Abstract
Ribosome stalling is an important incident enabling the cellular quality control machinery to detect aberrant mRNA. Saccharomyces cerevisiae Hbs1-Dom34 and Ski7 are homologs of the canonical release factor eRF3-eRF1, which recognize stalled ribosomes, promote ribosome release, and induce the decay of aberrant mRNA. Polyadenylated nonstop mRNA encodes aberrant proteins containing C-terminal polylysine segments which cause ribosome stalling due to electrostatic interaction with the ribosomal exit tunnel. Here we describe a novel mechanism, termed premature translation termination, which releases C-terminally truncated translation products from ribosomes stalled on polylysine segments. Premature termination during polylysine synthesis was abolished when ribosome stalling was prevented due to the absence of the ribosomal protein Asc1. In contrast, premature termination was enhanced, when the general rate of translation elongation was lowered. The unconventional termination event was independent of Hbs1-Dom34 and Ski7, but it was dependent on eRF3. Moreover, premature termination during polylysine synthesis was strongly increased in the absence of the ribosome-bound chaperones ribosome-associated complex (RAC) and Ssb (Ssb1 and Ssb2). On the basis of the data, we suggest a model in which eRF3-eRF1 can catalyze the release of nascent polypeptides even though the ribosomal A-site contains a sense codon when the rate of translation is abnormally low.
INTRODUCTION
In order to keep the expression of aberrant proteins low, cells need to distinguish between transcripts suitable for translation and transcripts that contain erroneous information. A large group of aberrant transcripts is characterized by the lack of in-frame stop codons. Such stop codon-free mRNAs (see Fig. 1A) can arise, for example, when polyadenylation occurs prematurely or through point mutations that disrupt the stop codon (1–3). It was estimated that nonstop mRNA molecules represent approximately 1% of all polyadenylated transcripts in Saccharomyces cerevisiae (4, 5). In addition, endonucleolytic cleavage generates a pool of mRNA molecules that not only are devoid of stop codons but also lack the poly(A) tail (see Fig. 1A). In order to distinguish those transcripts from polyadenylated nonstop mRNA, we refer to them as stop codon-less mRNA (see Fig. 1A). Proteins encoded by nonstop mRNA (nonstop proteins) as well as stop codon-less mRNA (stop codon-less proteins) may be detrimental for the cell because their C termini are significantly altered (see Fig. 1A). A specialized mRNA decay pathway, termed nonstop mRNA decay (NSD), ensures exosome-mediated degradation of stop codon-free transcripts from the 3′ end (1–3). Stop codon-free mRNA that escapes NSD is translationally repressed (6), and residual translation products are subject to proteasomal degradation (2, 7).
FIG 1.
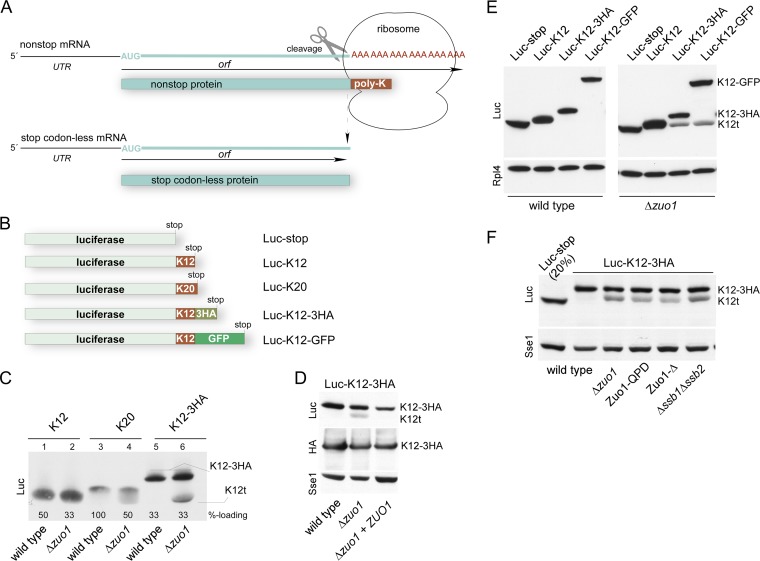
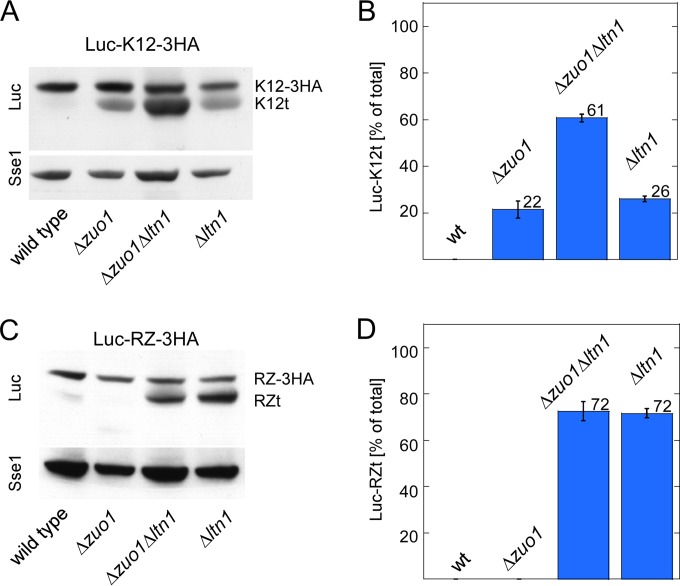
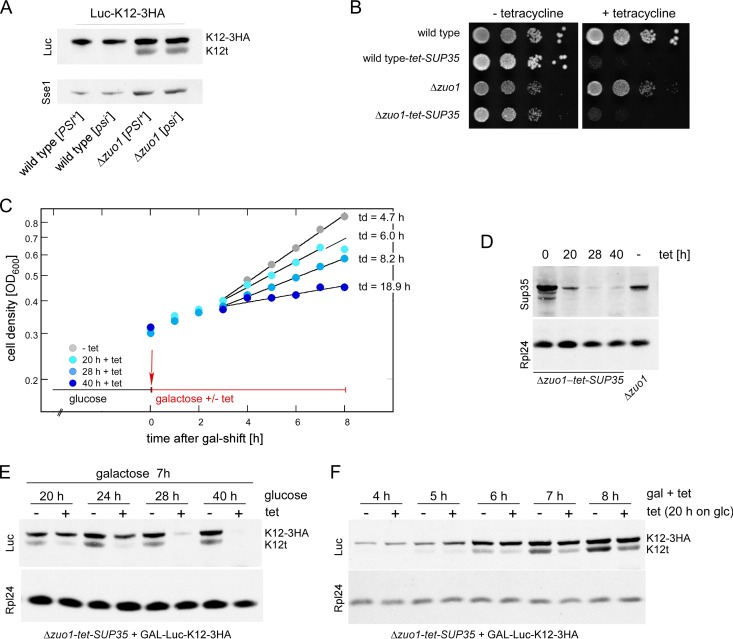
RAC/Ssb prevents expression of C-terminally truncated polylysine proteins. (A) Schematic representation of stop codon-free mRNAs and the corresponding translation products. Nonstop mRNA is characterized by the lack of in-frame stop codons between the AUG codon and poly(A) tail. If translation proceeds to the 3′ end of the nonstop mRNA, the translation product, which is termed nonstop protein, carries a polylysine tail (poly-K). Polylysine segments in the ribosomal tunnel cause stalling, which in turn induces endonucleolytic cleavage of the mRNA. The resulting cleavage product, termed stop codon-less mRNA, lacks a poly(A) tail, and accordingly, the corresponding stop codon-less protein lacks the polylysine segment. For details, see the introduction and Discussion. (B) Schematic representation of luciferase reporters containing C-terminal or internal polylysine segments. (C) Expression of polylysine reporters in a Δzuo1 strain. Total extracts of wild-type and Δzuo1 strains expressing luciferase reporters as indicated (compare to panel B) were analyzed via immunoblotting using an antibody directed against luciferase (Luc). Loading was adjusted to facilitate direct comparison of the luciferase band in the different strains (%-loading). Full-length Luc-K12-3HA (K12-3HA) and the luciferase fragment (K12t) are indicated (also in panels D to F). (D) Luc-K12t is a C-terminally truncated fragment of Luc-K12-3HA. Total extracts of wild-type strain, Δzuo1 strain, and Δzuo1 strain complemented with ZUO1 expressing Luc-K12-3HA were analyzed via immunoblotting using antibodies directed against luciferase (Luc), the HA epitope, and Sse1 as a loading control. (E) The amino acid sequence C terminal of the polylysine does not affect Luc-K12t expression. The total extracts of wild-type and Δzuo1 strains were separated on a single Tris-Tricine gel and subsequently analyzed via immunoblotting using antibodies directed against luciferase (Luc) and Rpl4 as a loading control. The immunoblot was cut to improve the clarity of the figure. (F) Luc-K12t is generated in strains with a nonfunctional RAC/Ssb system. The total extracts of the wild-type strain, Δzuo1 strain, Zuo1-QPD strain (J-domain inactive), Zuo1-Δ strain (ribosome binding deficient), and Δssb1 Δssb2 strain expressing Luc-K12-3HA were analyzed via immunoblotting using antibodies directed against luciferase (Luc) and Sse1 as a loading control. Fivefold-diluted total extract (see Materials and Methods) of the wild type expressing Luc-stop served as the size marker.
Because the poly(A) tail encodes lysines, nonstop proteins carry a positively charged C-terminal polylysine segment (see Fig. 1A) (8, 9). The polylysine segments interact with the negatively charged wall of the ribosomal tunnel, and this leads to a reduced rate of translation elongation, termed ribosomal stalling (9–11). Ribosomal stalling can also be caused by specific ribosome arrest peptides, as for example the arginine attenuator peptide, which causes Arg-regulated stalling of eukaryotic ribosomes (12, 13). Asc1, a nonessential WD protein of the small ribosomal subunit (14), is required for ribosomal stalling on positively charged nascent chain segments. The mechanism by which Asc1 affects the rate of translation elongation, however, is not understood (11, 15). Polylysine-induced ribosomal stalling provides an important signal for nonstop protein quality control. Ltn1, an E3 ligase required for the cotranslational polyubiquitination of nonstop proteins, is recruited to stalled ribosomes carrying nascent polylysine segments (11, 16–19). Moreover, polylysine segments connect nonstop to stop codon-less mRNA quality control, because stalling of ribosomes induces endonucleolytic cleavage of the mRNA in proximity to the stalled ribosome (2, 20–22). The 5′ fragment of nonstop mRNA cleavage possesses the properties of a stop codon-less mRNA (see Fig. 1A).
If a ribosome reaches an in-frame stop codon, the essential translation termination factor eRF3-eRF1 (Sup35-Sup45 in yeast) binds and induces hydrolytic cleavage of the peptidyl-tRNA at the ribosomal P-site (23–25). If a ribosome reaches the very 3′ end of a stop codon-free mRNA, its P-site is occupied with a peptidyl-tRNA; however, the A-site is empty. In this situation, Sup35-Sup45 does not bind efficiently, but instead Hbs1-Dom34, a complex homologous to Sup35-Sup45, interacts with ribosomes (2, 21, 26). Structural and functional data support the idea that Hbs1-Dom34 performs a role similar to that of Sup35-Sup45 if ribosomes stall with an empty A-site or close to the 3′ end of an mRNA molecule (2, 27, 28). Ski7 is yet another eRF3 homolog, which plays a role in the quality control of nonstop mRNA (29, 30). The current model indicates that Ski7, which interacts with the ribosome as well as with the exosome, recruits the exosome to stalled ribosomes during NSD (29, 31). Consistently, nonstop mRNA is stabilized in a Δski7 strain (29, 30, 32).
