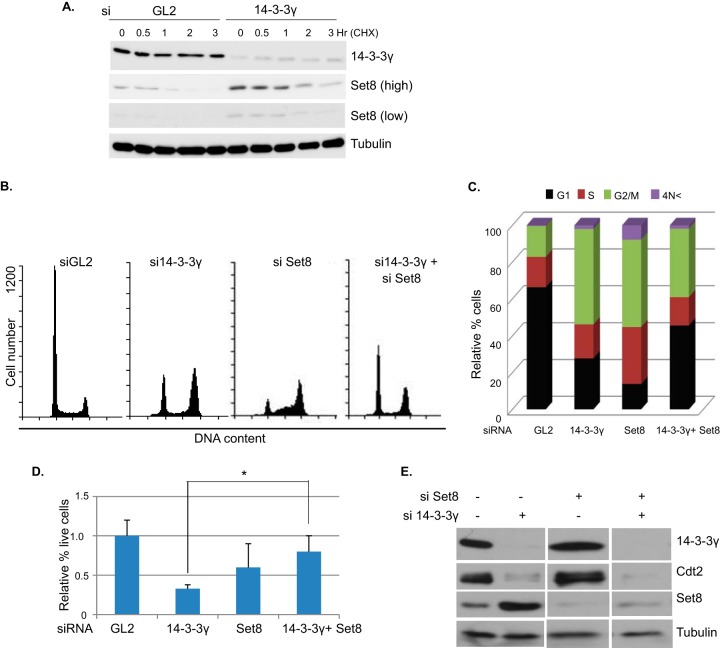
Since the posttranscriptional elevation of Set8 was most likely to be proximate to the decrease of Cdt2, we focused the following experiments on Set8. The half-life of Set8 was measured by treating cells with cycloheximide before and after depleting 14-3-3γ, to confirm that 14-3-3γ knockdown increases the half-life of Set8 (Fig. 8A). Conditional knockdown of Set8 or overexpression of CRL4Cdt2-resistant Set8 (Set8ΔPIP) causes cell cycle arrest in G2/M phase (5, 33). Since 14-3-3γ knockdown results in the accumulation of Set8 (a condition equivalent to the expression of Set8ΔPIP), we asked whether the G2/M arrest resulting from 14-3-3γ silencing could be rescued by decreasing the accumulation of Set8 by transfecting cells with an siRNA against Set8. As shown by the results in Fig. 8B and C, silencing 14-3-3γ or Set8 arrested cells in the G2/M stage of the cell cycle. However, knockdown of 14-3-3γ together with Set8 partly rescued the cell cycle progression (Fig. 8B and C). Consistent with this, knockdown of 14-3-3γ decreased cell proliferation, and coknockdown of Set8 partially rescued this defect (Fig. 8D). The efficiency of knockdown of the various proteins in these experiments was determined from a batch of cells grown and treated with siRNAs in parallel (Fig. 8E). Note that si-Set8 alone knocked down Set8 to levels that still allowed S phase progression and led to a block in G2 (Fig. 8B and C). Thus, the rescue of the G2 block by the combined knockdown of Set8 and 14-3-3γ is not secondary to the type of G1/S block that is seen after a more severe knockdown of Set8 (34). These results demonstrate that 14-3-3γ contributes to cell cycle progression by acting as a stabilizing factor for Cdt2 and thus preventing the accumulation of CRLCdt2 substrates like Set8.
FIG 8.
Preventing the accumulation of Set8 partly rescues the G2/M arrest seen when 14-3-3γ is depleted. (A) Half-life measurement of Set8 protein in siRNA-treated HeLa cells using cycloheximide (100 μg/ml) for different times. The lysates were analyzed by Western blotting with indicated antibodies. (B and C) FACS analysis of different cell cycle phases in HeLa cells after treatment with the siRNAs indicated for 60 h. 4N<, cells with rereplicated DNA. (D) HeLa cells were treated with the siRNAs indicated for 60 h. The relative numbers of viable cells are plotted as the averages of the results of the three independent experiments. Error bars show standard deviations. *, P < 0.05, two-tailed Student's t test. (E) HeLa cell lysates from experiments performed in parallel to those whose results are shown in panels B, C, and D were probed with the indicated antibodies.
