Abstract
In both animal and yeast cells, signaling pathways involving small guanosine triphosphatases (GTPases) regulate polarized organization of the actin cytoskeleton. In the budding yeast Saccharomyces cerevisiae, the Ras-like GTPase Bud1/Rsr1 and its guanosine 5’-diphosphate (GDP)/guanosine 5’-triphosphate (GTP) exchange factor Bud5 are involved in the selection of a specific site for growth, thus determining cell polarity. We found that Bud5 is localized at the cell division site and the presumptive bud site. Its localization is dependent on potential cellular landmarks, such as Bud3 and Axl2/Bud10 in haploid cells and Bud8 and Bud9 in diploid cells. Bud5 also physically interacts with Axl2/Bud10, a transmembrane glycoprotein, suggesting that a receptor-like transmembrane protein recruits a GDP/GTP exchange factor to connect an intrinsic spatial signal to oriented cell growth.
Yeast cells undergo oriented cell division by choosing a specific bud site on the cell cortex. Haploid a and α cells bud in an axial pattern in which both mother and daughter cells select a bud site immediately adjacent to their previous division site. Diploid a/α cells bud in a bipolar pattern: Mother cells select a bud site adjacent to their daughter or on the opposite end of the cell, whereas daughter cells always choose a bud site directed away from their mother (1–3). The axial pattern appears to depend on a transient cortical marker that may involve proteins such as septins, Bud3, Bud4, and Axl2 (4–12). The bipolar budding pattern appears to depend on persistent markers that are deposited at both poles of the cell. The genes that are specifically required for the bipolar pattern may encode or regulate the bipolar cortical cues (4, 13). A key issue in understanding the mechanism of polarity establishment is to determine how these potential landmarks are linked to the intracellular signaling pathways. A likely candidate that links these spatial cues to polarity establishment is the Bud1 GTPase module, which is composed of Bud1, its GTPase-activating protein (GAP) Bud2, and Bud5 (3, 4, 14–18). These proteins are required for both axial and bipolar budding patterns.
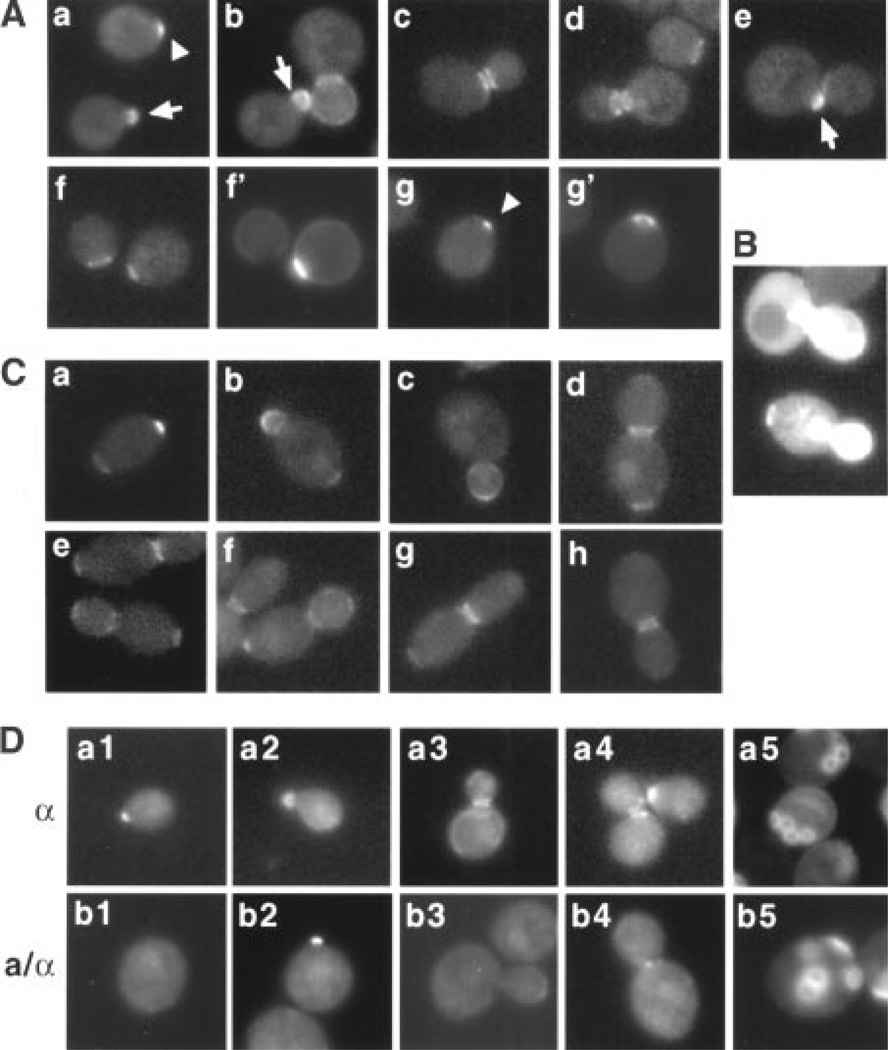
To examine Bud5 localization, we used a functional chromosomal BUD5-GFP (GFP, green fluorescent protein) fusion. Several aspects of the Bud5-GFP localization in haploid a and α cells were notable (Fig. 1A) (19). Bud5-GFP was present in a small patch in unbudded cells. After bud emergence, Bud5-GFP localized throughout the periphery of the bud. As the bud continued to grow, the Bud5-GFP signal at the bud periphery diminished, and the signal was observed as a