Yeast possesses a ribosome-bound chaperone triad consisting of the Hsp70 homologs Ssb1 and Ssb2 (collectively termed Ssb), Ssz1, and the J-domain protein Zuo1 (33). Zuo1 and Ssz1 form a stable complex, termed the ribosome-associated complex (RAC) (34). Ssb binds to ribosomes directly and interacts with nascent chains (34–38). RAC also binds to ribosomes and is required for the interaction of Ssb with nascent chains (39–41). RAC/Ssb is connected to diverse cellular functions, including cotranslational folding (38, 42, 43), translational fidelity (44, 45), and transcriptional regulation (46–49). Recently, we found that RAC/Ssb also plays a role in the quality control of nonstop proteins and of model proteins that possess a bona fide stop codon but contain a C-terminal polylysine segment (termed polylysine proteins) (32). C-terminal polylysine extensions reduce the level of expression of proteins in a wild-type strain (2, 7). Surprisingly, strains lacking RAC/Ssb express higher levels of correctly folded polylysine proteins than a wild-type strain does (32).
Here we have characterized the role of RAC/Ssb with respect to the quality control network maintaining low expression of nonstop and stop codon-less proteins. The results uncovered that the translation termination factor Sup35-Sup45 can release nascent polypeptides prematurely, even though the A-site contained a sense codon. Premature translation termination occurred during the translation of polylysine segments and was strongly enhanced in the absence of RAC/Ssb. The mechanism allows ribosome release of nascent polypeptides stalled due to the interaction of C-terminal polylysine segments with the ribosomal tunnel.
MATERIALS AND METHODS
Yeast strains and plasmids.
MH272-3f a/α (ura3/ura3 leu2/leu2 his3/his3 trp1/trp1 ade2/ade2) (50) was the parental wild-type strain for all haploid mutant strains used in this study. [PSI+] and [psi−] versions of the wild-type and Δzuo1 strains were previously described (44). Deletion strains lacking ZUO1 (Δzuo1), SKI7 (Δski7), HBS1 (Δhbs1), LTN1 (Δltn1), and the Δzuo1 Δski7 and Δzuo1 Δltn1 double deletion strains were described previously (32, 34, 51). Ssb is encoded by two closely related genes (SSB1 and SSB2), which are functionally interchangeable. Strains lacking SSB1 and SSB2 (Δssb1 Δssb2 [ssb1::kanMX4 ssb2::HIS3] and Δssb1 Δssb2 [ssb1::ADE2 ssb2::HIS3]) were previously described (36, 44). The Δssb1 Δssb2 strain (ssb1::kanMX4 ssb2::URA3) was constructed by replacing the ClaI/AgeI fragment within the SSB2 coding region with the URA3 gene in the Δssb1 (ssb1::kanMX4) (36) background. Different Δssb1 Δssb2 strains displayed the same phenotype and were used in the experiments according to the requirements for selection markers. A diploid heterozygous Δsup35 (sup35::ADE2/SUP35) strain was generated by replacing a DraIII/NsiI fragment of the SUP35 coding region with the ADE2 gene. The diploid strain was transformed with the pTET111-Sup35 plasmid and allowed to sporulate, and tetrads were dissected to obtain the haploid Δsup35 strain carrying pTET111-Sup35 (wild-type-tet-SUP35). The Δasc1 (asc1::HIS3) strain was generated by inserting the HIS3 gene into the AgeI sites of ASC1. This strain was used to generate the Δasc1 Δzuo1 and Δasc1 Δski7 double deletion strains. To preserve the snR24 snoRNA which is encoded in the intron of the ASC1 gene, the Δasc1 (asc1::TRP1) strain was generated by replacing a StyI/MscI fragment in the first exon of ASC1 with the TRP1 gene. This strain was used to generate a Δasc1 Δssb1 Δssb2 triple deletion strain. The presence (asc1::TRP1) or absence (asc1::HIS3) of the ASC1 intron sequence did not affect expression of proteins encoding polylysine segments (data not shown). An ADE2-expressing wild-type strain (MH272-3f ADE2+) was generated by reintegration of the wild-type ADE2 gene into the ade2 locus of the MH272-3fα strain. The Δdom34 strain was constructed by replacing DOM34 with the dom34::kanMX4 deletion cassette amplified by PCR from strain Y05329 (Euroscarf). Multiple deletion strains, such as Δzuo1 Δasc1 (asc1::HIS3), Δski7 Δasc1 (asc1::HIS3), Δssb1 Δssb2 Δasc1 (ssb1::kanMX4 ssb2::HIS3 asc1::TRP1), Δhbs1 Δski7, Δzuo1 Δhbs1, Δzuo1 Δdom34, and Δzuo1 Δhbs1 Δski7 strains and the Δzuo1 Δsup35 strain carrying pTET111-Sup35 (Δzuo1-tet-SUP35), were generated either by mating of the respective parental strains followed by dissection and tetrad analysis, or by transformation of the parental strains with the respective deletion cassettes amplified by PCR.
For constitutive expression of Zuo1, mutant versions of Zuo1, Ssb1, and luciferase reporter constructs, the respective genes were cloned into the low-copy-number plasmid pYCPlac33 (CEN URA3), the high-copy-number plasmid pYEPlac195 (2μ URA3), or the high-copy-number plasmid pYEPlac181 (2μ LEU2) (52). The plasmids pYEPlac195-Zuo1, pYCPlac33-Zuo1-QPD, and pYCPlac33-Zuo1-Δ282-311 were as described previously (34, 36, 40). pYCPlac33-Ssb1 is a derivative of pYEPlac195-Ssb1 (36), in which the SSB1 open reading frame (ORF) plus 300 bp of the 5′UTR and 3′UTR, respectively, was transferred into the BamHI/PstI sites of pYCPlac33. pYEPlac195-Luc-stop (Luc stands for luciferase), pYEPlac195-Luc-K12 (Luc-K12 stands for the luciferase reporter carrying 12 C-terminal lysines), pYEPlac195-Luc-K20 (Luc-K20 stands for the luciferase reporter carrying 20 C-terminal lysines), and pYEPlac195-Luc-nonstop were as described previously (32). Luciferase reporters were expressed from pYEPlac195 or pYEPlac181. Expression of luciferase reporters was under the control of the 5′ untranslated region (5′UTR) (300 bp) of the ZUO1 gene and the 3′UTR (266 bp) of the HIS3 gene. pYEPlac195-Luc-K12-3HA is based on pYEPlac195-Luc-K12-HA. Multimers of (AAG AAG AAA)4 followed by a PstI and XbaI restriction site and a hemagglutinin (HA) epitope encoding sequence (TATCCGTATGACGTCCCGGACTATGCA) were introduced in frame between the luciferase ORF and the HIS3 3′UTR in pYEPlac195-Luc-stop. To obtain pYEPlac195-Luc-K12-3HA, annealed oligonucleotides encoding two additional HA epitopes (separated by two codons) were inserted into the PstI/XbaI sites of pYEPlac195-Luc-K12-HA (see Fig. S1 in the supplemental material). The coding region of green fluorescent protein (GFP) was amplified by PCR and was inserted into the PstI/XbaI sites of pYEPlac195-Luc-K12-HA to result in pYEPlac195-Luc-K12-GFP. To generate pYEPlac195-Luc-GFP lacking the polylysine segment, the GFP coding region plus the HIS3 3′UTR was cut from pYEPlac195-Luc-K12-GFP and inserted into the PstI/HindIII sites of pYEPlac195-Luc-stop. For the construction of pYEPlac195-Luc-RZ-3HA, a PCR fragment was generated using a forward primer annealing 5′ of the unique SphI site, in the middle of the luciferase ORF, and a reverse primer fusing the hammerhead ribozyme (RZ) sequence in frame to the 3′ end of the luciferase orf ORF (compare the sequence in Fig. S1 to oligonucleotide pAV40 in reference 53). This PCR product was used to replace a SphI/PstI fragment in pYEPlac195-Luc-K12-3HA. The ribozyme-inactive variant pYEPlac195-Luc-RZ*-3HA was constructed according to a mutant of the hammerhead ribozyme, which carries a 3-bp substitution (compare the sequence in Fig. S1 to oligonucleotide pAV44 in reference 53). pYEPlac181-Luc-K12-3HA was generated by insertion of the BamHI/HindIII fragment (Luc-K12-3HA ORF plus the 5′UTR and 3′UTR) from pYEPlac195-Luc-K12-3HA into pYEPlac181. For galactose-inducible expression, pESC-URA-Luc-K12-3HA was constructed by replacing Luc-stop in pESC-URA-Luc-stop (32) with the SphI/HindIII fragment (Luc-K12-3HA ORF plus the HIS3 3′UTR) from pYEPlac195-Luc-K12-3HA. For tetracycline-repressible expression (Tet-off) we constructed pTET111 (CEN LEU2), which is a hybrid of the EcoRI/HindIII vector fragment of pYCPlac111 (52) and the tetracycline regulatory region of pCM190 (54). pTET111-Sup35 was generated by insertion of SUP35 plus 300 bp of the SUP35 3′UTR into the BamHI/NotI sites of pTET111.
Culture conditions.
Strains were grown at 30°C on selective minimal medium (SD) (0.67% yeast nitrogen base without amino acids [Difco] but with 2% glucose and the appropriate supplements). For galactose induction, cells were shifted to YPGal (2% peptone, 1% yeast extract, 2% galactose). Liquid cultures were incubated at 30°C and 200 rpm. For in vivo labeling of proteins with [35S]methionine, cells were grown in methionine-free SCD medium (0.67% yeast nitrogen base without amino acids [Difco], 2% glucose, 0.06% −Met/−Trp dropout [DO] supplement [Clontech], 0.02 mg/ml tryptophan). To slow down translation elongation, cells were grown in the presence of 150 ng/ml cycloheximide (11). Wild-type-tet-SUP35 and Δzuo1-tet-SUP35 strains harboring the Tet-off plasmid pTET111-Sup35 and the Gal-inducible plasmid pESC-URA-Luc-K12-3HA were grown in SD medium containing 10 μg/ml tetracycline to deplete Sup35 from cells. After the times indicated in the figures, the cells were collected and were immediately resuspended to an optical density at 600 nm (OD600) of ∼0.3 in YPGal medium containing 10 μg/ml tetracycline to induce Luc-K12-3HA expression.
RNA isolation and Northern blotting.
RNA isolation and Northern blot analysis were essentially performed as described previously (32). Total RNA (10 to 25 μg) was separated on a 1.2% agarose gel and transferred onto Hybond-N+ nylon membranes (Amersham) using 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) as a transfer buffer. Hybridization with radiolabeled probes was carried out in hybridization solution (5× SSC, 5× Denhardt's solution, 1% SDS, 0.1 mg/ml salmon sperm DNA). Probes were radiolabeled with the Ready-to-Go DNA labeling bead kit (Amersham). The probe for ACT1 was prepared as described previously (55). Luciferase mRNA was detected with a 550-bp probe covering positions 1100 to 1650 of the ORF (see Fig. S1 in the supplemental material).
Preparation of lysates, total protein extraction, and immunoblotting.
To prepare glass bead lysates, cells were harvested and resuspended (10 to 100 OD600 units/300 to 400 μl, depending on the experiment) in glass bead lysis buffer (20 mM HEPES-KOH [pH 7.4], 2 mM Mg acetate, 100 μg/ml cycloheximide, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 1× protease inhibitor mix; plus potassium acetate [K acetate] and dithiothreitol [DTT] as specified). Acid-washed glass beads (0.25 to 0.5 mm; Roth) corresponding to half of the volume of the cell suspension were added, and cell disruption was performed by six 20-s cycles in a Minilys homogenizer (Bertin Technologies) interrupted by 1-min intervals on ice. Cell debris was removed by centrifugation for 5 min at 3,000 × g. After a second 10-min clearing spin at 20,000 × g, glass bead lysates (lysates) were directly used for experiments.
