Summary
Mi-2/nucleosomal remodeling and deacetylase (NuRD) complexes are important epigenetic regulators of chromatin structure and gene expression. Mi-2/NuRD complexes are an assemblage of proteins that combine key epigenetic regulators necessary for (i) histone deacetylation and demethylation, (ii) binding to methylated DNA, (iii) mobilization of nucleosomes, and (iv) recruitment of additional regulatory proteins. Depending on their context in chromatin, Mi-2/NuRD complexes either activate or repress gene transcription. In this regard, they are important regulators of hematopoiesis and lymphopoiesis. Mi-2/NuRD complexes maintain pools of hematopoietic stem cells. Specifically, components of these complexes control multiple stages of B-cell development by regulating B-cell-specific transcription. With one set of components, they inhibit terminal differentiation of germinal center B cells into plasma B cells. They also mediate gene repression together with Blimp-1 during plasma cell differentiation. In cooperation with Ikaros, Mi-2/NuRD complexes also play important roles in T-cell development, including CD4 versus CD8 fate decisions and peripheral T-cell responses. Dysregulation of NuRD during lymphopoiesis promotes leukemogenesis. Here, we review general properties of Mi-2/NuRD complexes and focus on their functions in gene regulation and development of lymphocytes.
Keywords: B cells, T cells, transcription factors, gene regulation, hematopoiesis, lineage commitment/specification
Introduction
Epigenetics is the study of heritable changes in gene expression by mechanisms other than changes in DNA sequence. Epigenetic factors (i) modify the DNA itself (i.e. cytosine methylation), (ii) post-translationally modify histone tails on nucleosomes, and/or (iii) mobilize relative positioning of nucleosomes. Together, these modifications influence whether gene transcription is activated or repressed. It is fundamental to understand how these epigenetic marks are regulated because dysregulation can lead to aberrant gene expression and often tumorigenesis.
Chromatin remodeling complexes (CRCs) are multi-subunit assemblies of proteins that modulate nucleosome positioning. These complexes may also introduce or remove epigenetic marks from histones. One major CRC in eukaryotes is the Mi-2/nucleosome remodeling deacetylase (NuRD) complex (also known as Mi-2/NuRD) (1–3). Mi-2/NuRD was originally identified as a multiprotein complex including Mi-2β, an autoantigen that is targeted by the immune system in dermatomyositis (3). Further studies revealed that Mi-2/NuRD complexes not only mobilize nucleosomes via ATP-dependent chromatin remodeling, but also include subunits that deacetylate histones and bind methylated DNA (1–5). Relative to other types of CRCs, the combination of chromatin remodeling with histone deacetylases and demethylases are unique features of Mi-2/NuRD.
NuRD complexes facilitate the assembly of both open and closed chromatin, depending on the developmental context of the targeted gene (1, 3). In this regard, NuRD functions either as a co-activator or co-repressor of gene transcription. These activities are essential for Mi-2/NuRD complexes’ diverse functions, which include maintenance of genomic stability through DNA damage repair, DNA replication, chromatin assembly, and facilitating cell cycle progression (6–13).
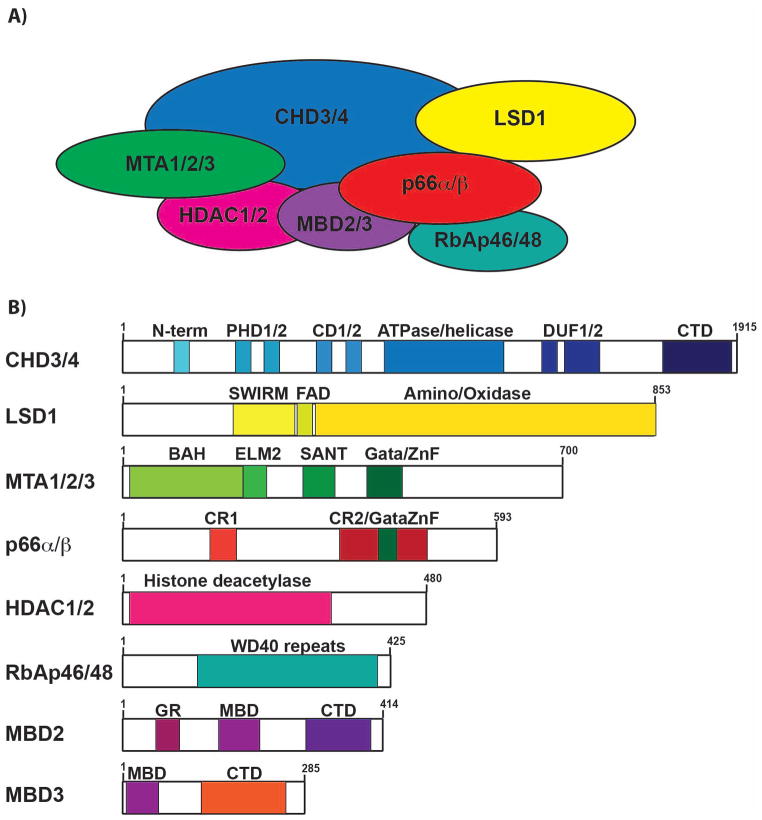
Mi-2/NuRD is a heterogeneous family of complexes with variable components. Each contains a catalytic core of either chromodomain helicase DNA-binding (CHD) domain 3 (also known as Mi-2α) or CHD4 (Mi-2β). In addition to the intrinsic activities of CHD3 and CHD4, these core subunits act as scaffolds for other proteins in these complexes, including histone deacetylases 1/2 (HDAC1/2), p66α (GATAD2A), p66β (GATAD2B), retinoblastoma binding protein 4 (RBBP4/RbAp48), and RBBP7 (RbAp46), metastasis-associated factors 1–3 (MTA1-3), and lysine-specific demethylase 1 (LSD1, KRDM1) (1–4, 14). The main components of NuRD aree defined in detail later in this review.
Here, we discuss how each component of Mi-2/NuRD functions to target or directly mediate epigenetic functional activities of the complexes. Although the majority of studies to date have implicated Mi-2/NuRD in transcriptional repression and chromatin compaction, recent work suggests that components of these complexes also participate in chromatin unfolding and active gene expression. We also address regulation of the development of B and T lymphocytes from hematopoietic stem cells by Mi-2/NuRD. Of special interest is the interaction of Mi-2/NuRD with transcription factors necessary for lymphopoiesis and for functions of mature lymphocytes.
Core subunits of Mi-2/NuRD complexes direct chromatin recognition and ATP-dependent nucleosome remodeling
Chromodomain Helicase DNA-binding domain proteins 3 and 4
Mi-2/NuRD complexes are assembled around catalytic core proteins consisting of helicase/ATPase subunits of the chromodomain helicase DNA-binding domain (CHD) family (CHD3, CHD4, and CHD5) (Fig. 1A). In lymphocytes, CHD3 (also known as Mi-2α) and CHD4 (Mi-2β) are the largest subunits (neuronal cells also express CHD5) (reviewed in 15) of Mi-2/NuRD complexes. In addition to their enzymatic activities, CHD3 and CHD4 function as scaffolds for the assembly of other proteins in Mi-2/NuRD complexes. CHD3 and CHD4 are mutually exclusive in individual NuRD complexes. Although both CHD3 and CHD4 are expressed in B and T cells (16), CHD4 is the predominant helicase that is expressed during hematopoiesis and is understood in more detail than CHD3 (17).
Fig. 1. The Mi-2/NuRD complex and its components.
(A) Schematic of the core components of the NuRD complex. (B) Protein domains of each of the proteins in the Mi-2/NuRD complex. Chromodomain helicase DNA binding protein (CHD3/4), Plant homeodomain (PHD), chromodomain (CD), domain of unknown function (DUF), C-terminal domain (CTD); Lysine-specific demethylase 1 (LSD1), Swi3, Rsc8 and Moira domain (SWIRM), flavin adenine dinucleotide (FAD); Metastasis-associated factors (MTA), bromo-adjacent homology domain (BAH), Egl27/MTA1 domain (ELM), SW13, ADA2, N-CoR and TFIIIB domain (SANT), and GATA-zinc finger domain (Gata-ZnF); conserved region (CR); Histone deacetylase (HDAC); Methyl-binding domain (MBD), glycine-arginine rich region (GR).
CHD3 and CHD4 functions are directed to target nucleosomes by other subunits that interact with histones and DNA. Each of these proteins includes two plant homeodomain (PHD) fingers, two tandem chromodomains (CD), a SWI/SNF2-like ATPase/helicase domains, a domain of unknown function (DUF), and a C-terminal domain (CTD) (Fig. 1B). Together, interactions between these domains provide a linked set of effectors for chromatin recognition and provide a platform for ATP hydrolysis and nucleosome mobilization (18).
PHD fingers are short zinc-binding motifs that bind proteins including histones. The PHD fingers of CHD4 (Fig. 1B) bind preferably to unmodified H3 histone tails and to repressive H3 marks including H3K9me (19, 20). Recognition of these motifs by the PHD fingers allows for bivalent interactions between CHD4 and single nucleosomes. Positive marks including H3K4me3 and H3 acetylation reduce CHD4 binding to chromatin. CHD4’s PHD fingers enhance its binding to unmodified H3 or repressive marks on H3 tails. Thus, interactions with CHD4 assist in localizing and stabilizing inactive genes and heterochromatin. This model was tested by introducing mutations into each of the PHD fingers of CHD4, which revealed their different affinities for binding H3 tails in vitro and functional contributions in transcriptional repression in B cells (21). PHD2 binds H3 tails strongly and is absolutely required for transcriptional repression in B cells, while PHD1 functions cooperatively with PHD2.
Chromodomains are highly conserved motifs present in many chromatin remodelers (reviewed in 22) (Fig. 1B). The tandem chromodomains of CHD4 bind to DNA and are necessary for its functional activities (23, 24). These mutations greatly reduced association of Mi-2/NuRD with chromatin and mutations in core residues of CD1 or CD2 of CHD4 blocked transcriptional repression in a B cell line (20, 21). Together, PHD fingers and CDs regulate the ATPase activity of human CHD4 (7).
The C-terminal domain (CTD) of CHD4 has not been studied in detail, but it is necessary for functional repression in B cells (21) (Fig. 1B). Deletion of the CTD, which may have a predominately α-helical structure, had no effect on the association of CHD4 with chromatin. However, transcriptional repression was lost in the absence of the CTD. This agrees with its proposed role as a docking site for co-repressor proteins or other subunits necessary for Mi-2/NuRD repressive functions.
