Abstract
The androgen receptor (AR) binds to and activates transcription of specific genes in response to its cognate steroid hormone, dihydrotestosterone. Transcriptional activation by the DNA-bound AR is accomplished with the help of a variety of coactivator proteins. For example, the p160 coactivators bind directly to AR and recruit additional coactivators such as the histone acetyltransferase p300 and the histone methyltransferase CARM1. The current study tested whether CARM1 can cooperate with other types of coactivator proteins. Recently it was shown that β-catenin can also bind directly to and serve as a coactivator for AR. Here it is shown that CARM1 binds to β-catenin and can function in synergy with β-catenin and p300 as coactivators for AR. The methyltransferase activity of CARM1 is important for its synergistic coactivator function with β-catenin. The synergistic coactivator function of β-catenin and CARM1 is not restricted to steroid receptors because these two coactivators can also act synergistically with another type of DNA binding transcriptional activator, LEF-1/TCF-4.
β-catenin is a critical component of at least two signaling pathways, one that regulates cell-cell adhesion and organization of the actin cytoskeleton, and another that mediates Wnt/ Wingless (Wg)1 signaling (1–3). β-catenin protein levels are usually maintained at a low level through active degradation. The glycogen synthase kinase complex constitutively phosphorylates β-catenin, targeting it for ubiquitination and degradation by proteasomes. Activation of the Wnt/Wg pathway leads to inhibition of glycogen synthase kinase activity and thus stabilizes β-catenin protein. As its level increases some β-catenin accumulates in the nucleus, where it binds to and enhances transcriptional activation by at least two different classes of enhancer element-binding transcriptional activator proteins: the lymphoid enhancer factor/T-cell factor (LEF/TCF) proteins (4, 5) and nuclear receptors such as the androgen receptor (AR) and retinoic acid receptor (6, 7). Thus, at the distal end of the Wnt/Wg and nuclear receptor signaling pathways β-catenin can act as a transcriptional coactivator protein.
Transcriptional coactivators are a diverse group of proteins that are recruited to a specific promoter by interaction with specific enhancer-binding proteins (2, 8, 9). Coactivators participate in chromatin remodeling and recruitment of RNA polymerase II to the promoter. Many coactivators function as complexes, in which one component (hereafter, a primary co-activator) anchors the complex to the enhancer-binding protein, while other components (hereafter, secondary coactivators) mediate the transcriptional activation process (10, 11). Binding of β-catenin to LEF/TCF causes displacement of core-pressor complexes containing histone deacetylases (5). In addition, β-catenin also recruits and acts synergistically as a coactivator with p300 or CBP (2, 12–14). These two related proteins acetylate histones and interact with components of the basal transcription machinery, such as TFIIB and the TATA-box binding protein, a component of TFIID that helps to recruit RNA polymerase (15–17). β-catenin can also bind directly to the TATA-box-binding protein (18).
To explore further the mechanism of coactivator function by β-catenin, we tested whether β-catenin can cooperate with another type of coactivator protein, the protein methyltransferase CARM1. CARM1 acts as a secondary coactivator for nuclear receptors, including steroid receptors such as AR (10). CARM1 is recruited to the promoter through its association with primary coactivators called p160 coactivators, which include GRIP1, steroid receptor coactivator-1, and activator of thyroid and retinoic acid receptors. GRIP1, CARM1, and p300 or CBP can form a ternary complex, and these three coactivators function synergistically to enhance the activity of nuclear receptors (19, 20). Furthermore, mutations that eliminated the methyltransferase activity of CARM1 also severely compromised the ability of CARM1 to cooperate with GRIP1 as coactivators for nuclear receptors (10, 20). CARM1 methylates specific arginine residues in the N-terminal tail of histone H3 in vitro (21). Chromatin immunoprecipitation assays indicated that methylation of the same arginine residue of histone H3 also occurs in vivo at specific promoters as part of the transcriptional activation process (22). These results suggest that histone methylation may cooperate with histone acetylation in chromatin remodeling.
