Abstract
Like all viruses, hepatitis B virus (HBV) replication and pathogenesis depends on the critical interplay between viral and host factors. In this review, we will focus on the recent progress in understanding the virus-host interactions at the level of the infected cell. These interactions include the requirement of cellular chaperones for the initiation of HBV reverse transcription, the role of the HBV X protein (HBx) in modifying viral and cellular transcription and signaling, the formation of the HBV episomal DNA and its epigenetic regulation in viral persistence, and the cellular factors involved viral entry, nucleocapsid maturation, and virion secretion.
Keywords: HBV, Reverse transcription, CCC DNA, pgRNA, HBx, Core protein, Surface protein, Phosphorylation
It is estimated that more than 350 million individuals are chronically infected with HBV; many of whom (ca. ¼) will eventually develop severe liver diseases, including chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (HCC), one of the most common forms of human cancer (Seeger et al., 2007). The estimated risk of HCC in chronic HBV carriers is approximately 100 times greater than in uninfected individuals. Currently, anti-HBV drugs such as interferon alpha and nucleoside analogs are either fraught with adverse reactions or are only effective in the short-term (Locarnini and Mason, 2006). Therefore, it is necessary to elucidate the intricate interactions between viral and host factors to facilitate the development of novel antiviral strategies.
HBV is an enveloped DNA virus that belongs to the Hepadnaviridae family. It contains a small (ca 3.2 kb), partially double-stranded (DS), relaxed-circular (RC) DNA genome that is reverse transcribed via an RNA intermediate, the pregenomic RNA (pgRNA) (Seeger and Mason, 2000; Seeger et al., 2007; Summers and Mason, 1982). The genome encodes four overlapping open reading frames (ORFs) that are translated to make the viral core protein, the surface proteins, a reverse transcriptase (RT), and HBx (Seeger et al., 2007).
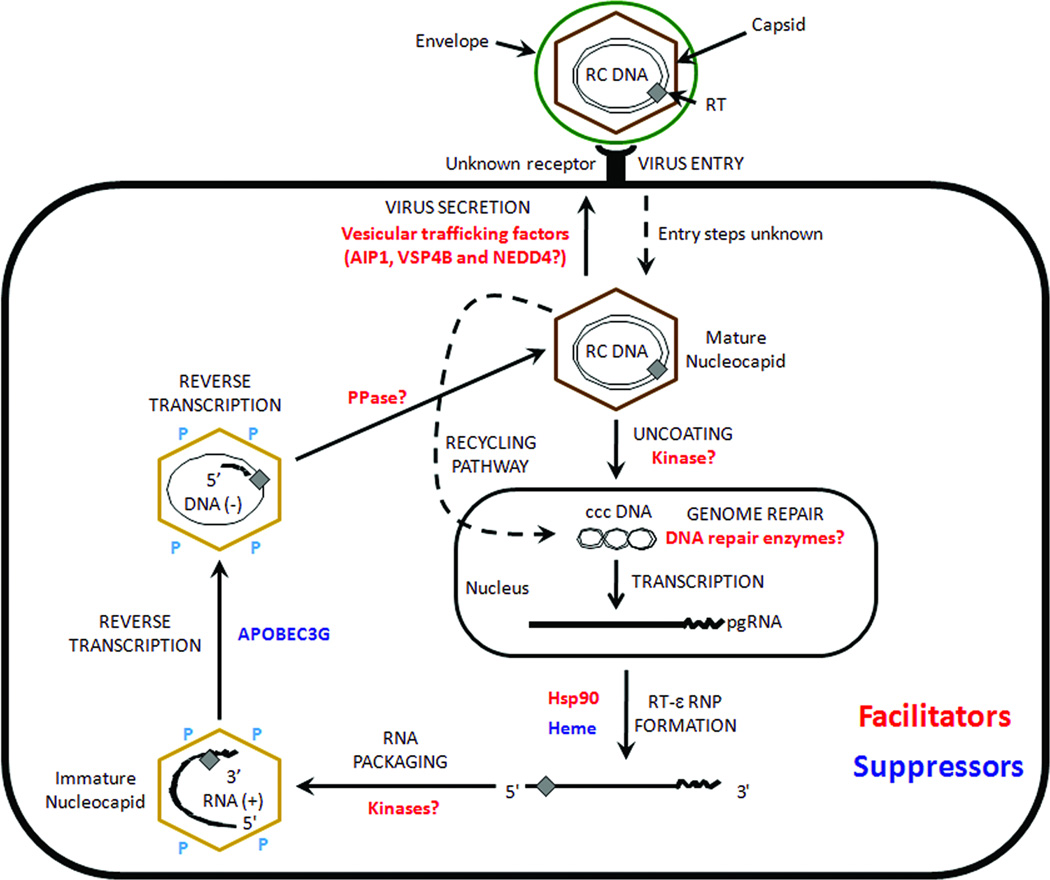
The HBV life cycle (Fig. 1) begins when the virus attaches to an unknown receptor on the host cell and is internalized. The virion RC DNA is delivered to the nucleus, where it is repaired to form a covalently closed-circular (CCC) DNA (Tuttleman et al., 1986). The episomal CCC DNA serves as the template for the transcription of the pgRNA and the other viral mRNAs by the host RNA polymerase II. The transcripts are then exported to the cytoplasm, where translation of the viral proteins occurs. RT binds to pgRNA and triggers assembly of the core proteins into immature, RNA-containing nucleocapsids (Bartenschlager and Schaller, 1992; Hirsch et al., 1990). The immature nucleocapsids then undergo a process of maturation where pgRNA is reversed transcribed by RT to make the mature RC DNA. A unique feature of hepadnavirus reverse transcription is the RT primed initiation of minus-strand DNA synthesis, which leads to the covalent linkage of RT to the 5’ end of the minus-strand DNA (Gerlich and Robinson, 1980; Wang and Seeger, 1992; Weber et al., 1994; Zoulim and Seeger, 1994). The mature, RC-DNA-containing nucleocapsids are then enveloped by the viral surface proteins and secreted as virions (secretion pathway) or alternatively, are recycled back to the nucleus to further amplify the pool of CCC DNA (recycling pathway) (Tuttleman et al., 1986; Wu et al., 1990).
Figure 1. The virus-cell interactions in the hepadnaviral lifecycle.
