Abstract
We present a custom, Boolean query generator utilizing common-table expressions (CTEs) that is capable of scaling with big datasets. The generator maps user-defined Boolean queries, such as those interactively created in clinical-research and general-purpose healthcare tools, into SQL. We demonstrate the effectiveness of this generator by integrating our work into the Informatics for Integrating Biology and the Bedside (i2b2) query tool and show that it is capable of scaling. Our custom generator replaces and outperforms the default query generator found within the Clinical Research Chart (CRC) cell of i2b2. In our experiments, sixteen different types of i2b2 queries were identified by varying four constraints: date, frequency, exclusion criteria, and whether selected concepts occurred in the same encounter. We generated non-trivial, random Boolean queries based on these 16 types; the corresponding SQL queries produced by both generators were compared by execution times. The CTE-based solution significantly outperformed the default query generator and provided a much more consistent response time across all query types (M=2.03, SD=6.64 vs. M=75.82, SD=238.88 seconds). Without costly hardware upgrades, we provide a scalable solution based on CTEs with very promising empirical results centered on performance gains. The evaluation methodology used for this provides a means of profiling clinical data warehouse performance.
Index Terms: Biomedical computing, data warehouses, biomedical informatics, health information management, data systems, large-scale systems
I. Introduction
Clinical data warehouses (CDWs) are a necessary component to any healthcare institution for operational and research purposes [1]. Informatics for Integrating Biology and the Bedside (i2b2) is an initiative sponsored by the NIH Roadmap National Centers for Biomedical Computing [2]. One of the initiative’s main products is the i2b2 Web Client – a query tool capable of supplying aggregate counts and basic analyses of patient populations from CDWs. i2b2 has been shown to be effective in estimating cohort sizes [2]–[4] and serves as a potential cost-effective back-end for performing genome-wide association studies [3], [5]. It is reported to be used by over half of all sites awarded a Clinical and Translational Science Award (CTSA), over 60 academic medical centers, and 10 international medical centers [3].
In the age of Google, users expect instantaneous query results. This client-side consumer assumption is often difficult to satisfy for informatics software due to open challenges with big data and the complexity of queries upon clinical data warehouses. For instance, i2b2’s performance is reasonable with our single hospital clinical data set containing approximately 540,000 patients across 4 million encounters. However, significant performance degradation was experienced with our larger state-wide Medicaid data set containing approximately 1.8 million patients across 160 million encounters. The poorest performing queries were those requiring concepts to occur in the same encounter, especially when additional constraints such as exclusion criteria or occurrences (frequency) were used.
Some institutions have been able to achieve reasonable performance on large datasets with significant investments in hardware upgrades. Because not every medical center can afford extensive and elaborate hardware environments and because the growth of biomedical data is outpacing Moore’s law for the growth of computational power [6], elegant software solutions that are capable of scaling are needed. The goal of this work is to provide a scalable query generator that greatly decreases the end-user’s wait time for obtaining aggregate counts. In addition, our experimental design provides a methodology for profiling and comparing the performance of clinical data warehouse query tools.
II. Background
Clinical data warehouses naturally expand as time elapses. An increased emphasis on adopting electronic heath records and other clinical information systems has lead to the expansion of CDWs in both volume and breadth [1]. Additionally, the inclusion of genomic data is increasingly important for translational science [5], which suggests that the scale of the modern CDW will continue to increase. In addition to scale, the use of the data is evolving; the Shared Health Research Information Network (SHRINE) is a federated query tool that was motivated by the CTSA to produce a tool to enable collaboration across multiple sites [7]. SHRINE is an i2b2-based solution where queries are distributed to multiple i2b2 warehouses within the SHRINE and result sets are aggregated for the end-user. The expansion of data in terms of raw size and scope of usage is further evidence that scalability is pivotal to the success of the individual informatics software solutions and consequently to the much larger goals of clinical and translational science.
Other query tools exist for performing clinical research, including BTRIS [8], DEDUCE [9], FURTHeR [10], STRIDE [11], and VISAGE [12]. Note that these are tools created with specific goals in mind, while i2b2 is essentially an extendable platform for open-source development. Regardless of the specific design, a tool must somehow allow the user to construct a query that pinpoints the data being requested. Boolean queries are important in clinical data warehousing because they provide the natural mapping for inclusion and exclusion criteria when selecting sets of patients, cohorts, and encounters. Additionally, this selection stage is critical for clinical research [1], [4].
Each of the listed tools has an interface that allows the user to actively construct a query, either visually or with the help of pull-down menus; the tool must somehow translate this construction into valid SQL so that the CDW can be interactively explored. We call this translation component the query generator, which plays a crucial role by allowing non-technical users to build queries without writing complex statements in a language such as SQL. Our query generator works by receiving an XML message from the tool and translates it into a SQL query that can be run against the CDW. FURTHeR uses i2b2 as a front-end and could use our query generator, since its queries are composed in an identical interface. The other tools would need to package their query requests in a similar XML message in order to receive a query from our generator.