In the current study we found a highly synergistic cooperation between CARM1 and β-catenin as coactivators for two different classes of enhancer-binding proteins. To investigate the mechanism of this synergy, we also tested the role of the methyltransferase activity of CARM1, whether CARM1 and β-catenin form a complex in vivo, and whether p300 can further contribute to the synergy between CARM1 and β-catenin. Our results further elucidate the mechanism of coactivator function by β-catenin and indicate that the coactivator role of CARM1 extends beyond its currently known cooperation with the p160 coactivators and nuclear receptors.
MATERIALS AND METHODS
Plasmids
The following luciferase reporter gene plasmids were described previously: for AR, MMTV-LUC (23); and for TCF-4, pGL3-OT containing three wild type TCF-4 binding elements and pGL3-OF with mutant TCF-4 binding sites (24). Mammalian expression vectors for the following proteins were described previously as follows: pSVAR0 encoding AR (25); pSG5.HA-CARM1 (10) and pSG5.HA-CARM1(E267Q) (20) encoding wild type and mutant CARM1, respectively; and pCMV-p300 (26) encoding p300. Mammalian expression vector pSG5.HA-bCAT was constructed by inserting the PCR-amplified chicken β-catenin coding region (27) into EcoRI and BamHI sites of the pSG5.HA vector (10). Bacterial expression vector pGEX-4T1.GRIP1C, encoding glutathione S-transferase fused to GRIP1 amino acids 1122–1462 was described previously (10).
Transient Transfection Assays
CV-1 cells (28) were transfected and luciferase assays were performed as described previously (29) except that 24-well or 96-well plates were seeded with 1 × 104 or 5 × 103 cells, respectively, and total DNA in each transfection was adjusted to 500 or 150 ng by addition of pSG5.HA vector. Where indicated, 20 nm dihydrotestosterone (DHT) was added after transfection to activate AR. Cells were harvested 48 h after transfection, and luciferase activity was determined. Luciferase activity is shown as the mean and standard deviation from four transfected cultures. Transfection efficiency was monitored by co-transfection with a β-galactosidase vector controlled by a Rous sarcoma virus promoter. However, because some of the coactivators used also enhance expression of this vector, the luciferase activity was not normalized to the β-galactosidase activity. Instead, multiple independent experiments were used to confirm results.
Protein-Protein Interactions
Binding of wild type or mutant CARM1, translated in vitro as 35S-labeled proteins, to the C-terminal region of GRIP1 fused to glutathione S-transferase were performed as described previously (30). For co-immunoprecipitation experiments, COS-7 cells (28) were seeded at a density of 1.5 × 106 per 100-mm culture dish and transfected with Targefect F-2 reagent (Targeting Systems) using 5 μg of each expression plasmid. After 48 h of growth in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum, cells were harvested with 1.0 ml of RIPA buffer (10 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% (v/v) Triton X-100, 0.1% (w/v) SDS). Cell lysate (0.45 ml) was incubated on a rotator with rabbit anti-β-catenin IgG or normal rabbit IgG (Santa Cruz Biotechnology) at a final concentration of 4 μg/ml for 12–16 h at 4 °C. After addition of 20 μl of a 50% slurry of protein A/G-agarose beads (Santa Cruz Biotechnology), the incubation was continued for 2 h. The immune complex was collected and washed three times by centrifugation with RIPA buffer. Immunoprecipitated proteins were analyzed by SDS-PAGE and immunoblot (31) using 0.1 μg/ml rat monoclonal antibody 3F10 (Roche Molecular Biochemicals) against the hemagglutinin (HA) epitope as primary antibody and horse-radish peroxidase-conjugated anti-rat IgG (Santa Cruz Biotechnology) as secondary antibody.