Following its localization to chromatin, CHD4 (and other CHD family members) mobilizes nucleosomes via its extensive ATPase/helicase domain (465 residues) (Fig. 1B). This domain, which is related to SNF2/RAD54 family of ATPases, is predicted to be a bi-lobed structure that functions as a molecular motor. Although the helicase domain of CHD4 has yet to be examined at atomic resolution, modes of action of related proteins, such as CHD1, have provided insights (24). Related DExx box-like helicase domains include subdomains that bind DNA, hydrolyze ATP and mobilize nucleosomes. This likely occurs by a sliding mechanism, but exact details have yet to be determined. However, Mi-2/NuRD function is absolutely dependent on the presence of an active ATPase/helicase domain. Mutation of K879 to alanine, which likely disrupts DNA binding by the ATPase/helicase domain, ablated CHD4’s ability to mediate transcriptional repression (21). Mutations predicted to disrupt ATP hydrolysis, including K757, E874 and R1159 to arginine, glutamine or alanine, respectively, each produced similarly complete loss of activity (21).
Metastasis-associated factors 1, 2, and 3
Metastasis-associated factors (MTA) were initially described as subunits of Mi-2/NuRD complexes (1–5). The MTA family includes MTA1, MTA2, and MTA3 (Fig. 1A), which are present in a mutually exclusive fashion within Mi-2/NuRD complexes (25, 26). MTA1, MTA2, and MTA3 each contain four highly conserved domains: the bromo-adjacent homology (BAH), Egl27/MTA1 (ELM), SW13, ADA2, N-CoR and TFIIIB (SANT), and GATA-zinc finger (ZF) domains (Fig. 1B). The BAH domains of MTA1, MTA2, and MTA3 are important for protein-protein interactions (27). The SANT domain of the MTA proteins contributes to the HDAC1-MTA2 interaction within Mi-2NuRD complexes (28). The ZF domains of the MTA family of proteins are important for interactions with transcription factors, such as friend of Gata-1 and 2 (FOG-1, FOG-2) (29), Bcl6 (30, 31), and Bcl11b (32). Additionally, the MTA-NuRD complexes bind to H3 peptides in vitro (33).
Studies have demonstrated that components including the MTA proteins contribute to specialized functions of different Mi-2/NuRD complexes. Mi-2/NuRD complexes including MTA1 primarily consist of HDAC1/2, RbAp46/48 and methyl-binding domain protein 3 (MBD3) and are associated with histone deacetylation and transcriptional Mi-2, and MBD3 and played primarily a housekeeping role and interact with p53 (4, 34, 35). MTA3-NuRD complexes are important for the initiation of hematopoiesis and also have reduced transcriptional repression abilities (26, 36). Interestingly, MTA1 and MTA2 may also functionally interact with each other (25) and a wide range of transcription factors. These factors include FOG-1 and FOG-2, which help recruit the NuRD complexes to GATA transcription factor family members to repress gene transcription in erythroid cells (29, 37). GATA-1 and NuRD complexes interact with another transcription factor, Ikaros, and the polycomb repressive complex 2 (PRC2) to suppress transcription of the Hes1 gene and promote erthyropoiesis in multipotent progenitors (38). MTA1 is a co-repressor that functions with estrogen receptor α (ERα), which plays important roles in breast cancer (39). It is unknown whether MTA1 plays similar roles in responses of lymphocytes to estrogen. MTA2 interacts with Yin-Yang-1 (YY1) and the immunophilin FKBP25 (25). The interaction with YY1 may account for some of this multifunctional protein’s mediation of transcriptional repression. MTA3 functions as a co-repressor when it binds Bcl-6, the ‘master regulator’ of germinal center lymphocyte development (30, 31). Overexpression of MTA3 results in lymphomagenesis. MTA3 expression is also regulated by estrogen. Interestingly, absence of MTA3 promotes aberrant regulation of Snail, which in turn represses E-cadherin and promotes breast cancer (26).
The methylation status of the MTA family proteins may determine functions of Mi-2/NuRD complexes. K532 of MTA1 can be methylated by G9a methyltransferase (6). Alternatively, this lysine can be demethylated by LSD1. When K532 of MTA1 is methylated, it promotes assembly of Mi-2/NuRD complexes. When demethylated, MTA1 is able to recognize acetylated H3K4 and H3K9, which in turn recruits the co-activator CRC NuRF-trithorax (6). Thus, lysine methylation of MTA1 is a component of a chromatin-based molecular switch that determines transcriptional outcomes. An open question is whether MTA2 and/or MTA3 are similarly regulated by post-translational modifications.
Methyl-binding domain proteins 2 and 3
Mi-2/NuRD complexes are associated with DNA methylation (5). Two members of the methyl-binding domain (MBD) protein family, MBD2 and MBD3, associate with Mi-2/NuRD complexes (4) (Fig. 1A). Members of the MBD family contain methyl-binding domains that bind to either 5-methylcytosine or 5-hydroxymethylcytosine, which may promote different epigenetic states (reviewed in 40). MBD2 binds to 5-methylcytosine, which can lead to transcriptional repression (41). MBD2 contains an N-terminal glycine-arginine (GR)-rich region, a methyl-binding domain (MBD), a transcriptional repression domain (TRD) adjacent to the MBD, and a coiled-coiled C-terminal domain (CTD) (41) (Fig. 1B). Arginine residues within the GR domain can be symmetrically dimethylated by the protein arginine methyltransferase enzyme, PRMT5, which attenuates its ability to repress transcription (42, 43). On the other hand, MBD3 cannot bind to 5-methylcytosine. Instead, MBD3 binds preferentially to 5-hydroxymethylcytosine via its truncated MBD (34, 44). MBD3 contains a truncated MBD region and a carboxy-terminal glutamic acid repeat domain (41) (Fig. 1B).
Further analysis of Mi-2/NuRD complexes identified versions of these complexes containing either MBD2 or MBD3 (45). Mice deficient in MBD2 do not have developmental defects (45). However, loss of MBD3 in mice results in embryonic lethality (45). This suggests that functions of MBD2 functions may be redundant with other methyl-binding proteins (e.g. MBD1, MBD4 and MeCP2), while those of MBD3 are unique. MBD2 and MBD3 bind to different proteins within Mi-2/NuRD complexes. MBD2 interacts with p66α, p66β, HDAC1, HDAC2 and CHD4 (46–49). MBD3 interacts with MTA2 and HDAC1 through its MBD, and with p66α and p66β (34, 49).
The Mi-2/NuRD complexes that include MBD3 are important for pluripotency in embryonic stem cells (ESCs) (50, 51). MBD3-deficient ESCs are unable to form functional NuRD complexes. Thus, they lack the ability to silence pre-implantation genes and cannot commit to a specific developmental lineage (50). Further analysis of MBD3-deficient ESCs demonstrated that these cells are unable to expand and differentiate. Together, the data indicate that Mi-2/NuRD complexes are required for all stages of embryogenesis and for pluripotency (51). Mi-2/NuRD complexes restrict expression of the key pluripotency genes, Oct4 and Nanog, which leads to a heterochromatic state in somatic cells (52).
Recent advances in chromatin immunoprecipitation-sequencing (ChIP-seq) have facilitated the analysis of MBD2 and MBD3 binding at gene targets across entire genomes (46). A recent study utilizing epitope-tagged MBD2 and MBD3 proteins with ChIP-seq confirmed that the MBD domain of MBD2 interacts preferentially with 5-methylcytosine in vivo (46). Interactions between its MBD and 5-methylcytosine directs MBD2 to directly bind to CpG dense, methylated regions. Interestingly, MBD2 and MBD3 each bind to a subset of unmethylated, active regulatory regions. The binding of MBD3 at these regions is independent of the presence of 5-hydroxymethylcytosine (46), which may reflect its presence in a large fraction of Mi-2/NuRD complexes. Additionally, another study reported that MBD3 binding is localized at promoters, gene bodies, and enhancers of active genes (53). The enrichment of MBD3 at these sites is associated with the histone marks H3K27me3 and H3K27ac. MBD3 binding is associated with CpG-rich promoters that bear the histone mark H3K4me3.
In parallel with its association with methylated DNA, Mi-2/NuRD complexes and MBD2 convert open chromatin with the euchromatic histone mark H3K9ac to a more tightly compacted heterochromatic state with the repressive H3K9me3 mark. Thus, MBD2 bound genes are generally expressed at low levels. In contrast, MBD3 was associated with histone marks of active promoters, including H3K4me2, H3K4me3, and H3K9ac (54). These studies indicate the importance of the localization of MBD2 and MBD3 and how they differentially regulate transcription.
Retinoblastoma-binding proteins 4 and 7
Retinoblastoma-binding proteins 4 and 7 (RBBP4 and RBBP7) are components of Mi-2/NuRD complexes that act as histone chaperones, which bind histones and facilitate nucleosome assembly (4) (Fig. 1A). RBBP4 and RBBP7 are also members of the Sin3A, CAF-1, and PRC2 repressive complexes (4, 55, 56). Both RBBP4 and RBBP7 contain six WD40 repeats, with short N-terminal and C-terminal domains flanking the WD40 repeats (Fig. 1B). Both RBBP4/7 proteins can interact with the N-terminus of FOG-1 (57), histone H3 tails (58), and histone H4 tails (59).
p66α/p66β
Mi-2/NuRD complexes also contain polypeptides that facilitate their assembly: p66α (GATAD2A) and p66β (GATAD2B) (5, 48, 49) (Fig. 1A). Both family members have conserved zinc finger domains (CR1 and CR2) (60) (Fig. 1B). The N-terminal domains of p66α and p66β are important for interacting with both MBD2 and MBD3 (48, 60). Importantly, MBD2-mediated gene repression is enhanced through interactions with p66α and p66β (48). The CR2 domain is responsible for interacting with histone tails and, together with MBD2 and MBD3, is important for targeting Mi-2/NuRD complexes to specific gene loci (48, 60). p66α and p66β interact with histone tails in heterochromatin, while acetylated histone tails decrease the interactions between p66α and p66β and histone tails (48). In this manner, p66α and p66β appear to be important for recruitment of Mi-2/NuRD complexes, which help maintain the state of repressed chromatin. Mice deficient in p66α exhibit embryonic lethality at day 9.5 and have defects related to DNA methylation and gene silencing (61).
Histone deacetylases 1 and 2
Histone deacetylases 1 (HDAC1) and 2 (HDAC2) are enzymes that mediate deacetylation of ε-amino groups of lysine residues (Fig. 1A). HDAC1 and HDAC2 are approximately 85% identical and belong to the class I type of histone deacetylases (62). Both proteins have a central catalytic deacetylase domain and a C-terminal retinoblastoma protein (Rb1)-binding motif (63) (Fig. 1B). In early studies, HDAC1/2 were detected in purified Mi-2NuRD complexes, which differentiated these CRCs from others that lack these subunits (1, 2, 4).