double ring encircling the mother-bud neck. Following cytokinesis, a single ring of Bud5-GFP persisted in both mother and newly born daughter cells. The Bud5-GFP ring then disappeared, and Bud5-GFP concentrated in a patch at the incipient bud site. Cal-cofluor staining of bud scars confirmed that the ring of Bud5-GFP localized at the division site and a new patch of Bud5-GFP appeared next to the previous division site (Fig. 1A). Thus, Bud5-GFP localized to the cell division site and the presumptive bud site, and its localization changed over the cell cycle. The localization of Bud5-GFP in haploid cells overlapped extensively with that of Axl2 throughout the cell cycle and also with those of Bud3 and Bud4 during M and early G1 phases (5–7, 12), suggesting that Bud5 may interact with these axial-specific components.
Fig. 1.
Localization of Bud5-GFP in haploid and diploid cells. (A) Localization of Bud5-GFP in haploid α cells. Approximately 150 cells were examined for each panel in Figs. 1 and 2, except where noted. Representative micrographs are shown. Bud5-GFP images in yeast cells (strain HPY307) at different stages of the cell cycle are shown (panels a through e). Panels f through g’ show two pairs of images of Bud5-GFP (f and g) and Calcofluor staining (f’ and g’) of unbudded cells. The arrowheads indicate the Bud5-GFP signal in a patch at the incipient bud site in G1 cells; the arrows indicate the Bud5-GFP signal in a small bud. (B) Overexpression of Bud5-GFP in haploid a cells. Bud5-GFP images of a bud5Δ (strain IH2423) cells expressing BUD5-GFP from a multicopy plasmid are shown. (C) Localization of Bud5-GFP in diploid a/α cells (strain HPY309). A small percentage (5%) of cells with small-sized buds also showed the Bud5-GFP signal at the opposite pole of the mother cell (panel b), in addition to buds. (D) Localization of Bud5-b1-GFP in haploid a and diploid a/α cells. Approximately 200 cells were examined for each panel. Strain HPY378 was used in panels a1 through a5, and strain HPY388 was used in panels b1 through b5. Calcofluor staining was shown in panels a5 and b5. Bud5-b1-GFP sometimes localized in a patch at the neck in a/α cells (panel b4), which was not observed in wild-type cells.
To examine whether localization of Bud5 was required for proper bud site selection, we determined the localization of Bud5-GFP and the budding pattern of haploid cells overexpressing BUD5-GFP from a multicopy plasmid. Bud5-GFP was often found at random locations in the cell periphery and also in the cytoplasm (Fig. 1B). These cells also exhibited partial random budding (20), suggesting that localization of Bud5 is important for proper bud site selection.