HBV binds to an unknown receptor on hepatocytes, is internalized, uncoated and delivers its RC DNA genome to the nucleus. The genome is repaired to form the CCC DNA that serves as the transcriptional template. Viral RNAs, including the pgRNA, are exported to the cytoplasm, where translation of the viral proteins occurs. The pgRNA is packaged together with RT into immature nucleocapsids composed of core protein. It is then reversed transcribed within the nucleocapsids into the mature RC DNA form. The mature, RC DNA-containing nucleocapsids are then enveloped by the viral envelope proteins and secreted extracellularly as virions or recycled to the nucleus to amplify the CCC DNA pool. Facilitators and suppressors of viral replication are highlighted in red and blue, respectively. RC DNA, relaxed circular DNA; CCC DNA, covalently closed circular DNA; pgRNA, pregenomic RNA; RT, reverse transcriptase; P, Phosphates, PPase, Phosphatase; Hsp 90, heat-shock protein 90.
In this short review, we will discuss the interactions between HBV viral factors and host proteins that are involved in HBV replication and pathogenesis. The review is organized based on the individual viral factors and their interactions with cellular proteins. In addition, systemic factors (e.g. cytokines) have also been implicated in the replication and pathogenesis of HBV infection; however, this is beyond the scope of the review and will not be discussed.
Surface Protein
The envelope of the HBV virion contains three surface proteins: large (L), middle (M), and small (S) proteins, all encoded by the S ORF. Additionally, the serum of infected individuals also contains subviral particles made up of cell-derived lipid envelope and HBV surface proteins without the genome or nucleocapsid. These particles can be found in as much as 10,000-fold excess of the infectious virus particles and may serve as a mechanism of evading the host immune system (Seeger et al., 2007). While the full complement of functions of surface proteins during HBV infection has not yet been characterized, the involvement of the surface protein in the virus’s entry into host cells and in the release of progeny virions will be discussed here.
The mechanism by which hepadnaviruses enter host cells is not completely understood. This entry mechanism has been primarily studied using duck hepatitis B virus (DHBV) as a model system. In 1994, a cellular factor that binds DHBV particles with high affinity was identified (Kuroki et al., 1994). This factor was shown to be carboxypeptidase D (gp180), which is now widely accepted to act as a primary receptor molecule for DHBV infection (Kuroki et al., 1995; Tong et al., 1995; Urban et al., 1998). However, it appears that additional factors are necessary for subsequent entry steps. Glycine decarboxylase has been identified as a candidate cofactor involved in some post-binding steps (Li et al., 2004; Li et al., 1999; Li et al., 1996). The receptor for human HBV has not yet been identified.
Only mature, DNA-containing nucleocapsids interact with the surface proteins to undergo subsequent envelopment and secretion from the host cell. However, it is currently unclear how the surface proteins select mature nucleocapsids for envelopment or which host factors may participate in this selection process. On the other hand, the budding and secretion of HBV virions from host cells has been suggested to involve the machinery of multivesicular body (MVB) formation, including the interaction of host factors γ2-adaptin, Nedd4 ubiquitin ligase, and Vps4 (Hartmann-Stuhler and Prange, 2001; Kian Chua et al., 2006; Rost et al., 2006). A recent report further supports the requirement of host MVB function: HBV S protein was found to colocalize with AIP1 and VSP4B, proteins involved in multivesicular body formation (Watanabe et al., 2007). Dominant negative mutants of AIP1 and VSP4B were able to inhibit the release of virions, but not subviral particles, suggesting that the release of mature virions may require the association of cellular factors discrete from those of other populations of viral particles (Watanabe et al., 2007).
Reverse Transcriptase
The HBV RT is a multifunctional protein that consists of four domains: terminal protein (TP), reverse transcriptase, RNase H, and a non-conserved spacer domain between the TP and RT domains. Initiation of reverse transcription and nucleocapsid assembly in HBV is carried out by RT (Bartenschlager and Schaller, 1992; Hirsch et al., 1990; Summers and Mason, 1982). The unique TP domain is used as a protein primer to initiate reverse transcription catalyzed by the RT domain (Pollack and Ganem, 1994; Wang and Seeger, 1992; Wang et al., 1994; Weber et al., 1994; Zoulim and Seeger, 1994). This protein priming reaction requires the recognition and binding of RT to a stem-loop structure located at the 5’ end of the pgRNA called epsilon (ε). As a result of the protein priming mechanism, the polymerase remains covalently linked via its TP domain to the 5’ end of the minus-strand DNA of the RC DNA genome. In addition to its role as a primer and polymerase for DNA synthesis, RT, via its specific recognition of the ε RNA, also triggers the assembly of the nucleocapsid, leading to the specific incorporation of both the pgRNA and RT into replication-competent nucleocapsids (Bartenschlager and Schaller, 1992; Pollack and Ganem, 1994).
Early biochemical studies on the hepadnavirus RT were thwarted by difficulties in obtaining an active recombinant protein due to problems of low expression, extreme instability, and insolubility, thus making the study of it structure and function very challenging (Hu and Seeger, 1996a). It was not until 1992 that an enzymatically active RT, from DHBV, was first produced using the rabbit reticulocyte lysate (RRL) in vitro translation system (Wang and Seeger, 1992). This then led to the discovery that a cellular molecular chaperone complex, consisting of heat shock protein 90 (Hsp90) and its cofactors, was required for the interaction between the viral RT and ε RNA (Hu and Seeger, 1996b; Hu et al., 1997). These observations suggested a model in which Hsp90 stabilizes a transient conformation of the viral polymerase to permit the formation of a ribonucleoprotein (RNP) complex between RT and ε RNA during viral assembly and reverse transcription.
Recent success in producing recombinant DHBV and HBV RT protein in Escherichia coli has allowed for the development of a defined biochemical reconstitution system that has greatly facilitated efforts to elucidate viral and cellular requirements for RNP formation and protein priming in hepadnaviruses (Hu and Anselmo, 2000; Hu et al., 2004; Hu et al., 2002). Together with Hsp90, several other chaperone proteins, including Hsp70, Hsp40, Hop, and p23 (all known components of the Hsp90 complex), are also necessary for the specific binding of DHBV and HBV RT to their cognate ε RNA in vitro (Hu et al., 2004; Hu et al., 2002). In the case of DHBV RT, the reconstituted RT-ε complex is then competent to carry out protein priming (Hu and Anselmo, 2000; Hu et al., 2002). However, despite the efficient reconstitution of HBV RT-ε interaction, protein priming activity could not be detected with the HBV RT protein, suggesting that additional cellular factors are required for HBV protein priming (Hu and Boyer, 2006; Hu et al., 2004). These results are consistent with the observation that the HBV RT translated in RRL had no detectable protein priming activity, even though the DHBV RT expressed in RRL was demonstrated early on to have protein priming activity (Wang and Seeger, 1992).