Lysine-specific demethylase 1
Lysine-specific demethylase 1 (LSD1) demethylates histone H3 at K4 and K9 sidechains, as well as lysines within other proteins (64–66) (Fig. 1A). Lysine demethylation promotes transcriptional repression. LSD1 contains an N-terminal SWIRM domain (a small α-helical domain named after the proteins SWI3, RSC8, and MOIRA in which the domain was first recognized). Also, LSD1 contains a C-terminal amine oxidase domain and a FAD binding motif (66) (Fig. 1B). The SWIRM domain mediates protein-protein interactions (65, 66). The amine oxidase domain is responsible for the lysine demethylase activity (66). LSD1 was initially identified as a component of Mi-2/NuRD complexes in mammalian breast cancer. Here, it regulates cell signaling pathways, including TGFβ. Mi-2/NuRD complexes that contain LSD1 suppress breast cancer metastasis (14). Additionally, these complexes interact with JARID1B to repress the chemokine CCL14, which in turn, prevents angiogenesis and metastasis (67). LSD1 is important for propagating embryonic stem cell differentiation by decommissioning enhancers of pluripotency during development (68). LSD1 also associates with other CRCs, including Co-REST (64).
Regulation of hematopoietic stem cells by CHD4 and Ikaros
CHD4 is expressed highly in hematopoietic stem cells (HSCs), early lymphoid progenitors, myeloid progenitors and erthyroid progenitors (69). In hematopoietic stem cells (HSCs), CHD4 is required for maintenance and multi-lineage differentiation in early hematopoiesis (70). CHD4 has been demonstrated to interact with the transcription factors Ikaros, Aiolos, and GATA-1 in hematopoietic progenitor cells (38, 69, 71–74). In mice, conditional depletion of CHD4 in HSCs by Mx-Cre promoted the expansion of both short-term HSCs (ST-HSCs) and long-term HSCs (LT-HSCs) (70). Additionally, in the absence of CHD4, progenitors shifted from a lymphoid to an erythroid bias in their differentiation (70). These stem cells resulted in fewer granulocyte (Mac-1+Gr-1+) and B-cell precursors (B220+CD19+), while increasing erythroid precursors (CD71hiTer119lo-int). Loss of CHD4 in HSCs also increased LT-HSC and ST-HSC populations, indicating that CHD4 is required for hematopoietic stem cell homeostasis. This conclusion was reflected by an increased number of cycling HSCs, which had higher rates of apoptosis. Microarray analysis of CHD4-deficient HSCs demonstrated that CHD4 is responsible for repressing 356 genes in these stem cells. Genes regulated by CHD4 can be grouped based on their functions. Group 1 genes were expressed in Lineage−Sca1+c-Kit+ (LSK) early progenitor cells, where they regulate self-renewal, homeostasis and cell cycle arrest. Group 2 genes were associated with lineage priming in HSCs and were myeloid-specific genes. These results indicate that the role of CHD4 in HSCs is to repress genes of myeloid and erythroid lineages, while promoting differentiation into lymphoid lineages.
HSCs isolated from mice lacking the zinc finger protein Ikaros (Ikzf1) have reduced long-term HSC populations and exhibit dysregulation of the receptor tyrosine kinases Flk-2/Flt3 and c-Kit (75). Ikaros is necessary for development of functional lymphoid-primed multipotent progenitors (LMPP), which have a lymphoid bias, but differentiate alternatively into myeloid cells. In the absence of Ikaros, mutant LMPPs were unable to differentiate into common lymphoid progenitors (CLPs). Instead, the mutant LMPPs differentiated into cells with a myeloid bias (76).
These studies indicate the importance of both CHD4 and Ikaros in determining a lymphoid bias in hematopoietic stem cells, while repressing transcription of genes associated with myeloid and erythroid lineages.
Mi-2 NuRD complexes and B-cell development
B cells develop from hematopoietic stem cells in the bone marrow (reviewed in 77). Different components of Mi-2/NuRD complexes regulate B-cell development in a temporally controlled fashion. Early roles of these complexes include repression of B-cell-specific genes, while later events include regulation of germinal center B-cell and plasma cell differentiation. These properties of Mi-2/NuRD complexes involve interactions with transcription factors that are themselves regulated temporally, including Ikaros, Bcl-6, and BLIMP-1 (Fig. 2).
Fig. 2. B cells develop from hematopoietic stem cells in the bone marrow.
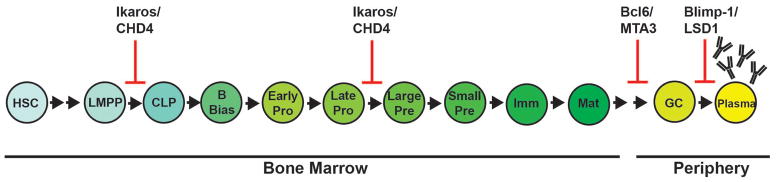
This schematic demonstrates a linear progression of B-cell development from hematopoietic stem cells (HSCs) in the bone marrow into functional B cells in the periphery. Lymphoid primed multipotent progenitors (LMPP) and common lymphoid progenitors (CLP) retain the potential to differentiate into other cell lineages. Expression of B-cell-specific transcription factors promote commitment to the B-cell lineage (as indicated above). Rearrangements of the DH to JH Igh loci occur in early pro-B cells and rearrangements of the VH to DJH locus occur in late pro-B cells. Rearrangements of the VL to JL of the Igk and Igl locus occur in large pre-B cells. After the small pre-B stage, B cells transition to an immature (Imm) B-cell stage and to a mature (Mat) B-cell stage in the bone marrow. B cells can migrate to the periphery and, upon antigen stimulation, can form germinal centers (GC) in peripheral lymphoid organs and can terminally differentiate into antibody-secreting plasma cells. Red arrows in this diagram indicate where components of the Mi-2/NuRD chromatin remodeling complex impede B cell development. Chromodomain helicase DNA-binding protein 4 (CHD4), Metastasis-associated factor 3 (MTA3), Lysine-specific demethylase 1 (LSD1).
Regulation of CD79a transcription by Mi-2/NuRD
The CD79a (mb-1) gene encodes Igα, a trans-membrane signaling component of the B-cell receptor in pre-B and mature B cells. Many transcription factors, including E2A, EBF1, Pax5, Runx1/CBFβ, Sp1, and Ets1, regulate transcriptional activation of the Cd79a gene. Activation of the Cd79a promoter occurs following the binding of transcription factors, which mediate a series of epigenetic changes including CpG demethylation (78, 79). This demethylation occurs progressively during early B-cell development. This process is initiated by binding of the transcription factor, early B-cell factor 1 (EBF1) (80). Demethylation of Cd79a promoter DNA occurs concurrently with increased accessibility, which facilitates the binding of Pax5. Pax5 recruits Ets family proteins to bind unmethylated DNA, which in turn activates functional transcription of the promoter (79).
More specifically, EBF1 recruits SWI/SNF CRCs to Cd79a promoters via direct interactions with Brahma-like gene 1 (Brg1) (80). This process is antagonized by Mi-2/NuRD complexes (Fig. 2). In the plasmacytoma cell line, μM.2, exogenous expression of EBF1 and Pax5 was sufficient for activating endogenous Cd79a gene transcription. However, knockdown of the CHD4 subunit of NuRD using shRNA potentiated effects of EBF1 and Pax5, resulting in higher amounts of Cd79a transcripts. Moreover, knockdown of CHD4 dramatically increased demethylation of CD79a promoters. Together, these data indicate that CHD4, and by inference, Mi-2/NuRD complexes, is present at the Cd79a promoter prior to its activation during B-cell development (80).
The domains of CHD4 necessary for repression were defined using a complementation system in the μM.2 plasmacytoma cell line. Point mutations, or complete domain deletions, of the PHD fingers, chromodomains, ATPase/helicase domain, and/or CTD of CHD4 were examined for repression of Cd79a transcription in μM.2 cells following depletion of endogenous CHD4 (20, 21). All domains of CHD4 except for the CTD are required for association of CHD4 with the Cd79a promoter. Overall, each of the domains of CHD4 is required for robust transcriptional repression of Cd79a (20, 21).
Similar to CHD4, MBD2 is necessary for repression of Cd79a transcription (21). In the μM.2 plasmacytoma cell line, Mbd2-specific shRNA potently increased Cd79a transcription, similar to effects of depleting CHD4 (Fig. 2). Depletion of MBD2 also increased demethylation of Cd79a promoter CpGs, reduced CHD4 occupancy and increased chromatin accessibility of Cd79a promoters. Thus, MBD2 is important for repression of Cd79a transcription, which involves its recruitment of Mi-2/NuRD complexes (21).
Regulation of B-cell development by Ikaros and CHD4
Ikaros (Ikzf1; also known as Lyf-1) is important for both B and T-cell development (81–83). Regulation of T-cell development by Ikaros is discussed later in this review. Ikaros is the founding member of the Ikaros family of proteins, which includes Helios (encoded by Ikzf2), Aiolos (Ikzf3), Eos (Ikzf4), and Pegasus (Ikzf5) (84). Two C-terminal zinc fingers (ZF5-6) are responsible for multimerization of Ikaros with itself and other family members (85). The conserved N-terminal DNA-binding domain of Ikaros contains up to four zinc fingers (ZF1-4) as a result of alternative splicing of Ikzf1 mRNA (85). The N-terminal zinc fingers have different DNA binding capabilities and are utilized in different combinations. Ikaros can also associate with HDAC1, HDAC2, RbAp48, and with SWI/SNF in T cells (69). Ikaros null mice exhibit extensive defects in hematopoietic stem cells and in the development of B and T lymphocytes and NK cells (81). Spleens from the Ikaros null mice had an increase of myeloid and erythroid cells, while fetal liver B-cell precursors, pro-B and pre-B cells from adult bone marrow, and mature B cells were absent (86).
Ikaros interacts with Mi-2/NuRD complexes in hematopoietic stem cells, B cells, and T cells, primarily through the CHD4 subunit (69, 72, 87, 88). Combined effects of Mi-2/NuRD and Ikaros localize genes within pericentromeric heterochromatin in developing lymphocytes (13, 69, 72, 89). Recruitment of Mi-2/NuRD was cell cycle-dependent, with localization occurring during late S phase (13). Moreover, recruitment of Mi-2/NuRD to heterochromatin involved co-localization with H3K9me3 marks and Heterochromatin Protein 1 (HP1). In the Ramos human B-cell line, Ikaros also associated with HP1γ in pericentromeric heterochromatin (13).