Total yeast extract for immunoblot analysis was prepared as described previously (56). Briefly, cells (1 to 1.6 OD600 units) were collected by centrifugation, resuspended in 200 μl of 0.1 M NaOH, and incubated for 5 min at room temperature. Cells were then collected by centrifugation, and the pellet was resuspended in 1× SDS sample buffer (20 mM Tris-HCl [pH 6.8], 1% SDS, 0.32 mM EDTA, 10% glycerol, 0.8% bromophenol blue, 5% β-mercaptoethanol), and were boiled for 5 min at 95°C. Loading of extracts derived from Δzuo1 and Δssb1 Δssb2 strains for SDS-polyacrylamide gel electrophoresis was adjusted as described previously (46). To avoid loss of samples, total extract of a wild-type strain, which did not express luciferase, was employed if total extracts were diluted prior to analysis. Enhanced chemiluminescence (ECL) detection of immunoblots was performed as described previously (57). The relative amount of Luc-K12-3HA and Luc-K12t (t stands for truncated) in different strain backgrounds was determined on the same exposure of a single immunoblot using AIDA Image Analyzer software (Raytest). To ensure that the intensity of the bands was within the linear range, dilution series of each sample were loaded and analyzed for each quantification.
Ribosome profiles and ribosome sedimentation assays.
Ribosome profiles and ribosome sedimentation assays were essentially performed as described previously (32, 58). Briefly, 100 μg/ml cycloheximide was added to early-log-phase cells prior to harvest. Cells (80 to 100 OD600 units) were resuspended in 400 μl glass bead lysis buffer complemented with 100 mM K acetate and 0.5 mM DTT, and lysates were prepared as described above. Samples corresponding to 10 A260 units were loaded onto a 15 to 55% linear sucrose gradient, centrifuged for 2.5 h at 200,000 × g (TH641 rotor; Sorvall), and fractionated from top to bottom with a density gradient fractionator monitoring A254 (Teledyne). Aliquots of the fractions and a total corresponding to 5% of the material loaded onto the gradient were separated on Tris-Tricine gels and subsequently analyzed by immunoblotting. Lysates for ribosome sedimentation experiments (20 to 30 OD600 units/300 μl buffer) were prepared in glass bead lysis buffer containing 120 mM K acetate and 2 mM DTT. Aliquots corresponding to 0.5 A260 unit in a total volume of 60 μl were loaded onto a 90-μl high-salt sucrose cushion (glass bead lysis buffer containing 25% sucrose, 800 mM K acetate, and 2 mM DTT). Ribosomal pellets were collected by ultracentrifugation (30 min, 350,000 × g, 4°C in a TLA-100 rotor [Beckman]). Aliquots of the total, supernatant, and ribosomal pellet were analyzed on Tris-Tricine gels followed by immunoblotting.
Determination of average ribosomal transit times.
Ribosomal transit times were essentially determined as described previously (59, 60). Briefly, cells were grown overnight in SCD (lacking methionine) medium at 30°C to mid-log phase. After addition of 0.5 μCi/ml [35S]Met, 10-ml aliquots were removed and rapidly mixed with 200 μg/ml cycloheximide at the time points indicated in the figures. After centrifugation, cell pellets (6 to 10 OD600 units) were resuspended in 150 μl glass bead lysis buffer containing 120 mM K acetate and 2 mM PMSF, and lysates were prepared as described above. Sixty microliters of each cleared lysate was loaded onto a 90-μl low-salt sucrose cushion (glass bead lysis buffer containing 25% sucrose, 120 mM K acetate, and 2 mM PMSF). After centrifugation for 25 min at 350,000 × g at 4°C (TLA-100 rotor; Beckman), ribosome-free supernatants (10 μl) and aliquots of the total lysates (4 μl) were spotted onto pieces (2 by 2 cm) of 3MM Whatman filter paper (Hartenstein). The filters were allowed to dry for 1 h, and after the hour of drying, 20 μl of 5% trichloroacetic acid (TCA) at 65°C was spotted on top of the original spot. Dried filters were then boiled in 5% TCA. The filters were then washed three times with cold 5% TCA and once with 96% ethanol. Dried filters were soaked in 500 μl Solvable solution (PerkinElmer) and incubated at 60°C for 1 h. The solution and filters were subjected to liquid scintillation counting (Tri-Carb 2800TR; PerkinElmer).
Miscellaneous.
Protease inhibitor mix (1×) contained 1.25 μg/ml leupeptin, 0.75 μg/ml antipain, 0.25 μg/ml chymostatin, 0.25 μg/ml elastinal, and 5 μg/ml pepstatin A. Polyclonal antibodies against Sse1, Rpl4, Rpl24, Rps9, and Sup35 were raised in rabbits (Eurogentec). Polyclonal rabbit antiluciferase antibody was purchased from Sigma (catalog no. L0159). Monoclonal mouse anti-HA antibody (clone 12CA5) was purchased from Roche. Total protein extracts, lysates, and ribosomal pellets were separated on 10% Tris-Tricine gels (61) prior to immunoblot analysis. Luciferase activity assays were performed as described previously (32). Sensitivity of strains toward cycloheximide was tested in liquid culture as previously described (44).
RESULTS
The absence of RAC/Ssb leads to enhanced expression of C-terminally truncated polylysine proteins.
We previously found that the expression of polylysine proteins is increased in the absence of ribosome-associated complex (RAC)/Ssb (32). We noticed that, upon prolonged separation on SDS-polyacrylamide gels, luciferase reporters carrying 12 or 20 C-terminal lysines (Luc-K12 and Luc-K20) (Fig. 1B) appeared fuzzy when expressed in the Δzuo1 strain but not when expressed in the wild-type strain (Fig. 1C, lanes 1 to 4). This suggested that C-terminally truncated luciferase species were generated during translation of the polylysine reporters. To analyze this phenomenon in more detail, we constructed Luc-K12-3HA (Fig. 1B; see Fig. S1 in the supplemental material). As expected, Luc-K12-3HA migrated with a higher molecular mass compared to Luc-K12 (Fig. 1C, lanes 1 and 5). However, when expressed in the Δzuo1 strain, an additional luciferase species, approximately the size of Luc-K12 appeared (Fig. 1C, lane 6). The molecular mass shift of this band, termed Luc-K12t, was due to a C-terminal truncation, because Luc-K12t was recognized with an antibody directed against luciferase, but it was not recognized with an antibody directed against the HA tag (Fig. 1D). Formation of the Luc-K12t fragment was suppressed when plasmid-borne ZUO1 was introduced into the Δzuo1 strain (Fig. 1D). We also constructed Luc-K12-GFP in which the 3HA tag was replaced with GFP (Fig. 1B). Luc-K12-GFP migrated with the expected molecular mass in wild-type and Δzuo1 strains (Fig. 1E). A fragment the size of Luc-K12t appeared in the Δzuo1 strain but not in the wild-type strain (Fig. 1E). Thus, neither the length nor the amino acid sequence C terminal of the polylysine affected the generation of Luc-K12t. Of note, the levels of expression of full-length Luc-K12-3HA or Luc-K12-GFP were similar in the wild-type and Δzuo1 strains (Fig. 1E). The combined data strongly suggested that the seemingly increased expression level of Luc-K12 and Luc-K20 in the Δzuo1 strain (32) was due to the comigration of full-length Luc-K12 or Luc-K20 with various protein species lacking only a few lysine residues at the C terminus (Fig. 1C and E). Luc-K12-3HA was next expressed in strains carrying mutant versions of Zuo1, which inactivate the function of Zuo1 in vivo. Zuo1-QPD carries a mutation in the J-domain of Zuo1, resulting in a failure to stimulate the ATPase activity of Ssb (36, 39). Zuo1-Δ carries an internal deletion, resulting in a failure of RAC to bind to the ribosome (40). Luc-K12t was detected in strains expressing Zuo1-QPD or Zuo1-Δ and was also expressed in a Δssb1 Δssb2 strain (Fig. 1F). Depending on loading and exposure of the immunoblot, a small amount of Luc-K12t was also detected in the wild-type strain (for example, Fig. 2A). However, the expression level of Luc-K12t was consistently higher in the absence of RAC/Ssb. We conclude that an active RAC/Ssb system was required to prevent the expression of C-terminally truncated species of proteins containing internal polylysine segments. Experiments below were performed employing either the Δzuo1 or Δssb1 Δssb2 mutations. The outcome was independent of which of the two deletion mutants was employed in the experiments (data not shown).
FIG 2.
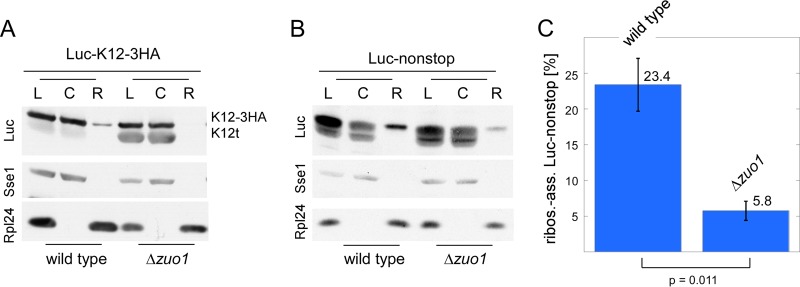
Luc-K12t and Luc-nonstop fragments are efficiently released from ribosomes. (A) The Luc-K12t fragment is efficiently released from ribosomes. Lysates (L) were separated into a cytosolic supernatant (C) and ribosomal pellet (R) by centrifugation through a high-salt sucrose cushion. The immunoblot was decorated with antibodies directed against luciferase (Luc), Sse1 (cytosolic marker) and Rpl24 (ribosomal marker). (B) The Luc-nonstop fragment is efficiently released from ribosomes. The experiment was performed as described above for panel A. (C) Quantification of 3 independent experiments as shown in panel B. The intensities of the cytosolic (C) plus ribosomal (R) luciferase bands were set to a total of 100%; the ribosome-associated Luc-nonstop fragment is given as a percentage of the total. Error bars represent the standard errors of the means. The P value was calculated via Student's t test.
Polylysine and nonstop proteins are efficiently released from ribosomes in the absence of RAC/Ssb.
One possibility for this observation was that ribosomes stalled on polylysine segments were permanently trapped in the absence of RAC/Ssb. If this were the case, we would expect Luc-K12t to be ribosome associated as a peptidyl-tRNA. To test for this possibility, we separated lysates derived from the wild-type or Δzuo1 strain expressing Luc-K12-3HA into a soluble supernatant and a ribosomal pellet under high-salt conditions (Fig. 2A). The analysis revealed that the Luc-K12t fragment was efficiently released from ribosomes (Fig. 2A). We also tested ribosome release of the Luc-nonstop fragment, which is encoded by a bona fide nonstop mRNA containing a poly(A) tail (Fig. 1A) (32). Consistent with the formation of C-terminally truncated protein fragments, Luc-nonstop runs as a fuzzy band when lysates are separated via SDS-PAGE (Fig. 2B) (32). The expression level of Luc-nonstop is only moderately increased in the absence of RAC/Ssb (Fig. 2B) (see Fig. S2A in the supplemental material) (32) (see below). The bulk of Luc-nonstop fragments was released from ribosomes under high-salt conditions; however, specifically, the fragments with the highest molecular mass were released only partly (Fig. 2B). A direct comparison between the wild-type and Δzuo1 strains revealed that ribosome release of Luc-nonstop was more efficient in the absence of RAC/Ssb (Fig. 2B and C). Thus, the absence of RAC/Ssb did not prevent high-salt ribosome release of proteins containing polylysine segments but rather enhanced ribosome release.
RAC/Ssb does not affect expression of stop codon-less proteins.
We speculated that Luc-K12t originated from translation of a stop codon-less 5′ fragment (termed 5′F-LUC-K12) derived from endonucleolytic cleavage of LUC-K12-3HA (Fig. 1A). If this model was correct, the Δzuo1 mutation might either enhance endonucleolytic cleavage of LUC-K12-3HA or stability/translation efficiency of 5′F-LUC-K12. We thus tried to detect 5′F-LUC-K12 in a Δzuo1 strain. However, the level of a potential 5′F-LUC-K12 was below the detection limit of the Northern blot analysis (data not shown).