DISCUSSION
The cell cycle transitions are driven by various E3 ubiquitin ligases that ubiquitinate and degrade specific target proteins (1). CRL4Cdt2 regulates G1/S and S phase progression of the cell cycle by degrading p21, Cdt1, and Set8 methyltransferase (4). p21 blocks the cell cycle at G1/S by inhibiting Cdk1 and also arrests cells at G2/M after DNA damage by blocking the interaction of Cdk1 with Cdk1-activating kinase (35). Cdt1 is involved in replication initiation, and its degradation by CRL4Cdt2 in S phase prevents replication relicensing (36). Set8 is targeted for destruction in S phase by CRL4Cdt2 in order to allow histone gene transcription and translation and progression through G2/M phase (5). Despite CRL4Cdt2 being a critical regulator of the cell cycle, little is known about the regulation of this E3 ligase.
In normally growing cells, Cdt2 (the substrate adaptor protein of the CRL4Cdt2 complex) is phosphorylated at threonine 464 by cyclin A/Cdk2 and cyclin B/Cdk1. Although Cdt2 is phosphorylated (at T464) throughout the cell cycle, hyperphosphorylation, involving additional sites, is seen in mitosis (Fig. 2E). The T464 phosphorylation in S phase prevents Cdt2 from interacting with another E3 ligase, SCFFbxO11, and thus protects it from ubiquitin-mediated degradation (24). In this report, we have identified that 14-3-3 proteins bind Cdt2 phosphorylated at T464 during S phase (Fig. 2F and G) to protect it from ubiquitin-mediated degradation (Fig. 2A to D, 3A to C, 4C and D, and 6B). The amino acid sequence around phospho-T464 of Cdt2, which is recognized and bound by 14-3-3 adaptors, does not match the consensus sequences that 14-3-3 proteins prefer (Fig. 2B), but this is something that has already been reported in many other contexts (27, 37, 38). Overexpressing 14-3-3γ overcomes FbxO11-mediated ubiquitination of Cdt2 (Fig. 5A), whereas silencing 14-3-3 proteins promotes interaction between Cdt2 and FbxO11 in cells (Fig. 5B). It is intriguing that the hyperphosphorylated Cdt2 in M phase is dissociated from 14-3-3 and this dissociation is accompanied by a decrease in Cdt2, perhaps due to destabilization of Cdt2. Further mapping of the sites on Cdt2 that are hyperphosphorylated in mitosis is required to prove this hypothesis and to test whether this mitotic regulation of Cdt2 is important for cell cycle progression.
Since the interaction of Cdt2 and 14-3-3 was discovered in a yeast two-hybrid screen, we assume either that yeast Cdk phosphorylated the Cdt2 in the bait on T464 or that in the artificial conditions of a yeast two-hybrid screen where both partner proteins are highly overexpressed, the residual interaction between 14-3-3 protein and Cdt2 is sufficient to reconstitute a functional transcriptional activator on the GAL4 binding sites upstream from the HIS3 promoter.
Both phosphomimetic Flag-Cdt2(T464D) and nonphosphorylatable Flag-Cdt2 (464A) have been reported not to coimmunoprecipitate with FbxO11 (24), but to our surprise, both are equally ubiquitinated when overexpressed in cells (Fig. 6B). We suggest that coimmunoprecipitation measures a more stringent interaction between Cdt2 and FbxO11 than is necessary for the catalytic interaction of FbxO11 with its substrate. In other words, even though Cdt2(T464A) and Cdt2(T464D) do not coimmunoprecipitate with FbxO11, they can still get ubiquitinated through transient interactions.
Compared to nonphosphorylatable Flag-Cdt2(464A), phosphomimetic Flag-Cdt2(T464D) is quite stable (Fig. 6A), because 14-3-3γ shields Flag-Cdt2(T464D) but not Flag-Cdt2(464A) from ubiquitination (Fig. 6B). These observations indicate that 14-3-3 shields Cdt2 from FbxO11 and, perhaps, other yet-to-be-identified E3 ligases.