A recent study using conditionally deleted Ikzf1 alleles revealed relationships between Ikaros and Mi-2/NuRD binding in pro-B cells in the bone marrow that was contrary to published observations in T cells (88) (Fig. 2). Floxed sequences encompassing ZF5-6 of Ikaros were conditionally deleted using mb-1 (Cd79a)-Cre during the transition from pre-proB to pro-B stage of B-cell development (87). Loss of functional Ikaros expression in these mice resulted in impaired transitioning of pro-B cells to pre-B cells. ChIP-seq was performed using pro-B cells from Cd79a-Cre Ikzf1fl/−Rag2−/− and Cd79a-Cre Ikzf1fl/+Rag2−/− mice to identify regulatory elements that are targets of Ikaros. Interestingly, the loss of Ikaros led to active histone marks at promoters and distal elements at gene targets that are normally repressed in pro-B cells. The association of CHD4 and Ikaros was also mapped using ChIP-seq in Cd79a-Cre Ikzf1fl/− and Cd79a-Cre Ikzf1fl/+ pro-B cells (87). Occupancy of promoters by CHD4 correlated strongly with that of Ikaros. However, there was little overlap between CHD4 and Ikaros binding at distal elements in pro-B cells. The binding of CHD4 at distal elements was similar in the presence or absence of Ikaros. This suggests that Ikaros is not required for retention of Mi-2/NuRD at binding sites. Additionally, the loss of Ikaros did not affect the abundance of H3K9ac at CHD4 or Ikaros binding sites in pro-B cells.
Regulation of terminal plasma cell differentiation by Bcl-6, MTA3 and Mi-2/NuRD Mi-2/NuRD complexes inhibit terminal differentiation of germinal center B cells through interactions of MTA3 and Bcl-6 (30)(Fig. 2). Bcl-6 is a master regulator of germinal center B cell differentiation, where it is expressed together with MTA3 (30). Co-immunoprecipitation studies demonstrated that Bcl-6 interacts with Mi-2/NuRD components including MBD3, HDAC1, MTA3, and CHD4. These interactions are dependent on the acetylation status of the central region of Bcl-6, which does not include the BTB-DOZ domain important for its interactions with the SMRT and NCoR co-repressors (90). Acetylation inactivates Bcl-6 function (90). MTA3-containing Mi-2/NuRD and Bcl-6 act as co-repressors of the plasma cell program in B cells. Depletion of MTA3 by shRNA in two human B cell lines, Raji and BJAB, led to derepression of plasma cell-specific transcripts including Prdm1 (Blimp-1), Ccl3 (MIP1-α), Sdc1 (Syndecan-1), and Xbp1 (XBP-1), and re-expression of CIITA. Enforced expression of Bcl-6 and MTA3 in terminally differentiated human plasma cell lines reprogrammed the cells to express markers of earlier stages of B cell development including CD19, CD20, and HLA-DR. Expression of these proteins was dependent on the presence of both MTA3 and Bcl-6, indicating that recruitment of Mi-2/NuRD complexes by Bcl-6 represses terminal differentiation of B cells into plasma cells (30).
MTA3 is expressed in centroblasts within germinal centers, as well as in germinal center B-cell lymphomas and follicular lymphomas (31). Gene expression profiling of lymphoma cells correlated MTA3 expression with germinal center B-cell-specific genes (Bcl6, Bach2, E2fs, Mybl1, Bcl7a, Fmr2, and Glet2), B-lineage transcription factors (Pax5, Ets-1, and Ebf1) and genes associated with proliferation (Cdca7, Rntre, Msf, Tcl6) (31).
Interaction of Blimp-1 and LSD1 during plasma cell differentiation
B-lymphocyte-induced maturation protein 1 (Blimp1) (encoded by the Prdm1 gene) is a transcriptional repressor that coordinates plasma cell differentiation (91, 92). Blimp-1 represses expression of c-myc, Pax5, CIITA, Id3, and Spib (reviewed in 93). Blimp-1 interacts with various co-repressors and epigenetic regulators, including HDAC1, HDAC2, G9a methyltransferase, PRMT5, and LSD1 (94–96). LSD1 interacts with Blimp-1 via its proline-rich domain (Fig. 2). LSD1 also interacts with HDAC1 and HDAC2. Depletion of LSD1 by shRNA in a plasma cell line restored expression of Pax5, CIITA, and Spib transcripts. LSD1 localized to target promoter sites of the Pax5, Ciita (CIITA), and Spib genes following Blimp-1. Further analysis showed that LSD1 binding is associated with accessible chromatin. Depletion of LSD1 changed the chromatin structure at these genes, with an increase in H3K4me2 and H3Ac. This links LSD1 with HDAC1/2 functional activity. Notably, LSD1 is required for formation of antibody-secreting cells. IgM secretion was decreased in the human B cell line SKW 6.4 in response to LSD1 depletion via shRNA. These results indicate that LSD1 is important for Blimp-1 mediated gene repression during plasma cell differentiation (97).
Control of T-cell development by Mi-2/NuRD complexes
T-cell precursors emigrate from the bone marrow to the thymus where they undergo positive and negative selection. This developmental process begins with CD4−CD8− double negative (DN) cells. To generate conventional T cells with αβ T-cell receptors (TCR), DN cells undergo TCR β chain rearrangements (at Tcrb genes), followed by TCR α chain rearrangements (at Tcra genes) and subsequent assembly of functional TCR proteins on the cell surface. Thymocytes progress from DN to double positive (DP) CD4+CD8+ cells, followed by differentiation to single positive (SP) cells that express one or the other co-receptor (CD4+ or CD8+) on the cell surface. CD4+ T cells can be polarized to different helper T-cell varieties (e.g. Th1 and Th2) that produce different cytokines following stimulation. CD8+ T cells are generally cytotoxic T cells that can kill target cells (reviewed in 98). Additional populations include regulatory T cells (Tregs) that oversee immune responses (99).
Multiple components of Mi-2/NuRD complexes function during early T-cell development in the thymus and in peripheral T-cell development by influencing Th1 versus Th2 cell fate decisions. Different components of the NuRD complex interact with the transcription factors Ikaros, Gata3, and Bcl11b throughout development (Fig. 3).
Fig. 3. T cells develop in the thymus from hematopoietic stem cells in the bone marrow.
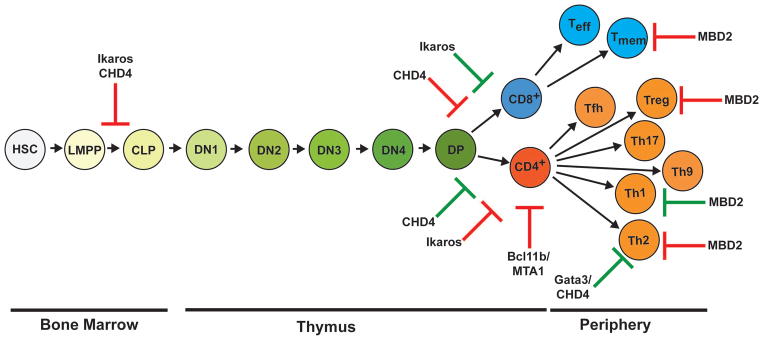
This schematic demonstrates a linear progression of T-cell development from hematopoietic stem cells (HSCs) in the bone marrow to differentiation in the thymus and final maturation in the periphery. Hematopoietic stem cells give rise to lymphoid primed multipotent progenitors (LMPP) and subsequently, common lymphoid progenitors (CLPs). CLPs that will differentiate into T cells egress to the thymus for development. T-cell progenitors start out as CD4/CD8 double negative (DN) cells. The four stages of double negative T cells (DN1-DN4) are characterized by expression of CD44, CD25, and CD117. Rearrangements of the Tcrb (TCRβ) locus and β selection occur during the DN3 stage of development. Rearrangements of the Tcra (TCRα) locus occur during the CD4/CD8 double positive (DP) stage of development. Additionally at the DP stage, positive and negative selection occurs. T cells that survive selection differentiate into CD4 and CD8 single positive (SP) T cells, which are able to egress to the periphery and perform many functions. Various cytokines allow for helper CD4+ T cells (Th) to differentiate and gain various functions; Th1, Th2, Th9, Th17, regulatory T cell (Treg), follicular T cell (Tfh). Cytotoxic CD8+ T cells terminally differentiate into effector CD8+ T cells (Teff) and central memory CD8+ T cells (Tmem). Red arrows in this diagram indicate where components of the Mi-2/NuRD chromatin remodeling complex impede T-cell development. Green arrows in this diagram indicate where components of the Mi-2/NuRD chromatin remodeling complex promote T-cell development. Chromodomain helicase DNA-binding protein 4 (CHD4), Metastasis-associated factor 1 (MTA1), Methyl-binding domain protein 2 (MBD2).
Interplay between CHD4 and Ikaros regulates T-cell development
Ikaros was first characterized as an important regulator of T cell development. Both Ikaros null mice and mice with an Ikaros dominant negative isoform have reductions in T cells (81, 86). Mice harboring dominant negative Ikaros lacking ZF1-4 have a complete absence of T-cell precursors, and thus, T cells (81). Mice with an Ikaros null mutation lack the C-terminal zinc fingers that are important for multimerization between Ikaros family members. These mice have an absence of fetal liver precursors and T cells. HSCs of adult mice with the null mutation give rise to thymic precursors; however, the resulting T cells are skewed toward CD4+ cells and clonal expansion is defective (86).
Ikaros interacts with Mi-2/NuRD complexes in T cells (69, 72, 100). Ikaros localizes to pericentromeric heterochromatin characterized by γ-satellite sequences and HP-1 in interphase nuclei (100). In resting T cells, Ikaros interacts with Brg1. Upon activation, Ikaros recruits Mi-2/NuRD to regions of heterochromatin (69, 101). In late S-phase, Ikaros-containing ‘toroidal’ structures co-localize with DNA replication clusters and heterochromatin (101). In activated T cells, Ikaros co-purifies with CHD3/4, HDAC1, HDAC2, and RbAp48 (69). Further analysis demonstrated that Ikaros interacts with SWI/SNF and Mi-2/NuRD in different complexes (69). Tandem affinity purification, followed by mass spectrometry of immature T-cell extracts (VL3-3M2) demonstrated that Ikaros predominantly interacts with components of the NuRD complex, including CHD3, CHD4, MTA1, MTA2, HDAC1, HDAC2, RbAp48, MBD3, and p66β (72). In addition to Mi-2/NuRD, Ikaros interacts with other CRCs and co-repressors, including Sin3 and CtBP (74, 102). Interestingly, both Ikaros and Helios can interact with CHD4 independently.
