Abstract
As the incidence of bone-marrow failure syndromes (BMFS) is 2-3x higher in East Asia than in the West, we examined peripheral blood or marrow cells of 100 Japanese patients for possible pathogenic mutations in the two main components of the telomere-synthesizing enzyme telomerase (hTERC RNA and hTERT protein) that have recently been implicated in the disease pathogenesis. We analyzed samples collected from 34 patients with acquired aplastic anemia (AA), 66 patients with myelodysplastic syndromes (MDS), and 120 healthy controls. In addition to two polymorphic germ-line sequence changes (n-771A/G and n-714 C insertion) in the promoter region of hTERC and eleven hTERT polymorphisms that were identified in both patients and healthy individuals, we found a novel germ-line C323T mutation in the hTERC RNA in an MDS patient only. This heterozygous C323T mutation abolished telomerase enzymatic activity and functioned in a haploinsufficiency manner to modulate telomerase activity in cells. In summary, this study reports a novel telomerase natural variant that abolishes telomerase function, which may lead to telomere shortening and marrow hypocellularity in patients with BMFS. This study also highlights the rarity of genetic alterations in BMFS patients in Japan, which suggests that other factors may play a more prominent role in the disease pathogenesis in East Asia.
Keywords: telomeres, telomerase, aplastic anemia, myelodysplastic syndromes, bone-marrow failure syndromes, hTERC, hTER, hTR, hTERT
INTRODUCTION
Dyskeratosis Congenita (DKC) is an inherited bone marrow failure syndrome (BMFS) typified by reticulated skin pigmentation, nails dystrophy and mucosal leucoplakia [1]. About 35% of the cases are X-linked recessive, 5% are autosomal dominant and the rest of the cases are with unidentifiable pattern of inheritance [2]. Whereas the gene responsible for X-linked recessive cases is DKC1 [3], those responsible for autosomal dominant cases are the telomerase hTERC and hTERT [4][5-7].
Telomerase is a ribonucleoprotein (RNP) complex with two main components: a protein (hTERT) with RNA-dependent DNA polymerase activity and an integral hTERC RNA, which provides a template to synthesize telomeric DNA repeats [8]. Telomeres are structural elements that seal and protect the ends of linear chromosomes from illegitimate recombination, end-to-end fusion, or being recognized as damaged DNA [8]. In human somatic cells, telomeres typically consist of more than 1000 simple repetitive DNA and associated proteins [8]. These repeats are gradually lost with cellular replication and aging, owing to the inability of DNA polymerase to fully replicate the 3’ end of DNA. Telomere attrition eventually leads to critically short telomeres, inducing cellular proliferative senescence and/or apoptosis possibly due to genomic instability [8]. It is thought that telomeres are shortened as a result of pathogenic mutations in DKC1 or telomerase gene components that lead to an impairment in the proliferative capacity of hematopoietic stem cells in patients with BMFS [4, 7, 9]. Furthermore, an association has been established between the degree of telomere shortening and that of disease severity and the age of onset [7, 9].
The existence of possibly cryptic DKC in patients who develop the disease later in life [10] and many cases of aplastic anemia (AA) with significantly shortened telomeres can also be attributed to mutations in telomerase gene components [11-13]. Several groups, including our own, have recently reported that some patients with paroxysmal nocturnal hemoglobinuria (PNH), myelodysplasia (MDS), in addition to those with AA or DKC, carry heterozygous mutations in the telomerase hTERC or hTERT gene [5, 14-18]. In vitro functional analyses of these mutations revealed that the mutations functioned either as dominant negatives or haploinsufficiency to attenuate telomerase enzymatic activity, which could explain the short telomeres in patients [6, 17, 19-21]. The largest controlled epidemiologic studies reported that the incidence of AA in the West is 2 per million per year and is about 2- to 3-fold higher in Asia [22]. The subjects of previous studies have mostly been those with Caucasian, Black or Hispanic ancestry. Screening for telomerase mutations among Asian populations has rarely been done [18, 23]. Therefore, we carried out an investigation to determine whether mutations in hTERC and hTERT genes are associated with the disease in our cohort of Japanese BMFS patients.