We show that two isoforms of the 14-3-3 family, 14-3-3ε and 14-3-3γ, can stabilize Cdt2 independent of each other when overexpressed (Fig. 3J). Such independent function would suggest redundancy between the two isoforms. Cdt2 knockout is lethal during early embryonic development of mice (13, 39). In this context, the lack of any phenotype in mice when 14-3-3γ is deleted (13, 39) may be explained if the Cdt2 levels in the mice are protected by 14-3-3ε protein. Why then, does the knockdown of the γ or ε isoform by itself cause a decrease in Cdt2 in cancer cell lines in culture? We propose that an increase in the level of FbxO11, activity of other E3 ligases that could act on Cdt2, activity of phosphatases that dephosphorylate phospho-T464 of Cdt2, or increased demand for 14-3-3 proteins make the cancer cell lines in culture highly dependent on adequate levels of both isoforms of 14-3-3 to stabilize the Cdt2.
CRL4Cdt2 degrades some of its substrates, like Set8, p21, Cdt1, and Cdc6, in a PCNA-dependent and replication-coupled manner during early S phase (4, 9). The 14-3-3 proteins bind Cdt2 in a phosphorylation-dependent manner (Fig. 2F and G) and maintain the latter's stability during S phase (Fig. 7E). How Cdt2 is recruited to chromatin and recognizes these PCNA-bound substrates on chromatin is not clear. A recent study has found that 14-3-3γ and 14-3-3ε are associated with nascent replicating chromatin (40). It will be interesting to study whether the 14-3-3 proteins mediate the recruitment of phosphorylated Cdt2 on chromatin to explain the mechanism of degradation of the PCNA-bound substrates by CRL4Cdt2 E3 ligase.
14-3-3 proteins are known to regulate the cell cycle by sequestering Cdc25A, Cdc25B, and Cdc25C phosphatases in the cytoplasm (27). Our results suggest that 14-3-3 proteins also regulate the cell cycle by protecting CRL4Cdt2 activity and, thus, restricting the level of Set8. Depletion of 14-3-3γ decreases cell proliferation by arresting cells in G2/M (Fig. 7A and 8B to D). Overexpression of CRL4Cdt2-resistant Set8 (Set8ΔPIP) also arrests cells in G2/M, perhaps due to undetectable levels of rereplication and attendant DNA damage (5, 8). The increase in Set8 levels by 14-3-3γ depletion is equivalent to the overexpression of Set8ΔPIP (Fig. 7B and F). That is why depletion of 14-3-3γ together with a partial Set8 depletion abrogates G2/M arrest and partly rescues the cell growth (Fig. 8B to D).
The transcriptional induction of p21 after 14-3-3γ depletion (Fig. 7C) is at odds with the report that 14-3-3ζ interacts with phosphorylated histone H3 at the p21 promoter and keeps the promoter in an induced state (41). The difference is probably due to the different roles of the γ isoform (tested here) and the ζ isoform. The mechanism of induction of p21 mRNA after 14-3-3γ knockdown is to be studied in the future. One possibility is that 14-3-3γ is repressing transcriptional activation of p53 in a Sirt2-dependent manner (42). Thus, the upregulation of p21 transcription (Fig. 7C) that we observe upon 14-3-3γ depletion may be a consequence of p53 accumulation. The other possibility is that the increase of Set8 causes DNA damage and activates p53 expression and that of its downstream transcriptional target p21 (5). If this is true, we expect the coknockdown of Set8 and 14-3-3γ to prevent the p21 induction. Alternatively, the p21 induction could be independent of p53 or Set8 and may indicate a new role of 14-3-3γ in repressing the p21 promoter.
Both 14-3-3γ and Cdt2 are associated with cell proliferation (13, 43). Cdt2 protein levels are high in several human malignancies, such as colon, breast, hepatic, and gastric cancers (15–18). It will be interesting to see whether increased expression of 14-3-3 proteins is important for maintaining the high levels of Cdt2 in these cancers. There are additional examples in the literature where the increase or decrease of 14-3-3 proteins leads to changes in cell proliferation or cell survival, and the question arises as to whether Cdt2 stabilization plays a role in these reported functions of 14-3-3 proteins. For example, 14-3-3γ is highly expressed in response to interleukin-3 (IL-3) in Ba/F3 immortalized bone marrow cells, and overexpression of 14-3-3γ overrides the dependency of these cells on IL-3 (43). Whether the IL-3 independence seen upon 14-3-3γ overexpression is mediated by the stabilization of Cdt2 is an example of the type of questions that will now need to be investigated.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Dutta laboratory for discussion, suggestions, and helpful comments. We are grateful to Michele Pagano (NYU Cancer Institute) for providing phospho-Cdt2 antibody.