Diploid a/α cells exhibit bipolar budding, as opposed to axial budding, and Bud5-GFP revealed distinct localization patterns in a/α cells, particularly during G1 and M phases (Fig. 1C) (19). Before bud emergence, Bud5-GFP was present at both poles: as a ring at one pole and in a small patch at the opposite pole. After bud emergence, Bud5-GFP localized throughout the periphery of the bud, as seen in haploid cells. At a later stage of the cell cycle, Bud5-GFP localized at the neck and at one or both poles of mother cell and bud, whereas a small percentage of cells showed a Bud5-GFP signal only at the neck. Such patterns of Bud5-GFP localization were not observed in haploid cells. Thus, localization of Bud5 to the potential bud site in a/α cells before the G1 phase was likely to contribute to the bipolar budding pattern of these cells.
We hypothesized that Bud5 was localized in distinct locations in different cell types by interacting with cell type–specific landmarks and predicted that cell type–specific alleles of BUD5 might fail to interact with one type of spatial cue but not with the other. Thus, we carried out site-directed mutagenesis and isolated a few bud5 mutants that were specifically defective in bipolar budding (19, 21), similar to a previously isolated bipolar-specific bud5 mutant (13). We localized one of the bipolar-specific Bud5 mutant proteins, Bud5-b1R17A,E18A [Arg17→Ala17 (R17A); Glu18→Ala18 (E18A)], and found that its localization was dramatically changed in diploid cells but not in haploid cells. Although Bud5-b1-GFP still appeared as a strong signal, filling the tiny bud in diploid cells, most of the signal was delocalized in unbudded cells and cells with medium- or large-sized buds (Fig. 1D).
Taken together, these data suggested that Bud5-GFP localized to distinct bud sites in each cell type, which correlated with the cell type–specific budding pattern. Localization of Bud5 was important for proper bud site selection because mislocalization of Bud5 by overexpression or specific mutation resulted in bud site selection defects.
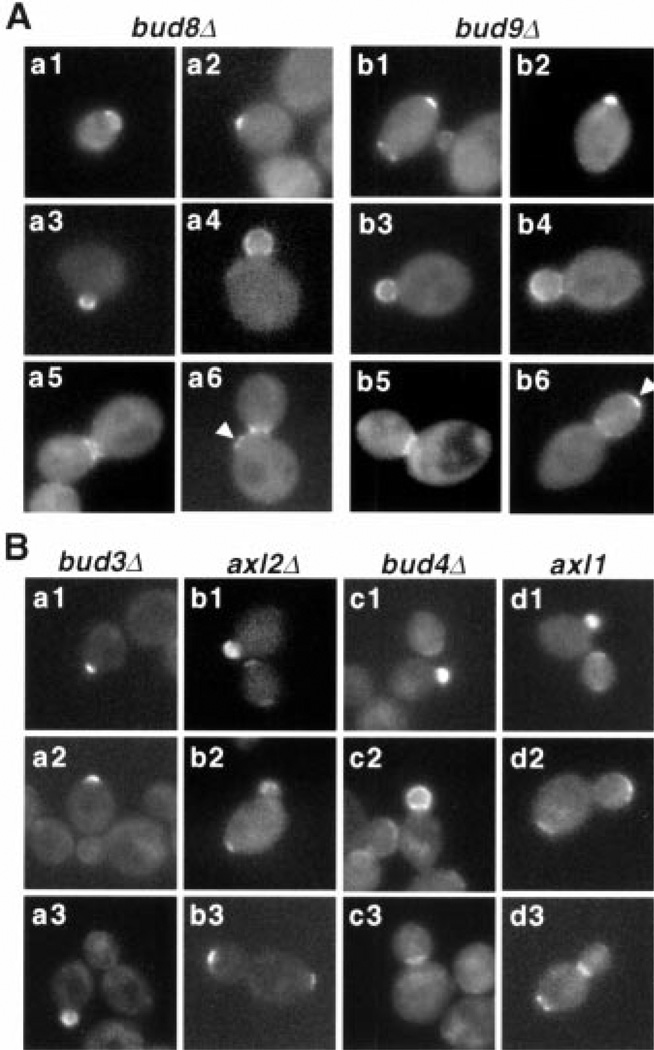
To understand how Bud5 localized to the distinct location in each cell type, we examined Bud5 localization in mutants defective in either axial or bipolar budding pattern. bud8 and bud9 mutants appear to have the most specific effects on bipolar bud site selection: bud8 mutants bud mostly around the pole proximal to the birth scar, whereas bud9 mutants bud almost exclusively around the pole distal to the birth scar (13). Thus, we determined Bud5 localization in diploid a/α cells homozygous for either bud8Δor bud9Δ(Fig. 2A and Web table 2) (19, 22). The most dramatic difference in Bud5-GFP localization in bud8Δmutants was found in unbudded cells and in cells with large-sized buds. Less than 1% of unbudded cells of bud8Δmutants showed both a ring and a patch at the opposite pole (compared to 70% in wild-type cells). The Bud5-GFP signal sometimes appeared all over the bud periphery, but it was never localized to the bud tip in bud8Δcells with large-sized buds, indicating that Bud5 failed to localize to the distal pole in bud8 mutants. These results support the idea that Bud5 may interact with Bud8, a distal pole marker, which has been shown to be localized to the bud tip (23–25).