Nucleoside analogs are highly effective in suppressing the activity of the HBV RT and thus, viral replication (Locarnini and Mason, 2006; Seeger et al., 2007). Treatment of chronic hepatitis B with these RT inhibitors has been associated with a rapid decrease in viremia. However, long-term treatment is required to maintain viral suppression, which leads to the development of drug-resistant HBV variants that renders the nucleoside analogs ineffective. Therefore, novel anti-HBV therapy needs to be developed. Recently, we discovered that iron protoporphyrin IX (heme), an endogenous component of hemoglobin and several other cellular enzymes, and several related porphyrin compounds, are potent inhibitors of hepadnavirus protein priming (Lin and Hu, 2007). Heme and its analogs disrupted the formation of the RT-ε complex, which is a prerequisite for protein priming. This block appears to be mediated by the binding of heme to the TP domain and to a lesser extent, the RT domain (Lin and Hu, 2007). These findings may represent a novel avenue for the development of anti-HBV drugs with an entirely new mechanism of action, which should complement the existing anti-HBV drugs very well. Furthermore, since the TP domain is unique to hepadnaviruses, drugs targeting TP should be highly selective for HBV.
The cellular deaminase protein, APOBEC3G, was recently shown to inhibit the replication of HBV, as well as a wide range of retroviruses (Cullen, 2006; Nguyen et al., 2007; Sheehy et al., 2002; Turelli et al., 2004). Initially, it was thought that the mechanism of inhibition of HBV replication was similar to that of retroviruses by inducing massive hypermutation on the viral minus-strand DNA via its deaminase activity (Lecossier et al., 2003; Rosler et al., 2005; Turelli et al., 2004). However, it turned out that the main mode of inhibition of HBV DNA synthesis is via a deaminase-independent mechanism (Nguyen et al., 2007; Rosler et al., 2005; Turelli et al., 2004). Although the details of the editing-independent mechanism are still not fully understood, we recently reported that APOBEC3G could block reverse transcription of HBV at a very early stage (Nguyen et al., 2007). APOBEC3G could potentially block this early step in DNA synthesis by inhibiting the polymerase function, or alternatively, it could affect the template function of the pgRNA via its known RNA binding activity.
Core Protein
Hepadnavirus nucleocapsids are composed of multiple (180 or 240) copies of one viral protein, the capsid or core protein, encoded by the C ORF. Core protein is subdivided into the N-terminal assembly domain (NTD) and the C-terminal functional domain (CTD), with the absolute requirement of the NTD for the formation of capsid particles (Nassal, 1992). The CTD is dispensable for assembly but is important in pgRNA packaging and DNA synthesis. It is arginine-rich and contains several serine/threonine phosphorylation sites (Nassal, 1992; Schlicht et al., 1989; Yu and Summers, 1994).
The phosphorylation state of DHBV nucleocapsids has been shown to correlate to the maturation stage of the virus. Immature nucleocapsids that contain RNA are phosphorylated at six CTD sites, while mature, DNA-containing nucleocapsids, either inside cells or in extracellular virions are completely dephosphorylated (Perlman et al., 2005; Pugh et al., 1989). Mutations made at these sites that mimic constitutively phosphorylated or un-phosphorylated states block viral replication at distinct stages. Phosphorylation of the HBV core protein has been shown to affect pgRNA packaging and DNA synthesis (Melegari et al., 2005; Yeh and Ou, 1991). In a similar manner, we recently reported that phosphorylation of the DHBV core protein is required for minus-strand DNA synthesis (Basagoudanavar et al., 2007), which suggests that phosphorylation of core protein plays an important role in the replication of all hepadnaviruses.
Purified HBV particles were found to demonstrate in vitro kinase activity almost 30 years ago (Albin and Robinson, 1980). Since the viral genome does not code for any proteins with kinase activity, it was predicted that the nucleocapsid packages a kinase of cellular origin during the assembly process (Albin and Robinson, 1980). The packaged kinase may play a role in the aforementioned CTD phosphorylation required for pgRNA packaging and minus-strand DNA synthesis. Additionally, the packaged kinase may participate in the uncoating process following viral entry, possibly initiating the uncoating process by phosphorylating the C-termini of the core subunits, thereby destabilizing the capsid (Barrasa et al., 2001). Over the last 3 decades, many kinases have been reported to be associated with viral capsids, including, but not limited to, a 46-kD kinase, a cdc2-like kinase, and SRPK1 and SRPK2 (Barrasa et al., 2001; Daub et al., 2002; Kau and Ting, 1998). Although these kinases have the ability to phosphorylate the core protein in vitro, no direct evidence for a role in viral replication has been convincingly demonstrated.
Similarly, because mature DHBV nucleocapsids are devoid of phosphorylation, a cellular phosphatase has been predicted to remove the phosphate groups during a late stage of reverse transcription, before the viral particles are enveloped and secreted (Basagoudanavar et al., 2007; Perlman et al., 2005; Pugh et al., 1989). It is clear that dynamic modification of the core protein by phosphorylation and dephosphorylation plays an indispensable role in the life cycle of hepadnaviruses. Identification of the host kinase(s) and phosphatase(s) responsible for this modification would undoubtedly provide new molecular targets for antiviral therapies, as would the identification of other cellular factors that interact with core protein during the different stages of HBV replication.
HBV X Protein
Despite all the recent progress in the field of HBV research, the function of one HBV protein, HBx, has remained elusive. HBx is encoded by the smallest HBV ORF and is 154 amino acids in size, with a molecular weight of about 17.5-kD. It has been suggested to affect viral replication, as well as host cell functions, by modulating a wide variety of cellular processes, including transcription, cell cycle progression, DNA damage repair, and apoptosis. However, its exact role in viral replication has yet to be established. Early studies have reported that HBx is essential for viral infection in vivo (Chen et al., 1993; Zoulim et al., 1994), yet it is dispensable for viral replication in cell culture (Blum et al., 1992). These differences in results obtained in vivo versus in vitro may suggest a critical role of HBx in the establishment of HBV infection in vivo, but not for viral replication per se.
Although it is not well defined, the subcellular localization of HBx seems to be mainly cytoplasmic, with a small fraction in the nucleus (Bouchard and Schneider, 2004). Thus it is believed that HBx may have a dual role in transcriptional regulation: cytoplasmic HBx could affect the regulation of signal transduction pathways while nuclear HBx may function at the promoter level (Doria et al., 1995).