RESULTS
Synergistic Enhancement of AR Function by β-Catenin and CARM1
β-catenin was previously shown to bind to and enhance transcriptional activation by AR (6). CARM1 binds to p160 coactivators, which target the coactivator complex to activated nuclear receptors (10). Because both β-catenin and CARM1 can function as coactivators with nuclear receptors, we tested their ability to cooperate as coactivators. Activation of the Wnt/Wg signaling pathway normally results in stabilization of β-catenin protein and thus increased cellular levels of β-catenin. An alternative method for increasing β-catenin protein levels is by exogenous expression from a β-catenin expression vector (2). Transient transfection of CV-1 cells with a luciferase reporter gene controlled by the mouse mammary tumor virus promoter (MMTV-LUC) and an expression vector encoding AR led to expression of luciferase, which depended on the presence of DHT, the activating hormone for AR (Fig. 1, compare lanes 1 and 9). In the presence of DHT co-expression of β-catenin enhanced reporter gene activity about 2-fold (lane 2), and co-expression of CARM1 with AR also modestly enhanced luciferase activity (lanes 6–8). When added together, CARM1 and β-catenin enhanced luciferase activity up to 33-fold (lanes 3–5) compared with AR alone (lane 1). The synergistic effect of β-catenin and CARM1 was completely dependent on the presence of DHT (lanes 9–12), demonstrating that they were not directly or non-specifically activating the reporter gene but were acting as coactivators for the hormone-activated AR.
FIG. 1. Synergistic coactivator function by β-catenin and CARM1 with AR.
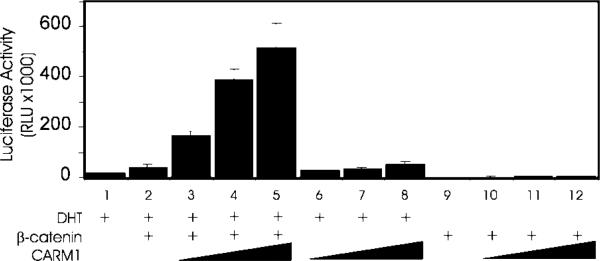
CV-1 cells were transfected in 24-well plates with 62.5 ng of MMTV-LUC reporter gene, 25 ng of AR expression vector, and the following coactivator expression vectors as indicated: β-catenin, 62.5 ng; and CARM1, 125, 188, or 250 ng. Transfected cells were grown with or without DHT as indicated, and luciferase activity was measured. Results shown are representative of five independent experiments.
Mechanism of Coactivator Synergy between CARM1 and β-Catenin
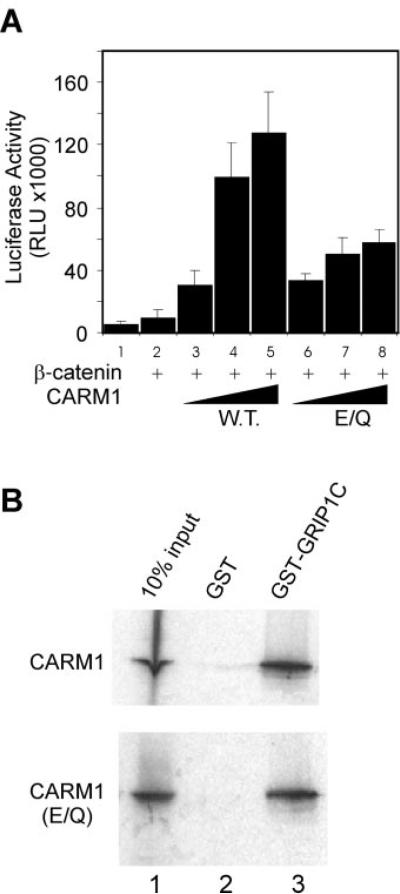
CARM1 is an arginine-specific protein methyltransferase that can methylate histone H3 in vitro (10, 21) and in vivo (22). Substitution of a CARM1 mutant lacking histone H3 methyltransferase activity for wild type CARM1 in the transient transfection assays resulted in a substantial reduction in the ability of CARM1 to function synergistically as a coactivator with β-catenin (Fig. 2A). Mutant and wild type proteins were expressed at similar levels (20). Furthermore, the mutant and wild type CARM1 proteins bound equally in vitro to the C-terminal region of GRIP1 (Fig. 2B), indicating the structural integrity of the mutant CARM1 protein. Thus, the reduction in coactivator activity observed with the CARM1 mutant indicates that the methyltransferase activity of CARM1 plays an important role in its coactivator function; however, part of the coactivator function was independent of the methyltransferase activity, indicating that CARM1 enhances AR function by additional mechanisms, such as protein-protein interaction with the transcription machinery.