Fig. 2.
Localization of Bud5-GFP in various mutants. (A) Localization of Bud5-GFP in a/α bud8Δ/bud8Δ(strain HPY373) (panels a1 through a6) and a/α bud9Δ/bud9Δ(strain HPY359) (panels b1 through b6) cells. Arrowheads indicate the Bud5-GFP signal next to the proximal pole of the mother cell (panel a6) and at the bud tip (panel b6). (B) Localization of Bud5-GFP in a bud3Δ (strain HPY352), a axl2Δ(strain HPY357), a bud4Δ (strain HPY353), and a axl1 (strain HPY458) cells.
The localization of Bud5-GFP in bud9Δcells was quite similar to that of wild-type cells during most of the cell cycle, except at a later stage (Fig. 2A). Although the percentage of large-budded cells with Bud5-GFP localization at the neck or distal pole of buds was similar to that of wild-type cells, no cells with the Bud5-GFP signal at the proximal pole of the mother cell were observed in bud9Δcells. These results support the idea that a proximal pole marker is absent or defective in bud9 mutants (13, 24); thus, Bud5 fails to localize at the proximal pole of the mother cells in bud9Δmutants.
Bud5-GFP localization was significantly altered in a distinct manner in mutants defective in axial budding pattern (Fig. 2B and Web table 2) (19). The most dramatic difference in Bud5-GFP localization was found in a bud3Δ and a axl2Δcells in G1 and M phases. The number of unbudded cells with a Bud5-GFP ring was greatly reduced in bud3Δ and axl2Δmutants. Most axl2Δcells with a medium- or large-sized bud lost a Bud5-GFP ring at the neck. Instead, they showed the Bud5-GFP signal at the bud tip and/or the pole of the mother cell. Almost half of the bud3Δ cells with a medium- or large-sized bud did not show any localized Bud5-GFP signal. Bud5-GFP was similarly mislocalized in a bud4Δ mutants, but to a lesser extent. Because the localization of Bud3 and Bud4 is interdependent (12), the defect of Bud5 localization in bud4 mutants could be due to the fact that Bud3 is inefficiently localized in bud4 mutants. Bud5-GFP localization in a axl1 mutants, which are also defective in axial budding (26), was very similar to that in wild-type a/α cells. Bud5 localization in these mutants that are defective in axial budding pattern is different from that in wild-type a/α cells, except in axl1 mutants. The different patterns of Bud5 localization in these mutants, which belong to the same group based on their phenotypes, may reflect the distinct roles of each protein in axial budding and underlie the slight differences in bud site selection seen in these mutants. Together, these data suggest a functional interaction between Bud5 and Axl2 or Bud3 for axial bud site selection.
