Abstract
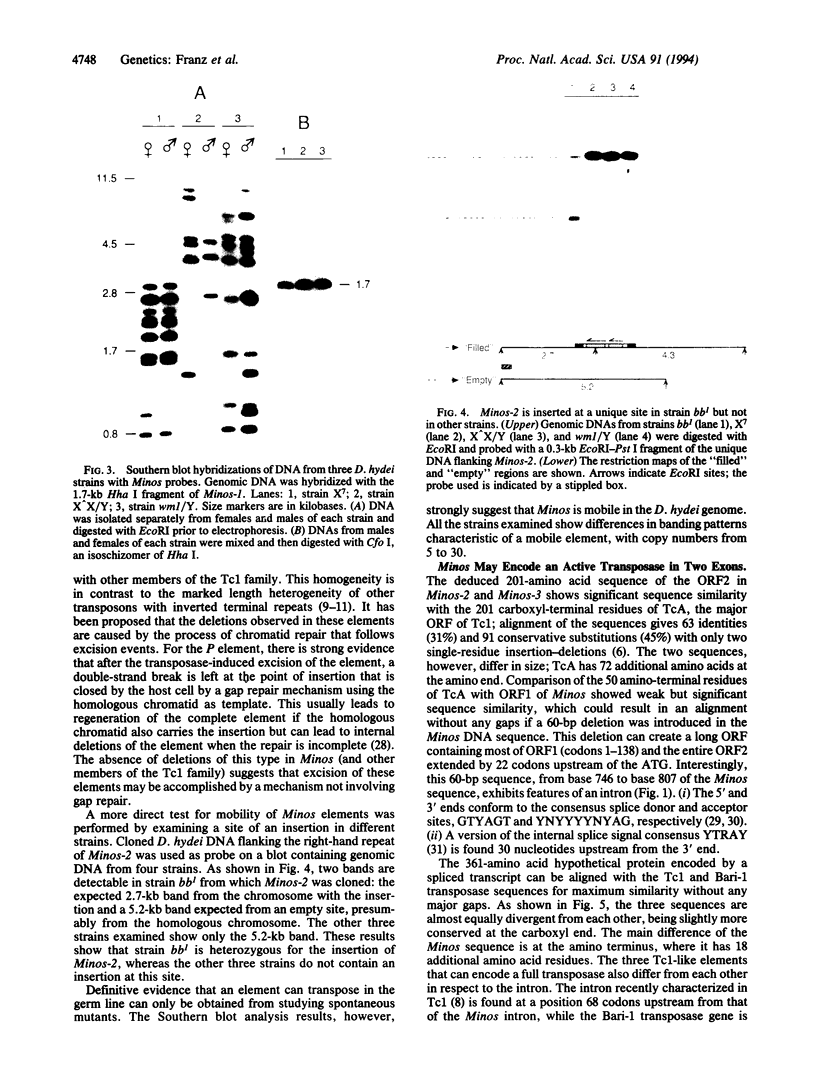
Elements related to the Tc1-like Minos mobile element have been cloned from Drosophila hydei and sequenced. Southern blot and sequence analyses show that (i) the elements are actively transposing in the Drosophila hydei germ line, (ii) they are characterized by a striking degree of sequence and size homogeneity, and (iii) like Tc1, they insert at a TA dinucleotide that is probably duplicated during the process. The nucleotide sequences of two elements, Minos-2 and Minos-3, differ at only one position from each other and contain two nonoverlapping open reading frames that are separated by a putative 60-nucleotide intron. The amino-terminal part of the Minos putative transposase shows sequence similarity to the paired DNA-binding domain. Forced transcription of a modified Minos element that was introduced into the Drosophila melanogaster germ line by P element-mediated transformation resulted in the production of accurately spliced polyadenylylated RNA molecules. It is proposed that Minos-2 and/or Minos-3 may encode an active transposase containing an amino-terminal DNA-binding domain that is distantly related to the paired DNA-binding domain.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Beck H. New compound (1) chromosomes and the production of large quantities of X/O males in Drosophila hydei. Genet Res. 1975 Dec;26(3):313–317. doi: 10.1017/s0016672300016116. [DOI] [PubMed] [Google Scholar]
- Brezinsky L., Wang G. V., Humphreys T., Hunt J. The transposable element Uhu from Hawaiian Drosophila--member of the widely dispersed class of Tc1-like transposons. Nucleic Acids Res. 1990 Apr 25;18(8):2053–2059. doi: 10.1093/nar/18.8.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley H. L., Potter S. S. Distinct characteristics of loop sequences of two Drosophila foldback transposable elements. Nucleic Acids Res. 1985 Jan 25;13(2):485–500. doi: 10.1093/nar/13.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caizzi R., Caggese C., Pimpinelli S. Bari-1, a new transposon-like family in Drosophila melanogaster with a unique heterochromatic organization. Genetics. 1993 Feb;133(2):335–345. doi: 10.1093/genetics/133.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D., Anderson P. Insertion and excision of Caenorhabditis elegans transposable element Tc1. Mol Cell Biol. 1988 Feb;8(2):737–746. doi: 10.1128/mcb.8.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons S. W., Yesner L., Ruan K. S., Katzenberg D. Evidence for a transposon in Caenorhabditis elegans. Cell. 1983 Jan;32(1):55–65. doi: 10.1016/0092-8674(83)90496-8. [DOI] [PubMed] [Google Scholar]
- Engels W. R., Johnson-Schlitz D. M., Eggleston W. B., Sved J. High-frequency P element loss in Drosophila is homolog dependent. Cell. 1990 Aug 10;62(3):515–525. doi: 10.1016/0092-8674(90)90016-8. [DOI] [PubMed] [Google Scholar]
- Engler J. A., van Bree M. P. The nucleotide sequence and protein-coding capability of the transposable element IS5. Gene. 1981 Aug;14(3):155–163. doi: 10.1016/0378-1119(81)90111-6. [DOI] [PubMed] [Google Scholar]
- Franz G., Kunz W., Grimm C. Determination of the region of rDNA involved in polytenization in salivary glands of Drosophila hydei. Mol Gen Genet. 1983;191(1):74–80. doi: 10.1007/BF00330892. [DOI] [PubMed] [Google Scholar]
- Franz G., Savakis C. Minos, a new transposable element from Drosophila hydei, is a member of the Tc1-like family of transposons. Nucleic Acids Res. 1991 Dec 11;19(23):6646–6646. doi: 10.1093/nar/19.23.6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D. IS1 insertion generates duplication of a nine base pair sequence at its target site. Cell. 1978 Mar;13(3):419–426. doi: 10.1016/0092-8674(78)90316-1. [DOI] [PubMed] [Google Scholar]
- Gruss P., Walther C. Pax in development. Cell. 1992 May 29;69(5):719–722. doi: 10.1016/0092-8674(92)90281-g. [DOI] [PubMed] [Google Scholar]