FIG. 2. Role of the methyltransferase activity of CARM1 in coactivator synergy with β-catenin.
A, CV-1 cells were transfected in 96-well plates with 20 ng of MMTV-LUC, 1 ng of AR expression vector, and the following coactivator expression vectors as indicated: β-catenin, 25 ng; CARM1 wild type (WT) or E267Q mutant (E/Q), 20, 50, or 100 ng. Transfected cultures were grown with DHT, and lucifer-ase activity was determined. Results are representative of two independent experiments. B, CARM1 wild type or E267Q (E/Q) mutant protein was translated in vitro in the presence of [35S]methionine and tested for binding to beads containing a C-terminal fragment of GRIP1 (amino acids 1122–1462) fused to glutathione S-transferase (GST-GRIP1C) or to glutathione S-transferase (GST) alone. Bound proteins were eluted (lanes 2–3) and analyzed by SDS-polyacrylamide gel electrophoresis and autoradiography. In the lane 1, 10% of the input protein was loaded on the gel.
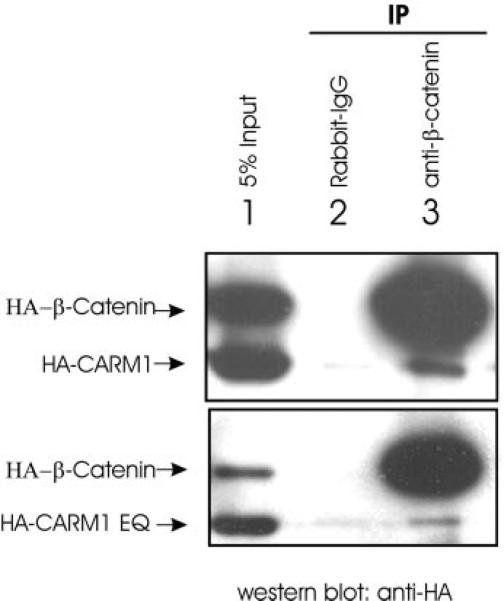
To explore the mechanism of synergy between β-catenin and CARM1, we tested whether these two proteins can form a complex in vivo. When HA-tagged CARM1 and HA-tagged β-catenin were co-expressed in COS-7 cells, both proteins were detected by immunoblot using antibodies against the HA epitope (Fig. 3, lane 1). Antiserum against β-catenin precipitated a large amount of β-catenin and a small amount of CARM1 (lane 3), whereas normal rabbit IgG failed to precipitate either protein (lane 2). Both wild type CARM1 (top panel) and the methyltransferase-deficient mutant of CARM1 (bottom panel) interacted with β-catenin, and results of six independent co-immunoprecipitation experiments indicated that there was no appreciable difference in the interaction of wild type and mutant CARM1 with β-catenin. Because β-catenin can bind directly to AR and CARM1 can bind to β-catenin in vivo, β-catenin nin may recruit CARM1 to hormone-activated AR bound to its target promoters.
FIG. 3. Co-immunoprecipitation of β-catenin and CARM1.
COS-7 cells were transfected with expression vectors for HA-tagged β-catenin and CARM1 wild type (top panel) or EQ mutant (bottom panel). After cell lysates were immunoprecipitated (IP) with antiserum against β-catenin or normal rabbit IgG, immunoprecipitates were resolved by SDS-PAGE, and CARM1 and β-catenin were visualized by immunoblot using antibodies against the HA-epitope. A portion of the original cell lysate was reserved and loaded as input. Results shown are representative of six independent experiments.