The most commonly published enhancements or extensions of i2b2 have been in the form of project forks and plugins that provide novel functionality [13]–[15] or in-depth examinations of i2b2’s use cases [4], [16], [17]. In this paper, we present our research in developing a scalable query generator and present our results on associated performance gains when it is used as an alternative to the internal generator of i2b2. We note that there exists related work [18] directed toward enhancing performance of i2b2, which focuses on pre-computing aggregate values and obtaining inexact counts via simulation. This related work could also be layered on top of our solution, which, in contrast, focuses on improvements in query generation.
i2b2 consists of a web client and desktop client that increasingly share similar functionality. In both clients, the user interface consists of a series of panels (also called groups). Users select concepts from an ontology and drag them into the panels to form basic Boolean queries; concepts within a panel represent a logical OR, while concepts across panels represent a logical AND (see details in the METHODS section). Additionally, panels can be negated and constrained by time, frequency, and whether the concepts occurred in the same encounter — that is, a single billable visit to a provider or inpatient stay at a hospital. The basic internal structure of i2b2 is that of a modular hive, where each hive cell is designed to provide a certain function [19]. Our contribution concerns the Clinical Research Chart (CRC) cell, which acts as the core data repository handler. When the user submits a query, the CRC cell converts the panels that the user has composed through the interface into an SQL statement that is executed against the local data repository. Our work replaces the query generation logic found within the CRC cell, so that more elegant SQL statements are produced and are followed by performance gains.
The default query generator that is packaged with i2b2 generates queries that use a procedural, multiple transaction approach. The panels assembled by the user are converted into multiple SQL statements. This approach takes patient records that match the concepts in the first panel and inserts them into a temporary table. The process continues and the records that also match the second panel’s concepts are updated in the temporary table. This process continues for all panels where the subsequent set of patients in the temporary table to be updated is never larger than the previous and the resulting temporary table will hold only those patient records that match the query the user constructed. The major issue is that this technique is heavily dependent on hard disk access- and write-speeds coupled with the large amounts of temporary storage on the database required for storing and logging these temporary result sets.
III. Methods
As an alternative to the procedural approach, we developed a query generator that produces a single relational query using Common Table Expressions (CTEs), which were first introduced in Microsoft SQL Server 2005 [20] and later integrated into Oracle and other database management systems. Fig. 1 shows the general framework for a CTE.
Fig. 1.
The general framework of a CTE containing N common tables that join N select statements. In each common table, a select statement determines the result set and previous common tables can be referenced in subsequent ones. The last SELECT statement dictates what the final result set will be.
CTEs are temporary, named result sets that reside in memory as complex derived views of the underlying data and can be referenced multiple times within a single SQL statement (including self-reference); additionally, they can be chained together to create a more complex final query. Due to the derived nature of CTEs, there is no need to write these temporary result sets to disk, thus reducing I/O requirements to complete each transaction.
Suppose we have multiple tables that contain different complementary pieces of information about a patient such as their personal information, physician visits, and prescriptions. Often, researchers need to correlate information in these different tables to select a set of patient records that satisfy certain criteria. This correlation is generally accomplished using database table joins performed on fields common across all tables. A straightforward approach to join multiple tables is to form a sequence of joins, where the result of a join between two tables is used in turn to perform a join with the next table. As such, the result set of a join can also be treated as a table consisting of fields from multiple tables that are selected in the join. Instead of storing these intermediate results as explicit temporary tables, CTEs enable us to use named result sets or common tables that provide the same functionality.
A. Transformation of i2b2 queries to CTE-based queries
A high-level overview of the translation process can be found in Fig. 2 and Fig. 3 represents an example of the innermost workings. To make things less abstract, we use an example to demonstrate how queries from the i2b2 interface are transformed into the corresponding CTE-based SQL queries. As mentioned earlier, the i2b2 interface provides panels to formulate Boolean queries on patient records involving certain concepts. In our approach, each panel in the i2b2 user-interface is mapped to a common table where the joins between CTEs facilitate logical ANDs and the inner-expression within each CTE facilitates logical ORs and NOTs by using nested leftjoins. Left-join predicates capture additional constraints such as dates, frequency, concept modifiers, and can be used to support additional query features such as lab value constraints, provider details, and text searches.