This work is supported by R01 grants CA60499 and CA166054 to A. Dutta.
Footnotes
Published ahead of print 25 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00838-14.
REFERENCES
- 1.Teixeira LK, Reed SI. 2013. Ubiquitin ligases and cell cycle control. Annu. Rev. Biochem. 82:387–414. 10.1146/annurev-biochem-060410-105307. [DOI] [PubMed] [Google Scholar]
- 2.Deshaies RJ, Joazeiro CA. 2009. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78:399–434. 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 3.Hua Z, Vierstra RD. 2011. The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 62:299–334. 10.1146/annurev-arplant-042809-112256. [DOI] [PubMed] [Google Scholar]
- 4.Havens CG, Walter JC. 2011. Mechanism of CRL4(Cdt2), a PCNA-dependent E3 ubiquitin ligase. Genes Dev. 25:1568–1582. 10.1101/gad.2068611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A. 2010. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol. Cell 40:9–21. 10.1016/j.molcel.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. 2008. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 22:2496–2506. 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arias EE, Walter JC. 2006. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat. Cell Biol. 8:84–90. 10.1038/ncb1346. [DOI] [PubMed] [Google Scholar]
- 8.Centore RC, Havens CG, Manning AL, Li JM, Flynn RL, Tse A, Jin J, Dyson NJ, Walter JC, Zou L. 2010. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol. Cell 40:22–33. 10.1016/j.molcel.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clijsters L, Wolthuis R. 2014. PIP-box-mediated degradation prohibits re-accumulation of Cdc6 during S phase. J. Cell Sci. 127:1336–1345. 10.1242/jcs.145862. [DOI] [PubMed] [Google Scholar]
- 10.Hall JR, Lee HO, Bunker BD, Dorn ES, Rogers GC, Duronio RJ, Cook JG. 2008. Cdt1 and Cdc6 are destabilized by rereplication-induced DNA damage. J. Biol. Chem. 283:25356–25363. 10.1074/jbc.M802667200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim Y, Starostina NG, Kipreos ET. 2008. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 22:2507–2519. 10.1101/gad.1703708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Senga T, Sivaprasad U, Zhu W, Park JH, Arias EE, Walter JC, Dutta A. 2006. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J. Biol. Chem. 281:6246–6252. 10.1074/jbc.M512705200. [DOI] [PubMed] [Google Scholar]
- 13.Liu CL, Yu IS, Pan HW, Lin SW, Hsu HC. 2007. L2dtl is essential for cell survival and nuclear division in early mouse embryonic development. J. Biol. Chem. 282:1109–1118. 10.1074/jbc.M606535200. [DOI] [PubMed] [Google Scholar]
- 14.Mackintosh C, Ordonez JL, Garcia-Dominguez DJ, Sevillano V, Llombart-Bosch A, Szuhai K, Scotlandi K, Alberghini M, Sciot R, Sinnaeve F, Hogendoorn PC, Picci P, Knuutila S, Dirksen U, Debiec-Rychter M, Schaefer KL, de Alava E. 2012. 1q gain and CDT2 overexpression underlie an aggressive and highly proliferative form of Ewing sarcoma. Oncogene 31:1287–1298. 10.1038/onc.2011.317. [DOI] [PubMed] [Google Scholar]
- 15.Baraniskin A, Birkenkamp-Demtroder K, Maghnouj A, Zollner H, Munding J, Klein-Scory S, Reinacher-Schick A, Schwarte-Waldhoff I, Schmiegel W, Hahn SA. 2012. MiR-30a-5p suppresses tumor growth in colon carcinoma by targeting DTL. Carcinogenesis 33:732–739. 10.1093/carcin/bgs020. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Ng EK, Ng YP, Wong CY, Yu J, Jin H, Cheng VY, Go MY, Cheung PK, Ebert MP, Tong J, To KF, Chan FK, Sung JJ, Ip NY, Leung WK. 2009. Identification of retinoic acid-regulated nuclear matrix-associated protein as a novel regulator of gastric cancer. Br. J. Cancer 101:691–698. 10.1038/sj.bjc.6605202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan HW, Chou HY, Liu SH, Peng SY, Liu CL, Hsu HC. 2006. Role of L2DTL, cell cycle-regulated nuclear and centrosome protein, in aggressive hepatocellular carcinoma. Cell Cycle 5:2676–2687. 10.4161/cc.5.22.3500. [DOI] [PubMed] [Google Scholar]