HBx has been shown to transactivate a variety of viral and cellular promoters (Bouchard and Schneider, 2004; Seeger et al., 2007). Its ability to regulate a wide range of genes suggests that HBx could upregulate the expression of HBV gene by transactivating its own promoters, as well as modify the environment by transactivating cellular genes in infected hepatocytes to facilitate viral replication. Since HBx does not directly bind DNA, its ability to activate transcription of host genes is thought to occur indirectly by interaction with nuclear transcription factors or by activation of different signal transduction pathways. In support of its role in transcriptional regulation, HBx was reported to associate with several components of the basal transcriptional machinery, including TFIIB, TFIIH, and RBP5, a subunit of mammalian RNA polymerase (Cheong et al., 1995). It has also been demonstrated to bind to transcription factors including CREB, ATF-2, and AP-2 to modify their activities (Maguire et al., 1991; Seto et al., 1990). It was recently demonstrated that HBx could interact and cooperate with CREB-binding protein (CBP)/p300 to synergistically enhance CREB activity (Cougot et al., 2007). Thus, the ability of HBx to interact with these cellular factors may provide a mechanism for its participation in transcriptional regulation.
Aside from its transactivating capabilities, a number of different cytoplasmic signal transduction cascades appear to be affected by HBx, including Ras-Raf-mitogen-activated protein kinase (Ras-Raf-MAPK), extracellular signal-regulated kinase (ERK), the stress-activated protein kinases/NH2-terminal-Jun kinase (SAPK/JNK), protein kinase B (PKB/Akt), and the Janus kinase/STAT (JAK/STAT) (Benn et al., 1996; Bouchard and Schneider, 2004). Recently, Bouchard et al. (Bouchard et al., 2001) demonstrated that HBx could stimulate cellular calcium signaling pathways, resulting in the release of Ca2+ ions into the cytosol. This led to the activation of focal adhesion kinase (FAK) and proline-rich tyrosine kinase 2 (Pyk2), and the subsequent activation of Src tyrosine kinases. This, in turn, resulted in the induction of downstream signaling pathways such as Ras-Raf-MAPK pathway. Such a model for HBx function is attractive because Ca2+ affects so many cellular processes, including transcription, cell cycle control, and apoptosis, many of which have been suggested to be affected by HBx.
HBx has also been implicated in the development of HCC, yet its link to the progression of HCC remains controversial (Kim et al., 1991; Lee et al., 1990; Madden et al., 2001; Yu et al., 1999). It is generally viewed that HBx can promote cellular transformation by either disrupting cell signaling pathways involved in transcription, apoptosis, and cell proliferation (Benn and Schneider, 1995; Bouchard and Schneider, 2004; Bouchard et al., 2001; Chirillo et al., 1997). Alternatively, HBx could stimulate viral replication (Leupin et al., 2005) which may lead to an enhanced immune response against the virus. This will result in the continual destruction and regeneration of hepatocytes due to chronic liver inflammation and thus increase the chance of genetic mutations (Chisari et al., 1989).
HBx has been found to be overexpressed in human HCC and its overexpression in hepatocyte cultures and transgenic mice resulted in a tumorigenic phenotype in some model systems, although this remains controversial (Kim et al., 1991; Yu et al., 1999). Some evidence suggested that HBx interacts with the Ras-Raf-MAPK pathway to promote cell growth and activity (Wang et al., 1997). In addition, multiple pathways of DNA repairs are also comprised by HBx. Several groups have demonstrated that an interaction between HBx and the DNA repair protein, damaged DNA-binding proteins (DDB) 1 could stimulate viral replication (Leupin et al., 2005; Sitterlin et al., 2000). More recently, it was found that both HBx and telomerase were highly expressed in hepatoma tissue and liver cirrhosis tissues (Zhang et al., 2005) and that HBx could upregulate the expression and activity of hTERT, the catalytic subunit of telomerase (Zhang et al., 2005). These findings suggest that HBx expression may play a role in the development of HCC by modulating telomerase activity, given that telomerase activation has been associated with late stage tumors (Kim et al., 1994).
Covalently Closed Circular DNA
The HBV CCC DNA exists as an episome in the nucleus with an estimated copy number of 5–50 molecules per infected cell (Locarnini and Mason, 2006; Wu et al., 1990; Zhang et al., 2003). It is organized as a minichromosome (Newbold et al., 1995) and is regulated by epigenetic changes (see below). It serves as the sole intranuclear transcriptional template for the synthesis of viral RNAs. As such, a critical step in HBV infection is the establishment and maintenance of a pool of viral CCC DNA. However, very little is known about the factors that may be involved in CCC DNA formation. The only known viral protein to regulate CCC DNA formation is the viral large surface (LS) protein (Gao and Hu, 2007; Lenhoff and Summers, 1994; Summers et al., 1990), which was shown to suppress CCC DNA amplification by using a negative-feedback mechanism. When CCC DNA levels are low, viral transcription and protein expression (including LS protein) are low, and the mature, RC-containing, nucleocapsids enter the recycling pathway where the RC DNA is converted to CCC DNA, thus increasing the pool of CCC DNA. As the CCC DNA level increases, LS protein will accumulate which will result the preferential secretion of the mature nucleocapsids, over its recycling back to the nucleus.
Similarly, virtually nothing is known about the potential host factors involved in CCC DNA formation. The observation that the liver of HBV transgenic mice failed to accumulate detectable levels of CCC DNA, despite the presence of high levels of RC DNA precursor, led to the suggestion that CCC formation may require species-specific host factors (Guidotti et al., 1995). However, HBV transgenic mouse that lack HNF1α synthesized low levels of nuclear CCC DNA, suggesting that the physiological state of the host hepatocytes may somehow modulate CCC DNA formation (Raney et al., 2001). Also, since the conversion of RC DNA to CCC DNA can be thought of as a DNA damage response, it has been speculated that host cell DNA repair enzymes may, in part, be involved in this step. However no specific host factors that participate in CCC DNA formation have thus far been identified. Alternatively, it is possible that the viral RT may solely be responsible for this conversion step.
Biochemically, the formation of CCC DNA from RC DNA involves the completion of the plus-strand DNA, removal of the RT protein that is covalently linked to the 5’ end of the minus-strand DNA (Gao and Hu, 2007), and the ligation of the two DNA strands. Recent identification of a protein-free RC DNA from which the RT protein has been removed may provide a valuable window into the mechanism of RC DNA processing in CCC DNA formation (Gao and Hu, 2007; Guo et al., 2007).