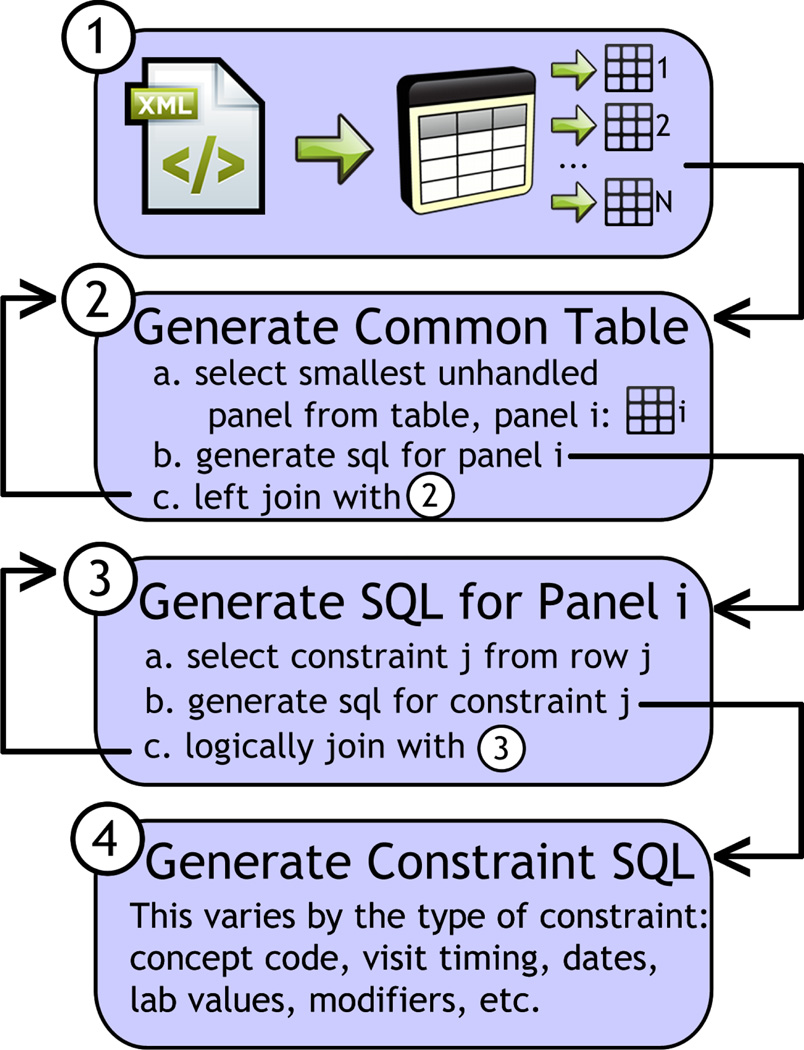
Fig. 2.
Overview of the CTE-generation process. In stage 1, the incoming XML message containing the query definition is placed inside a table where groups of rows represent panels. Joined common tables are generated (stage 2) from generating SQL for panels (stage 3) by generating logically joined SQL for constraints (stage 4).
Fig. 3.

An example of stage 4 from Fig. 2 showing the logic needed to generate the SQL for a date constraint. This is conditionally based on the query definition’s values for the “to” and “from” of the date constraint received from the user interface; for example, if both values are present, a between clause is needed. This returns a SQL clause comparing “start_date”, which is found in the source database table.
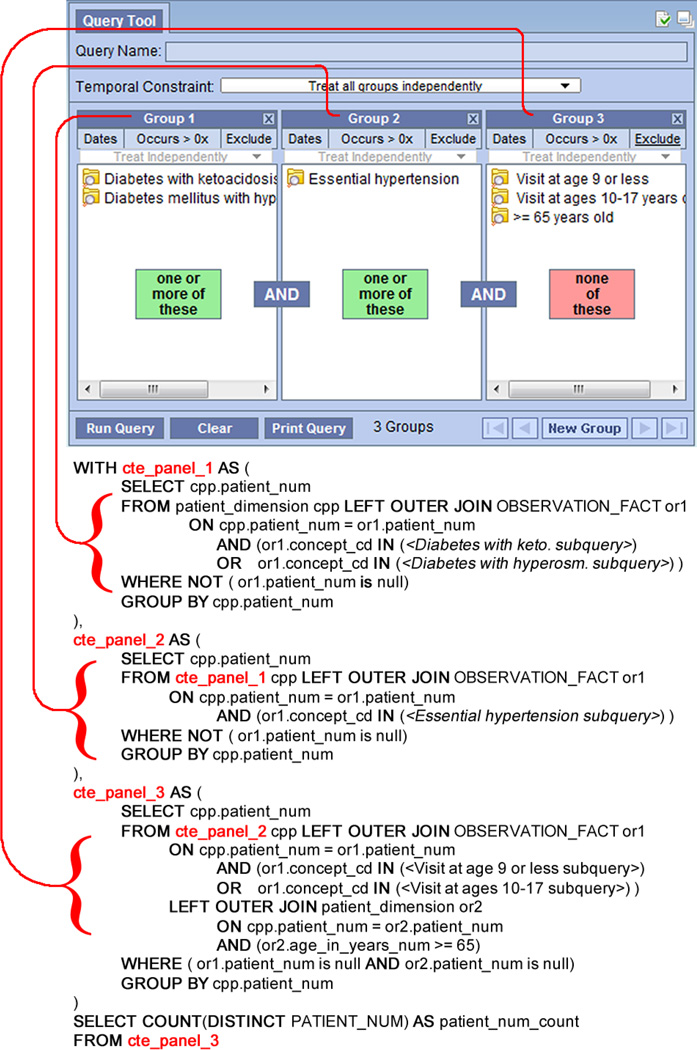
Fig. 4 shows a query that has 3 panels, where the first panel is a logical OR upon two different diabetes-related concepts and the second panel simply selects the hypertension concept. The last panel is slightly more complicated; it is the logical OR between those patients who had a visit at an age of less than 9 years, or between 10–17 years, or older than 65 years today. This panel is also negated, so only those patients who are outside of those age ranges will be selected. We show the corresponding CTE-based SQL query below the panel diagram. To conserve space, sub-queries for concept children in the ontology have been replaced by placeholders. We use a special table of inter-concept hierarchical relationships to help manage our ontology so that child concepts can be returned quickly. In short, this mapping generates a single SQL statement that can be completed in one transaction with minimal need for temporary disk space or logging.