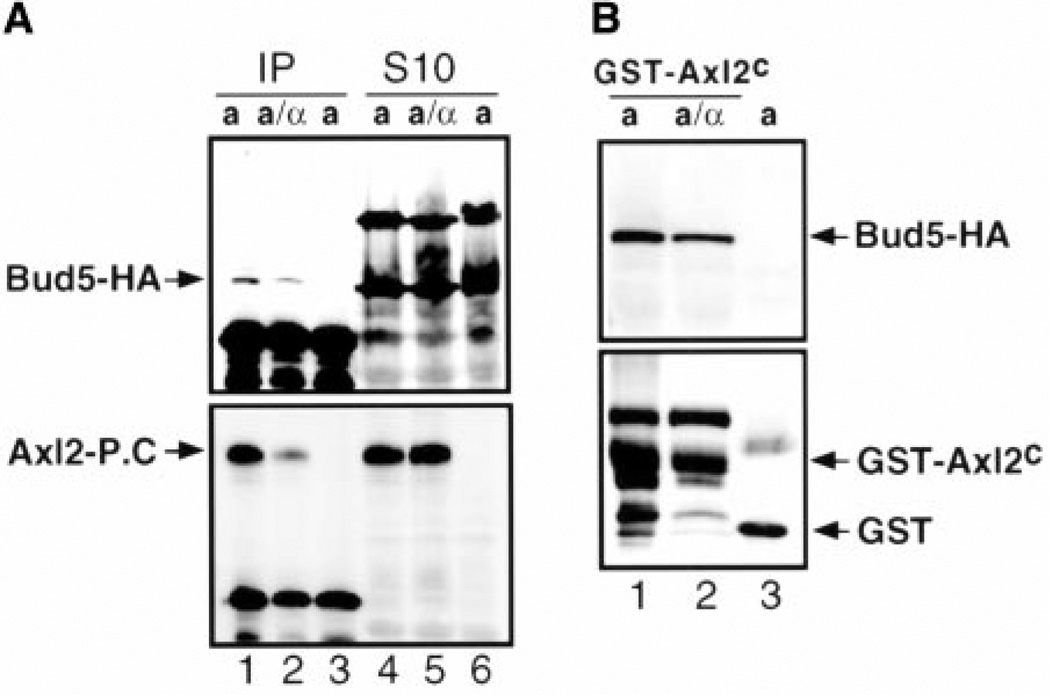
To investigate whether Bud5 physically interacts with these potential axial landmarks, we performed immunoprecipitation experiments, using yeast cells expressing hemagglutinin epitope–tagged Bud5 (Bud5-HA), protein C epitope–tagged Axl2 (Axl2–protein C), and Bud3 (19). When Axl2–protein C was immunoprecipitated with antibodies recognizing the protein C epitope, Bud5-HA was also coprecipitated (Fig. 3A), suggesting that Bud5 interacts with Axl2 (27).
Fig. 3.
Bud5 interacts with Axl2. (A) Bud5-HA coimmunoprecipitates with Axl2–protein C. The top and bottom panels show immunoblots carried out with monoclonal antibodies against the hemagglutinin epitope and the protein C epitope, respectively. Lanes 1 and 2 show the eluents from immunoprecipitation (IP) experiments with extracts from a and a/α cells expressing Axl2–protein C (Axl2-P.C), respectively; lanes 4 and 5 show the soluble fractions (S10) from a and a/α cells expressing Axl2–protein C, respectively. Lanes 3 and 6 show the control immunoprecipitation experiment and S10 fraction from cells expressing untagged Axl2, respectively. (B) Bud5 copurifies with the cytoplasmic domain of Axl2. The top and bottom panels show immunoblots carried out with monoclonal antibodies against the hemagglutinin epitope and with polyclonal antibodies against GST, respectively. Lane 1 shows a cells expressing GST-Axl2C, lane 2 shows a/α cells expressing GST-Axl2C, and lane 3 shows a cells expressing GST alone.
Axl2 is a transmembrane glycoprotein with a predicted intracellular domain at the COOH-terminus (6, 7). We hypothesized that Bud5 interacts with the cytoplasmic domain of Axl2. Thus, the putative cytoplasmic domain of Axl2 was expressed as a glutathione S-transferase fusion protein (GST-Axl2C) and was purified from a yeast strain that also expressed Bud5-HA (19). Bud5-HA copurified with GST-Axl2C, but not with GST (Fig. 3B), suggesting that Bud5 interacts with the cytoplasmic domain of Axl2.