It is widely accepted that the clearance of the HBV CCC DNA is essential to resolving chronic HBV infections. Although effective suppression of RC DNA synthesis by inhibition of RT can lead to a significant reduction in CCC DNA levels in patients with chronic HBV infection due either to cell death or intrinsic CCC DNA instability (Hu and Nguyen, 2004), most often, the infection is not resolved owing to the persistence of the HBV CCC DNA in the infected liver (Locarnini and Mason, 2006). This accounts for the rapid reappearance of HBV replication upon cessation of antiviral therapy (Locarnini and Mason, 2006). Thus, the successful eradication of chronic HBV infection requires either the permanent clearance of the viral CCC DNA from the infected cell (i.e. curing) or the elimination of the infected cell itself (i.e. killing).
Viral DNA Integration
HBV DNA has been reported to be integrated into host chromosomes in tissues of HCC patients or in cell lines derived from HCC (Seeger and Mason, 2000). Although HBV replication cannot be sustained by integration of the viral DNA into the host genome due to the disruption of the circular genome, it may still contribute to the development of HCC by several mechanisms. It was hypothesized that HBV could act as an insertional mutagen causing the activation of a proto-oncogene. In the woodchuck model, woodchuck hepatitis virus (WHV) DNA was found to integrate close to members of the myc proto-oncogene family and the resulting oncogene deregulation appeared to be a major cause of liver cancer (Fourel et al., 1990). In humans, HBV integration is random and plays a direct role in carcinogenesis in only a few cases, where the viral DNA is inserted next to important growth regulatory genes (Dejean et al., 1986; Wang et al., 1990). Viral DNA integration may also promote the general instability of chromosomal DNA in the infected cells, which may be exacerbated by the chronic inflammation in the infected liver (Hagen et al., 1994; Summers and Mason, 2004).
Regulation of HBV Transcription
Transcription of the HBV genes is controlled by four viral promoters and two enhancer elements. In vitro studies have demonstrated that the transcription of the HBV genome is regulated by a variety of transcription factors, such as the liver-enriched transcription factors C/EBP, retinoid X receptor alpha (RXRα), peroxisome proliferator-activated receptor alpha (PPARα), hepatocyte nuclear factor (HNF) 3, HNF4 and the ubiquitous transcription factors, Sp1, NFκB, p53 (Seeger et al., 2007). Studies have also indicated that members of the nuclear receptor family (i.e. HNF4) may be involved in the downregulation of some viral promoters (Yu and Mertz, 1997).
Recently, Pollicono et al. (Pollicino et al., 2006) reported that HBV replication seems to be regulated by the acetylation status of histones H3 and H4 bound to CCC DNA minichromosome. Furthermore, it appears that cellular acetyltransferases, p300 and CBP, and cellular deacetylases, HDAC1, were recruited in vitro and in vivo onto the HBV CCC DNA. Using histone deacetylase inhibitors, they saw an increase in HBV transcripts, which was paralleled by an increase of HBV replicative intermediates in the cytoplasm, as well as an increase in secreted HBV viral particles. These results suggested that HBV transcription can be modulated by epigenetic changes to the CCC DNA and provide new insight into how cellular host factors might regulate HBV replication. One potential mechanism by which host transcription factors can regulate transcription of CCC DNA may be through modifying the CCC DNA-bound histones via the recruitment of acetyltransferases and deacetylases to the viral promoters. Similarly, the role of HBx in the epigenetic regulation of CCC DNA can also be envisioned since it was reported to interact with p300 (Cougot et al., 2007).
Conclusion
The past two decades have seen tremendous advances in our understanding of the HBV life cycle. However, we have only just begun to unravel the complex interplay between the virus and host cell and its impact on viral replication and pathogenesis. The role of HBx during viral replication in vivo and its involvement in the development of HCC are still unknown. How is CCC DNA converted from RC DNA? Does this process involve cellular DNA repair enzymes? What is the cellular receptor(s) involved in HBV entry? What is the identity of the kinase(s) or phosphatase(s) that regulate core phosphorylation/dephosphorylation and thus viral DNA synthesis? How are mature HBV nucleocapsids selected for virion secretion? Are host proteins involved in this process? What is the identity of the putative cellular factor(s) required for HBV RT priming following RT-ε interaction? The recent discoveries that APOBEC3G and heme could inhibit HBV replication points to the existence of cellular factors that could counteract viral replication and therefore serve as natural antiviral defense mechanisms. Ultimately, understanding these complex virus-host interactions during an HBV infection will be critical to the development of novel antiviral therapies in the future.