Fig. 4.
An example of a patient-count query translated into common table expression.
The CTE-based approach described above is in stark contrast to the procedural approach, which generates multiple SQL statements. In turn these statements invoke costly inserts, updates, and hard-disk temporary table commits which require disk space and are limited by disk-access speeds. Additionally, the multiple statement procedural approach prevents the SQL optimizer from generating an optimal join strategy due to its limited knowledge of the overall query sequence. In contrast, our relational approach presents the optimizer with a single query that can be optimized effectively.
The left outer-join provides an elegant method for performing both the logical OR and NOT operations by using a compound WHERE predicate with the help of the NOT operand. For example, when a panel contains two or more concepts from different tables, a new alias or1, or2, …, orN is added to each table in subsequent joins. The final WHERE predicate for a panel then looks like WHERE not (or1.patient_num is NULL AND or2.patient_num is NULL … AND orN.patient_num is NULL).
Note that this will return patients where at least one of the concepts in the OR panel returns true. The logical NOT of a panel is found simply by removing the NOT keyword before the compound WHERE predicate. That is, the left join will then only return those patient records that fail to return null for all of the concepts in the OR list. The records that match at least one of the concepts will be excluded. The modified query generator also combines all exclusion panels into one CTE, where it is logically consistent to do so.
For any query that can be logically composed in the i2b2 user interface, the query generator can construct a corresponding CTE-based query. Although lab values were not present in our CDW at the time of this testing, the current version of the software supports searching for explicitly entered values such as glucose >126 mg/dL. We also support the use of patient sets or previous queries as a panel concept. Additionally, it is feasible to expand functionality to support queries with temporal relationships between panels such as before, after, or within a certain number of days, by retrieving the encounter date to further refine the joins between common tables.
B. Experiments
We created a random query generator to generate randomized XML request messages that the CRC cell would have normally received from the user interface. The query generators processed these XML messages so that they could translate the user supplied i2b2 query into actual SQL queries that can be run on the warehouse. We chose to generate random queries because our i2b2 user-base is small and may not accurately represent the usage at other institutions. For example, the occurrence constraint might be used more at institutions with large longitudinal datasets as opposed to institutions with smaller datasets with transient populations. The goal was to stress test i2b2 by capturing every possible type of query that users might design; we determined that there were 16 types of queries possible by having binary choices on the four user interface constraints (date, frequency, negation/exclusion, and whether the concepts occurred in the same encounter) (See Table I). For each of the 16 types of queries, 40 queries were randomly selected. Each of these 40 random queries had between one and four panels, each of which had randomized constraints and contained between one and three randomly chosen concepts. Concepts that did not appear in the dataset were not considered to prevent trivial empty set calculations. Here concepts are arbitrary placeholders that represent sets of patients, allowing us to focus on how each query generator interprets the logical constraints to construct a resulting aggregate patient set. This approach to test query formulation might not represent clinical reality, but does represent faithfully the effort it would take to logically combine sets of these sizes for each generator. In all our queries, each of the four possible constraints of query timing, exclusion, date, and occurrence (frequency) need only apply to at least one panel in the query to be considered that query type. Date ranges were randomly assigned. The start date and end date of a date range was restricted to exist between the minimum and maximum date of our dataset, and the end date was required to be after the start date. Of these 640 queries, a small number were unable to be correctly processed due to erroneous SQL being generated by the stock query generator.
TABLE I.