MATERIALS
Patients and healthy controls
We examined mononuclear cells (MNC) of peripheral blood or bone marrow from 100 BMFS patients with acquired AA (n=34) or with MDS (RA) (n=66) diagnosed between 1993 to 2005 at the Nippon Medical School and its affiliated hospital. These patients were diagnosed with AA based on the blood count criteria of the International Study of AA and agranulocytosis with severity determined by the criteria of Camitta et al [24]. We excluded AA patients who had achieved complete remission or good response to immunosuppressive therapy. Good response was defined as a resolution of all blood transfusion requirements and a more than 2 g/dL increase in hemoglobin as compared with pretreatment levels. Most AA patients (91.1%) received ATG and cyclosporine A combination therapy and showed either only partial or no response to treatment (Table 1). For a diagnosis of MDS, patients were subclassified according to the French-American-British (FAB) nomenclature [25]. MDS (RA) classification might include several heterogeneous BMF or other hematologic diseases. However, we excluded patients who had developed MDS (RAEB) or MDS leukemia for more than three years from original diagnosis. Most of selected patients with MDS (RA) were treated with blood transfusion or anabolic steroid. As normal controls, we analyzed blood samples from 120 healthy individuals. Our volunteers provided informed consent prior to genetic testing as approved by our institutional review board.
Table 1.
Patients clinical background
| AA (n=34) | MDS (RA) (n=66) | ||
|---|---|---|---|
| Sex | Male | 14 | 34 |
| Female | 20 | 32 | |
| Age (Range) | 13–77 | 19–90 | |
| Family history | + | 0 | 2 |
| - | 34 | 64 | |
| Chromosome abnormality | trisomy 8 | 0 | 4 |
| 7q- | 1 | 1 | |
| del (20) | 0 | 3 | |
| complex | 0 | 2 | |
| Treatment | |||
| Immunosuppressive therapy | |||
| Partial Response | 17 | 5 | |
| No Response | 14 | 9 | |
| Others | |||
| Blood transfusion only | 0 | 27 | |
| Metenolone | 2 | 22 | |
| Stem cell transplantation | 1 | 3 |
Mutational analysis
Mononuclear cells (MNC) from bone marrow or peripheral blood were isolated by density gradient centrifugation using lymphocyte separation medium (Organon, Durham, NC). The genomic DNA of MNC was extracted with the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). Polymerase chain reaction (PCR) amplification of telomerase gene components (hTERC and hTERT) was carried out essentially as described previously [16, 17]. The Advantage GC2 PCR amplification kit (BD Biosciences Clontech, CA, USA) and the TaKaRa Ex Taq DNA polymerase (Takara, Shiga, Japan) were used to amplify the genes from genomic DNA. PCR products were purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany) and sequenced bidirectionally using the Big Dye Termination 3.1 kit and the ABI Prism 310 system (Perkin-Elmer Cetus, CA, USA). Specific sequences of primers used for sequencing are available upon request. To validate the sequencing results, PCR products were inserted into the pCR2.1-TOPO vector using the TOPO TA cloning kit (Invitrogen, CA, USA). Recombinant plasmids isolated from 8 to 12 white colonies were sequenced.
In vivo reconstitution of telomerase enzymatic activity
Wild-type or mutant pcDNA3-hTERC DNAs (2 micrograms) were transfected into VA13+hTERT cells (at approximately 70% confluency) in 6-well polystyrene dishes using SuperFect transfection reagent (Qiagen, Hilden, Germany) according to the manufacturer's instructions. In certain cases, two different versions of the hTERC gene, each on a separate vector and at 1 μg, were co-expressed simultaneously in the VA13+hTERT cells. To monitor transfection efficiency, a plasmid (peGFP-N1) (Strategene, CA, USA) expressing green fluorescent protein was transfected in a parallel transfection reaction. The eGFP protein expression level was examined under fluorescence microscope. Approximately 48 hours after transfection, cells were scraped from the dish in the presence of 1 mL cold phosphate-buffered saline. Cellular extracts were then prepared in 1X CHAPS lysis buffer as suggested by the manufacturer (Chemicon International, CA, USA). Telomerase activity of the cellular extract from 2×104 cells was assayed using the TRAPeze Telomerase Detection Kit following the manufacturer's directions (Chemicon International, CA, USA), except that PCR was performed as follows: 95°C for 2 min; 25 cycles of 94°C for 10 sec, 50°C for 30 sec, 72°C for 30 sec; and 72°C for 5 min. Products were analyzed on a 12% native polyacrylamide gel and examined by phosphor imaging (Molecular Dynamics, GE Healthcare Bio-Sciences Corp., NJ, USA).