References
- Albin C, Robinson WS. Protein kinase activity in hepatitis B virus. J Virol. 1980;34(1):297–302. doi: 10.1128/jvi.34.1.297-302.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrasa MI, Guo JT, Saputelli J, Mason WS, Seeger C. Does a cdc2 kinase-like recognition motif on the core protein of hepadnaviruses regulate assembly and disintegration of capsids? J Virol. 2001;75(4):2024–2028. doi: 10.1128/JVI.75.4.2024-2028.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R, Schaller H. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. Embo J. 1992;11(9):3413–3420. doi: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basagoudanavar SH, Perlman DH, Hu J. Regulation of hepadnavirus reverse transcription by dynamic nucleocapsid phosphorylation. J Virol. 2007;81(4):1641–1649. doi: 10.1128/JVI.01671-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benn J, Schneider RJ. Hepatitis B virus HBx protein deregulates cell cycle checkpoint controls. Proc Natl Acad Sci U S A. 1995;92(24):11215–11219. doi: 10.1073/pnas.92.24.11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benn J, Su F, Doria M, Schneider RJ. Hepatitis B virus HBx protein induces transcription factor AP-1 by activation of extracellular signal-regulated and c-Jun N-terminal mitogen-activated protein kinases. J Virol. 1996;70(8):4978–4985. doi: 10.1128/jvi.70.8.4978-4985.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum HE, Zhang ZS, Galun E, von Weizsacker F, Garner B, Liang TJ, Wands JR. Hepatitis B virus X protein is not central to the viral life cycle in vitro. J Virol. 1992;66(2):1223–1227. doi: 10.1128/jvi.66.2.1223-1227.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard MJ, Schneider RJ. The enigmatic X gene of hepatitis B virus. J Virol. 2004;78(23):12725–12734. doi: 10.1128/JVI.78.23.12725-12734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard MJ, Wang LH, Schneider RJ. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science. 2001;294(5550):2376–2378. doi: 10.1126/science.294.5550.2376. [DOI] [PubMed] [Google Scholar]
- Chen HS, Kaneko S, Girones R, Anderson RW, Hornbuckle WE, Tennant BC, Cote PJ, Gerin JL, Purcell RH, Miller RH. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J Virol. 1993;67(3):1218–1226. doi: 10.1128/jvi.67.3.1218-1226.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong JH, Yi M, Lin Y, Murakami S. Human RPB5, a subunit shared by eukaryotic nuclear RNA polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. Embo J. 1995;14(1):143–150. doi: 10.1002/j.1460-2075.1995.tb06984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirillo P, Pagano S, Natoli G, Puri PL, Burgio VL, Balsano C, Levrero M. The hepatitis B virus X gene induces p53-mediated programmed cell death. Proc Natl Acad Sci U S A. 1997;94(15):8162–8167. doi: 10.1073/pnas.94.15.8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisari FV, Klopchin K, Moriyama T, Pasquinelli C, Dunsford HA, Sell S, Pinkert CA, Brinster RL, Palmiter RD. Molecular pathogenesis of hepatocellular carcinoma in hepatitis B virus transgenic mice. Cell. 1989;59(6):1145–1156. doi: 10.1016/0092-8674(89)90770-8. [DOI] [PubMed] [Google Scholar]
- Cougot D, Wu Y, Cairo S, Caramel J, Renard CA, Levy L, Buendia MA, Neuveut C. The hepatitis B virus X protein functionally interacts with CREB-binding protein/p300 in the regulation of CREB-mediated transcription. J Biol Chem. 2007;282(7):4277–4287. doi: 10.1074/jbc.M606774200. [DOI] [PubMed] [Google Scholar]
- Cullen BR. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J Virol. 2006;80(3):1067–1076. doi: 10.1128/JVI.80.3.1067-1076.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub H, Blencke S, Habenberger P, Kurtenbach A, Dennenmoser J, Wissing J, Ullrich A, Cotten M. Identification of SRPK1 and SRPK2 as the major cellular protein kinases phosphorylating hepatitis B virus core protein. J Virol. 2002;76(16):8124–8137. doi: 10.1128/JVI.76.16.8124-8137.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejean A, Bougueleret L, Grzeschik KH, Tiollais P. Hepatitis B virus DNA integration in a sequence homologous to v-erb-A and steroid receptor genes in a hepatocellular carcinoma. Nature. 1986;322(6074):70–72. doi: 10.1038/322070a0. [DOI] [PubMed] [Google Scholar]
- Doria M, Klein N, Lucito R, Schneider RJ. The hepatitis B virus HBx protein is a dual specificity cytoplasmic activator of Ras and nuclear activator of transcription factors. Embo J. 1995;14(19):4747–4757. doi: 10.1002/j.1460-2075.1995.tb00156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel G, Trepo C, Bougueleret L, Henglein B, Ponzetto A, Tiollais P, Buendia MA. Frequent activation of N-myc genes by hepadnavirus insertion in woodchuck liver tumours. Nature. 1990;347(6290):294–298. doi: 10.1038/347294a0. [DOI] [PubMed] [Google Scholar]
- Gao W, Hu J. Formation of hepatitis B virus covalently closed circular DNA: removal of genome-linked protein. J Virol. 2007;81(12):6164–6174. doi: 10.1128/JVI.02721-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich WH, Robinson WS. Hepatitis B virus contains protein attached to the 5' terminus of its complete DNA strand. Cell. 1980;21(3):801–809. doi: 10.1016/0092-8674(80)90443-2. [DOI] [PubMed] [Google Scholar]
- Guidotti LG, Matzke B, Schaller H, Chisari FV. High-level hepatitis B virus replication in transgenic mice. J Virol. 1995;69(10):6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Jiang D, Zhou T, Cuconati A, Block TM, Guo JT. Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation. J Virol. 2007;81(22):12472–12484. doi: 10.1128/JVI.01123-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen TM, Huang S, Curnutte J, Fowler P, Martinez V, Wehr CM, Ames BN, Chisari FV. Extensive oxidative DNA damage in hepatocytes of transgenic mice with chronic active hepatitis destined to develop hepatocellular carcinoma. Proc Natl Acad Sci U S A. 1994;91(26):12808–12812. doi: 10.1073/pnas.91.26.12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann-Stuhler C, Prange R. Hepatitis B virus large envelope protein interacts with gamma2-adaptin, a clathrin adaptor-related protein. J Virol. 2001;75(11):5343–5351. doi: 10.1128/JVI.75.11.5343-5351.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch RC, Lavine JE, Chang LJ, Varmus HE, Ganem D. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as wel as for reverse transcription. Nature. 1990;344(6266):552–555. doi: 10.1038/344552a0. [DOI] [PubMed] [Google Scholar]
- Hu J, Anselmo D. In vitro reconstitution of a functional duck hepatitis B virus reverse transcriptase: posttranslational activation by Hsp90. J Virol. 2000;74(24):11447–11455. doi: 10.1128/jvi.74.24.11447-11455.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Boyer M. Hepatitis B virus reverse transcriptase and epsilon RNA sequences required for specific interaction in vitro. J Virol. 2006;80(5):2141–2150. doi: 10.1128/JVI.80.5.2141-2150.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Flores D, Toft D, Wang X, Nguyen D. Requirement of heat shock protein 90 for human hepatitis B virus reverse transcriptase function. J Virol. 2004;78(23):13122–13131. doi: 10.1128/JVI.78.23.13122-13131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Nguyen D. Therapy for chronic hepatitis B: the earlier, the better? Trends Microbiol. 2004;12(10):431–433. doi: 10.1016/j.tim.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Hu J, Seeger C. Expression and characterization of hepadnavirus reverse transcriptases. Methods Enzymol. 1996a;275:195–208. doi: 10.1016/s0076-6879(96)75013-9. [DOI] [PubMed] [Google Scholar]