- 18.Ueki T, Nishidate T, Park JH, Lin ML, Shimo A, Hirata K, Nakamura Y, Katagiri T. 2008. Involvement of elevated expression of multiple cell-cycle regulator, DTL/RAMP (denticleless/RA-regulated nuclear matrix associated protein), in the growth of breast cancer cells. Oncogene 27:5672–5683. 10.1038/onc.2008.186. [DOI] [PubMed] [Google Scholar]
- 19.Lin JJ, Milhollen MA, Smith PG, Narayanan U, Dutta A. 2010. NEDD8-targeting drug MLN4924 elicits DNA rereplication by stabilizing Cdt1 in S phase, triggering checkpoint activation, apoptosis, and senescence in cancer cells. Cancer Res. 70:10310–10320. 10.1158/0008-5472.CAN-10-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lydeard JR, Schulman BA, Harper JW. 2013. Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep. 14:1050–1061. 10.1038/embor.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, Cullis CA, Doucette A, Garnsey JJ, Gaulin JL, Gershman RE, Lublinsky AR, McDonald A, Mizutani H, Narayanan U, Olhava EJ, Peluso S, Rezaei M, Sintchak MD, Talreja T, Thomas MP, Traore T, Vyskocil S, Weatherhead GS, Yu J, Zhang J, Dick LR, Claiborne CF, Rolfe M, Bolen JB, Langston SP. 2009. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458:732–736. 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 22.Ishii T, Shiomi Y, Takami T, Murakami Y, Ohnishi N, Nishitani H. 2010. Proliferating cell nuclear antigen-dependent rapid recruitment of Cdt1 and CRL4Cdt2 at DNA-damaged sites after UV irradiation in HeLa cells. J. Biol. Chem. 285:41993–42000. 10.1074/jbc.M110.161661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abbas T, Mueller AC, Shibata E, Keaton M, Rossi M, Dutta A. 2013. CRL1-FBXO11 promotes Cdt2 ubiquitylation and degradation and regulates Pr-Set7/Set8-mediated cellular migration. Mol. Cell 49:1147–1158. 10.1016/j.molcel.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossi M, Duan S, Jeong YT, Horn M, Saraf A, Florens L, Washburn MP, Antebi A, Pagano M. 2013. Regulation of the CRL4(Cdt2) ubiquitin ligase and cell-cycle exit by the SCF(Fbxo11) ubiquitin ligase. Mol. Cell 49:1159–1166. 10.1016/j.molcel.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hermeking H. 2003. The 14-3-3 cancer connection. Nat. Rev. Cancer 3:931–943. 10.1038/nrc1230. [DOI] [PubMed] [Google Scholar]
- 26.Reinhardt HC, Yaffe MB. 2013. Phospho-Ser/Thr-binding domains: navigating the cell cycle and DNA damage response. Nat. Rev. Mol. Cell Biol. 14:563–580. 10.1038/nrm3640. [DOI] [PubMed] [Google Scholar]
- 27.Hermeking H, Benzinger A. 2006. 14-3-3 proteins in cell cycle regulation. Semin. Cancer Biol. 16:183–192. 10.1016/j.semcancer.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Margolis SS, Perry JA, Forester CM, Nutt LK, Guo Y, Jardim MJ, Thomenius MJ, Freel CD, Darbandi R, Ahn JH, Arroyo JD, Wang XF, Shenolikar S, Nairn AC, Dunphy WG, Hahn WC, Virshup DM, Kornbluth S. 2006. Role for the PP2A/B56delta phosphatase in regulating 14-3-3 release from Cdc25 to control mitosis. Cell 127:759–773. 10.1016/j.cell.2006.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shibata E, Dar A, Dutta A. 2014. CRL4Cdt2 E3 ubiquitin ligase and proliferating cell nuclear antigen (PCNA) cooperate to degrade thymine DNA glycosylase in S phase. J. Biol. Chem. 289:23056–23064. 