We therefore constructed a reporter containing the self-cleaving hammerhead ribozyme (RZ) sequence (Fig. 3A; see Fig. S1 in the supplemental material). A catalytically inactive version, termed RZ*, served as a control (21, 53, 62, 63). Autocatalytic cleavage of RZ generates a stop codon-less 5′ fragment (5′F-LUC-RZ) (Fig. 3A; also see Fig. S1) comprised of the complete luciferase ORF. This 5′F-LUC-RZ fragment resembles the potential 5′F-LUC-K12 fragment, because both lack a stop codon, 3′UTR, and poly(A) tail (Fig. 3A; also see Fig. S1). Northern blot analysis revealed that autocatalytic cleavage of LUC-RZ-3HA was efficient (Fig. 3B). However, cleavage was not complete (Fig. 3B), and a small amount of full-length Luc-RZ-3HA protein was detected in immunoblots (Fig. 3C). In comparison, LUC-RZ*-3HA mRNA remained uncleaved and was efficiently translated (Fig. 3B and C). Of note, stop codon-less 5′F-LUC-RZ mRNA was readily detected via Northern blotting (Fig. 3B), while the corresponding stop codon-less protein (Luc-RZt) was not (Fig. 3C). This is consistent with a model in which low expression of stop codon-less mRNA is maintained not only via nonstop mRNA decay (NSD) but also via translational repression (6, 32).
FIG 3.
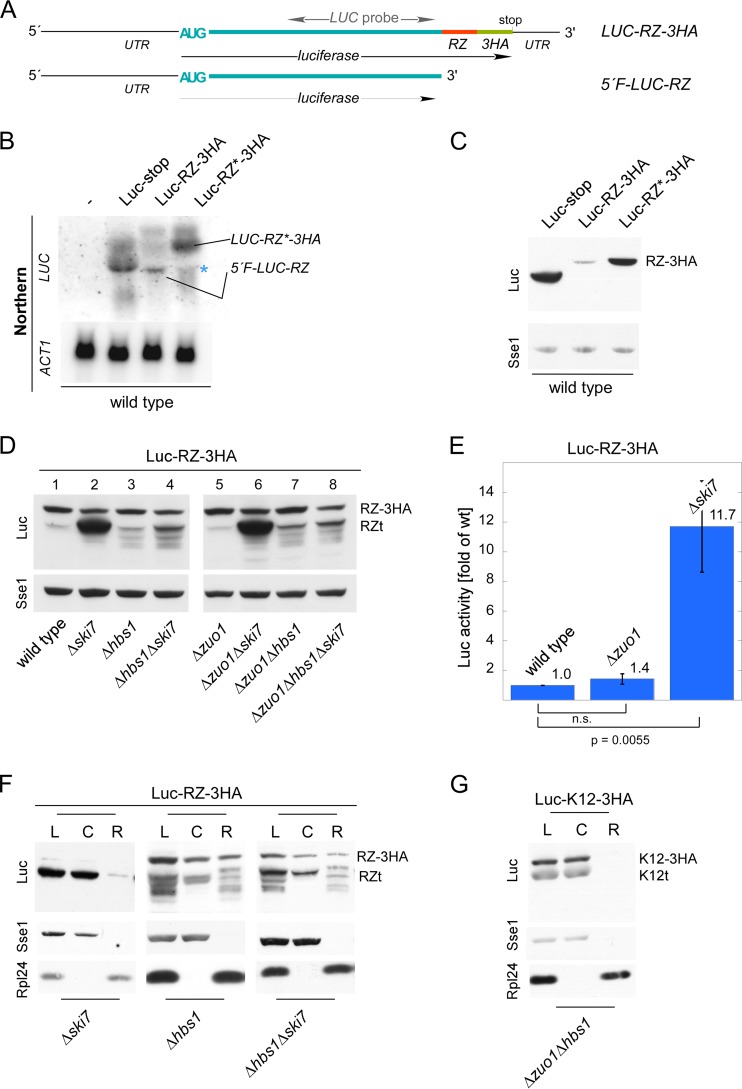
Expression of stop codon-less proteins is independent of RAC/Ssb. (A) Luc-RZ-3HA reporter. Luciferase was fused in frame to the hammerhead ribozyme (RZ) sequence, followed by a sequence encoding three consecutive HA tags (LUC-RZ-3HA). RZ undergoes autocatalytic cleavage, generating a stop codon-less 5′ fragment (5′F-LUC-RZ). The position of the Northern probe employed in panel B is indicated (see Fig. S1 in the supplemental material). (B) The hammerhead ribozyme sequence induces mRNA cleavage. Total RNA isolated from the wild-type strain expressing Luc-stop, Luc-RZ-3HA, or Luc-RZ*-3HA was analyzed via Northern blotting using a probe recognizing luciferase (LUC). The wild type without a luciferase reporter served as a control for probe specificity. Actin (ACT1) served as a loading control. The positions of LUC-RZ-3HA/LUC-RZ*-3HA and 5′F-LUC-RZ are indicated. The asterisk indicates a background band. (C) The 5′F-LUC-RZ fragment is not efficiently translated. Total extracts of the wild type expressing luciferase reporters as indicated were analyzed via immunoblotting with antibodies recognizing luciferase (Luc), and as a loading control, Sse1. The product of LUC-RZ-3HA/LUC-RZ*-3HA translation (RZ-3HA) is indicated. (D) The Δhbs1 and Δski7 mutations, but not the Δzuo1 mutation, affect expression of 5′F-LUC-RZ. The indicated strains harboring the Luc-RZ-3HA reporter were analyzed as described above for panel C. The products of LUC-RZ-3HA translation (RZ-3HA) and of 5′F-LUC-RZ translation (RZt) are indicated. (E) Luc-RZt is catalytically active. Luciferase activity was determined in lysates of the indicated strains expressing Luc-RZ-3HA. Luciferase activity in the wild-type strain was set at 1. The averages of 4 independent experiments are shown; error bars represent the standard errors of the means. P values were calculated via Student's t test. n.s., not significant. (F) Ribosome release of Luc-RZt requires Hbs1. Lysates (L) derived from strains expressing Luc-RZ-3HA were separated into a cytosolic supernatant (C) and ribosomal pellet (R). Analysis was performed as described in the legend to Fig. 2A. Because of the high level of expression of Luc-RZt (see panel D also) in the Δski7 strain, only 25% of the aliquots was loaded. (G) Ribosome release of Luc-K12t does not require Hbs1. Lysate (L) derived from the Δzuo1Δhbs1 strain expressing Luc-K12-3HA was separated into a cytosolic supernatant (C) and a ribosomal pellet (R). Analysis was performed as described in the legend to Fig. 2A.
We next analyzed the Luc-RZ-3HA reporter in strains carrying mutations in Δski7 or Δhbs1, because these mutations reportedly affect expression of stop codon-less mRNA (21, 53). The levels of expression of the residual uncleaved LUC-RZ-3HA mRNA were similar in all strains (Fig. 3D, lanes 1 to 4, RZ-3HA). However, expression of 5′F-LUC-RZ was strongly affected in the mutant strains (Fig. 3D, lanes 1 to 4, RZt). The Δski7 mutation led to high expression of Luc-RZt (Fig. 3D, lane 2), which was enzymatically active (Fig. 3E), and was released from ribosomes (Fig. 3F). In the Δhbs1 strain, additional luciferase fragments of lower molecular mass appeared (Fig. 3D, lane 3). The fragments of this Luc-RZ ladder were ribosome associated, suggesting a “ribosome jam” at the 3′ end of the mRNA in the absence of Hbs1 (Fig. 3F) or Dom34 (Fig. 4A and B). Interestingly, the effect of the Δhbs1 mutation was dominant over the Δski7 mutation, because in the Δhbs1 Δski7 double deletion strain, Luc-RZt expression was similar to Luc-RZt expression in the Δhbs1 strain (Fig. 3D, compare lanes 2 to 4), and the Luc-RZ ladder remained ribosome associated (Fig. 3F). Thus, Hbs1-Dom34 was sufficient for ribosome release of Luc-RZt in the wild-type and Δski7 strains. These results are fully consistent with the function of Hbs1-Dom34 in the dissociation of ribosomes stalled at the 3′ end of stop codon-less mRNA (21, 64). With respect to Ski7, the data are consistent with a role of Ski7 for the efficient decay of stop codon-less mRNA (29, 53). However, Ski7 was not required for efficient ribosome release of Luc-RZt. Analysis in the Δzuo1 background (Fig. 3D, lanes 5 to 8) revealed that Luc-RZt expression and activity in the wild-type and Δzuo1 strains were similar (Fig. 3D, lanes 1 and 5, and Fig. 3E). Moreover, expression of Luc-RZt and the Luc-RZ ladder in the Δski7, Δhbs1, or Δhbs1 Δski7 strains was unaffected by the Δzuo1 mutation (Fig. 3D, compare lanes 1 to 4 to lanes 5 to 8). We also analyzed expression of Luc-nonstop and Luc-K12-3HA fragments in strains lacking Δzuo1, Δski7, Δhbs1, or combinations of these mutations (see Fig. S2B to D in the supplemental material). Upon expression of these reporters, stop codon-less mRNA species are produced by endonucleolytic cleavage (see introduction; also see Fig. S2). Consistently, truncated luciferase species originated from two independent processes. One was enhanced in the absence of RAC/Ssb, and the other was enhanced in the absence of Ski7 and was abolished in the absence of Hbs1-Dom34 (see Fig. S2B to S2D; see below). The combined data strongly suggested that RAC/Ssb did not affect expression of stop codon-less mRNA. To test this possibility further, we expressed the Luc-K12-3HA reporter in a Δzuo1 Δhbs1 strain (Fig. 3G). The analysis revealed that the Δhbs1 mutation did not affect ribosome release of Luc-K12t (Fig. 3G). Thus, the Δhbs1 mutation prevented ribosome release of Luc-RZt in the Δhbs1 Δski7 strain (Fig. 3F) but did not prevent ribosome release of Luc-K12t in the Δzuo1 Δhbs1 strain (Fig. 3G). The combined data strongly suggested that enhanced production of truncated protein species in the absence of RAC/Ssb was not connected to translation of a stop codon-less mRNA fragments derived by endonucleolytic cleavage.
FIG 4.
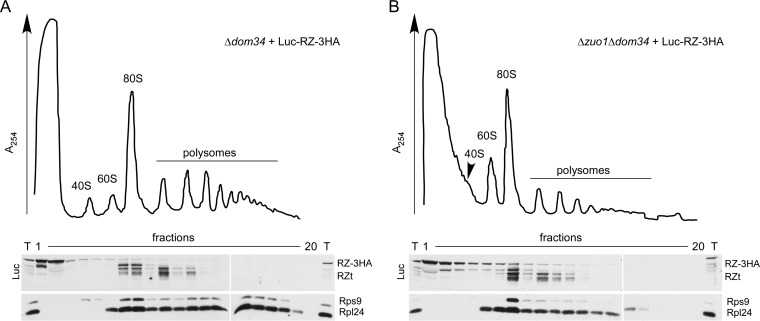
Luc-RZt species generated in the absence of Hbs1-Dom34 are associated with ribosomes. (A and B) Ribosome profiles of Δdom34 (A) and Δzuo1 Δdom34 (B) strains expressing Luc-RZ-3HA. Lysates were separated on 15 to 55% sucrose gradients and subsequently fractionated monitoring A254. Fractions were analyzed via immunoblotting using antibodies directed against luciferase (Luc), Rps9 (40S subunit), and Rpl24 (60S subunit). The bulk of full-length Luc-RZ-3HA was recovered in the cytosolic fractions, and Luc-RZt species with a higher molecular mass were recovered in the monosome fractions, while Luc-RZt species with a lower molecular mass were recovered in the polysome fractions.