Average query response time (in seconds) per query type.
| Query Type | Modified | Stock | |||||
|---|---|---|---|---|---|---|---|
| Timing | Neg. | Date | Freq. | Mean | s.d. | Mean | s.d. |
| Any | F | F | F | 0.73 | 0.34 | 0.47 | 0.78 |
| Any | T | F | F | 1.36 | 0.84 | 8.44 | 3.75 |
| Any | F | T | F | 0.59 | 0.37 | 0.50 | 0.55 |
| Any | F | F | T | 1.22 | 0.97 | 1.11 | 1.25 |
| Any | T | T | F | 1.27 | 0.96 | 7.00 | 5.63 |
| Any | T | F | T | 6.01 | 17.56 | 7.87 | 7.13 |
| Any | F | T | T | 1.21 | 0.63 | 1.50 | 1.12 |
| Any | T | T | T | 1.68 | 4.55 | 5.76 | 6.00 |
| Same | F | F | F | 1.31 | 2.43 | 2.49 | 4.97 |
| Same | T | F | F | 2.03 | 2.23 | 376.82 | 365.96 |
| Same | F | T | F | 0.61 | 0.50 | 1.12 | 1.47 |
| Same | F | F | T | 1.00 | 0.53 | 0.75 | 0.75 |
| Same | T | T | F | 2.50 | 6.38 | 332.04 | 293.49 |
| Same | T | F | T | 5.52 | 13.41 | 304.55 | 426.57 |
| Same | F | T | T | 1.28 | 1.35 | 1.12 | 1.19 |
| Same | T | T | T | 4.44 | 10.82 | 236.37 | 516.81 |
The requested result type was restricted to a simple patient count. The dataset used for this testing contains 10 years of Kentucky State Medicaid claims data, covering 1.8 million patients across 160 million encounters with 660 million facts. The Medicaid data contains only demographics, diagnosis billing codes (excluding procedures), and visit details such as age at visit, length of stay, and visit type. For comparison purposes, the i2b2 environment at the University of Kentucky is housed on SQL Server 2008 R2 on a Dell PowerEdge M910 blade server with 32 processor cores and 128GB RAM; storage is provided by a EMC CX3-40 SAN housing four 7.2K RPM hard drives with 4Gbps transfer capacity. Version 1.6.04 of i2b2 was used during the testing process. Both generators had access to the pre-calculated frequency of each concept. This allows the panels to be ordered starting with the smallest first, since an intersection is no larger than its smallest member.
IV. Results and Discussion
Because i2b2 users’ queries are placed into a processing queue, we note that the stress test results based on our query types are especially important in a multiple-client setting. Excessive delays incurred by poorly formed queries from novice users can affect the usability of i2b2 for other more experienced users. With the development and launch of SHRINE [7], query response times should be consistent and have as little impact on the server’s resources as possible, which also improves the local user’s experience.
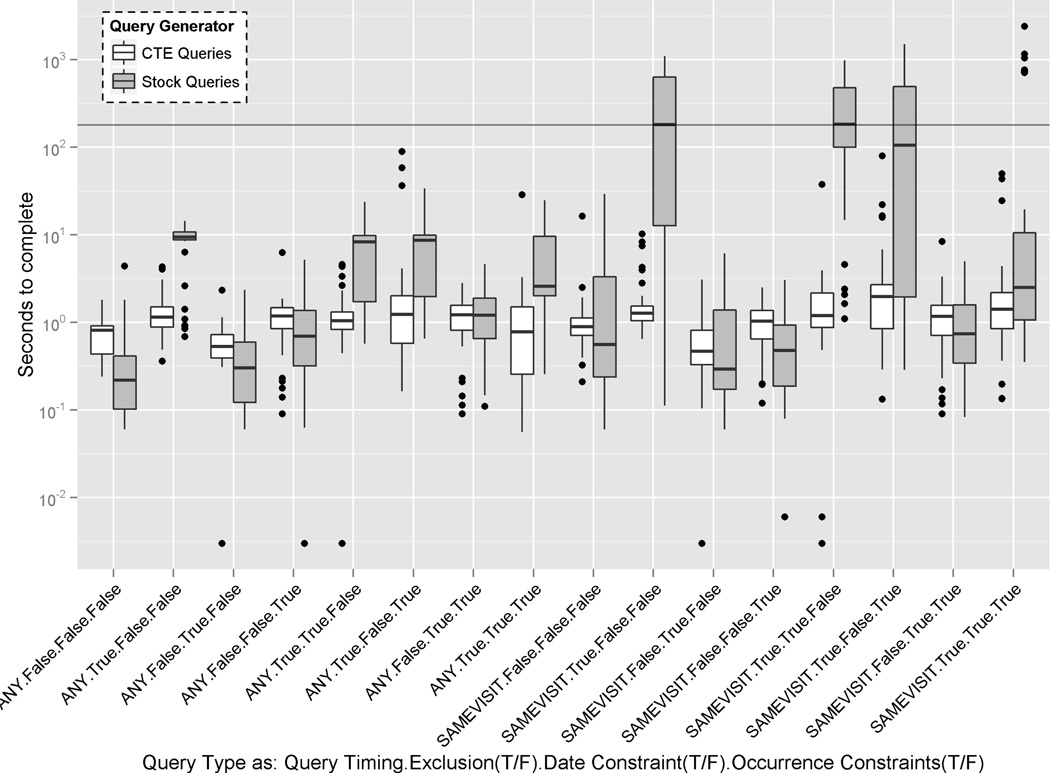
Table I shows the mean and standard deviation of time in seconds for each of the 16 query types discussed in the previous section. Bolded values indicate the approach that provided the best performance in either average query response time or standard deviation. Table II shows the global average response time if all executed queries are considered. This gives an indication of the resources both techniques might use when deployed. In addition to these summary statistics, we chose to plot our testing results in a box plot to illustrate the variance in performance within and across all 16 query types. Fig. 5 shows the 16 query types by the four variables discussed in the last section: query encounter timing, exclusion, date, and occurrence (frequency) constraints.