Both Bud5 and Axl2 are equally abundant in all cell types (Fig. 3A) (6). Bud5 is required for both axial and bipolar budding, but Axl2 is required only for axial budding (6, 7, 15). It has been proposed that Axl2 acts as an anchor in the plasma membrane that directs new growth components to the axial budding site (6, 7). Thus, Bud5 may interact productively with Axl2 only in haploid cells and have other partners in diploid cells. However, we found that Bud5 still interacted with Axl2 in the extracts prepared from diploid a/α cells (Fig. 3A and B) (28). It remains to be determined whether Axl2 regulates the GDP/GTP exchange factor (GEF) activity of Bud5 differently in different cell types and whether Bud5 interacts with potential bipolar landmarks in a/α cells. Likely candidates include Bud8 and Bud9, which are also transmembrane proteins with a short intracellular domain (24), and Rax2, which has recently been reported as a persistent cortical marker of diploid cells (25). Consistent with this idea, we found that localization of Bud5 was altered in diploid bud8 and bud9 mutants.
Bud1 interacts with specific proteins required for polarity establishment, such as Cdc24, a GEF for Cdc42 (18, 29). The coupling of Bud1 and Cdc42 GTPase cycles is crucial for actin cytoskeleton organization at the proper bud site. However, Bud1 is not localized to the incipient budding site, but rather is distributed uniformly around the plasma membrane (30), suggesting that it is not sufficient to direct positioning of bud site assembly proteins. Instead, localization of its regulators may be crucial in linking the cell type–specific cues to polarity establishment. Indeed, its GAP Bud2 (31) and GEF Bud5 are localized to the presumptive bud sites that are distinct in different cell types (19). In particular, the physical association between Bud5 and Axl2 reported here suggests that Bud5 is probably involved in linking an axial-specific landmark to polarity establishment (Web fig. 1) (19). Axl2 is predicted to possess a type I membrane topology similar to that of integrins (6, 7). In mammalian cells, an integrin, LFA-1, interacts with cytohesin, a GEF for adenosine 5’-diphosphate ribosylation factor GTPase, which regulates cell adhesion (32). Thus, interaction between an exchange factor and a receptor-like transmembrane protein may be a conserved mechanism linking a spatial signal to cell polarity in both yeast and mammalian cells.
Supplementary Material
Acknowledgments
We thank I. Herskowitz, A. Simcox, R. Hill, R. Tabtiang, E. Oakley, P. Wilson, and H. Chamberlin for comments on the manuscript; J. Pringle and B. Glick for discussions; and J. Pringle, M. Snyder, D. Kellogg, J. Chant, S. Sanders, and A. Straight for strains and plasmids. Supported by an NIH grant and a Basil OÕConnor Starter Scholar Research Award from the March of Dimes Birth Defects Foundation to H.-O.P.
References and Notes
- 1.Freifelder D. J. Bacteriol. 1960;80:567. doi: 10.1128/jb.80.4.567-568.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hicks JB, Strathern JN, Herskowitz I. Genetics. 1977;85:373. doi: 10.1093/genetics/85.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chant J, Herskowitz I. Cell. 1991;65:1203. doi: 10.1016/0092-8674(91)90015-q. [DOI] [PubMed] [Google Scholar]
- 4.Chant J, Pringle JR. J. Cell Biol. 1995;129:751. doi: 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chant J, Mischke M, Mitchell E, Herskowitz I, Pringle JR. J. Cell Biol. 1995;129:767. doi: 10.1083/jcb.129.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roemer T, Madden K, Chang J, Snyder M. Genes Dev. 1996;10:777. doi: 10.1101/gad.10.7.777. [DOI] [PubMed] [Google Scholar]
- 7.Halme A, Michelitch M, Mitchell EL, Chant J. Curr. Biol. 1996;6:570. doi: 10.1016/s0960-9822(02)00543-2. [DOI] [PubMed] [Google Scholar]