Conditional knockout mice revealed important roles of CHD4 in thymocyte development (Fig. 3). Floxed sequences encompassing the ATPase/helicase domain of CHD4 were conditionally deleted using Lck-Cre, which mediates excision after positive selection (17). In this system, loss of CHD4 expression resulted in a marked reduction in thymic cellularity, including decreased DP and CD4+ SP thymocytes and increased CD8+ SP T cells. Therefore, CHD4 regulates the transition from the DN to the DP stage of development that is subsequent to β selection. Surprisingly, overall expression of the TCR in Chd4-deleted T cells was not reduced. Further characterization of the mutant T cells demonstrated that the CD8+ SP thymocytes were actually DP-like but failed to express appropriate levels of CD4. T cells derived from the CHD4-deficient mice had proliferation defects: reductions were noted both in the numbers of activated T cells and of T cells that were able to undergo successive cell divisions. ChIP analysis of Cd4 regulatory elements detected the direct association of CHD4 with Cd4 proximal enhancers (17). Thus, CHD4 may regulate Cd4 genes by physically associating with the HEB (Tcf12) transcription factor and p300 at regulatory elements.
Analysis of T cells deficient in both CHD4 and Ikaros revealed antagonistic activities of the two proteins in T cells (103) (Fig. 3). Loss of Ikaros or CHD4 alone led to activation or repression, respectively, of Cd4 genes during T-cell development. In contrast, mice deficient in both CHD4 and Ikaros exhibited relatively normal expression of CD4 (103). These data suggest that Ikaros directs inappropriate repression of the Cd4 locus in the absence of CHD4. Analysis of the Cd4 silencer demonstrated that both Ikaros and CHD4 bind to the silencer and that CHD4 counteracts effects of Ikaros (103). The binding of CHD4 to the Cd4 silencer in DP thymocytes recruits HAT proteins, including MOZ, which mediates silencer inactivation (and thus, gene activation) concurrently with changes in its chromatin structure and histone acetylation.
Ikaros and Mi-2/NuRD also impact upon CD8+ T-cell development (104, 105). Ikaros associates with the CD8a loci throughout T cell development, where it increases chromatin accessibility by binding to Cd8a regulatory elements (105) (Fig. 3). Analysis of mice with reduced Ikzf1 (Ikaros) and Ikzf3 (Aiolos) alleles detected increased CD4+ thymocytes due to inactivation of Cd8a transcription, suggesting that the Ikaros family proteins drive Cd8a expression. Additionally, Cd8a transcription is regulated through CHD4 recruitment. ChIP-seq experiments demonstrated that Cd8a loci bear bivalent chromatin marks (H3K4me3 and H3K27me3) in DN thymocytes. Mi-2/NuRD complexes are associated with CD8a loci in DN1 and DN2 populations. However, the associations are reduced dramatically in DN3 and DN4 populations and are almost undetectable on these genes in DP and CD8+ SP thymocytes. At the DN3 stage, Cd8a loci become hypersensitive to DNaseI, indicating that CHD4 eviction is coinciding with altered chromatin accessibility necessary for transcription (104).
ChIP-seq experiments demonstrated the importance of CHD4 and Ikaros binding during lymphocyte differentiation (88). In an elegant study by Zhang and colleagues (88), ChIP-seq was used to detect localization of CHD4 to genes in the presence or absence of Ikaros in DP thymocytes. DP thymocytes contain Ikaros, Aiolos, CHD4, and other components of Mi-2/NuRD complexes. In the presence of Ikaros, Mi-2/NuRD was targeted by Ikaros to transcriptionally active genes involved in lymphocyte development. The binding of Ikaros to chromatin inhibits accessibility and nucleosomal remodeling. In the absence of Ikaros, Aiolos recruited Mi-2/NuRD to new gene targets. The absence of Ikaros led to increased Mi-2/NuRD binding activity, but at fewer sites associated with RNA polymerase II and lymphoid-specific gene transcription. Instead, Mi-2/NuRD binding was redistributed to sites of permissive chromatin, or “poised” genes, that are not normally associated with Ikaros binding. These inappropriately activated genes include those genes associated with metabolism and proliferation. In this manner, loss of Ikaros and dysregulation of Mi-2/NuRD binding promotes leukemogenesis (88).
Ikaros and CHD4 also collaborate to regulate V(D)J recombination at Tcra genes (106) (Fig. 3). The DN3-like thymoma JE131 arose spontaneously in an Ikaros null mouse (107). Vα to Jα rearrangements are greatly increased by enforced expression of Ikaros in these cells (106). ChIP experiments demonstrated that Ikaros binds directly to the Tcra enhancer. Knockdown of CHD4 using shRNA greatly potentiated Ikaros-dependent V-J recombination. Together, the data indicate that Mi-2/NuRD complexes repress V(D)J recombination of Tcra loci, but other aspects of this mechanism remain to be determined.
Mi-2/NuRD complexes determine CD4+ T-helper cell polarization
In addition to their roles in early T-cell development, components of Mi-2/NuRD complexes influence the balance between Th1 and Th2 CD4+ T-cell responses (polarization) in mice (Fig. 3). Following stimulation of their TCRs and co-receptors, Th1 cells generate pro-inflammatory responses by secreting interferon-γ (IFNγ) and interleukin-2 (IL-2). In contrast, activated Th2 cells secrete IL-4, IL-5, IL-13, which help generate IgE and atopy, and IL-10, which has anti-inflammatory properties. Differential expression of MBD2, MTA2, and CHD4 skew polarization towards Th2 responses (108–111). In part, these proteins function by augmenting/inhibiting transcription factors that mediate helper T-cell polarization: T-bet and Gata3 are master regulators of Th1 versus Th2 responses, respectively.
MBD2 has been implicated directly in Th2 responses. MBD2 knockout mice have normal lymphoid development and distribution of lymphocyte subsets (110). Mbd2−/− T cells cultured under Th1 (IFNγ) or Th2 (IL-4) polarizing conditions demonstrated greater activation of IFNγ and IL-4, respectively, compared to their wildtype littermates (110). These data indicate that MBD2 functions in cytokine gene silencing. Additionally, under Th1 polarizing conditions, Mbd2−/− T cells had expressed IL-4 ectopically. Further analysis of Mbd2−/− T cells showed that loss of MBD2 renders the transcription factor Gata3, which is essential for IL-4 expression in normal T cells, dispensable for IL-4 cytokine production. Expression of MBD2 by itself silences Il4 gene transcription. During activation of Il4 loci, Gata3 displaces MBD2 prior to CpG demethylation by competing with MBD2 for DNA binding (110).
The effects of MBD2 on Th1 versus Th2 polarization were tested in infection models using the protozoa Leishmania major (L. major) and the intestinal helminth, Trichuris muris (T. muris) (109). L. major causes a robust Th1 response by inducing IFNγ; in contrast, T. muris causes a robust Th2 response by inducing IL-4 and IL-13. Mbd2−/− mice infected with L. major in the footpad were able to control the infection compared to their wildtype littermates. Control of the infection was mediated by robust IFNγ secretion. In contrast, when Mbd2−/− mice were infected orally with T. muris, they demonstrated greater susceptibility to the helminth compared to their wild-type littermates. The Mbd2−/− mice had a significant increase of eggs in the intestine, excessive IFNγ responses and increased IgG2a antibodies (production of IgG2a is stimulated by IFNγ). MBD2 and T-bet inversely regulate IFNγ expression. MBD2 induces T-bet expression and represses Gata3 expression. In response to infections, Mbd2−/− Th2 T cells express IFNγ in the absence of T-bet. This observation is similar to responses of Mbd2−/− Th1 cells, which express IL-4 in the absence of Gata3. Thus, a role of MBD2 in polarization by each of these master regulators is evident.
In addition to MBD2-containing Mi-2/NuRD complexes, Gata3 also requires CHD4 to regulate Th2 responses in mice (108) (Fig. 3). CHD4 assembles complexes with Gata3 that activate Th2 cytokine production and repress IFNγ expression. Gata3 and CHD4 are localized at target genes together with the HAT p300 or HDAC2. The Gata3/CHD4/p300 and Gata3/CHD4/HDAC2 complexes are functionally distinct. Gata3, CHD4, and p300 are recruited to Th2 cytokine loci, including Il4, Il5, and Il13 to induce their transcription. In Th2 CD4+ T cells, Gata3, CHD4, and HDAC2 localize to the gene encoding T-bet, Tbx21, to repress T-bet-dependent IFNγ expression. Gata3, CHD4, MTA1, MTA2, p66, and MBD3 co-purified with each other from the Th2 CD4+ T-cell line, D10G4.1. When CHD4 expression was ablated through Chd4 specific shRNAs, polarization to Th2 was decreased. Transcription of Il4 and Il5 were repressed without altering Gata3 expression, while Ifng (IFNγ) transcripts were activated. The effects of CHD4 were also examined in a Th2-dominated model of allergic inflammation (108). Mice that were challenged in an allergic model of asthma with transfer of CHD4-deficient T cells exhibited (i) decreased inflammatory cells in bronchial alveolar lavage, (ii) modest mononuclear cell infiltration, and (iii) lower levels of mucus production compared to CHD4-sufficient T cells. These results indicate that CHD4, along with Gata3, is important for activating Th2 cytokine production and repressing Th1 cytokine production (108).
Other regulatory functions of Mi-2/NuRD components in T-cell development
Tregs are a subpopulation of T cells that modulate immune responses by maintaining tolerance and suppressing autoimmune disease (99). MBD2 plays a role in Treg development by binding to regulatory regions of Foxp3, the transcription factor that directs Treg polarization (112) (Fig. 3). MBD2 binds a unique CpG-rich island of the Foxp3 upstream enhancer. This region is unmethylated in natural Tregs that arise in the thymus, but it is methylated in naive CD4+ T cells, activated T cells, and peripheral TGFβ-induced Tregs. The CpG-rich island is bound by DNMT1, DNMT3b, MeCP2, and MBD2, which initiate and maintain DNA methylation. Demethylation of this site by the DNA methyltransferase inhibitor 5-azacytidine induced Foxp3 expression. Conversely, expression of IL-6 repressed Foxp3 transcription (112). IL-6 induces Stat-3-dependent upregulation of DNMT1. In turn, DNMT1 increases Foxp3 DNA methylation (113). Another study investigated the importance of a second regulatory region in Foxp3 genes: the Treg-specific demethylation region (TSDR) (114). In Mbd2−/− mice, Tregs were decreased with reduced suppressive functions. While TSDR regions in T cells from WT mice were completely demethylated, Mbd2−/− T cells were >75% methylated. Re-introduction of MBD2 into Mbd2−/− cells restored TSDR demethylation, Foxp3 expression and Treg suppressive functions. Further analysis of Mbd2−/− T cells revealed that they had reduced binding by the Ten-Eleven Translocase 2 (Tet2) oxidase, which has been implicated in DNA demethylation, at the TSDR (114). Details of the mechanism have yet to be determined, but alternative binding of methylated DNA by other methyl-binding proteins may be involved.