Northern blotting analysis
Wild-type or mutant pcDNA3-hTERC vector (2 micrograms) was transfected into VA13+hTERT cells (at approximately 70% confluency) in 6-well polystyrene dishes using SuperFect transfection reagent. Approximately 48 hr after transfection, Trizol reagent was used to extract total cellular RNA as suggested by the manufacturer (Invitrogen, CA, USA). Northern blot analysis was performed essentially as described [26].
Immunoprecipitation-Northern blotting analysis
FLAG-tagged hTERT protein was expressed in vitro from the pCR3-FLAG-hTERT vector using the TnT quick-coupled transcription-translation system (Promega) in the presence of 200 ng of in vitro-transcribed, gel-purified CR4-CR5 fragment of hTERC RNA spanning nucleotides 239 to 332 at 37°C for 2 h. The resulting telomerase complexes were affinity-enriched on anti-FLAG agarose beads (Sigma, St. Louis, MO). To detect hTERT-bound telomerase RNAs, Northern blotting was performed on the enriched telomerase preparations as described above.
RESULTS
Telomerase mutational analysis
We selected 100 BMFS patients who were diagnosed with either AA, mainly those who showed either only partial or no response to immunosuppressive therapy, or with MDS RA (Table 1). Genomic DNA from peripheral blood cells or marrow stem cells was extracted in order to amplify the hTERT and hTERC genes for sequencing. Even though we did not find any pathogenic mutations in the hTERT gene, we identified eleven polymorphic sequence changes (Table 2). These sequence polymorphisms were identified in both intronic and exonic regions of the gene and did not result in amino acid substitutions in the corresponding protein. Two of the polymorphic sequence changes (IVS6 -93 G/A and codon837 CTC/CTG) have not been reported previously.
Table 2.
Polymorphisms of hTERT
| Exon | Substitution | No(%) |
|---|---|---|
| 2 | Codon305 GCA/GCG(Ala/Ala) | 25 |
| 3 | IVS3+130C/T | 24 |
| 4 | IVS3-24C/T | 14 |
| 5 | Codon699 GCC/GCT(Ala/Ala) | 3 |
| 7 | IVS6-93G/A | 2 |
| Codon837 CTC/CTG(Leu/Leu) | 1 | |
| 9 | Codon840 CTG/CTA(Leu/Leu) | 3 |
| IVS9+11C/T | 3 | |
| 13 | IVS13+45C/T | 5 |
| 14 | Codon1013 CAC/CAT(His/His) | 13 |
| 15 | IVSE14-94C/T | 2 |
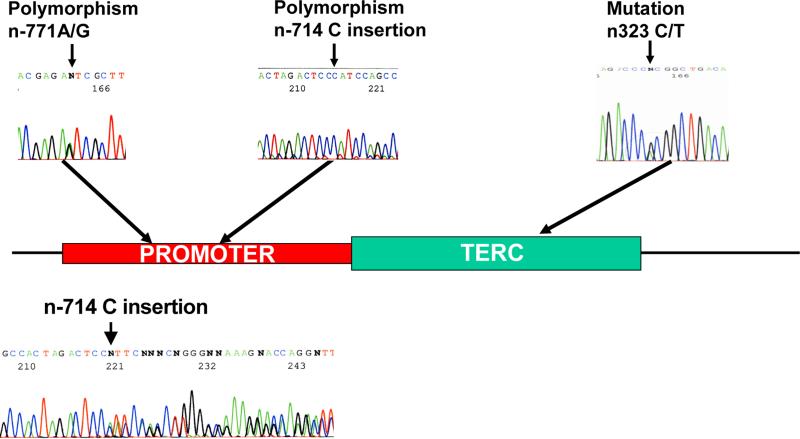
In regard to the hTERC gene, we identified two novel heterozygous sequence polymorphisms in its promoter (n-771A/G and n-714 C insertion) at about a similar frequency in both patients and healthy controls (Table 3 and Fig. 1). On the contrary, we identified a novel heterozygous germ-line mutation in the hTERC gene (C322T) in an MDS patient only (Table 3 and Fig. 1). This patient was a 72 yr-old man, who was clinically diagnosed with MDS RA. His bone marrow was slightly hypocellular but showed no sign of dysplasia or chromosomal abnormality. He showed a good and sustained response to metenolone and did not have a family history for the disease. Since this is an archival case, primary specimen (blood) collected from this patient did not yield an adequate amount of genomic DNA for measuring telomere lengths. The patient has deceased from ischemic heart disease.