Ltn1 affects expression of polylysine and stop codon-less proteins.
Consistent with earlier observations (11, 16), the expression level of Luc-K12t was increased in a Δltn1 strain (Fig. 5A). The effects of Δltn1 and Δzuo1 on Luc-K12t expression were additive (Fig. 5A and B). This is consistent with our previous data indicating that RAC/Ssb and Ltn1 affect independent steps of polylysine protein quality control (32). Even though Luc-RZt does not contain a stretch of polybasic amino acids, Luc-RZt expression was strongly enhanced in the Δltn1 strain (Fig. 5C and D). However, Luc-RZt expression was not further enhanced in the Δzuo1 Δltn1 double deletion strain (Fig. 5C and D). Thus, Ltn1 was involved not only in the efficient degradation of nascent proteins containing polylysine segments but also in the degradation of stop codon-less proteins. A similar observation was recently reported for a different stop codon-less reporter protein (65). In contrast to Ltn1, the role of RAC/Ssb was confined to the expression of the polylysine reporter.
FIG 5.
RAC/Ssb and the ribosome-bound E3-ligase Ltn1 affect expression of Luc-K12t and Luc-RZt by distinct mechanisms. (A) The Δzuo1 and Δltn1 mutations synergistically enhance expression of Luc-K12t. Total extracts of the indicated strains expressing Luc-K12-3HA were analyzed via immunoblotting using antibodies recognizing luciferase (Luc) and, as a loading control, Sse1. The positions of Luc-K12-3HA (K12-3HA) and Luc-K12t (K12t) are indicated. (B) The effects of the Δzuo1 and Δltn1 mutations on Luc-K12t expression are additive. For each strain, the total intensity of the luciferase bands (Luc-K12-3HA plus Luc-K12t) was set at 100%. The intensity of the Luc-K12t band is given as a percentage of the total (see Materials and Methods). The results of 3 independent experiments are shown; error bars represent the standard errors of the means. wt, wild type. (C) The Δltn1, but not the Δzuo1, mutation enhances expression of Luc-RZt. Total extracts of the indicated strains expressing Luc-RZ-3HA were analyzed as described above for panel A. The positions of Luc-RZ-3HA (RZ-3HA) and Luc-RZt (RZt) are indicated. (D) The Δzuo1 mutation does not affect Luc-RZt expression in a Δltn1 strain. Analysis was performed as described above for panel B. The results of 3 independent experiments are shown; error bars represent the standard errors of the means.
Expression of C-terminally truncated proteins correlates with a slowdown of translation.
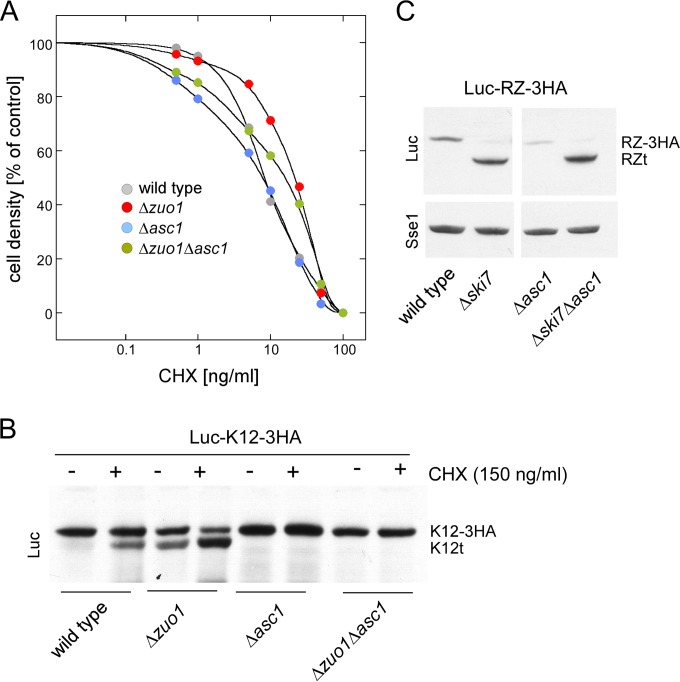
Cycloheximide (CHX) reduces the overall elongation rate on eukaryotic ribosomes (66, 67). It was previously shown that C-terminally truncated polybasic reporters are generated when yeast grows in the presence of sublethal doses of CHX (11). We reasoned that a low rate of translation might also be responsible for the effect of RAC/Ssb on polylysine protein expression. Consistent with previous data (35), the Δzuo1 and Δssb1 Δssb2 mutations did not affect CHX sensitivity in our strain background (Fig. 6A and data not shown). This allowed us to compare the effect of CHX on Luc-K12t expression in the presence or absence of RAC/Ssb. The results revealed that Luc-K12t expression was indeed enhanced in the wild-type and Δzuo1 strains in the presence of CHX (Fig. 6B). We further tested the possibility that Luc-K12t expression was related to the rate of translation employing the Δasc1 mutation, which causes a defect in ribosomal stalling (15) (see introduction). Indeed, the Δasc1 mutation prevented CHX-induced expression of Luc-K12t in the wild type as well as in the absence of RAC/Ssb (Fig. 6B and data not shown). However, the Δasc1 mutation did not influence Luc-RZt expression (Fig. 6C). Thus, Asc1 affected expression of a protein containing an internal stretch of polylysines but did not affect expression of a stop codon-less protein.
FIG 6.
The ribosomal protein Asc1 is involved in the expression of Luc-K12t. (A) Cycloheximide sensitivity of yeast is not significantly altered in the absence of Asc1 or RAC/Ssb. Strains were inoculated to the same OD600 into medium containing the indicated concentrations of cycloheximide (CHX). Cultures were then grown until the no-CHX control had reached an OD600 of 0.9 (100%). The OD600 of the cultures containing CHX is given in the no-CHX control. (B) Luc-K12t expression depends on Asc1. Strains harboring the Luc-K12-3HA reporter were grown in the absence (−) or presence (+) of sublethal doses of CHX. Total extracts were analyzed on an immunoblot decorated with luciferase antibody (Luc). The positions of Luc-K12-3HA (K12-3HA) and Luc-K12t (K12t) are indicated. (C) Luc-RZt expression is independent of Asc1. Total extracts of strains harboring the Luc-RZ-3HA reporter were analyzed via immunoblotting using antibodies recognizing luciferase (Luc) and Sse1. The positions of Luc-RZ-3HA (RZ-3HA) and Luc-RZt (RZt) are indicated.
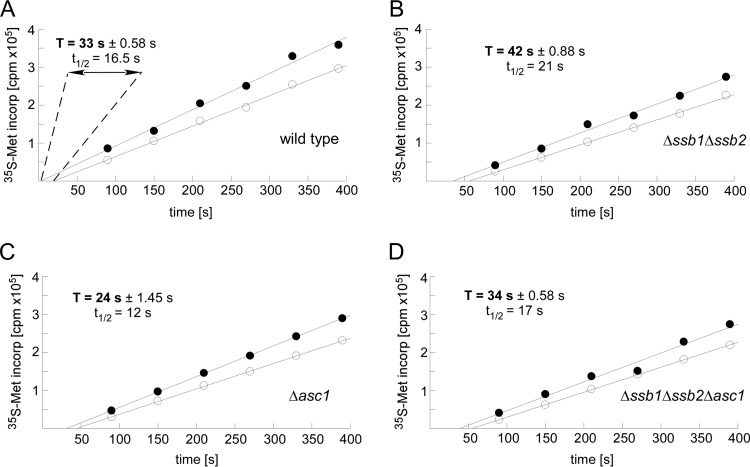
On the basis of the above, we analyzed the effects of RAC/Ssb and Asc1 on the average ribosomal transit times. The average transit time is defined as the time required for a ribosome, after initiation, to decode an mRNA and release the completed polypeptide chain, i.e., the time of elongation plus termination (59, 68, 69). In the absence of RAC/Ssb, the average transit time was increased by approximately 30% compared to that of the wild-type strain (Fig. 7A and B). In contrast, in the absence of Asc1, the average transit time was reduced by approximately 30% (Fig. 7A and C). When both RAC/Ssb and Asc1 were absent, the transit time was similar to that of the wild type (Fig. 7A and D). Thus, RAC/Ssb and Asc1 exerted opposing effects on ribosomal transit times, i.e., the combined rate of translation elongation and termination (Fig. 7A to D).
FIG 7.
Ribosomal transit times depend on RAC/Ssb and Asc1. (A to D) Determination of average ribosomal transit times. [35S]Met incorporation into total protein (nascent chains plus completed polypeptides [filled circles]) and completed polypeptides released from ribosomes (open circles) was determined as described in Materials and Methods. Half average transit times (t1/2) were determined from the displacement in time between the two lines obtained by linear regression analysis. The average transit time is calculated by doubling the difference in the x-axis intercepts of the two lines. For each strain, the experiment was performed in triplicate. The standard error of the mean is given for each experiment. Student's t test was used to calculate P values based on 3 independent experiments: wild type versus Δssb1 Δssb2 strain, P = 0.0012; wild type versus Δasc1 strain, P = 0.0052; Δssb1 Δssb2 strain versus Δasc1 strain, P = 0.00052; Δssb1 Δssb2 strain versus Δssb1 Δssb2 Δasc1 strain, P = 0.0019; Δasc1 strain versus Δssb1 Δssb2 Δasc1 strain, P = 0.0035. Ribosomal transit times in wild-type and Δssb1 Δssb2 Δasc1 strains did not differ significantly.
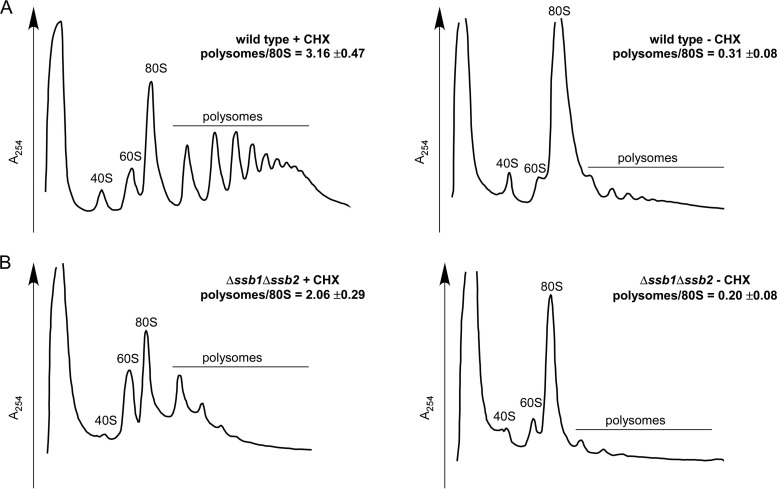
Defects that enhance ribosomal transit times due to a reduced rate of translation elongation result in a stabilization of polysomes in the absence of CHX. This is not the case if ribosomal transit times are enhanced due to defects in translation termination (59, 70, 71). We thus tested whether polysomes were stabilized in the absence of RAC/Ssb. As previously reported (33, 35), the polysome content of the Δssb1 Δssb2 strain was lower than that of the wild-type strain (Fig. 8A and B, left panels). In both strains, however, the polysome content dropped approximately 10-fold when CHX was omitted from the preparation (Fig. 8A and B, right panels). These data strongly suggested that translation elongation was not significantly impaired in the Δssb1 Δssb2 strain and that ribosomal transit times were increased mainly due to a defect in translation termination (see Discussion). This is consistent with the additive effect of the Δssb1 Δssb2 and Δasc1 mutations in respect to ribosomal transit times, caused by a translation elongation defect in the absence of Asc1 (15) (Fig. 7) and a translation termination defect in the absence of RAC/Ssb (Fig. 7 and 8).
FIG 8.