10.1074/jbc.M114.574210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dar A, Shibata E, Dutta A. 2013. Deubiquitination of Tip60 by USP7 determines the activity of the p53-dependent apoptotic pathway. Mol. Cell. Biol. 33:3309–3320. 10.1128/MCB.00358-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim Y, Kipreos ET. 2007. Cdt1 degradation to prevent DNA re-replication: conserved and non-conserved pathways. Cell Div. 2:18. 10.1186/1747-1028-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeda DY, Parvin JD, Dutta A. 2005. Degradation of Cdt1 during S phase is Skp2-independent and is required for efficient progression of mammalian cells through S phase. J. Biol. Chem. 280:23416–23423. 10.1074/jbc.M501208200. [DOI] [PubMed] [Google Scholar]
- 33.Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, Torres-Padilla ME, Heard E, Reinberg D. 2009. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol. Cell. Biol. 29:2278–2295. 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jorgensen S, Eskildsen M, Fugger K, Hansen L, Larsen MS, Kousholt AN, Syljuasen RG, Trelle MB, Jensen ON, Helin K, Sorensen CS. 2011. SET8 is degraded via PCNA-coupled CRL4(CDT2) ubiquitylation in S phase and after UV irradiation. J. Cell Biol. 192:43–54. 10.1083/jcb.201009076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abbas T, Dutta A. 2009. p21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer 9:400–414. 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arias EE, Walter JC. 2005. Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev. 19:114–126. 10.1101/gad.1255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henriksson ML, Francis MS, Peden A, Aili M, Stefansson K, Palmer R, Aitken A, Hallberg B. 2002. A nonphosphorylated 14-3-3 binding motif on exoenzyme S that is functional in vivo. Eur. J. Biochem. 269:4921–4929. 10.1046/j.1432-1033.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- 38.Zhang L, Wang H, Liu D, Liddington R, Fu H. 1997. Raf-1 kinase and exoenzyme S interact with 14-3-3zeta through a common site involving lysine 49. J. Biol. Chem. 272:13717–13724. 10.1074/jbc.272.21.13717. [DOI] [PubMed] [Google Scholar]
- 39.Steinacker P, Schwarz P, Reim K, Brechlin P, Jahn O, Kratzin H, Aitken A, Wiltfang J, Aguzzi A, Bahn E, Baxter HC, Brose N, Otto M. 2005. Unchanged survival rates of 14-3-3gamma knockout mice after inoculation with pathological prion protein. Mol. Cell. Biol. 25:1339–1346. 10.1128/MCB.25.4.1339-1346.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alabert C, Bukowski-Wills JC, Lee SB, Kustatscher G, Nakamura K, de Lima Alves F, Menard P, Mejlvang J, Rappsilber J, Groth A. 2014. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 16:281–293. 10.1038/ncb2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simboeck E, Sawicka A, Zupkovitz G, Senese S, Winter S, Dequiedt F, Ogris E, Di Croce L, Chiocca S, Seiser C. 2010. A phosphorylation switch regulates the transcriptional activation of cell cycle regulator p21 by histone deacetylase inhibitors. J. Biol. Chem. 285:41062–41073. 10.1074/jbc.M110.184481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin YH, Kim YJ, Kim DW, Baek KH, Kang BY, Yeo CY, Lee KY. 2008. Sirt2 interacts with 14-3-3 beta/gamma and down-regulates the activity of p53. Biochem. Biophys. Res. Commun. 368:690–695. 10.1016/j.bbrc.2008.01.114. [DOI] [PubMed] [Google Scholar]
- 43.Ajjappala BS, Kim YS, Kim MS, Lee MY, Lee KY, Ki HY, Cha DH, Baek KH. 2009. 14-3-3 gamma is stimulated by IL-3 and promotes cell proliferation. J. Immunol. 182:1050–1060. 10.4049/jimmunol.182.2.1050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.