Table 3.
Polymorphisms and mutation of hTERC
| AA, MDS (n=100) | control (n=120) | |
|---|---|---|
| Polymorphisms | ||
| n-771A/G | 11 (11.0%) | 18 (15.0%) |
| n-714C insertion | 12 (12.0%) | 20 (16.7%) |
| Mutation | ||
| n323C/T | 1 (1.0%) | 0 (0%) |
Figure 1.
Schematic depiction of the hTERC gene with naturally occurring sequence variations that occur in its promoter and coding sequence. Electrophereograms showing the heterozygous nature of these sequence variations are also shown. In the case of the n-714 C insertion, hTERC PCR product was subcloned into pCR2.1-TOPO expression vector before sequencing. Sequence analysis of PCR product of this sequence variant showing the heterozygous nature of the mutation is also shown in the bottom.
Functional analysis of the hTERC C323T mutation
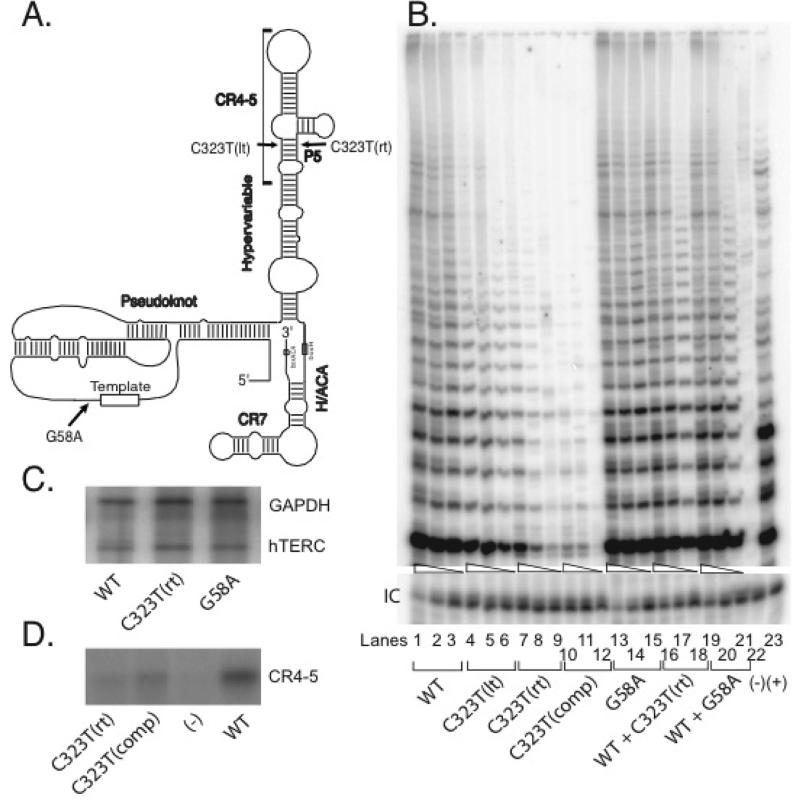
The novel C323T mutation changes a cytosine to a uracil in the hTERC RNA and is located on one strand of the P5 stem of the predicted hTERC RNA secondary structure (Fig. 2A) [27]. As such, we hypothesize that it may disrupt the basepairing interaction of this stem structure, which may lead to defective telomerase enzymatic function. In order to test this hypothesis, we introduced this hTERC natural variant C323T [or C323T(rt)] into a mammalian expression plasmid encoding the full-length (451-base) hTERC sequence. Because this specific nucleotide change might disrupt the basepairing interaction of the predicted P5 stem structure (Fig. 2A), we also created additional mutants designed to test the importance of this basepair. We designed a mutant denoted as C323T(lt) in which a guanine nucleotide located at position 246, on the opposite strand from the natural mutation, was mutated to an adenine (Fig. 2A). We also created a compensatory mutant [denoted as C323T(comp)] in which the natural C323T(rt) base mutation was accompanied by a complementary mutation on the opposite strand [C323T(lt)], which would theoretically restore the predicted intramolecular basepairing interactions of the stem structure.