Synergistic Enhancement of AR Function by the Three Coactivators β-Catenin, CARM1, and p300
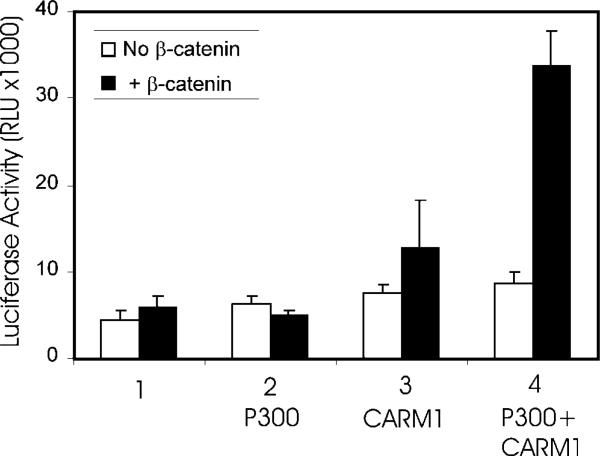
The related histone acetyltransferases p300 and CBP have been shown previously to cooperate with β-catenin as coactivators (2, 12–14). With nuclear receptors, CARM1 and p300 have also been shown to act synergistically as secondary coactivators in cooperation with the primary p160 coactivators (19, 20). We thus tested for synergistic cooperation among β-catenin, CARM1, and p300. Our previous studies demonstrated that use of low levels of nuclear receptor expression vectors was conducive for observing synergy among three different coactivators (20, 30). Under these conditions, none of the three coactivators individually caused a significant enhancement of reporter gene activation by AR (Fig. 4). Co-expression of p300 or CARM1 with β-catenin also had little or no effect. However, co-expression of all three coactivators together caused a 7-fold enhancement of AR activity. Thus, as demonstrated for primary p160 coactivators (19), the primary coactivator function of β-catenin may involve recruitment of secondary coactivators such as CARM1 and p300 or CBP.
FIG. 4. Synergistic enhancement of AR function by β-catenin, CARM1, and p300.
CV-1 cells were transfected in 96-well plates with 20 ng of MMTV-LUC, 0.1 ng of AR expression vector, and the following coactivator expression vectors as indicated: β-catenin, 25 ng; CARM1, 50 ng; and p300, 50 ng. Transfected cultures were grown with DHT, and luciferase activity was determined. Results are representative of four independent experiments.
Synergy between CARM1 and β-Catenin with another Class of Enhancer-binding Proteins
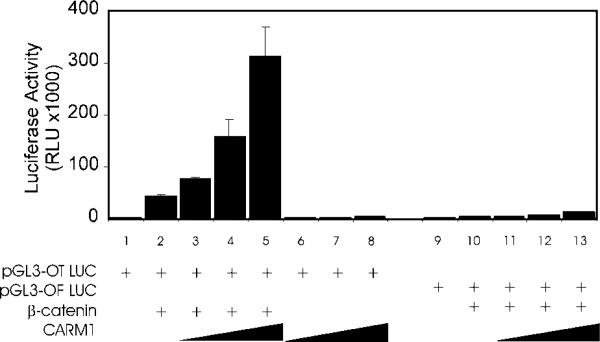
β-catenin serves as a coactivator for the LEF/TCF family of enhancer-binding proteins as well as AR (4, 5). After observing the synergistic coactivator function of CARM1 and β-catenin with AR, we tested whether these two coactivators could also cooperate to enhance transcriptional activation by LEF-1/TCF-4. By immunoblot, we determined that CV-1 cells contain endogenous TCF-4 (data not shown). When luciferase reporter gene pGL3-OT, which contains three tandem wild type TCF-4 binding elements, was transiently transfected into CV-1 cells, a low level of luciferase activity was observed (Fig. 5, lane 1). This activity was enhanced 16-fold by co-expression of β-catenin (lane 2), and a further enhancement up to 130-fold was observed when CARM1 was co-expressed with β-catenin (lanes 3–5). However, CARM1 alone failed to enhance the reporter gene activity (lanes 6–8). When reporter gene pGL3-OF, which contains mutant TCF-4 binding elements, was compared with pGL3-OT, co-expression of β-catenin and CARM1 caused a modest enhancement of pGL3-OF activity of up to 4-fold (lanes 9–13) as opposed to their enhancement up to 130-fold of pGL3-OT activity (lanes 1–5). Thus, the ability of β-catenin and CARM1 to enhance reporter gene activity depended on the recruitment of the enhancer binding protein, TCF-4, to the promoter.