- Hu J, Seeger C. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proceedings of the National Academy of Sciences of the United States of America. 1996b;93(3):1060–1064. doi: 10.1073/pnas.93.3.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Toft D, Anselmo D, Wang X. In vitro reconstitution of functional hepadnavirus reverse transcriptase with cellular chaperone proteins. J Virol. 2002;76(1):269–279. doi: 10.1128/JVI.76.1.269-279.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Toft DO, Seeger C. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. Embo J. 1997;16(1):59–68. doi: 10.1093/emboj/16.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau JH, Ting LP. Phosphorylation of the core protein of hepatitis B virus by a 46-kilodalton serine kinase. J Virol. 1998;72(5):3796–3803. doi: 10.1128/jvi.72.5.3796-3803.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kian Chua P, Lin MH, Shih C. Potent inhibition of human Hepatitis B virus replication by a host factor Vps4. Virology. 2006;354(1):1–6. doi: 10.1016/j.virol.2006.07.018. [DOI] [PubMed] [Google Scholar]
- Kim CM, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351(6324):317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266(5193):2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Kuroki K, Cheung R, Marion PL, Ganem D. A cell surface protein that binds avian hepatitis B virus particles. J Virol. 1994;68(4):2091–2096. doi: 10.1128/jvi.68.4.2091-2096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroki K, Eng F, Ishikawa T, Turck C, Harada F, Ganem D. gp180, a host cell glycoprotein that binds duck hepatitis B virus particles, is encoded by a member of the carboxypeptidase gene family. J Biol Chem. 1995;270(25):15022–15028. doi: 10.1074/jbc.270.25.15022. [DOI] [PubMed] [Google Scholar]
- Lecossier D, Bouchonnet F, Clavel F, Hance AJ. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science. 2003;300(5622):1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- Lee TH, Finegold MJ, Shen RF, DeMayo JL, Woo SL, Butel JS. Hepatitis B virus transactivator X protein is not tumorigenic in transgenic mice. J Virol. 1990;64(12):5939–5947. doi: 10.1128/jvi.64.12.5939-5947.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhoff RJ, Summers J. Coordinate regulation of replication and virus assembly by the large envelope protein of an avian hepadnavirus. J Virol. 1994;68(7):4565–4571. doi: 10.1128/jvi.68.7.4565-4571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leupin O, Bontron S, Schaeffer C, Strubin M. Hepatitis B virus X protein stimulates viral genome replication via a DDB1-dependent pathway distinct from that leading to cell death. J Virol. 2005;79(7):4238–4245. doi: 10.1128/JVI.79.7.4238-4245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Tong S, Lee HB, Perdigoto AL, Spangenberg HC, Wands JR. Glycine decarboxylase mediates a postbinding step in duck hepatitis B virus infection. J Virol. 2004;78(4):1873–1881. doi: 10.1128/JVI.78.4.1873-1881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Tong S, Wands JR. Identification and expression of glycine decarboxylase (p120) as a duck hepatitis B virus pre-S envelope-binding protein. J Biol Chem. 1999;274(39):27658–27665. doi: 10.1074/jbc.274.39.27658. [DOI] [PubMed] [Google Scholar]
- Li JS, Tong SP, Wands JR. Characterization of a 120-Kilodalton pre-S-binding protein as a candidate duck hepatitis B virus receptor. J Virol. 1996;70(9):6029–6035. doi: 10.1128/jvi.70.9.6029-6035.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Hu J. Inhibition of Hepadnavirus Reverse Transcriptase-{epsilon} RNA Interaction by Porphyrin Compounds. J Virol. 2007 doi: 10.1128/JVI.02147-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locarnini S, Mason WS. Cellular and virological mechanisms of HBV drug resistance. J Hepatol. 2006;44(2):422–431. doi: 10.1016/j.jhep.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Madden CR, Finegold MJ, Slagle BL. Hepatitis B virus X protein acts as a tumor promoter in development of diethylnitrosamine-induced preneoplastic lesions. J Virol. 2001;75(8):3851–3858. doi: 10.1128/JVI.75.8.3851-3858.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire HF, Hoeffler JP, Siddiqui A. HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science. 1991;252(5007):842–844. doi: 10.1126/science.1827531. [DOI] [PubMed] [Google Scholar]
- Melegari M, Wolf SK, Schneider RJ. Hepatitis B virus DNA replication is coordinated by core protein serine phosphorylation and HBx expression. J Virol. 2005;79(15):9810–9820. doi: 10.1128/JVI.79.15.9810-9820.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassal M. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J Virol. 1992;66(7):4107–4116. doi: 10.1128/jvi.66.7.4107-4116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbold JE, Xin H, Tencza M, Sherman G, Dean J, Bowden S, Locarnini S. The covalently closed duplex form of the hepadnavirus genome exists in situ as a heterogeneous population of viral minichromosomes. J Virol. 1995;69(6):3350–3357. doi: 10.1128/jvi.69.6.3350-3357.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DH, Gummuluru S, Hu J. Deamination-independent inhibition of hepatitis B virus reverse transcription by APOBEC3G. J Virol. 2007;81(9):4465–4472. doi: 10.1128/JVI.02510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman DH, Berg EA, O'Connor PB, Costello CE, Hu J. Reverse transcription-associated dephosphorylation of hepadnavirus nucleocapsids. Proc Natl Acad Sci U S A. 2005;102(25):9020–9025. doi: 10.1073/pnas.0502138102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack JR, Ganem D. Site-specific RNA binding by a hepatitis B virus reverse transcriptase initiates two distinct reactions: RNA packaging and DNA synthesis. J Virol. 1994;68(9):5579–5587. doi: 10.1128/jvi.68.9.5579-5587.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollicino T, Belloni L, Raffa G, Pediconi N, Squadrito G, Raimondo G, Levrero M. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology. 2006;130(3):823–837. doi: 10.1053/j.gastro.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Pugh J, Zweidler A, Summers J. Characterization of the major duck hepatitis B virus core particle protein. J Virol. 1989;63(3):1371–1376. doi: 10.1128/jvi.63.3.1371-1376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raney AK, Eggers CM, Kline EF, Guidotti LG, Pontoglio M, Yaniv M, McLachlan A. Nuclear covalently closed circular viral genomic DNA in the liver of hepatocyte nuclear factor 1 alpha-null hepatitis B virus transgenic mice. J Virol. 2001;75(6):2900–2911. doi: 10.1128/JVI.75.6.2900-2911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosler C, Kock J, Kann M, Malim MH, Blum HE, Baumert TF, von Weizsacker F. APOBEC-mediated interference with hepadnavirus production. Hepatology. 2005;42(2):301–309. doi: 10.1002/hep.20801. [DOI] [PubMed] [Google Scholar]