Figure 2.
(A) Schematic depiction of the predicted secondary structure of hTERC as proposed by Chen et al [27]. The 8-base template sequence (rectangle) and other structural features are indicated, including the pseudoknot, CR4-CR5, box H/ACA, and CR7 domains, as well as the hypervariable paired region. The inconsequential G58A polymorphism, disease-associated C323T mutation [aka C323T(rt)] and C323T(lt) engineered variant are shown. (B) Telomerase enzymatic activities as determined in VA13+hTERT cells for the naturally occurring hTERC mutation and its derivatives. A representative TRAP gel showing the relative telomerase enzymatic activities obtained from the substitution mutations and compensatory mutation. The compensatory mutation [C323T(comp)] was created in order to restore the P5 stem structure. Serial fivefold dilutions of the transfected cell lysates (indicated by triangles) were assayed for each sample to ensure linearity of the assay. Lane 22 shows a negative control composed of wild-type cell lysate denatured at 95°C for 5 minutes prior to assay. Lane 23 shows PCR products amplified from the non-hTERC control TSR8 DNA template supplied in the TRAP kit. “IC” indicates PCR products amplified from an unrelated DNA template, which is included as an internal control for PCR amplification efficiency in each reaction. (C) Northern blot analysis of selected naturally occurring hTERC sequence variants and wild-type sequence expressed in transfected VA13+hTERT cells. Cellular GAPDH mRNA was assayed simultaneously. (D) Northern blot analysis of affinity-enriched telomerase complexes assembled using RNA fragments representing the CR4-5 domain of hTERC (Fig. 2A) with either the wild-type or mutated sequences and in vitro expressed hTERT catalytic protein. The negative control (lane 3) was a reaction that lacks the hTERT-expressing construct.
Biological activities of these hTERC variants were assessed by transient transfection of each vector into the VA13+hTERT cell line, a human lung-derived line that lacks endogenous hTERC but has been engineered to express stably the hTERT protein. These cells cannot ordinarily produce functional telomerase, but can assemble the active enzymatic complex when transfected transiently with a vector that expresses a functional hTERC copy. We prepared extracts of the cells approximately 48 hrs after transfection with the various hTERC constructs, and estimated the steady-state levels of exogenous hTERC expression by Northern blot analysis. We then tested the extracts for telomerase enzymatic activity by measuring their ability to add telomeric DNA repeats onto a synthetic DNA primer in vitro, using a semi-quantitative, PCR-based telomere repeat amplification protocol (TRAP).
As compared to cell lysates that carry either the wild-type hTERC RNA or the one with a known inconsequential G58A polymorphic sequence change [16, 19] (Fig. 2B, lanes 1-3 and 13-15), we found that in each of the cases, the mutations located within the P5 stem drastically reduced telomerase enzymatic activity (lanes 4-12). More specifically, mutations located on the individual strands of the P5 stem [i.e., either the natural C323T(rt) mutation or the C323T(lt) mutation alone] abolished telomerase activity to about the same degree (Fig. 2B, lanes 4-9). Surprisingly, we found that the compensatory mutation C323T(comp) did not restore telomerase activity (lanes 10-12). Rather, this version of the RNA seemed to further reduce telomerase function in cells to an almost undetectable level. These results were not attributable to differences in hTERC RNA synthesis, processing, or stability, as Northern blotting verified that each construct produced comparable steady-state levels of hTERC expression in the transfected cells (Fig. 2C). As the disease-associated mutation is located within the highly conserved CR4-5 domain of hTERC that has been implicated as one of the hTERT-interacting sites, we asked whether our RNA mutants can also affect hTERT binding activity. Telomerase complexes were reconstituted in vitro using rabbit reticulocyte lysates to express a FLAG-tagged version of the hTERT protein in the presence of a synthetic hTERC RNA spanning nucleotides 239 to 332 of the CR4-5 domain. Anti-FLAG antibody was used to immunoprecipitate telomerase RNP complexes, which was used to probe for the CR4-5 RNA fragment. As shown in Fig. 2D, both the C323T(rt) and the C323T(comp) RNAs showed substantially impaired binding to hTERT protein. Taken together, these findings indicate that the disease-associated hTERC variant and its derivatives are functionally defective and that their defects may result from altering the conserved secondary structure and/or primary sequence of the RNA that abolishes its ability to interact with the hTERT catalytic protein component of telomerase.