Driving CD8+ memory versus effector functions also has been attributed to MBD2 (115). Mbd2−/− mice were infected with the acute Armstrong strain of lymphocytic choriomenengitis virus (LCMV). Deficiency in MBD2 resulted in normal viral clearance and numbers of primary effector CD8+ T cells. However, Mbd2−/− mice had reduced numbers of antigen-specific CD8+ memory cells and the appearance of robustly functional CD127high memory cells was delayed. When mice were challenged with the chronic Clone 13 strain of LCMV, the memory CD8+ T cells from Mbd2−/− mice were not protective. Memory CD8+ T cells isolated from Mbd2−/− mice exhibited decreased CD127, CD27, CD62L, CD44, Ly6c, and bcl2. Together, these data suggest defects in gene activation. Additionally, Mbd2−/− T cells produced less IFNγ than wildtype T cells. The impaired precursor and memory cell induction in Mbd2−/− mice is CD8+ T-cell intrinsic. These studies confirm the importance of MBD2 for the development of CD8+ T-memory cells (115) (Fig. 3).
Bcl11b represses transcription via MTA1 and Mi-2/NuRD in T cells
Bcl11b (also known as Ctip2) is a transcription factor associated with transcriptional repression in T cells (reviewed in 116) (Fig. 3). Bcl11b is required for positive selection and survival of thymocytes, where it constitutes one of the earliest developmental checkpoints (117–119). Bcl11b has a C2H2 zinc finger and mediates transcriptional repression via its heterologous DNA binding domain. In Jurkat and MOLT4 T cells, endogenous Bcl11b associates with multiple components of Mi-2/NuRD complexes, including: MTA1, MTA2, CHD3, CHD4, MBD3, HDAC1, HDAC2, RbAp46, and RpAp48 (32). In co-immunoprecipitation assays, MTA1 and MTA2 directly interacted with Bcl11b through its N-terminus. Transcriptional repression mediated by Bcl11b is dependent on MTA1. When MTA1 was depleted by siRNA, transcriptional repression was reduced. Likewise, when MTA1 was overexpressed in T cells, transcriptional repression was enhanced. ChIP experiments revealed that MTA1 is specifically recruited to Bcl11b target promoters (32). Targets of Bcl11b include activation of NF-κB, Il-2 transcription and repression of the long-terminal repeat of HIV-1 in T cells (120–122).
Inactivation of MTA2 leads to an autoimmune-like disease
Effects of MTA2 deficiency were tested in Mta2 knockout mice (111). Loss of MTA2 caused embryonic and perinatal lethality. Mta2−/− embryos had defects in axial skeleton, skin and craniofacial structures; however, some Mta2−/− mice survived to adulthood. Surviving Mta2−/− adult mice had smaller body sizes, developed erosive skin lesions and female infertility. Mta2−/− mice developed serum autoantibodies that are commonly associated with systemic lupus erythematosus (SLE), a systemic autoimmune disease. T cells from Mta2−/− mice appeared normal as they developed in the thymus and migrated to the periphery; however, the mutant T cells hyperproliferated in response to T-cell stimulation by anti-CD3 and anti-CD28. The activated mutant T cells produced more IL-2, IL-4, and IFNγ in the absence of MTA2. However, MTA2 deficiency did not affect Th1 polarization. These results indicate that when MTA2 is present in CD4+ T cells, MTA2 suppresses both IL-4 and IFNγ cytokine production in Th2 responses. ChIP assays demonstrate that MTA2 binds to the Il4 promoter and to another site downstream of its transcriptional start site (111).
Conclusions
The Mi-2/NuRD chromatin remodeling complex is a multi-subunit complex that couples the processes of (i) histone deacetylation, (ii) histone demethylation, (iii) nucleosomal mobilization, (iv) recruitment of regulatory proteins, and (v) recruitment to methylated DNA. The composition of these complexes helps determine functions in different cell types and at different developmental stages throughout hematopoiesis. In HSCs, expression of CHD4 and the transcription factor, Ikaros, bias differentiation, in part, by repressing the myeloid- and erythroid-specific developmental programs. Prior to B-lineage specification and commitment, Mi-2/NuRD components CHD4 and MBD2 repress transcription of the B-cell-specific gene, Cd79a. Progression of B-cell development requires Ikaros, which co-localizes at promoters with CHD4 in pro-B cells. Additionally, Mi-2/NuRD complexes influences terminal differentiation of B cells through interactions with MTA3 (through its interaction with Bcl6) and LSD1 (with its interaction with Blimp1). Mi-2/NuRD complexes are essential for normal T-cell development and peripheral CD4+ and CD8+ responses. The interplay between CHD4 and Ikaros determines CD4+ versus CD8+ cell fates and determines how CHD4 binds to specific gene targets. In addition, Ikaros and CHD4 help propagate V(D)J recombination in thymocytes. Mi-2/NuRD complexes drive Th1 and Th2 polarization in CD4 T cells via activities of CHD4, MTA2 and MBD2. Additionally, MBD2 is important for regulatory T-cell development and CD8+ memory formation. Through its interaction with Bcl11b, Mi-2/NuRD controls transcription of T-cell specific genes.
One question that remains unanswered with the NuRD complex is how it acts uniquely as both a transcriptional activator and repressor depending on the context of expression. Solution of this problem will require greater understanding of epigenetic mechanisms involving Mi-1/NuRD, including cross-talk between transcription factors, various chromatin-remodeling complexes, and signaling. By understanding the epigenetic regulation that NuRD imposes during hematopoiesis and lymphopoiesis, new cancer therapeutics can be developed, in particular those for hematological malignancies. The Mi-2/NuRD complex is an attractive target for chemotherapy because of its involvement in epigenetic regulation and control of proliferation. Moreover, further investigation concerning the regulation of Mi-2/NuRD complexes is important for our understanding of epigenetics in general.
Acknowledgments
This work was supported by NIH grants AI081878 and AI098417. CD was supported by a Pre-doctoral Emphasis Pathway in Tumor Immunology Fellowship from the Cancer Res Institute. We would like to thank Kara Lukin and Sarah Greaves for editorial and technical assistance with the manuscript.
Footnotes
The authors have no financial conflicts of interest to disclose.
References
- 1.Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–21. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 2.Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–61. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–89. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–35. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–6. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 6.Nair SS, Li DQ, Kumar R. A core chromatin remodeling factor instructs global chromatin signaling through multivalent reading of nucleosome codes. Mol Cell. 2013;49:704–18. doi: 10.1016/j.molcel.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watson AA, Mahajan P, Mertens HD, Deery MJ, Zhang W, Pham P, et al. The PHD and chromo domains regulate the ATPase activity of the human chromatin remodeler CHD4. J Mol Biol. 2012;422:3–17. doi: 10.1016/j.jmb.2012.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morra R, Lee BM, Shaw H, Tuma R, Mancini EJ. Concerted action of the PHD, chromo and motor domains regulates the human chromatin remodelling ATPase CHD4. FEBS Lett. 2012;586:2513–21. doi: 10.1016/j.febslet.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moshkin YM, Chalkley GE, Kan TW, Reddy BA, Ozgur Z, van Ijcken WF, et al. Remodelers organize cellular chromatin by counteracting intrinsic histone-DNA sequence preferences in a class-specific manner. Mol Cell Biol. 2012;32:675–88. doi: 10.1128/MCB.06365-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morey L, Brenner C, Fazi F, Villa R, Gutierrez A, Buschbeck M, et al. MBD3, a component of the NuRD complex, facilitates chromatin alteration and deposition of epigenetic marks. Mol Cell Biol. 2008;28:5912–23. doi: 10.1128/MCB.00467-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sims JK, Wade PA. Mi-2/NuRD complex function is required for normal S phase progression and assembly of pericentric heterochromatin. Mol Biol Cell. 2011;22:3094–102. doi: 10.1091/mbc.E11-03-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwintkiewicz J, Padilla-Banks E, Jefferson WN, Jacobs IM, Wade PA, Williams CJ. Metastasis-associated protein 3 (MTA3) regulates G2/M progression in proliferating mouse granulosa cells. Biol Reprod. 2012;86:1–8. doi: 10.1095/biolreprod.111.096032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helbling Chadwick L, Chadwick BP, Jaye DL, Wade PA. The Mi-2/NuRD complex associates with pericentromeric heterochromatin during S phase in rapidly proliferating lymphoid cells. Chromosoma. 2009;118:445–57. doi: 10.1007/s00412-009-0207-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138:660–72. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 15.Kolla V, Zhuang T, Higashi M, Naraparaju K, Brodeur GM. Role of CHD5 in human cancers: 10 years later. Cancer Res. 2014;74:652–8. doi: 10.1158/0008-5472.CAN-13-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–4. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 17.Williams CJ, Naito T, Arco PG, Seavitt JR, Cashman SM, De Souza B, et al. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity. 2004;20:719–33. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Watson AA, Mahajan P, Mertens HD, Deery MJ, Zhang W, Pham P, et al. The PHD and chromo domains regulate the ATPase activity of the human chromatin remodeler CHD4. Journal of molecular biology. 2012;422:3–17. doi: 10.1016/j.jmb.2012.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Musselman CA, Mansfield RE, Garske AL, Davrazou F, Kwan AH, Oliver SS, et al. Binding of the CHD4 PHD2 finger to histone H3 is modulated by covalent modifications. Biochem J. 2009;423:179–87. doi: 10.1042/BJ20090870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Musselman CA, Ramirez J, Sims JK, Mansfield RE, Oliver SS, Denu JM, et al. Bivalent recognition of nucleosomes by the tandem PHD fingers of the CHD4 ATPase is required for CHD4-mediated repression. Proc Natl Acad Sci U S A. 2012;109:787–92. doi: 10.1073/pnas.1113655109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramirez J, Dege C, Kutateladze TG, Hagman J. MBD2 and multiple domains of CHD4 are required for transcriptional repression by Mi-2/NuRD complexes. Mol Cell Biol. 2012;32:5078–88. doi: 10.1128/MCB.00819-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones DO, Cowell IG, Singh PB. Mammalian chromodomain proteins: their role in genome organisation and expression. BioEssays. 2000;22:124–37. doi: 10.1002/(SICI)1521-1878(200002)22:2<124::AID-BIES4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 23.Bouazoune K, Mitterweger A, Langst G, Imhof A, Akhtar A, Becker PB, et al. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. EMBO J. 2002;21:2430–40. doi: 10.1093/emboj/21.10.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hauk G, McKnight JN, Nodelman IM, Bowman GD. The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol Cell. 2010;39:711–23. doi: 10.1016/j.molcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao YL, Yang WM. The metastasis-associated proteins 1 and 2 form distinct protein complexes with histone deacetylase activity. J Biol Chem. 2003;278:42560–8. doi: 10.1074/jbc.M302955200. [DOI] [PubMed] [Google Scholar]