Polysomes do not accumulate in the absence of RAC/Ssb. (A and B) Ribosome profiles of wild-type (A) and Δssb1 Δssb2 (B) strains in the absence (−) or presence (+) of cycloheximide (CHX). Strains were grown at 30°C and then shifted to 20°C for 2.5 h prior to harvest. The temperature shift was performed to enhance effects due to the absence of RAC/Ssb. The Δssb1 Δssb2 mutation causes severe cold sensitivity (33). Cells were then harvested in the presence or absence of cycloheximide (CHX) as indicated. Lysates were separated on 15 to 55% sucrose gradients and subsequently fractionated monitoring A254. The polysome/80S ratio represents the mean of 3 independent experiments, and the standard error of the mean is indicated. Quantifications were performed using ImageJ software.
The translation termination factor Sup35 is required for premature translation termination.
The combined data strongly suggested that Luc-K12t was derived from premature termination on ribosomes engaged in the translation of full-length LUC-K12-3HA mRNA. Because ribosome release of Luc-K12t was independent of Hbs1-Dom34 (Fig. 3G), we speculated that Sup35-Sup45 might execute release in this situation. If this was the case, we reasoned that limiting the concentration of functional Sup35-Sup45 would affect expression of Luc-K12t more severely than expression of full-length Luc-K12-3HA, because Sup35-Sup45 is expected to bind with only low affinity to ribosomes containing a sense codon in the A-site (23, 72). Like many laboratory strains, MH272-3f carries the [PSI+] element, which traps a fraction of Sup35 in prion-like aggregates (23, 44). We initially tested whether increasing the concentration of functional Sup35 by curing cells from [PSI+] would enhance the expression level of Luc-K12t in the wild-type or Δzuo1 strain (Fig. 9A). However, neither Luc-K12-3HA expression nor Luc-K12t expression was affected by the [PSI+] status (Fig. 9A). To test for the possibility that severe reduction of Sup35 affects Luc-K12t expression, we constructed wild-type-tet-SUP35 and Δzuo1-tet-SUP35 strains, in which expression of Sup35 was regulated via a Tet-off promoter (Fig. 9B). To determine the time interval during which Sup35 concentrations were limiting but not lethal, the Δzuo1-tet-SUP35 strain was grown for increasing periods of time in the presence of tetracycline (Fig. 9C). The doubling time of the Δzuo1-tet-SUP35 strain started to drop after >20 h and ceased after >40 h growth on tetracycline (Fig. 9C). Consistently, Sup35 levels were significantly reduced after >28 h on tetracycline (Fig. 9D). To analyze expression under Sup35-limiting conditions, Luc-K12-3HA was expressed under the control of the inducible GAL1 promoter in the Δzuo1-tet-SUP35 strain. The GAL-Luc-K12-3HA reporter allowed us to compare the effect of Sup35 on canonical termination resulting in Luc-K12-3HA expression (stop codon in the A-site) with the effect of Sup35 on premature termination resulting in Luc-K12t expression (sense codon in the A-site). First, we analyzed expression of Luc-K12-3HA and Luc-K12t in the presence of decreasing Sup35 concentrations, i.e., after different lengths of time of growth on tetracycline (Fig. 9E). Luc-K12t was severely reduced after only 20 h on tetracycline, while Luc-K12-3HA was not (Fig. 9E). After 28 h on tetracycline, both Luc-K12t and Luc-K12-3HA were no longer expressed (Fig. 9E). We then analyzed a time course experiment of reporter induction after a short period of Sup35 depletion, when growth of Δzuo1-tet-SUP35 was still only mildly affected (Fig. 9C, 20 h on tetracycline). Under these conditions, Luc-K12-3HA expression was unaffected for up to 8 h, while Luc-K12t expression was already severely reduced (Fig. 9F). The combined data indicate that termination within the coding region of Luc-K12-3HA was affected by low concentration of Sup35 more severely compared to termination at a bona fide stop codon.
FIG 9.
Premature translation termination requires the translation termination factor Sup35. (A) Luc-K12-3HA was expressed in [PSI+] and [psi−] wild-type and Δzuo1 strains. Aliquots of total extracts were analyzed via immunoblotting using antibodies directed against luciferase (Luc) and Sse1. (B) Depletion of Sup35 is lethal. Δsup35 or Δzuo1 Δsup35 strains complemented with SUP35 on a Tet-off plasmid (wild-type-tet-SUP35 and Δzuo1-tet-SUP35, respectively) fail to grow in the presence of tetracycline (10 μg/ml). Tenfold serial dilutions of the indicated strains were spotted onto YPD plates with or without tetracycline. Growth was assessed after 3 days at 30°C. (C) Depletion of Sup35 for short periods of time leads to slow growth. Δzuo1-tet-SUP35 (see panel B) harboring GAL-Luc-K12-3HA was grown in glucose-containing liquid medium with or without tetracycline for the indicated times. Luc-K12-3HA expression was then induced by shifting cells to galactose-containing medium with tetracycline (+ tet) or without tetracycline (− tet). Growth was monitored after the galactose shift. The doubling times (td) are given. (D) Tetracycline treatment leads to Sup35 depletion. Δzuo1-tet-SUP35 (see panels B and C) was grown in the presence of tetracycline for the time periods indicated. The Δzuo1 strain served as a control for normal Sup35 levels. Total extracts were analyzed via immunoblotting using antibodies recognizing Sup35 and, as a loading control, Rpl24. (E) Sup35 is required for the expression of Luc-K12-3HA and Luc-K12t. The Δzuo1-tet-SUP35 strain harboring GAL-Luc-K12-3HA (see panel C) was grown on glucose-containing medium with or without tetracycline (tet) for the indicated periods of time and then shifted to galactose-containing medium with or without tetracycline for 7 h to induce Luc-K12-3HA expression. Aliquots of total extracts were analyzed as described above for panel D. (F) Sup35 depletion severely affects Luc-K12t expression. The Δzuo1-tet-SUP35 strain harboring GAL-Luc-K12-3HA (see panel C) was grown on glucose-containing medium with or without tetracycline for 20 h and then shifted to galactose-containing medium with or without tetracycline for the indicated periods of time. Aliquots of total extracts were analyzed as described above for panel D.
DISCUSSION
When translation comes to a halt, for example because polylysine segments interact with the wall of the ribosomal tunnel, the mRNA molecule is prone to endonucleolytic cleavage close to the site where the ribosome stalls (11, 20, 21, 28). The mechanism of endonucleolytic cleavage is not understood. The actual endonucleolytic activity might be provided by the exosome, an unknown nuclease recruited by Hbs1-Dom34, or possibly by the ribosome itself (2, 22). In any case, the resulting stop codon-less 5′ mRNA fragment is rapidly degraded by the exosome, but it is translated in Δski7 strains in which exosome-mediated degradation of mRNA from the 3′ end is defective (53). Efficient ribosome release of the stop codon-less mRNA and the corresponding C-terminally truncated translation product requires Hbs1-Dom34 (21). In the context of this study, we confirm the roles of Ski7 and Hbs1-Dom34 for the expression and ribosome release of polypeptides encoded by stop codon-less mRNA. In addition, we define a distinct mechanism that also leads to expression and ribosome release of C-terminally truncated proteins. This mechanism does not involve endonucleolytic cleavage of the translated mRNA but is based on premature translation termination on a full-length mRNA molecule (Fig. 10). Premature termination (termed ribosome drop-off) was previously suggested to occur in the mammalian system; the mechanism, however, remained obscure (5, 73). Another example for translation termination at sense codons is provided by a group of viral proteins containing specific peptide sequences, termed 2A peptides (72, 74).
FIG 10.
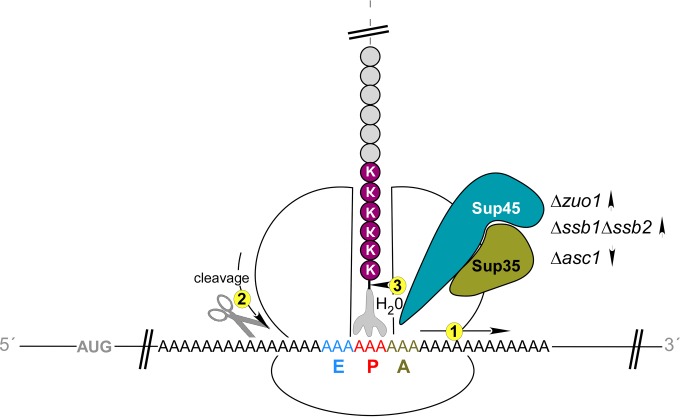
Consequences of ribosomal stalling on polylysine segments. Polylysine segments (KKKKKK) electrostatically interact with the interior of the ribosomal tunnel. This leads to a slowdown of translation elongation, which is termed ribosomal stalling. In this situation, translation is not completely blocked; however, expression of the full-length polypeptide is significantly reduced compared to a control lacking the polylysine segment (1). At least two events compete with translation elongation on stalled ribosomes. First, endonucleolytic cleavage of the mRNA can occur close to where the ribosome pauses (2). Second, the translation termination factor Sup35-Sup45 can erroneously induce premature translation termination, even though the A-site contains a sense codon (3). Mutations that reduce the overall rate of translation elongation/termination (Δzuo1 or Δssb1 Δssb2) enhance Sup35-Sup45-mediated premature termination; mutations that enhance the rate of translation elongation/termination (Δasc1) decrease premature termination. For details and references, see the Discussion.
Several lines of evidence indicate that the frequency of premature termination on polylysine segments was connected to a reduced rate of translation. First, polylysine segments are known to slow down the rate of elongation (10). Second, premature termination was enhanced in the presence of CHX, which also slows down elongation. Third, the Δasc1 mutation suppressed premature termination, consistent with the enhanced rate of translation in the Δasc1 strain (75; this study) and with previous data indicating that Asc1 is required for ribosomal stalling (15). A straightforward explanation for these observations is that a slowdown of translation elongation increases the time window for Sup35-Sup45 to erroneously interact with the ribosome and terminate translation. In vitro data indicate that, in principle, such an activity can be executed by Sup35-Sup45, which slowly releases peptides from the P-site-bound peptidyl-tRNA without a termination codon at the A-site (64, 76). In vivo, eRF3-eRF1 was indeed found to catalyze termination on ribosomes stalled on the 2A peptide even though the ribosomal A site contained a proline-encoding sense codon (77). Of note, the polylysine segment of the Luc-K12-3HA reporter is encoded by alternating AAG and AAA, both of which differ in only a single nucleotide from the universal stop codons TAG and TAA. In combination with the polylysine-induced stalling, this might result in a relatively high frequency of premature termination. In the cell, premature termination via Sup35-Sup45 may provide a mechanism to release ribosomes stalled on the poly(A) tail of nonstop proteins and also upon read-through through the stop codon and the 3′UTR.
A central finding of this study was that the ribosome-bound chaperones RAC/Ssb strongly affect premature termination on polylysine segments. We envisage that RAC/Ssb may impact on this process either by controlling the rate of elongation or via a more direct effect on the actual termination process. The first possibility would be in agreement with the long standing hypothesis that binding of nascent chains to Hsp70 homologs might further their translocation through the tunnel in general (35), and in the case of polylysine segments RAC/Ssb may thus help to overcome stalling. The function of RAC/Ssb during translocation through the ribosomal tunnel would then resemble the function of other Hsp70 homologs during the translocation of polypeptides across cellular membranes (78). Interestingly, it was recently shown in the mammalian system that Hsp70 is depleted from the vicinity of the exit tunnel during stress and that this depletion correlates with translational pausing at an early stage of the elongation process (79, 80). Moreover, a recent in vivo study in yeast identified physiologically relevant cotranslational folding substrates of RAC/Ssb. The analysis revealed that Ssb was preferentially associated with nascent chains with a predicted low translation rate (38). Thus, a similar set of proteins might require the RAC/Ssb system to facilitate translocation through the ribosomal tunnel and subsequently fold. Alternatively, slow translation of specific polypeptide segments in the absence of RAC/Ssb may also hamper cotranslational folding, because polypeptides may not be able to fold until a domain is fully synthesized (81). Of note, folding of the luciferase reporters, with or without a C-terminal polylysine tail, was independent of RAC/Ssb (32; this study).