hTERC C323T natural mutation functions as haploinsufficiency to modulate telomerase function
As the natural hTERC C323T variant was identified in an individual who is a heterozygous carrier for the gene, the altered allele might modulate normal telomerase function through either a haploinsufficiency or dominant negative fashion. In order to address this, we performed TRAP assays on cell lysates prepared from VA13+hTERT cells that had been cotransfected with plasmids to express the wild-type hTERC sequence and either the disease-associated hTERC C323T variant or the polymorphic G58A variant. As shown in Fig 2B, little to no effects were observed between cells that carried only the wildtype hTERC vector and those that carried both the wild-type and the individual mutated hTERC copy (lanes 16-21), consistent with the idea that the natural variant functions in a haploinsufficiency manner to modulate wildtype telomerase function [5, 19].
DISCUSSION
This study shows that a novel variant telomerase RNA allele found in a Japanese patient with MDS is unable to support a normal level of telomerase enzymatic activity. This is consistent with the hypothesis that defects in telomerase function and telomere maintenance contribute to the pathogenesis of BMFS. This patient has deceased from ischemic heart disease. It has been documented that short telomeres may contribute to the pathophysiology of atherosclerosis and to the development of ischemic heart disease [28-30]. Some studies have implicated short telomere lengths to atherogenesis [31] and indicated that telomere lengths can serve as an effective marker for biological aging at a cellular level, such as aging cells of the vascular tissues [28-30].
The novel hTERC mutation reported in the current study is located on one side of the predicted P5 stem of the hTERC RNA secondary structure (Fig. 2A) that when altered drastically reduces telomerase enzymatic activity (Fig. 2B). More importantly, a combined substitution of both sides of this stem almost completely abolished telomerase function. It is noteworthy that the C323T variant allele described here is one of three known disease-associated alleles identified so far in this region (i.e., G322A, C323T, and G305A), and each seems to effectively abolish telomerase function [20]. Therefore, it is possible that this region may serve as one of the hotspots for the natural process of mutagenesis that can result in defective telomerase function and telomere shortening effect observed in patients with hematologic disorders.
Our study indicates that this P5 region of the hTERC structure contributes to optimal telomerase function, and this intricately basepaired structure and/or its primary sequence serve as a critical feature for its biological activity. Indeed, it is striking that a high proportion of the seemingly minor point mutations examined in our earlier study, which aimed at comprehensively analyzing the structure and function of the entire hTERC molecule, severely compromised telomerase function by perturbing RNA structural formation [32]. However, in those cases, we found that compensatory mutations could fully restore telomerase activity, highlighting the importance of the normal basepairing pattern of the RNA. In the current study, we describe for the first time a unique region (P5) of the telomerase RNA molecule that seems to be highly sensitive to sequence alteration. As this region is part of the conserved CR4-5 region (Fig. 2A) that has been shown to serve as one of the sites for the catalytic hTERT protein to assemble onto the RNA [20, 33, 34], we show here that indeed both the disease-associated mutant C323T(rt) and the compensatory C323T(comp) mutant substantially impair hTERT binding. Our data, therefore, support the idea that even a minute sequence change in this region can perturb this critical telomerase ribonucleoprotein interaction and hence abrogate its enzymatic activity.
Together with earlier reports [4, 17, 20, 26, 32], our data presented here are consistent with the idea that defects in telomerase function and telomere maintenance contribute to the pathogenesis of BMFS in a subset of these patients. As we found only one patient with a potential pathogenic mutation in the hTERC gene and none with pathogenic hTERT mutations among a cohort of 220 Japanese men and women, our study revealed that natural sequence variations in telomerase gene components rarely occurred in the Japanese population. This is consistent with a recent finding of no mutations in the hTERC gene in 35 Japanese MDS patients and 134 healthy volunteers [23]. A separate group examined 96 Japanese children with acquired AA and 76 healthy controls and similarly found no mutations in the hTERC gene [20]. However, this study revealed two nonsynonymous mutations in the hTERT gene among several inconsequential polymorphic sequence changes in this gene. Collectively, these studies showed that mutational frequencies of telomere-synthesizing genes among Japanese BMFS patients were lower than what had been reported for other ethnic groups [5, 7, 16, 17] and that this genetic difference could not explain the higher incidence of the disease in Asian populations.