Fig. 5. Synergistic coactivator function by β-catenin and CARM1 with TCF-4.
CV-1 cells were transfected in 96-well plates with 25 ng of pGL3-OT (containing wild type TCF-4 binding sites) or pGL3-OF (containing mutant TCF-4 binding sites) luciferase reporter plasmid and the following coactivator expression vectors as indicated: β-catenin, 25 ng; and CARM1, 25, 50, or 100 ng. Luciferase activities shown are representative of four independent experiments.
DISCUSSION
β-catenin was previously shown to function as a coactivator for enhancer-binding proteins from two different protein families: AR (6) and LEF-1/TCF-4 (4, 5). β-catenin binds directly to AR and TCF-4, and over-expression of β-catenin alone enhances the ability of these enhancer-binding proteins to activate transcription of their target genes. Thus, β-catenin acts as a primary coactivator and presumably helps to activate transcription by recruiting secondary coactivators, which remodel chromatin and/or recruit and activate RNA polymerase II. The fact that β-catenin binds p300 and CBP (2, 12–14) and can act cooperatively with these proteins as coactivators explains part of the mechanism by which β-catenin helps the enhancer-binding protein to transmit the activating signal to the transcription machinery. The current study showed that CARM1 can also bind to β-catenin and cooperate with β-catenin and p300 to enhance the ability of enhancer binding proteins to activate transcription of their target genes. These findings suggest that CARM1 is another part of the mechanism for activation of transcription employed by β-catenin.
That CARM1 is acting as a coactivator was shown by its inability to enhance reporter gene function unless the activated enhancer-binding protein was present and unless the promoter of the reporter gene contained binding sites for the enhancer-binding protein. That CARM1 is acting as a secondary coactivator was shown by its inability to enhance reporter gene activity unless the primary coactivator, in this case β-catenin, was present to recruit CARM1 to the promoter.
The E267Q mutation in CARM1 eliminated the methyltransferase activity (20) and dramatically reduced the coactivator function of CARM1 with β-catenin (Fig. 2A); however, the mutation did not affect the expression of CARM1 (20) or the binding of CARM1 to the C-terminal domain of GRIP1 (Fig. 2B) or to β-catenin (Fig. 3). The selective elimination of methyltransferase activity in this mutant allows us to conclude that the methyltransferase activity was important for the full coactivator function of CARM1 with β-catenin. Histone H3 is a good substrate for CARM1 in vitro and is a target of CARM1 methylation during gene activation in vivo (10, 21, 22). However, other proteins may be methylated in addition to or instead of histone H3 when CARM1 is recruited to some promoters. The fact that the elimination of the methyltransferase activity of CARM1 reduced but did not eliminate the coactivator function of CARM1 indicates that CARM1 uses another mechanism, in addition to protein methylation, to transmit the activating signal to the transcription machinery. Other domains of CARM1 may contribute to transcriptional activation through protein-protein interactions with components of the transcription machinery. By analogy, when CBP and p300 act as coactivators, they also use protein-protein interactions as well as their protein acetyltransferase activity to transmit the activating signal (15, 16).
CARM1 was previously shown to function as a secondary coactivator for nuclear receptors by cooperating with p160 primary coactivators (10, 19). The current study broadens the scope of the potential coactivator function of CARM1 in the cell: CARM1 can function as a coactivator for another class of enhancer-binding protein, TCF-4; and CARM1 can function in cooperation with a different class of primary coactivator, β-catenin. In future work it will be important to test even more broadly for combinatorial functional interactions of CARM1 with other types of coactivators and enhancer-binding proteins.