- 26.Fujita N, Jaye DL, Kajita M, Geigerman C, Moreno CS, Wade PA. MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell. 2003;113:207–19. doi: 10.1016/s0092-8674(03)00234-4. [DOI] [PubMed] [Google Scholar]
- 27.Oliver AW, Jones SA, Roe SM, Matthews S, Goodwin GH, Pearl LH. Crystal structure of the proximal BAH domain of the polybromo protein. Biochem J. 2005;389:657–64. doi: 10.1042/BJ20050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Humphrey GW, Wang Y, Russanova VR, Hirai T, Qin J, Nakatani Y, et al. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J Biol Chem. 2001;276:6817–24. doi: 10.1074/jbc.M007372200. [DOI] [PubMed] [Google Scholar]
- 29.Roche AE, Bassett BJ, Samant SA, Hong W, Blobel GA, Svensson EC. The zinc finger and C-terminal domains of MTA proteins are required for FOG-2-mediated transcriptional repression via the NuRD complex. J Mol Cell Cardiol. 2008;44:352–60. doi: 10.1016/j.yjmcc.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujita N, Jaye DL, Geigerman C, Akyildiz A, Mooney MR, Boss JM, et al. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell. 2004;119:75–86. doi: 10.1016/j.cell.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 31.Jaye DL, Iqbal J, Fujita N, Geigerman CM, Li S, Karanam S, et al. The BCL6-associated transcriptional co-repressor, MTA3, is selectively expressed by germinal centre B cells and lymphomas of putative germinal centre derivation. J Pathol. 2007;213:106–15. doi: 10.1002/path.2199. [DOI] [PubMed] [Google Scholar]
- 32.Cismasiu VB, Adamo K, Gecewicz J, Duque J, Lin Q, Avram D. BCL11B functionally associates with the NuRD complex in T lymphocytes to repress targeted promoter. Oncogene. 2005;24:6753–64. doi: 10.1038/sj.onc.1208904. [DOI] [PubMed] [Google Scholar]
- 33.Wu M, Wang L, Li Q, Li J, Qin J, Wong J. The MTA family proteins as novel histone H3 binding proteins. Cell Biosci. 2013;3:1–14. doi: 10.1186/2045-3701-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saito M, Ishikawa F. The mCpG-binding domain of human MBD3 does not bind to mCpG but interacts with NuRD/Mi2 components HDAC1 and MTA2. J Biol Chem. 2002;277:35434–9. doi: 10.1074/jbc.M203455200. [DOI] [PubMed] [Google Scholar]
- 35.Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408:377–81. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 36.Li X, Jia S, Wang S, Wang Y, Meng A. Mta3-NuRD complex is a master regulator for initiation of primitive hematopoiesis in vertebrate embryos. Blood. 2009;114:5464–72. doi: 10.1182/blood-2009-06-227777. [DOI] [PubMed] [Google Scholar]
- 37.Hong W, Nakazawa M, Chen YY, Kori R, Vakoc CR, Rakowski C, et al. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J. 2005;24:2367–78. doi: 10.1038/sj.emboj.7600703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ross J, Mavoungou L, Bresnick EH, Milot E. GATA-1 utilizes Ikaros and polycomb repressive complex 2 to suppress Hes1 and to promote erythropoiesis. Mol Cell Biol. 2012;32:3624–38. doi: 10.1128/MCB.00163-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh RR, Barnes CJ, Talukder AH, Fuqua SA, Kumar R. Negative regulation of estrogen receptor alpha transactivation functions by LIM domain only 4 protein. Cancer Res. 2005;65:10594–601. doi: 10.1158/0008-5472.CAN-05-2268. [DOI] [PubMed] [Google Scholar]
- 40.Fournier A, Sasai N, Nakao M, Defossez PA. The role of methyl-binding proteins in chromatin organization and epigenome maintenance. Brief Funct Genomics. 2012;11:251–64. doi: 10.1093/bfgp/elr040. [DOI] [PubMed] [Google Scholar]
- 41.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–47. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le Guezennec X, Vermeulen M, Brinkman AB, Hoeijmakers WA, Cohen A, Lasonder E, et al. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol Cell Biol. 2006;26:843–51. doi: 10.1128/MCB.26.3.843-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan CP, Nakielny S. Control of the DNA methylation system component MBD2 by protein arginine methylation. Mol Cell Biol. 2006;26:7224–35. doi: 10.1128/MCB.00473-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15:710–23. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baubec T, Ivanek R, Lienert F, Schubeler D. Methylation-dependent and -independent genomic targeting principles of the MBD protein family. Cell. 2013;153:480–92. doi: 10.1016/j.cell.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 47.Gnanapragasam MN, Scarsdale JN, Amaya ML, Webb HD, Desai MA, Walavalkar NM, et al. p66Alpha-MBD2 coiled-coil interaction and recruitment of Mi-2 are critical for globin gene silencing by the MBD2-NuRD complex. Proc Natl Acad Sci U S A. 2011;108:7487–92. doi: 10.1073/pnas.1015341108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brackertz M, Gong Z, Leers J, Renkawitz R. p66alpha and p66beta of the Mi-2/NuRD complex mediate MBD2 and histone interaction. Nucleic Acids Res. 2006;34:397–406. doi: 10.1093/nar/gkj437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brackertz M, Boeke J, Zhang R, Renkawitz R. Two highly related p66 proteins comprise a new family of potent transcriptional repressors interacting with MBD2 and MBD3. J Biol Chem. 2002;277:40958–66. doi: 10.1074/jbc.M207467200. [DOI] [PubMed] [Google Scholar]
- 50.Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–92. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- 51.Kaji K, Nichols J, Hendrich B. Mbd3, a component of the NuRD co-repressor complex, is required for development of pluripotent cells. Development. 2007;134:1123–32. doi: 10.1242/dev.02802. [DOI] [PubMed] [Google Scholar]
- 52.Luo M, Ling T, Xie W, Sun H, Zhou Y, Zhu Q, et al. NuRD blocks reprogramming of mouse somatic cells into pluripotent stem cells. Stem Cells. 2013;31:1278–86. doi: 10.1002/stem.1374. [DOI] [PubMed] [Google Scholar]
- 53.Shimbo T, Du Y, Grimm SA, Dhasarathy A, Mav D, Shah RR, et al. MBD3 localizes at promoters, gene bodies and enhancers of active genes. PLoS Genet. 2013;9:e1004028. doi: 10.1371/journal.pgen.1004028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gunther K, Rust M, Leers J, Boettger T, Scharfe M, Jarek M, et al. Differential roles for MBD2 and MBD3 at methylated CpG islands, active promoters and binding to exon sequences. Nucleic Acids Res. 2013;41:3010–21. doi: 10.1093/nar/gkt035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 57.Lejon S, Thong SY, Murthy A, AlQarni S, Murzina NV, Blobel GA, et al. Insights into association of the NuRD complex with FOG-1 from the crystal structure of an RbAp48.FOG-1 complex. J Biol Chem. 2011;286(2):1196–203. doi: 10.1074/jbc.M110.195842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Migliori V, Muller J, Phalke S, Low D, Bezzi M, Mok WC, et al. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat Struct Mol Biol. 2012;19:136–44. doi: 10.1038/nsmb.2209. [DOI] [PubMed] [Google Scholar]
- 59.Murzina NV, Pei XY, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, et al. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077–85. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feng Q, Cao R, Xia L, Erdjument-Bromage H, Tempst P, Zhang Y. Identification and functional characterization of the p66/p68 components of the MeCP1 complex. Mol Cell Biol. 2002;22:536–46. doi: 10.1128/MCB.22.2.536-546.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marino S, Nusse R. Mutants in the mouse NuRD/Mi2 component P66alpha are embryonic lethal. PLoS One. 2007;2:e519. doi: 10.1371/journal.pone.0000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Allen HF, Wade PA, Kutateladze TG. The NuRD architecture. Cell Mol Life Sci. 2013;70:3513–24. doi: 10.1007/s00018-012-1256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thiagalingam S, Cheng KH, Lee HJ, Mineva N, Thiagalingam A, Ponte JF. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann N Y Acad Sci. 2003;983:84–100. doi: 10.1111/j.1749-6632.2003.tb05964.x. [DOI] [PubMed] [Google Scholar]
- 64.Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–5. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 65.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–9. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 66.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 67.Li Q, Shi L, Gui B, Yu W, Wang J, Zhang D, et al. Binding of the JmjC demethylase JARID1B to LSD1/NuRD suppresses angiogenesis and metastasis in breast cancer cells by repressing chemokine CCL14. Cancer Res. 2011;71:6899–908. doi: 10.1158/0008-5472.CAN-11-1523. [DOI] [PubMed] [Google Scholar]
- 68.Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–5. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–55. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 70.Yoshida T, Hazan I, Zhang J, Ng SY, Naito T, Snippert HJ, et al. The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev. 2008;22:1174–89. doi: 10.1101/gad.1642808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O’Neill DW, Schoetz SS, Lopez RA, Castle M, Rabinowitz L, Shor E, et al. An ikaros-containing chromatin-remodeling complex in adult-type erythroid cells. Mol Cell Biol. 2000;20:7572–82. doi: 10.1128/mcb.20.20.7572-7582.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sridharan R, Smale ST. Predominant interaction of both Ikaros and Helios with the NuRD complex in immature thymocytes. J Biol Chem. 2007;282:30227–38. doi: 10.1074/jbc.M702541200. [DOI] [PubMed] [Google Scholar]