ACKNOWLEDGEMENTS
This work was supported in part by grants from the American Cancer Society (RSG-06-162-01-GMC) and the AA & MDS International Foundation, Inc. to H.L. We wish to thank the patients and their families for their supports and participation in the study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Dokal I. Dyskeratosis congenita in all its forms. Br J Haematol. 2000;110:768–79. doi: 10.1046/j.1365-2141.2000.02109.x. [DOI] [PubMed] [Google Scholar]
- 2.Marrone A, Walne A, Dokal I. Dyskeratosis congenita: telomerase, telomeres and anticipation. Curr Opin Genet Dev. 2005;15:249–57. doi: 10.1016/j.gde.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Heiss NS, Knight SW, Vulliamy TJ, et al. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet. 1998;19:32–8. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 4.Vulliamy T, Marrone A, Goldman F, et al. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432–5. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 5.Vulliamy TJ, Walne A, Baskaradas A, et al. Mutations in the reverse transcriptase component of telomerase (TERT) in patients with bone marrow failure. Blood Cells Mol Dis. 2005;34:257–63. doi: 10.1016/j.bcmd.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Armanios M, Chen JL, Chang YP, et al. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc Natl Acad Sci U S A. 2005;102(44):15960–4. doi: 10.1073/pnas.0508124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vulliamy TJ, Marrone A, Knight SW, et al. Mutations in dyskeratosis congenita: their impact on telomere length and the diversity of clinical presentation. Blood. 2006;107:2680–5. doi: 10.1182/blood-2005-07-2622. [DOI] [PubMed] [Google Scholar]
- 8.Cong YS, Wright WE, Shay JW. Human telomerase and its regulation. Microbiol Mol Biol Rev. 2002;66:407–25. doi: 10.1128/MMBR.66.3.407-425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vulliamy TJ, Knight SW, Mason PJ, et al. Very short telomeres in the peripheral blood of patients with X-linked and autosomal dyskeratosis congenita. Blood Cells Mol Dis. 2001;27:353–7. doi: 10.1006/bcmd.2001.0389. [DOI] [PubMed] [Google Scholar]
- 10.Fogarty PF, Yamaguchi H, Wiestner A, et al. Late presentation of dyskeratosis congenita as apparently acquired aplastic anaemia due to mutations in telomerase RNA. Lancet. 2003;362:1628–30. doi: 10.1016/S0140-6736(03)14797-6. [DOI] [PubMed] [Google Scholar]
- 11.Ball SE, Gibson FM, Rizzo S, et al. Progressive telomere shortening in aplastic anemia. Blood. 1998;91:3582–92. [PubMed] [Google Scholar]
- 12.Brummendorf TH, Maciejewski JP, Mak J, et al. Telomere length in leukocyte subpopulations of patients with aplastic anemia. Blood. 2001;97:895–900. doi: 10.1182/blood.v97.4.895. [DOI] [PubMed] [Google Scholar]
- 13.Lee JJ, Kook H, Chung IJ, et al. Telomere length changes in patients with aplastic anaemia. Br J Haematol. 2001;112:1025–30. doi: 10.1046/j.1365-2141.2001.02669.x. [DOI] [PubMed] [Google Scholar]
- 14.Chen JL, Greider CW. Telomerase RNA structure and function: implications for dyskeratosis congenita. Trends Biochem Sci. 2004;29:183–92. doi: 10.1016/j.tibs.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Vulliamy T, Marrone A, Dokal I, et al. Association between aplastic anaemia and mutations in telomerase RNA. Lancet. 2002;359:2168–70. doi: 10.1016/S0140-6736(02)09087-6. [DOI] [PubMed] [Google Scholar]
- 16.Yamaguchi H, Baerlocher GM, Lansdorp PM, et al. Mutations of the human telomerase RNA gene (TERC) in aplastic anemia and myelodysplastic syndrome. Blood. 2003;102:916–8. doi: 10.1182/blood-2003-01-0335. [DOI] [PubMed] [Google Scholar]
- 17.Yamaguchi H, Calado RT, Ly H, et al. Mutations in TERT, the gene for telomerase reverse transcriptase in aplastic anemia. N Engl J Med. 2005;352:1413–24. doi: 10.1056/NEJMoa042980. [DOI] [PubMed] [Google Scholar]