- Halling S. M., Kleckner N. A symmetrical six-base-pair target site sequence determines Tn10 insertion specificity. Cell. 1982 Jan;28(1):155–163. doi: 10.1016/0092-8674(82)90385-3. [DOI] [PubMed] [Google Scholar]
- Harris L. J., Baillie D. L., Rose A. M. Sequence identity between an inverted repeat family of transposable elements in Drosophila and Caenorhabditis. Nucleic Acids Res. 1988 Jul 11;16(13):5991–5998. doi: 10.1093/nar/16.13.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heierhorst J., Lederis K., Richter D. Presence of a member of the Tc1-like transposon family from nematodes and Drosophila within the vasotocin gene of a primitive vertebrate, the Pacific hagfish Eptatretus stouti. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):6798–6802. doi: 10.1073/pnas.89.15.6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Detection of Caenorhabditis transposon homologs in diverse organisms. New Biol. 1992 Apr;4(4):382–388. [PubMed] [Google Scholar]
- Ingolia T. D., Craig E. A., McCarthy B. J. Sequence of three copies of the gene for the major Drosophila heat shock induced protein and their flanking regions. Cell. 1980 Oct;21(3):669–679. doi: 10.1016/0092-8674(80)90430-4. [DOI] [PubMed] [Google Scholar]
- Keller E. B., Noon W. A. Intron splicing: a conserved internal signal in introns of Drosophila pre-mRNAs. Nucleic Acids Res. 1985 Jul 11;13(13):4971–4981. doi: 10.1093/nar/13.13.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori I., Benian G. M., Moerman D. G., Waterston R. H. Transposable element Tc1 of Caenorhabditis elegans recognizes specific target sequences for integration. Proc Natl Acad Sci U S A. 1988 Feb;85(3):861–864. doi: 10.1073/pnas.85.3.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hare K., Rubin G. M. Structures of P transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell. 1983 Aug;34(1):25–35. doi: 10.1016/0092-8674(83)90133-2. [DOI] [PubMed] [Google Scholar]
- Pohlman R. F., Fedoroff N. V., Messing J. The nucleotide sequence of the maize controlling element Activator. Cell. 1984 Jun;37(2):635–643. doi: 10.1016/0092-8674(84)90395-7. [DOI] [PubMed] [Google Scholar]
- Rosenzweig B., Liao L. W., Hirsh D. Target sequences for the C. elegans transposable element Tc1. Nucleic Acids Res. 1983 Oct 25;11(20):7137–7140. doi: 10.1093/nar/11.20.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G. M., Spradling A. C. Genetic transformation of Drosophila with transposable element vectors. Science. 1982 Oct 22;218(4570):348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Stalder R., Caspers P., Olasz F., Arber W. The N-terminal domain of the insertion sequence 30 transposase interacts specifically with the terminal inverted repeats of the element. J Biol Chem. 1990 Mar 5;265(7):3757–3762. [PubMed] [Google Scholar]
- Streck R. D., Macgaffey J. E., Beckendorf S. K. The structure of hobo transposable elements and their insertion sites. EMBO J. 1986 Dec 20;5(13):3615–3623. doi: 10.1002/j.1460-2075.1986.tb04690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D., Tautz C., Webb D., Dover G. A. Evolutionary divergence of promoters and spacers in the rDNA family of four Drosophila species. Implications for molecular coevolution in multigene families. J Mol Biol. 1987 Jun 5;195(3):525–542. doi: 10.1016/0022-2836(87)90181-1. [DOI] [PubMed] [Google Scholar]
- Teem J. L., Abovich N., Kaufer N. F., Schwindinger W. F., Warner J. R., Levy A., Woolford J., Leer R. J., van Raamsdonk-Duin M. M., Mager W. H. A comparison of yeast ribosomal protein gene DNA sequences. Nucleic Acids Res. 1984 Nov 26;12(22):8295–8312. doi: 10.1093/nar/12.22.8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos J. C., van Luenen H. G., Plasterk R. H. Characterization of the Caenorhabditis elegans Tc1 transposase in vivo and in vitro. Genes Dev. 1993 Jul;7(7A):1244–1253. doi: 10.1101/gad.7.7a.1244. [DOI] [PubMed] [Google Scholar]