While we do not want to exclude an effect of RAC/Ssb on the rate of elongation, our data suggest that RAC/Ssb significantly impacts also on translation termination. Elongation factor mutants cause polysome accumulation (59, 70), while translation termination factor mutants do not (59, 71). We did not observe a stabilization of polysomes in the absence of RAC/Ssb; thus, in this respect, loss of RAC/Ssb more closely resembled a situation in which termination does not occur properly. Also, it is difficult to envisage how the stalling defect caused by the Δasc1 mutation (see above) could be overcome by the Δssb1 Δssb2 mutation. We rather favor a model in which translation elongation is enhanced in the Δasc1 and Δssb1 Δssb2 Δasc1 strains. However, due to mixed effects on elongation and termination, ribosomal transit time in the Δssb1 Δssb2 Δasc1 strain resembled that of the wild type. This model is also consistent with the finding that Δzuo1 and Δssb1 Δssb2 strains are highly sensitive to aminoglycosides, which affect both elongation and termination, but do not display increased sensitivity toward the elongation inhibitor CHX (23, 33, 35). Of note, growth inhibition curves, as performed in this work, take into account the fact that Δzuo1 and Δssb1Δssb2 strains display slow growth at 30°C. We found that, due to the underlying slow growth phenotype of the mutant strains, CHX sensitivity on solid media was difficult to assess. On the basis of these observations, we speculate that RAC/Ssb might affect termination via modulating the activity or specificity of the translation termination factors. Indeed, the Sup35 subunit of the yeast termination factor is connected to Ssb via multiple lines of evidence. Sup35 and Ssb physically interact (82). Also, RAC/Ssb is involved in the formation of the [PSI+] factor, which is the prion version of Sup35 (44, 82–84). Moreover, high levels of Ssb enhance the efficiency of translation termination (45), while the absence of RAC/Ssb results in enhanced read-through at stop codons (23, 44). At first glance, it seems puzzling that loss of RAC/Ssb enhances Sup35-Sup45-mediated polypeptide release at a sense codon, while at the same time loss of RAC/Ssb enhances the frequency of read-through (23, 44). However, this dual role of RAC/Ssb might simply indicate a general decrease of Sup35-Sup45 performance in the absence of RAC/Ssb.
A third possibility is that RAC/Ssb influences translation termination indirectly. In yeast cells lacking RAC/Ssb, the protein kinase SNF1 is hyperactive, due to phosphorylation of a specific threonine residue on the Snf1 subunit (46). SNF1 is the yeast homolog of the mammalian AMP-activated protein kinase, which regulates the activity of many proteins, including translation factors (85, 86), in response to glucose availability (87). Interestingly, mutations within a specific threonine residue of Sup35, which can be phosphorylated in vitro, severely affected the in vivo function (88). The negative effect of the Δasc1 mutation on premature termination of Luc-K12-3HA might also be connected to a deregulation of signaling pathways. Asc1 and its mammalian homolog RACK1 are not only part of the small ribosomal subunit but also highly conserved adaptor proteins, which interact with components of various signaling pathways and in this way affect the activity of important kinases (14). It is thus possible that the effect of the Δasc1 mutation on ribosomal stalling involves modulation of the activity of ribosomal proteins and/or of translation factors via phosphorylation (2, 89–91).
In summary, Sup35-mediated premature termination at a sense codon of a polylysine segment is a previously unrecognized type of translational error. How exactly Sup35 induces hydrolytic cleavage of a peptidyl tRNA in this situation is unclear. We also do not know whether premature termination occurs, although with lower frequency, independent of polylysine stalling. In fact, we still lack substantial information on the communication between stop codon decoding and peptide release, even when a canonical stop codon is contained in the ribosomal A-site (23, 24). However, our findings indicate that premature termination provides yet another possibility for the cell to recycle ribosome-nascent chain complexes.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by SFB 746, DFG RO 1028/5-1, and by the Excellence Initiative of the German federal and state governments (BIOSS-2) (to S.R.).
Footnotes
Published ahead of print 25 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00799-14.
REFERENCES
- 1.Klauer AA, van Hoof A. 2012. Degradation of mRNAs that lack a stop codon: a decade of nonstop progress. Wiley Interdiscip. Rev. RNA 3:649–660. 10.1002/wrna.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inada T. 2013. Quality control systems for aberrant mRNAs induced by aberrant translation elongation and termination. Biochim. Biophys. Acta 1829:634–642. 10.1016/j.bbagrm.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 3.Parker R. 2012. RNA degradation in Saccharomyces cerevisiae. Genetics 191:671–702. 10.1534/genetics.111.137265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graber JH, Cantor CR, Mohr SC, Smith TF. 1999. Genomic detection of new yeast pre-mRNA 3′-end-processing signals. Nucleic Acids Res. 27:888–894. 10.1093/nar/27.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akimitsu N. 2008. Messenger RNA surveillance systems monitoring proper translation termination. J. Biochem. 143:1–8. 10.1093/jb/mvm204. [DOI] [PubMed] [Google Scholar]
- 6.Inada T, Aiba H. 2005. Translation of aberrant mRNAs lacking a termination codon or with a shortened 3′-UTR is repressed after initiation in yeast. EMBO J. 24:1584–1595. 10.1038/sj.emboj.7600636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodrigo-Brenni MC, Hegde RS. 2012. Design principles of protein biosynthesis-coupled quality control. Dev. Cell 23:896–907. 10.1016/j.devcel.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 8.Ito-Harashima S, Kuroha K, Tatematsu T, Inada T. 2007. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 21:519–524. 10.1101/gad.1490207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimitrova LN, Kuroha K, Tatematsu T, Inada T. 2009. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J. Biol. Chem. 284:10343–10352. 10.1074/jbc.M808840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu J, Deutsch C. 2008. Electrostatics in the ribosomal tunnel modulate chain elongation rates. J. Mol. Biol. 384:73–86. 10.1016/j.jmb.2008.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, Dunn JG, Rouskin S, Inada T, Frost A, Weissman JS. 2012. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151:1042–1054. 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhushan S, Meyer H, Starosta AL, Becker T, Mielke T, Berninghausen O, Sattler M, Wilson DN, Beckmann R. 2010. Structural basis for translational stalling by human cytomegalovirus and fungal arginine attenuator peptide. Mol. Cell 40:138–146. 10.1016/j.molcel.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Wei J, Wu C, Sachs MS. 2012. The arginine attenuator peptide interferes with the ribosome peptidyl transferase center. Mol. Cell. Biol. 32:2396–2406. 10.1128/MCB.00136-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams DR, Ron D, Kiely PA. 2011. RACK1, a multifaceted scaffolding protein: structure and function. Cell Commun. Signal. 9:22. 10.1186/1478-811X-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuroha K, Akamatsu M, Dimitrova L, Ito T, Kato Y, Shirahige K, Inada T. 2010. Receptor for activated C kinase 1 stimulates nascent polypeptide-dependent translation arrest. EMBO Rep. 11:956–961. 10.1038/embor.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bengtson MH, Joazeiro CA. 2010. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467:470–473. 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Defenouillere Q, Yao Y, Mouaikel J, Namane A, Galopier A, Decourty L, Doyen A, Malabat C, Saveanu C, Jacquier A, Fromont-Racine M. 2013. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc. Natl. Acad. Sci. U. S. A. 110:5046–5051. 10.1073/pnas.1221724110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao S, von der Malsburg K, Hegde RS. 2013. Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol. Cell 50:637–648. 10.1016/j.molcel.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verma R, Oania RS, Kolawa NJ, Deshaies RJ. 2013. Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. eLife 2:e00308. 10.7554/eLife.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doma MK, Parker R. 2006. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440:561–564. 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsuboi T, Kuroha K, Kudo K, Makino S, Inoue E, Kashima I, Inada T. 2012. Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol. Cell 46:518–529. 10.1016/j.molcel.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Harigaya Y, Parker R. 2010. No-go decay: a quality control mechanism for RNA in translation. Wiley Interdiscip. Rev. RNA 1:132–141. [DOI] [PubMed] [Google Scholar]
- 23.Rospert S, Rakwalska M, Dubaquié Y. 2005. Polypeptide chain termination and stop codon readthrough on eukaryotic ribosomes. Rev. Physiol. Biochem. Pharmacol. 155:1–30. 10.1007/3-540-28217-3_1. [DOI] [PubMed] [Google Scholar]
- 24.Dever TE, Green R. 2012. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb. Perspect. Biol. 4:a013706. 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franckenberg S, Becker T, Beckmann R. 2012. Structural view on recycling of archaeal and eukaryotic ribosomes after canonical termination and ribosome rescue. Curr. Opin. Struct. Biol. 22:786–796. 10.1016/j.sbi.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 26.Atkinson GC, Baldauf SL, Hauryliuk V. 2008. Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components. BMC Evol. Biol. 8:290. 10.1186/1471-2148-8-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shoemaker CJ, Green R. 2011. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc. Natl. Acad. Sci. U. S. A. 108:E1392–E1398. 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Passos DO, Doma MK, Shoemaker CJ, Muhlrad D, Green R, Weissman J, Hollien J, Parker R. 2009. Analysis of Dom34 and its function in no-go decay. Mol. Biol. Cell 20:3025–3032. 10.1091/mbc.E09-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295:2262–2264. 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 30.Frischmeyer PA, van Hoof A, O'Donnell K, Guerrerio AL, Parker R, Dietz HC. 2002. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295:2258–2261. 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 31.Araki Y, Takahashi S, Kobayashi T, Kajiho H, Hoshino S, Katada T. 2001. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J. 20:4684–4693. 10.1093/emboj/20.17.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiabudini M, Conz C, Reckmann F, Rospert S. 2012. RAC/Ssb is required for translational repression induced by polylysine segments within nascent chains. Mol. Cell. Biol. 32:4769–4779. 10.1128/MCB.00809-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peisker K, Chiabudini M, Rospert S. 2010. The ribosome-bound Hsp70 homolog Ssb of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1803:662–672. 10.1016/j.bbamcr.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 34.Gautschi M, Lilie H, Fünfschilling U, Mun A, Ross S, Lithgow T, Rücknagel P, Rospert S. 2001. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc. Natl. Acad. Sci. U. S. A. 98:3762–3767. 10.1073/pnas.071057198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. 1992. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 71:97–105. 10.1016/0092-8674(92)90269-I. [DOI] [PubMed] [Google Scholar]
- 36.Gautschi M, Mun A, Ross S, Rospert S. 2002. A functional chaperone triad on the yeast ribosome. Proc. Natl. Acad. Sci. U. S. A. 99:4209–4214. 10.1073/pnas.062048599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfund C, Lopez-Hoyo N, Ziegelhoffer T, Schilke BA, Lopez-Buesa P, Walter WA, Wiedmann M, Craig EA. 1998. The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J. 17:3981–3989. 10.1093/emboj/17.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willmund F, Del Alamo M, Pechmann S, Chen T, Albanese V, Dammer EB, Peng J, Frydman J. 2013. The cotranslational function of ribosome-associated hsp70 in eukaryotic protein homeostasis. Cell 152:196–209. 10.1016/j.cell.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang P, Gautschi M, Walter W, Rospert S, Craig EA. 2005. The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nat. Struct. Mol. Biol. 12:497–504. 10.1038/nsmb942. [DOI] [PubMed] [Google Scholar]