- 73.Rodriguez P, Bonte E, Krijgsveld J, Kolodziej KE, Guyot B, Heck AJ, et al. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 2005;24:2354–66. doi: 10.1038/sj.emboj.7600702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koipally J, Renold A, Kim J, Georgopoulos K. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J. 1999;18:3090–100. doi: 10.1093/emboj/18.11.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nichogiannopoulou A, Trevisan M, Neben S, Friedrich C, Georgopoulos K. Defects in hemopoietic stem cell activity in Ikaros mutant mice. J Exp Med. 1999;190:1201–14. doi: 10.1084/jem.190.9.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yoshida T, Ng SY, Zuniga-Pflucker JC, Georgopoulos K. Early hematopoietic lineage restrictions directed by Ikaros. Nat Immunol. 2006;7:382–91. doi: 10.1038/ni1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hardy RR, Kincade PW, Dorshkind K. The protean nature of cells in the B lymphocyte lineage. Immunity. 2007;26:703–14. doi: 10.1016/j.immuni.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 78.Maier H, Ostraat R, Gao H, Fields S, Shinton SA, Medina KL, et al. Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated with mb-1 transcription. Nat Immunol. 2004;5:1069–77. doi: 10.1038/ni1119. [DOI] [PubMed] [Google Scholar]
- 79.Maier H, Colbert J, Fitzsimmons D, Clark DR, Hagman J. Activation of the early B-cell-specific mb-1 (Ig-alpha) gene by Pax-5 is dependent on an unmethylated Ets binding site. Mol Cell Biol. 2003;23:1946–60. doi: 10.1128/MCB.23.6.1946-1960.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gao H, Lukin K, Ramirez J, Fields S, Lopez D, Hagman J. Opposing effects of SWI/SNF and Mi-2/NuRD chromatin remodeling complexes on epigenetic reprogramming by EBF and Pax5. Proc Natl Acad Sci U S A. 2009;106:11258–63. doi: 10.1073/pnas.0809485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Georgopoulos K, Bigby M, Wang JH, Molnar A, Wu P, Winandy S, et al. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–56. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- 82.Georgopoulos K, Moore DD, Derfler B. Ikaros, an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science. 1992;258:808–12. doi: 10.1126/science.1439790. [DOI] [PubMed] [Google Scholar]
- 83.Hahm K, Ernst P, Lo K, Kim GS, Turck C, Smale ST. The lymphoid transcription factor LyF-1 is encoded by specific, alternatively spliced mRNAs derived from the Ikaros gene. Mol Cell Biol. 1994;14:7111–23. doi: 10.1128/mcb.14.11.7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.John LB, Ward AC. The Ikaros gene family: transcriptional regulators of hematopoiesis and immunity. Mol Immunol. 2011;48:1272–8. doi: 10.1016/j.molimm.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 85.Sun L, Liu A, Georgopoulos K. Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J. 1996;15:5358–69. [PMC free article] [PubMed] [Google Scholar]
- 86.Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M, et al. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–49. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- 87.Schwickert TA, Tagoh H, Gultekin S, Dakic A, Axelsson E, Minnich M, et al. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat Immunol. 2014;15:283–93. doi: 10.1038/ni.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang J, Jackson AF, Naito T, Dose M, Seavitt J, Liu F, et al. Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat Immunol. 2012;13:86–94. doi: 10.1038/ni.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koipally J, Georgopoulos K. Ikaros-CtIP interactions do not require C-terminal binding protein and participate in a deacetylase-independent mode of repression. J Biol Chem. 2002;277:23143–9. doi: 10.1074/jbc.M202079200. [DOI] [PubMed] [Google Scholar]
- 90.Bereshchenko OR, Gu W, Dalla-Favera R. Acetylation inactivates the transcriptional repressor BCL6. Nat Genet. 2002;32:606–13. doi: 10.1038/ng1018. [DOI] [PubMed] [Google Scholar]
- 91.Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 92.Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–20. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 93.Shapiro-Shelef M, Calame K. Regulation of plasma-cell development. Nat Rev Immunol. 2005;5:230–42. doi: 10.1038/nri1572. [DOI] [PubMed] [Google Scholar]
- 94.Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, et al. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8:623–30. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- 95.Gyory I, Wu J, Fejer G, Seto E, Wright KL. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat Immunol. 2004;5:299–308. doi: 10.1038/ni1046. [DOI] [PubMed] [Google Scholar]
- 96.Yu J, Angelin-Duclos C, Greenwood J, Liao J, Calame K. Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol Cell Biol. 2000;20:2592–603. doi: 10.1128/mcb.20.7.2592-2603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Su ST, Ying HY, Chiu YK, Lin FR, Chen MY, Lin KI. Involvement of histone demethylase LSD1 in Blimp-1-mediated gene repression during plasma cell differentiation. Mol Cell Biol. 2009;29:1421–31. doi: 10.1128/MCB.01158-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Germain RN. T-cell development and the CD4-CD8 lineage decision. Nat Rev Immunol. 2002;2:309–22. doi: 10.1038/nri798. [DOI] [PubMed] [Google Scholar]
- 99.Vignali DA, Collison LW, Workman CJ. How regulatory T cells work. Nat Rev Immunol. 2008;8:523–32. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–54. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 101.Avitahl N, Winandy S, Friedrich C, Jones B, Ge Y, Georgopoulos K. Ikaros sets thresholds for T cell activation and regulates chromosome propagation. Immunity. 1999;10:333–43. doi: 10.1016/s1074-7613(00)80033-3. [DOI] [PubMed] [Google Scholar]
- 102.Koipally J, Georgopoulos K. Ikaros interactions with CtBP reveal a repression mechanism that is independent of histone deacetylase activity. J Biol Chem. 2000;275:19594–602. doi: 10.1074/jbc.M000254200. [DOI] [PubMed] [Google Scholar]
- 103.Naito T, Gomez-Del Arco P, Williams CJ, Georgopoulos K. Antagonistic interactions between Ikaros and the chromatin remodeler Mi-2beta determine silencer activity and Cd4 gene expression. Immunity. 2007;27:723–34. doi: 10.1016/j.immuni.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 104.Harker N, Garefalaki A, Menzel U, Ktistaki E, Naito T, Georgopoulos K, et al. Pre-TCR signaling and CD8 gene bivalent chromatin resolution during thymocyte development. J Immunol. 2011;186:6368–77. doi: 10.4049/jimmunol.1003567. [DOI] [PubMed] [Google Scholar]
- 105.Harker N, Naito T, Cortes M, Hostert A, Hirschberg S, Tolaini M, et al. The CD8alpha gene locus is regulated by the Ikaros family of proteins. Mol Cell. 2002;10:1403–15. doi: 10.1016/s1097-2765(02)00711-6. [DOI] [PubMed] [Google Scholar]
- 106.Collins B, Clambey ET, Scott-Browne J, White J, Marrack P, Hagman J, et al. Ikaros promotes rearrangement of TCR alpha genes in an Ikaros null thymoma cell line. Eur J Immunol. 2013;43:521–32. doi: 10.1002/eji.201242757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kathrein KL, Lorenz R, Innes AM, Griffiths E, Winandy S. Ikaros induces quiescence and T-cell differentiation in a leukemia cell line. Mol Cell Biol. 2005;25:1645–54. doi: 10.1128/MCB.25.5.1645-1654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hosokawa H, Tanaka T, Suzuki Y, Iwamura C, Ohkubo S, Endoh K, et al. Functionally distinct Gata3/Chd4 complexes coordinately establish T helper 2 (Th2) cell identity. Proc Natl Acad Sci U S A. 2013;110:4691–6. doi: 10.1073/pnas.1220865110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hutchins AS, Artis D, Hendrich BD, Bird AP, Scott P, Reiner SL. Cutting edge: a critical role for gene silencing in preventing excessive type 1 immunity. J Immunol. 2005;175:5606–10. doi: 10.4049/jimmunol.175.9.5606. [DOI] [PubMed] [Google Scholar]
- 110.Hutchins AS, Mullen AC, Lee HW, Sykes KJ, High FA, Hendrich BD, et al. Gene silencing quantitatively controls the function of a developmental trans-activator. Mol Cell. 2002;10:81–91. doi: 10.1016/s1097-2765(02)00564-6. [DOI] [PubMed] [Google Scholar]
- 111.Lu X, Kovalev GI, Chang H, Kallin E, Knudsen G, Xia L, et al. Inactivation of NuRD component Mta2 causes abnormal T cell activation and lupus-like autoimmune disease in mice. J Biol Chem. 2008;283:13825–33. doi: 10.1074/jbc.M801275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lal G, Zhang N, van der Touw W, Ding Y, Ju W, Bottinger EP, et al. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J Immunol. 2009;182:259–73. doi: 10.4049/jimmunol.182.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lal G, Bromberg JS. Epigenetic mechanisms of regulation of Foxp3 expression. Blood. 2009;114:3727–35. doi: 10.1182/blood-2009-05-219584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang L, Liu Y, Han R, Beier UH, Thomas RM, Wells AD, et al. Mbd2 promotes foxp3 demethylation and T-regulatory-cell function. Mol Cell Biol. 2013;33:4106–15. doi: 10.1128/MCB.00144-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kersh EN. Impaired memory CD8 T cell development in the absence of methyl-CpG-binding domain protein 2. J Immunol. 2006;177:3821–6. doi: 10.4049/jimmunol.177.6.3821. [DOI] [PubMed] [Google Scholar]
- 116.Rothenberg EV. Transcriptional drivers of the T-cell lineage program. Curr Opin Immunol. 2012;24:132–8. doi: 10.1016/j.coi.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wakabayashi Y, Watanabe H, Inoue J, Takeda N, Sakata J, Mishima Y, et al. Bcl11b is required for differentiation and survival of αβ T lymphocytes. Nat Immunol. 2003;4:533–9. doi: 10.1038/ni927. [DOI] [PubMed] [Google Scholar]
- 118.Albu DI, Feng D, Bhattacharya D, Jenkins NA, Copeland NG, Liu P, et al. BCL11B is required for positive selection and survival of double-positive thymocytes. J Exp Med. 2007;204:3003–15. doi: 10.1084/jem.20070863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ikawa T, Hirose S, Masuda K, Kakugawa K, Satoh R, Shibano-Satoh A, et al. An essential developmental checkpoint for production of the T cell lineage. Science. 2010;329:93–6. doi: 10.1126/science.1188995. [DOI] [PubMed] [Google Scholar]
- 120.Cismasiu VB, Duque J, Paskaleva E, Califano D, Ghanta S, Young HA, et al. BCL11B enhances TCR/CD28-triggered NF-kappaB activation through up-regulation of Cot kinase gene expression in T-lymphocytes. Biochem J. 2009;417:457–66. doi: 10.1042/BJ20080925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cismasiu VB, Ghanta S, Duque J, Albu DI, Chen HM, Kasturi R, et al. BCL11B participates in the activation of IL2 gene expression in CD4+ T lymphocytes. Blood. 2006;108:2695–702. doi: 10.1182/blood-2006-05-021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cismasiu VB, Paskaleva E, Suman Daya S, Canki M, Duus K, Avram D. BCL11B is a general transcriptional repressor of the HIV-1 long terminal repeat in T lymphocytes through recruitment of the NuRD complex. Virology. 2008;380:173–81. doi: 10.1016/j.virol.2008.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]