While we have shown here that β-catenin, CARM1, and p300 can function synergistically as coactivators for AR (Fig. 4), we have shown previously that p160 coactivators (such as GRIP1 and steroid receptor coactivator-1), CARM1, and p300 can also function synergistically as coactivators for AR and other nuclear receptors (20). Both p160 coactivators and β-catenin bind directly to AR and can thereby recruit their respective coactivator complexes to the promoter. Which of these coactivator complexes is used in any given situation presumably will depend on the relative cellular levels of p160 coactivator and β-catenin and the activation status of signaling pathways that regulate the activity of β-catenin (1–3) or p160 coactivators (32). Alternatively, it is conceivable that β-catenin and p160 coactivators could form a single coactivator complex with CARM1 and p300, which is more effective in mediating transcriptional activation than either of the ternary complexes mentioned above. Thus, future studies should test for physical interaction and functional synergy between p160 coactivators and β-catenin.
The wnt pathway and β-catenin are pivotal for driving growth in many types of cancer and during normal development (2, 33, 34). In the growth of epithelial appendages such as prostate glands, mammary glands, hair, and feathers, the androgen and estrogen pathways also have profound effects on new growth. The work presented here sets the stage for further exploration of whether sex hormone-regulated growth is mediated by the β-catenin pathway.
Acknowledgment
We thank Dr. Bert Vogelstein (Johns Hopkins University) for kindly providing the pGL3-OT and pGL3-OF plasmids.
Footnotes
This work was supported by United States Public Health Service Grants DK43093 (to M. R. S.), AR42177 (to C.-M. C.), and CA83716 (to R. B. W.) from the National Institutes of Health. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: Wg, Wingless; AR, androgen receptor; CARM, coactivator-associated arginine methyltransferase; CBP, CREB-binding protein; DHT, dihydrotestosterone; GRIP, glucocorticoid receptor-interacting protein; HA, hemagglutinin epitope tag; LEF, lymphoid enhancer factor; LUC, luciferase; MMTV, mouse mammary tumor virus; TCF, T-cell factor.
REFERENCES
- 1.Hecht A, Kemler R. EMBO Rep. 2000;1:24–28. doi: 10.1093/embo-reports/kvd012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morin PJ. Bioessays. 1999;21:1021–1030. doi: 10.1002/(SICI)1521-1878(199912)22:1<1021::AID-BIES6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 3.Eastman Q, Grosschedl R. Curr. Opin. Cell Biol. 1999;11:233–240. doi: 10.1016/s0955-0674(99)80031-3. [DOI] [PubMed] [Google Scholar]
- 4.Liu T, DeCostanzo AJ, Liu X, Wang H, Hallagan S, Moon RT, Malbon CC. Science. 2001;292:1718–1722. doi: 10.1126/science.1060100. [DOI] [PubMed] [Google Scholar]
- 5.Billin AN, Thirlwell H, Ayer DE. Mol. Cell. Biol. 2000;20:6882–6890. doi: 10.1128/mcb.20.18.6882-6890.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Truica CI, Byers S, Gelmann EP. Cancer Res. 2000;60:4709–4713. [PubMed] [Google Scholar]
- 7.Easwaran V, Pishvaian, Salimuddin M, Byers S. Curr. Biol. 1999;9:1415–1418. doi: 10.1016/s0960-9822(00)80088-3. [DOI] [PubMed] [Google Scholar]
- 8.Cheung P, Allis CD, Sassone-Corsi P. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 9.Glass CK, Rosenfeld MG. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 10.Chen D, Ma H, Hong H, Koh SS, Huang S-M, Schurter BT, Aswad DW, Stallcup MR. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 11.Stallcup MR. Oncogene. 2001;20:3014–3020. doi: 10.1038/sj.onc.1204325. [DOI] [PubMed] [Google Scholar]