TABLE II.
Average, maximum, and standard deviation of all query response times (in seconds).
| Modified Query Gen. | Stock Query Gen. | |
|---|---|---|
| Avg. Time to Complete | 2.03 | 75.82 |
| Max. Time to Complete | 89.48 | 2407.64 |
| Standard Deviation | 6.64 | 238.88 |
Fig. 5.
Logarithmic-scale plot of the execution times of the randomly generated queries across the 16 query combinations that were tested. The grey horizontal line represents the default amount of time (180 seconds) that the user interface allows queries to run before timing out.
The modified queries perform fairly consistently across all 16 query types and only show a large variance within a population of queries in three cases: (1) a query with both an exclusion and frequency constraint where the concepts must occur in the same encounter, (2) a query with both an exclusion and frequency constraint where the concepts do not have to occur in the same encounter, and (3) a query with exclusion, date, and frequency constraints where the concepts must occur in the same encounter. In all three cases, the CTE queries performed better on average. The variation seen in these queries is relatively small (10–20 seconds) when compared to the stock query generator’s largest variation (293–516 seconds).
As Fig. 5 shows, the stock queries performed very inconsistently with highly varying minima, medians, and maxima. Furthermore, the inter-quartile range for the stock queries is much higher (almost twice the size) than those for CTE-based queries in 15 of the 16 types of queries. A non-parametric binomial test of proportions shows that significantly more than half (59%, p<0.0001) of the queries required less computing time for our modified approach; for these queries, the mean difference between stock and modified execution times was 125.66 seconds (s.d. 297.61), which indicates a significant, positive change in user experience. For the queries where stock outperformed our modified generator, the mean difference of execution times was only 1.21 seconds (s.d. 5.54).
The testing also reveals several use-cases in which the stock procedural approach performs much more poorly than to be expected, such as a query with same encounter timing paired with an exclusion constraint. We feel this is a typical use case that many users are interested in. For example, this could be a query that is looking for patients who have diabetes but are not recorded to have hypertension in the same encounter. This test case and a few others in the same encounter timing query types present a great deal of variance and represent an unpredictable impact on server resources and the user’s experience.
There are query types in which the stock approach outperformed the modified CTEs, such as the simplest case where no constraints are chosen at all. In these cases, both methods produced queries that finished in less than one or two seconds on average, likely resulting in no substantial difference in the user’s experience. Additionally, all of the modified CTEs performed well under the 180 second timeout feature of the web-client user-interface, which would provide each user with a better experience in either concurrent-user systems or in systems that support i2b2 SHRINE.
Our solutions were implemented for Microsoft SQL Server, but i2b2 is released for both Microsoft and Oracle systems. Publishing an Oracle-compatible package is left as future work; common-table expressions and all other tuning features can be leveraged in an Oracle environment. We are confident we would see performance gains in a similar Oracle environment because there are principal differences in our query techniques. We generate relational queries that require only a single database transaction and thus are more friendly to the internal optimizer; this is partly why we see a high degree of parallelism. The stock technique generates procedural queries that require multiple transactions, which paralyzes the optimizer and inhibits parallelism. These philosophical differences in techniques are expected to carry performance differences regardless of architecture. We also acknowledge that testing across varying hardware configurations may provide insight into the relationship between memory and disk usage for these approaches. We currently use our approach in our production i2b2 environment and provide this as an open-source [21] proof of concept for SQL Server.
The key benefit of this work is that it can improve the users’ experience by reducing response times and ensuring consistent performance regardless of the logical constraints of their queries. In i2b2, this is demonstrated by our queries returning far more quickly than the default 180 seconds that queries are allowed to run in the user-interface. Because a queue for queries submitted by the users is used to delegate resources, returning result sets quickly is crucial. Otherwise, queries that take far too long to complete can block activity from other users, which would be especially critical to address in a federated query network such as SHRINE.
V. Conclusion
We have shown that stable and scalable Boolean queries can be constructed using common table expressions. This improvement is purely algorithmic and the result of elegant relational programming. Hardware improvements can also provide performance gains but require a costly investment. The improvements we have presented can be installed and integrated into an i2b2 system in a short amount of time and can provide performance gains with no investment of additional financial resources. Because of their low cost, these types of graceful programming solutions are crucial. Furthermore, without scalable algorithms, clinical and translational science cannot be performed at a national level. Translational work must ultimately bridge and transcend across individual institutions, which is only possible with truly scalable architecture. Our contribution enables efficient local querying, which in turn, enables efficient federated querying.