- Rost M, Mann S, Lambert C, Doring T, Thome N, Prange R. Gamma-adaptin, a novel ubiquitin-interacting adaptor, and Nedd4 ubiquitin ligase control hepatitis B virus maturation. J Biol Chem. 2006;281(39):29297–29308. doi: 10.1074/jbc.M603517200. [DOI] [PubMed] [Google Scholar]
- Schlicht HJ, Bartenschlager R, Schaller H. The duck hepatitis B virus core protein contains a highly phosphorylated C terminus that is essential for replication but not for RNA packaging. J Virol. 1989;63(7):2995–3000. doi: 10.1128/jvi.63.7.2995-3000.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64(1):51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger C, Mason WS, Zoulim F. Hepadnaviruses. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2977–3029. [Google Scholar]
- Seto E, Mitchell PJ, Yen TS. Transactivation by the hepatitis B virus X protein depends on AP-2 and other transcription factors. Nature. 1990;344(6261):72–74. doi: 10.1038/344072a0. [DOI] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418(6898):646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Sitterlin D, Bergametti F, Transy C. UVDDB p127-binding modulates activities and intracellular distribution of hepatitis B virus X protein. Oncogene. 2000;19(38):4417–4426. doi: 10.1038/sj.onc.1203771. [DOI] [PubMed] [Google Scholar]
- Summers J, Mason WS. Replication of the genome of a hepatitis B--like virus by reverse transcription of an RNA intermediate. Cell. 1982;29(2):403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- Summers J, Mason WS. Residual integrated viral DNA after hepadnavirus clearance by nucleoside analog therapy. Proc Natl Acad Sci U S A. 2004;101(2):638–640. doi: 10.1073/pnas.0307422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers J, Smith PM, Horwich AL. Hepadnavirus envelope proteins regulate covalently closed circular DNA amplification. J Virol. 1990;64(6):2819–2824. doi: 10.1128/jvi.64.6.2819-2824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Li J, Wands JR. Interaction between duck hepatitis B virus and a 170-kilodalton cellular protein is mediated through a neutralizing epitope of the pre-S region and occurs during viral infection. J Virol. 1995;69(11):7106–7112. doi: 10.1128/jvi.69.11.7106-7112.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli P, Mangeat B, Jost S, Vianin S, Trono D. Inhibition of hepatitis B virus replication by APOBEC3G. Science. 2004;303(5665):1829. doi: 10.1126/science.1092066. [DOI] [PubMed] [Google Scholar]
- Tuttleman JS, Pourcel C, Summers J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell. 1986;47(3):451–460. doi: 10.1016/0092-8674(86)90602-1. [DOI] [PubMed] [Google Scholar]
- Urban S, Breiner KM, Fehler F, Klingmuller U, Schaller H. Avian hepatitis B virus infection is initiated by the interaction of a distinct pre-S subdomain with the cellular receptor gp180. J Virol. 1998;72(10):8089–8097. doi: 10.1128/jvi.72.10.8089-8097.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GH, Seeger C. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell. 1992;71(4):663–670. doi: 10.1016/0092-8674(92)90599-8. [DOI] [PubMed] [Google Scholar]
- Wang GH, Zoulim F, Leber EH, Kitson J, Seeger C. Role of RNA in enzymatic activity of the reverse transcriptase of hepatitis B viruses. J Virol. 1994;68(12):8437–8442. doi: 10.1128/jvi.68.12.8437-8442.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HD, Trivedi A, Johnson DL. Hepatitis B virus X protein induces RNA polymerase III-dependent gene transcription and increases cellular TATA-binding protein by activating the Ras signaling pathway. Mol Cell Biol. 1997;17(12):6838–6846. doi: 10.1128/mcb.17.12.6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chenivesse X, Henglein B, Brechot C. Hepatitis B virus integration in a cyclin A gene in a hepatocellular carcinoma. Nature. 1990;343(6258):555–557. doi: 10.1038/343555a0. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Sorensen EM, Naito A, Schott M, Kim S, Ahlquist P. Involvement of host cellular multivesicular body functions in hepatitis B virus budding. Proc Natl Acad Sci U S A. 2007;104(24):10205–10210. doi: 10.1073/pnas.0704000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Bronsema V, Bartos H, Bosserhoff A, Bartenschlager R, Schaller H. Hepadnavirus P protein utilizes a tyrosine residue in the TP domain to prime reverse transcription. J Virol. 1994;68(5):2994–2999. doi: 10.1128/jvi.68.5.2994-2999.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TT, Coates L, Aldrich CE, Summers J, Mason WS. In hepatocytes infected with duck hepatitis B virus, the template for viral RNA synthesis is amplified by an intracellular pathway. Virology. 1990;175(1):255–261. doi: 10.1016/0042-6822(90)90206-7. [DOI] [PubMed] [Google Scholar]
- Yeh CT, Ou JH. Phosphorylation of hepatitis B virus precore and core proteins. J Virol. 1991;65(5):2327–2331. doi: 10.1128/jvi.65.5.2327-2331.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu DY, Moon HB, Son JK, Jeong S, Yu SL, Yoon H, Han YM, Lee CS, Park JS, Lee CH, Hyun BH, Murakami S, Lee KK. Incidence of hepatocellular carcinoma in transgenic mice expressing the hepatitis B virus X-protein. J Hepatol. 1999;31(1):123–132. doi: 10.1016/s0168-8278(99)80172-x. [DOI] [PubMed] [Google Scholar]
- Yu M, Summers J. Multiple functions of capsid protein phosphorylation in duck hepatitis B virus replication. J Virol. 1994;68(7):4341–4348. doi: 10.1128/jvi.68.7.4341-4348.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Mertz JE. Differential regulation of the pre-C and pregenomic promoters of human hepatitis B virus by members of the nuclear receptor superfamily. J Virol. 1997;71(12):9366–9374. doi: 10.1128/jvi.71.12.9366-9374.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Dong N, Zhang H, You J, Wang H, Ye L. Effects of hepatitis B virus X protein on human telomerase reverse transcriptase expression and activity in hepatoma cells. J Lab Clin Med. 2005;145(2):98–104. doi: 10.1016/j.lab.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Zhang YY, Zhang BH, Theele D, Litwin S, Toll E, Summers J. Single-cell analysis of covalently closed circular DNA copy numbers in a hepadnavirus-infected liver. Proc Natl Acad Sci U S A. 2003;100(21):12372–12377. doi: 10.1073/pnas.2033898100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J Virol. 1994;68(3):2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoulim F, Seeger C. Reverse transcription in hepatitis B viruses is primed by a tyrosine residue of the polymerase. J Virol. 1994;68(1):6–13. doi: 10.1128/jvi.68.1.6-13.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]