- 18.Liang J, Yagasaki H, Kamachi Y, et al. Mutations in telomerase catalytic protein in Japanese children with aplastic anemia. Haematologica. 2006;91:656–8. [PubMed] [Google Scholar]
- 19.Marrone A, Stevens D, Vulliamy T, et al. Heterozygous telomerase RNA mutations found in dyskeratosis congenita and aplastic anemia reduce telomerase activity via haploinsufficiency. Blood. 2004;104:3936–42. doi: 10.1182/blood-2004-05-1829. [DOI] [PubMed] [Google Scholar]
- 20.Ly H, Calado RT, Allard P, et al. Functional characterization of telomerase RNA variants found in patients with hematologic disorders. Blood. 2005;105:2332–9. doi: 10.1182/blood-2004-09-3659. [DOI] [PubMed] [Google Scholar]
- 21.Xin ZT, Beauchamp AD, Calado RT, et al. Functional characterization of natural telomerase mutations found in patients with hematologic disorders. Blood. 2007;109:524–32. doi: 10.1182/blood-2006-07-035089. [DOI] [PubMed] [Google Scholar]
- 22.Issaragrisil S, Kaufman DW, Anderson T, et al. The epidemiology of aplastic anemia in Thailand. Blood. 2006;107:1299–1307. doi: 10.1182/blood-2005-01-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohyashiki K, Shay JW, Ohyashiki JH. Lack of mutations of the human telomerase RNA gene (hTERC) in myelodysplastic syndrome. Haematologica. 2005;90:691. [PubMed] [Google Scholar]
- 24.Camitta BM, Thomas ED, Nathan DG, et al. Severe aplastic anemia: a prospective study of the effect of early marrow transplantation on acute mortality. Blood. 1976;48:63–70. [PubMed] [Google Scholar]
- 25.Bennett JM, Catovsky D, Daniel MT, et al. Proposals for the classification of the myelodysplastic syndromes. Br J Haematol. 1982;51:189–99. [PubMed] [Google Scholar]
- 26.Ly H, Schertzer M, Jastaniah W, et al. Identification and functional characterization of 2 variant alleles of the telomerase RNA template gene (TERC) in a patient with dyskeratosis congenita. Blood. 2005;106:1246–52. doi: 10.1182/blood-2005-01-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–14. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 28.Samani NJ, Boultby R, Butler RB, et al. Telomere shortening in atherosclerosis. Lancet. 2002;358:472–3. doi: 10.1016/S0140-6736(01)05633-1. [DOI] [PubMed] [Google Scholar]
- 29.Cawthon R, Smith K, O'Brien E, et al. Association between telomere length in blood and mortality in people aged 60 years or older. Lancet. 2003;361:393–5. doi: 10.1016/S0140-6736(03)12384-7. [DOI] [PubMed] [Google Scholar]
- 30.Brouilette SW, Moore JS, McMahon AD, et al. Telomere length, risk of coronary heart disease, and statin treatment in the West of Scotland Primary Prevention Study: a nested case-control study. Lancet. 2007;369:107–14. doi: 10.1016/S0140-6736(07)60071-3. [DOI] [PubMed] [Google Scholar]
- 31.Minamino T, Miyauchi H, Yoshida T, et al. Endothelial cell senescence in human atherosclerosis. Role of telomere in endothelial dysfunction. Circulation. 2002;105:1541–4. doi: 10.1161/01.cir.0000013836.85741.17. [DOI] [PubMed] [Google Scholar]
- 32.Ly H, Blackburn EH, Parslow TG. Comprehensive Structure-Function Analysis of The Core Domain of Human Telomerase RNA. Mol. Cell. Biol. 2003;23:6849–56. doi: 10.1128/MCB.23.19.6849-6856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen JL, Opperman KK, Greider CW. A critical stem-loop structure in the CR4-CR5 domain of mammalian telomerase RNA. Nucl. Acids Res. 2002;30:592–7. doi: 10.1093/nar/30.2.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moriarty TJ, Fluard S, Dupuis S, Autexier C. Functional Multimerization of Human Telomerase Requires an RNA Interaction Domain in the N Terminus of the Catalytic Subunit. Mol. Cell. Biol. 2002;22:1253–65. doi: 10.1128/MCB.22.4.1253-1265.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]