Our experiments also describe an evaluation methodology that provides a means of profiling and comparing CDW performance using one or more algorithmic approaches across a range of query types, and for tuning CDWs for the types of queries that are most common to local user needs. The process of enumerating the possible query types and randomly generating queries per type gives a good indication of the global performance profile and may help avoid local institutional biases.
Acknowledgments
This work was supported by HRSA grant D1BRH20410, the National Center for Research Resources, UL1RR033173, and the National Center for Advancing Translational Sciences, UL1TR000117. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or HRSA.
Biographies

Daniel R. Harris received a B.S. degree in computer science and mathematics from the University of Kentucky, Lexington, KY in 2008, where he is currently a Ph.D. student of the Department of Computer Science.
He is also currently an Information Technology Manager at the Center for Clinical and Translational Science at the University of Kentucky. His research interests include biomedical informatics, information visualization, data and text mining, and database systems.

Darren W. Henderson received a B.S. degree in Biology from the University of Kentucky, Lexington, KY in 2011.
He is currently a Database Administrator for the Center for Clinical and Translational Science at the University of Kentucky. He has worked in this capacity for 6 years. His interests include performance tuning and effective clinical data warehousing.

Ramakanth Kavuluru received a B.Tech degree in computer science and engineering from the Jawaharlal Nehru Technological University, India, in 2002. He received an M.S degree in computer science from the Western Kentucky University, USA, in 2004 and a Ph.D degree in computer science from the University of Kentucky, USA, in 2009.
He currently works as an Assistant Professor in the Division of Biomedical Informatics in the Department of Biostatistics at the University of Kentucky; he also holds a joint appointment in the Department of Computer Science. Earlier, he worked as a Postdoctoral Research Scientist in the Kno.e.sis Center at the Wright State University. His current research interests include biomedical and health infomatics, text mining, Semantic Web techniques, knowledge-based approaches to machine learning and data mining, and information security and privacy. Earlier, for his doctoral thesis, he pursued research in analysis of security measures for pseudorandom sequences in the context of stream ciphers.
Dr. Kavuluru is a member of the American Medical Informatics Association, ACM SIGBio, and the International Society for Computational Biology.

Arnold J. Stromberg received his B.S. in Mathematical Sciences from Stanford University in 1983 and his M.S. (1988) and Ph.D. (1989) in Statistics are from the University of North Carolina at Chapel Hill.
He is Professor and Chair of the Department of Statistics at the University of Kentucky. He is also Director of Data Analysis for the U.K. Microarray Core Facility. He is Co-Director of Biomedical Informatics Core for the U.K. Center for Clinical and Translational Science. He and his research assistant provide statistical services for many federal grants. His research interests include statistical bioinformatics, outlier detection and statistical process control.

Todd R. Johnson received a B.S. degree in Computer and Information Science from the Ohio State University in 1984, a M.S. Degree in Computer and Information Science from the Ohio State University in 1986, and a Ph.D. in Computer and Information Science (Artificial Intelligence with minors in cognitive science and theory of computation) from the Ohio State University in 1991.
He is a Professor and the Director of the Division of Biomedical Informatics in the Department of Biostatistics, College of Public Health at the University of Kentucky. He is also the Director of the Biomedical Informatics Core at the Center for Clinical and Translational Science at the University of Kentucky. His research areas include clinical informatics, clinical research informatics, patient safety, decision making, human-centered design of information technology and medical devices, and information visualization.
Contributor Information
Daniel R. Harris, Department of Computer Science and the Center for Clinical and Translation Science at the University of Kentucky, Lexington, KY, USA
Darren W. Henderson, Center for Clinical and Translation Science at the University of Kentucky, Lexington, KY, USA
Ramakanth Kavuluru, Division of Biomedical Informatics, Department of Biostatistics, College of Public Health, University of Kentucky, Lexington, KY, USA.