- 12.Sun Y, Kolligs FT, Hottiger MO, Mosavin R, Fearon ER, Nabel GJ. Proc. Natl. Acad. Sci. U. S. A. 2000;97:12613–12618. doi: 10.1073/pnas.220158597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hecht A, Vleminckx K, Stemmler MP, van Roy F, Kemler R. EMBO J. 2000;19:1839–1850. doi: 10.1093/emboj/19.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyagishi M, Fujii R, Hatta M, Yoshida E, Araya N, Nagafuchi A, Ishihara S, Nakajima T, Fukamizu A. J. Biol. Chem. 2000;275:35170–35175. doi: 10.1074/jbc.C000258200. [DOI] [PubMed] [Google Scholar]
- 15.Korzus E, Torchia J, Rose DW, Xu L, Kurokawa R, McInerney EM, Mullen T-M, Glass CK, Rosenfeld MG. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 16.Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 17.Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 18.Hecht A, Litterst CM, Huber O, Kemler R. J. Biol. Chem. 1999;274:18017–18025. doi: 10.1074/jbc.274.25.18017. [DOI] [PubMed] [Google Scholar]
- 19.Chen D, Huang S-M, Stallcup MR. J. Biol. Chem. 2000;275:40810–40816. doi: 10.1074/jbc.M005459200. [DOI] [PubMed] [Google Scholar]
- 20.Lee Y-H, Koh SS, Zhang X, Cheng X, Stallcup MR. Mol. Cell. Biol. 2002;22:3621–3632. doi: 10.1128/MCB.22.11.3621-3632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schurter BT, Koh SS, Chen D, Bunick GJ, Harp JM, Hanson L, Henschen-Edman A, Mackay DR, Stallcup MR, Aswad DW. Biochem. 2001;40:5747–5756. doi: 10.1021/bi002631b. [DOI] [PubMed] [Google Scholar]
- 22.Ma H, Baumann CT, Li H, Strahl BD, Rice R, Jelinek MA, Aswad DW, Allis CD, Hager GL, Stallcup MR. Curr. Biol. 2001;11:1981–1985. doi: 10.1016/s0960-9822(01)00600-5. [DOI] [PubMed] [Google Scholar]
- 23.Umesono K, Evans RM. Cell. 1989;57:1139–1146. doi: 10.1016/0092-8674(89)90051-2. [DOI] [PubMed] [Google Scholar]
- 24.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 25.Brinkmann AO, Faber PW, van Rooij HCJ, Kuiper GGJM, Ris C, Klaassen P, van der Korput JAGM, Voorhorst MM, van Laar JH, Mulder E, Trapman J. J. Steroid Biochem. Molec. Biol. 1989;34:307–310. doi: 10.1016/0022-4731(89)90098-8. [DOI] [PubMed] [Google Scholar]
- 26.Puri PL, Sartorelli V, Yang XJ, Hamamori Y, Ogryzko VV, Howard BH, Kedes L, Wang JY, Graessmann A, Nakatani Y, Levrero M. Mol. Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 27.Lu J, Chuong CM, Widelitz RB. Gene. 1997;196:201–207. doi: 10.1016/s0378-1119(97)00228-x. [DOI] [PubMed] [Google Scholar]
- 28.Gluzman Y. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- 29.Ma H, Hong H, Huang S-M, Irvine RA, Webb P, Kushner PJ, Coetzee GA, Stallcup MR. Mol. Cell. Biol. 1999;19:6164–6173. doi: 10.1128/mcb.19.9.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koh SS, Chen D, Lee Y-H, Stallcup MR. J. Biol. Chem. 2001;276:1089–1098. doi: 10.1074/jbc.M004228200. [DOI] [PubMed] [Google Scholar]
- 31.Ding XF, Anderson CM, Ma H, Hong H, Uht RM, Kushner PJ, Stallcup MR. Mol. Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 32.Lopez GN, Turck CW, Schaufele F, Stallcup MR, Kushner PJ. J. Biol. Chem. 2001;276:22177–22182. doi: 10.1074/jbc.M010718200. [DOI] [PubMed] [Google Scholar]
- 33.Barker N, Clevers H. Bioessays. 2000;22:961–965. doi: 10.1002/1521-1878(200011)22:11<961::AID-BIES1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 34.Chuong CM, Chodankar R, Widelitz RB, Jiang TX. Curr. Opin. Genet. Dev. 2000;10:449–456. doi: 10.1016/s0959-437x(00)00111-8. [DOI] [PMC free article] [PubMed] [Google Scholar]