- 40.Peisker K, Braun D, Wölfle T, Hentschel J, Fünfschilling U, Fischer G, Sickmann A, Rospert S. 2008. Ribosome-associated complex binds to ribosomes in close proximity of Rpl31 at the exit of the polypeptide tunnel in yeast. Mol. Biol. Cell 19:5279–5288. 10.1091/mbc.E08-06-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leidig C, Bange G, Kopp J, Amlacher S, Aravind A, Wickles S, Witte G, Hurt E, Beckmann R, Sinning I. 2013. Structural characterization of a eukaryotic chaperone-the ribosome-associated complex. Nat. Struct. Mol. Biol. 20:23–28. 10.1038/nsmb.2447. [DOI] [PubMed] [Google Scholar]
- 42.Koplin A, Preissler S, Ilina Y, Koch M, Scior A, Erhardt M, Deuerling E. 2010. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J. Cell Biol. 189:57–68. 10.1083/jcb.200910074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Albanese V, Reissmann S, Frydman J. 2010. A ribosome-anchored chaperone network that facilitates eukaryotic ribosome biogenesis. J. Cell Biol. 189:69–81. 10.1083/jcb.201001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rakwalska M, Rospert S. 2004. The ribosome-bound chaperones RAC and Ssb1/2p are required for accurate translation in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:9186–9197. 10.1128/MCB.24.20.9186-9197.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hatin I, Fabret C, Namy O, Decatur W, Rousset JP. 2007. Fine tuning of translation termination efficiency in Saccharomyces cerevisiae involves two factors in close proximity to the exit tunnel of the ribosome. Genetics 177:1527–1537. 10.1534/genetics.107.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Plehwe U, Berndt U, Conz C, Chiabudini M, Fitzke E, Sickmann A, Petersen A, Pfeifer D, Rospert S. 2009. The Hsp70 homolog Ssb is essential for glucose sensing via the SNF1 kinase network. Genes Dev. 23:2102–2115. 10.1101/gad.529409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prunuske AJ, Waltner JK, Kuhn P, Gu B, Craig EA. 2012. Role for the molecular chaperones Zuo1 and Ssz1 in quorum sensing via activation of the transcription factor Pdr1. Proc. Natl. Acad. Sci. U. S. A. 109:472–477. 10.1073/pnas.1119184109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richly H, Rocha-Viegas L, Ribeiro JD, Demajo S, Gundem G, Lopez-Bigas N, Nakagawa T, Rospert S, Ito T, Di Croce L. 2010. Transcriptional activation of Polycomb-repressed genes by ZRF1. Nature 468:1124–1128. 10.1038/nature09574. [DOI] [PubMed] [Google Scholar]
- 49.Ribeiro JD, Morey L, Mas A, Gutierrez A, Luis NM, Mejetta S, Richly H, Benitah SA, Keyes WM, Di Croce L. 2013. ZRF1 controls oncogene-induced senescence through the INK4-ARF locus. Oncogene 32:2161–2168. [DOI] [PubMed] [Google Scholar]
- 50.Heitman J, Movva NR, Hiestand PC, Hall MN. 1991. FK 506-binding protein proline rotamase is a target for the immunosuppressive agent FK 506 in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 88:1948–1952. 10.1073/pnas.88.5.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Belyi Y, Tartakovskaya D, Tais A, Fitzke E, Tzivelekidis T, Jank T, Rospert S, Aktories K. 2012. Elongation factor 1A is the target of growth inhibition in yeast caused by Legionella pneumophila glucosyltransferase Lgt1. J. Biol. Chem. 287:26029–26037. 10.1074/jbc.M112.372672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gietz RD, Sugino A. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527–534. 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 53.Meaux S, Van Hoof A. 2006. Yeast transcripts cleaved by an internal ribozyme provide new insight into the role of the cap and poly(A) tail in translation and mRNA decay. RNA 12:1323–1337. 10.1261/rna.46306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gari E, Piedrafita L, Aldea M, Herrero E. 1997. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13:837–848. . [DOI] [PubMed] [Google Scholar]
- 55.Carvin CD, Kladde MP. 2004. Effectors of lysine 4 methylation of histone H3 in Saccharomyces cerevisiae are negative regulators of PHO5 and GAL1–10. J. Biol. Chem. 279:33057–33062. 10.1074/jbc.M405033200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kushnirov VV. 2000. Rapid and reliable protein extraction from yeast. Yeast 16:857–860. . [DOI] [PubMed] [Google Scholar]
- 57.Raue U, Oellerer S, Rospert S. 2007. Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. J. Biol. Chem. 282:7809–7816. 10.1074/jbc.M611436200. [DOI] [PubMed] [Google Scholar]
- 58.Ashe MP, De Long SK, Sachs AB. 2000. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell 11:833–848. 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saini P, Eyler DE, Green R, Dever TE. 2009. Hypusine-containing protein eIF5A promotes translation elongation. Nature 459:118–121. 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fan H, Penman S. 1970. Regulation of protein synthesis in mammalian cells. II. Inhibition of protein synthesis at the level of initiation during mitosis. J. Mol. Biol. 50:655–670. [DOI] [PubMed] [Google Scholar]
- 61.Schägger H, von Jagow G. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368–379. 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 62.Haseloff J, Gerlach WL. 1988. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature 334:585–591. 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- 63.Düvel K, Valerius O, Mangus DA, Jacobson A, Braus GH. 2002. Replacement of the yeast TRP4 3′ untranslated region by a hammerhead ribozyme results in a stable and efficiently exported mRNA that lacks a poly(A) tail. RNA 8:336–344. 10.1017/S1355838202021039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shoemaker CJ, Eyler DE, Green R. 2010. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 330:369–372. 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matsuda R, Ikeuchi K, Nomura S, Inada T. 2014. Protein quality control systems associated with no-go and nonstop mRNA surveillance in yeast. Genes Cells 19:1–12. 10.1111/gtc.12106. [DOI] [PubMed] [Google Scholar]
- 66.Ogg SC, Walter P. 1995. SRP samples nascent chains for the presence of signal sequences by interacting with ribosomes at a discrete step during translation elongation. Cell 81:1075–1084. 10.1016/S0092-8674(05)80012-1. [DOI] [PubMed] [Google Scholar]
- 67.Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO. 2010. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 6:209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nielsen PJ, McConkey EH. 1980. Evidence for control of protein synthesis in HeLa cells via the elongation rate. J. Cell Physiol. 104:269–281. 10.1002/jcp.1041040302. [DOI] [PubMed] [Google Scholar]
- 69.Sivan G, Kedersha N, Elroy-Stein O. 2007. Ribosomal slowdown mediates translational arrest during cellular division. Mol. Cell. Biol. 27:6639–6646. 10.1128/MCB.00798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anand M, Chakraburtty K, Marton MJ, Hinnebusch AG, Kinzy TG. 2003. Functional interactions between yeast translation eukaryotic elongation factor (eEF) 1A and eEF3. J. Biol. Chem. 278:6985–6991. 10.1074/jbc.M209224200. [DOI] [PubMed] [Google Scholar]
- 71.Stansfield I, Eurwilaichitr L, Akhmaloka Tuite MF. 1996. Depletion in the levels of the release factor eRF1 causes a reduction in the efficiency of translation termination in yeast. Mol. Microbiol. 20:1135–1143. 10.1111/j.1365-2958.1996.tb02634.x. [DOI] [PubMed] [Google Scholar]
- 72.Yan F, Doronina VA, Sharma P, Brown JD. 2010. Orchestrating ribosomal activity from inside: effects of the nascent chain on the peptidyltransferase centre. Biochem. Soc. Trans. 38:1576–1580. 10.1042/BST0381576. [DOI] [PubMed] [Google Scholar]
- 73.Akimitsu N, Tanaka J, Pelletier J. 2007. Translation of nonSTOP mRNA is repressed post-initiation in mammalian cells. EMBO J. 26:2327–2338. 10.1038/sj.emboj.7601679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luke G, Escuin H, De Felipe P, Ryan M. 2010. 2A to the fore – research, technology and applications. Biotechnol. Genet. Eng. Rev. 26:223–260. 10.5661/bger-26-223. [DOI] [PubMed] [Google Scholar]
- 75.Gerbasi VR, Weaver CM, Hill S, Friedman DB, Link AJ. 2004. Yeast Asc1p and mammalian RACK1 are functionally orthologous core 40S ribosomal proteins that repress gene expression. Mol. Cell. Biol. 24:8276–8287. 10.1128/MCB.24.18.8276-8287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kurata S, Shen B, Liu JO, Takeuchi N, Kaji A, Kaji H. 2013. Possible steps of complete disassembly of post-termination complex by yeast eEF3 deduced from inhibition by translocation inhibitors. Nucleic Acids Res. 41:264–276. 10.1093/nar/gks958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Doronina VA, Wu C, de Felipe P, Sachs MS, Ryan MD, Brown JD. 2008. Site-specific release of nascent chains from ribosomes at a sense codon. Mol. Cell. Biol. 28:4227–4239. 10.1128/MCB.00421-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goloubinoff P, De Los Rios P. 2007. The mechanism of Hsp70 chaperones: (entropic) pulling the models together. Trends Biochem. Sci. 32:372–380. 10.1016/j.tibs.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 79.Shalgi R, Hurt JA, Krykbaeva I, Taipale M, Lindquist S, Burge CB. 2013. Widespread regulation of translation by elongation pausing in heat shock. Mol. Cell 49:439–452. 10.1016/j.molcel.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu B, Han Y, Qian SB. 2013. Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Mol. Cell 49:453–463. 10.1016/j.molcel.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Frydman J. 2001. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 70:603–647. 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- 82.Allen KD, Wegrzyn RD, Chernova TA, Muller S, Newnam GP, Winslett PA, Wittich KB, Wilkinson KD, Chernoff YO. 2005. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]. Genetics 169:1227–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chernoff YO, Newnam GP, Kumar J, Allen K, Zink AD. 1999. Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone ssb in formation, stability, and toxicity of the [PSI] prion. Mol. Cell. Biol. 19:8103–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shorter J, Lindquist S. 2008. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J. 27:2712–2724. 10.1038/emboj.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cherkasova V, Qiu H, Hinnebusch AG. 2010. Snf1 promotes phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2 by activating Gcn2 and inhibiting phosphatases Glc7 and Sit4. Mol. Cell. Biol. 30:2862–2873. 10.1128/MCB.00183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leprivier G, Remke M, Rotblat B, Dubuc A, Mateo AR, Kool M, Agnihotri S, El-Naggar A, Yu B, Somasekharan SP, Faubert B, Bridon G, Tognon CE, Mathers J, Thomas R, Li A, Barokas A, Kwok B, Bowden M, Smith S, Wu X, Korshunov A, Hielscher T, Northcott PA, Galpin JD, Ahern CA, Wang Y, McCabe MG, Collins VP, Jones RG, Pollak M, Delattre O, Gleave ME, Jan E, Pfister SM, Proud CG, Derry WB, Taylor MD, Sorensen PH. 2013. The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell 153:1064–1079. 10.1016/j.cell.2013.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hedbacker K, Carlson M. 2008. SNF1/AMPK pathways in yeast. Front. Biosci. 13:2408–2420. 10.2741/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fabret C, Cosnier B, Lekomtsev S, Gillet S, Hatin I, Le Marechal P, Rousset JP. 2008. A novel mutant of the Sup35 protein of Saccharomyces cerevisiae defective in translation termination and in GTPase activity still supports cell viability. BMC Mol. Biol. 9:22. 10.1186/1471-2199-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gibson TJ. 2012. RACK1 research - ships passing in the night? FEBS Lett. 586:2787–2789. 10.1016/j.febslet.2012.04.048. [DOI] [PubMed] [Google Scholar]
- 90.Gandin V, Senft D, Topisirovic I, Ronai ZA. 2013. RACK1 function in cell motility and protein synthesis. Genes Cancer 4:369–377. 10.1177/1947601913486348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Regmi S, Rothberg KG, Hubbard JG, Ruben L. 2008. The RACK1 signal anchor protein from Trypanosoma brucei associates with eukaryotic elongation factor 1A: a role for translational control in cytokinesis. Mol. Microbiol. 70:724–745. 10.1111/j.1365-2958.2008.06443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.