Arnold J. Stromberg, Department of Statistics, College of Arts and Sciences, University of Kentucky, Lexington, KY, USA
Todd R. Johnson, Division of Biomedical Informatics, Department of Biostatistics, College of Public Health, University of Kentucky, Lexington, KY, USA
References
- 1.Marsolo K. Pediatric Biomedical Informatics. Springer; 2012. Research patient data warehousing; pp. 93–108. [Google Scholar]
- 2.Murphy SN, Weber G, Mendis M, Gainer V, Chueh HC, Churchill S, Kohane I. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) Journal of the American Medical Informatics Association. 2010;17(2):124–130. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohane IS, Churchill SE, Murphy SN. A translational engine at the national scale: informatics for integrating biology and the bedside. Journal of the American Medical Informatics Association. 2012;19(2):181–185. doi: 10.1136/amiajnl-2011-000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deshmukh V, Meystre S, Mitchell J. Evaluating the informatics for integrating biology and the bedside system for clinical research. BMC Medical Research Methodology. 2009;9(1):70. doi: 10.1186/1471-2288-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy S, Churchill S, Bry L, Chueh H, Weiss S, Lazarus R, Zeng Q, Dubey A, Gainer V, Mendis M, et al. Instrumenting the health care enterprise for discovery research in the genomic era. Genome Research. 2009;19(9):1675–1681. doi: 10.1101/gr.094615.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.National Human Genome Research Institute. Dna sequencing costs. [accessed 27-July-2012];2012 [Online]. [Online]. Available: http://www.genome.gov/sequencingcosts/
- 7.Weber GM, Murphy SN, McMurry AJ, MacFadden D, Nigrin DJ, Churchill S, Kohane IS. The shared health research information network (shrine): a prototype federated query tool for clinical data repositories. Journal of the American Medical Informatics Association. 2009;16(5):624–630. doi: 10.1197/jamia.M3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cimino JJ. Data Integration in the Life Sciences. Springer; 2012. The biomedical translational research information system: clinical data integration at the national institutes of health; pp. 92–92. [Google Scholar]
- 9.Horvath MM, Winfield S, Evans S, Slopek S, Shang H, Ferranti J. The deduce guided query tool: providing simplified access to clinical data for research and quality improvement. Journal of Biomedical Informatics. 2011;44(2):266–276. doi: 10.1016/j.jbi.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Livne OE, Schultz ND, Narus SP. Federated querying architecture with clinical & translational health it application. Journal of medical systems. 2011;35(5):1211–1224. doi: 10.1007/s10916-011-9720-3. [DOI] [PubMed] [Google Scholar]
- 11.Lowe HJ, Ferris TA, Hernandez PM, Weber SC. in AMIA Annual Symposium Proceedings. Vol. 2009. American Medical Informatics Association; 2009. Stride–an integrated standards-based translational research informatics platform; p. 391. [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang G-Q, Siegler T, Saxman P, Sandberg N, Mueller R, Johnson N, Hunscher D, Arabandi S. Visage: a query interface for clinical research. AMIA Summits on Translational Science Proceedings. 2010;2010:76. [PMC free article] [PubMed] [Google Scholar]
- 13.Segagni D, Ferrazzi F, Larizza C, Tibollo V, Napolitano C, Priori SG, Bellazzi R. R engine cell: integrating r into the i2b2 software infrastructure. Journal of the American Medical Informatics Association. 2011;18(3):314–317. doi: 10.1136/jamia.2010.007914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segagni D, Gabetta M, Tibollo V, Zambelli A, Priori SG, Bellazzi R. Data Integration in the Life Sciences. Springer; 2012. Onco-i2b2: improve patients selection through case-based information retrieval techniques; pp. 93–99. [Google Scholar]
- 15.Wattanasin N, Porter A, Ubaha S, Mendis M, Phillips L, Mandel J, Ramoni R, Mandl K, Kohane I, Murphy SN. AMIA Annual Symposium Proceedings. Vol. 2012. American Medical Informatics Association; 2012. Apps to display patient data, making smart available in the i2b2 platform; p. 960. [PMC free article] [PubMed] [Google Scholar]
- 16.Ganslandt T, Mate S, Helbing K, Sax U, Prokosch H. Unlocking data for clinical research–the german i2b2 experience. Methods of Information in Medicine. 2008;47(2):117–123. doi: 10.4338/ACI-2010-09-CR-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gainer V, Hackett K, Mendis M, Kuttan R, Pan W, Phillips LC, Chueh HC, Murphy S. Using the i2b2 hive for clinical discovery: an example. AMIA Annual Symposium Proceedings. 2007;959 [PubMed] [Google Scholar]
- 18.Weber GM. AMIA Summit on Translational Bioinformatics Proceedings. Vol. 2012. American Medical Informatics Association; 2012. Supercharging i2b2; p. 182. [Google Scholar]
- 19.Murphy SN, Mendis M, Hackett K, Kuttan R, Pan W, Phillips LC, Gainer V, Berkowicz D, Glaser JP, Kohane I, et al. AMIA Annual Symposium Proceedings. Vol. 2007. American Medical Informatics Association; 2007. Architecture of the open-source clinical research chart from informatics for integrating biology and the bedside; p. 548. [PMC free article] [PubMed] [Google Scholar]
- 20.Microsoft. Sql server 2005 beta 2 transact-sql enhancements. [accessed 27-July-2012];2004 [Online]. [Online]. Available: http://msdn.microsoft.com/en-us/library/ms345144.aspx#sql05b2tsqlen_recquercte. [Google Scholar]
- 21.Division of Biomedical Informatics, University of Kentucky. i2b2 common-table expression query generator. [accessed 26-October-2012];2012 [Online]. [Online]. Available: https://code.google.com/p/i2b2-cte-query-generator-ukbmi/ [Google Scholar]