- 8.Haarer BK, Pringle JR. Mol. Cell. Biol. 1987;7:3678. doi: 10.1128/mcb.7.10.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ford SK, Pringle JR. Dev. Genet. 1991;12:281. doi: 10.1002/dvg.1020120405. [DOI] [PubMed] [Google Scholar]
- 10.Kim HB, Haarer BK, Pringle JR. J. Cell Biol. 1991;112:535. doi: 10.1083/jcb.112.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flescher EG, Madden K, Snyder M. J. Cell Biol. 1993;122:373. doi: 10.1083/jcb.122.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanders S, Herskowitz I. J. Cell Biol. 1996;134:413. doi: 10.1083/jcb.134.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zahner J, Harkins HI, Pringle JR. Mol. Cell. Biol. 1996;16:1857. doi: 10.1128/mcb.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bender A, Pringle JR. Proc. Natl. Acad. Sci. U.S.A. 1989;86:9976. doi: 10.1073/pnas.86.24.9976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chant J, Corrado K, Pringle JR, Herskowitz I. Cell. 1991;65:1213. doi: 10.1016/0092-8674(91)90016-r. [DOI] [PubMed] [Google Scholar]
- 16.Powers S, Gonzales E, Christensen T, Cubert J, Broek D. Cell. 1991;65:1225. doi: 10.1016/0092-8674(91)90017-s. [DOI] [PubMed] [Google Scholar]
- 17.Park H-O, Chant J, Herskowitz I. Nature. 1993;365:269. doi: 10.1038/365269a0. [DOI] [PubMed] [Google Scholar]
- 18.Park H-O, Bi E, Pringle J, Herskowitz I. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4463. doi: 10.1073/pnas.94.9.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.For supplementary data, as well as more information about strains, plasmids, and methods, see Science Online (www.sciencemag.org/cgi/content/full/1060360/DC1).
- 20.These cells showed ~60% random budding (n = 200), as compared to 8% random budding of the wild-type strain (n = 200).
- 21.α cells expressing Bud5-b1-GFP budded in an axial manner (85%; n = 200), whereas a/α cells expressing Bud5-b1-GFP budded in a random manner (93%; n= 200),
- 22.Localization of Bud5-GFP in haploid bud8Δ and bud9Δ strains was indistinguishable from that in haploid wild-type cells.
- 23.Taheri N, Kohler T, Braus GH, Mosch H-U. EMBO J. 2000;19:6686. doi: 10.1093/emboj/19.24.6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harkins HA, et al. Mol. Biol. Cell doi: 10.1091/mbc.12.8.2497. , in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen T, et al. Science. 2000;290:1975. doi: 10.1126/science.290.5498.1975. [DOI] [PubMed] [Google Scholar]
- 26.Fujita A, et al. Nature. 1994;372:567. doi: 10.1038/372567a0. [DOI] [PubMed] [Google Scholar]
- 27.In a similar approach, we were unable to detect coimmunoprecipitation of Bud5 and Bud3. Bud3 may not interact with Bud5 but rather may indirectly influence localization of Bud5. Alternatively, Bud5 may interact with Bud3 very transiently, which cannot be easily coimmunoprecipitated.
- 28.Although a slightly less amount of Bud5 was co-immunoprecipitated or copurified with Axl2 from a/α cells compared to a cells, Axl2 was also recovered less efficiently from a/α cells by immunoprecipitation and GST column chromatography. We suspect that an unknown protein interacts with Axl2 in a/α cells, resulting in poor recovery of Axl2. One caveat of both approaches is that Axl2 is overexpressed when either a multicopy plasmid or the GAL promoter is used; hence it may not reflect the exact scenario occurring in the cell.
- 29.Zheng Y, Bender A, Cerione RA. J. Biol. Chem. 1995;270 doi: 10.1074/jbc.270.2.626. [DOI] [PubMed] [Google Scholar]
- 30.Michelitch M, Chant J. Curr. Biol. 1996;6:446. doi: 10.1016/s0960-9822(02)00512-2. [DOI] [PubMed] [Google Scholar]
- 31.Park H-O, Sanson A, Herskowitz I. Genes Dev. 1999;13:1912. doi: 10.1101/gad.13.15.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geiger C, et al. EMBO J. 2000;19:2525. doi: 10.1093/emboj/19.11.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.