Abstract
BACKGROUND
We report a genome-wide association study (GWAS) of nicotine dependence defined on the basis of scores on the Fagerström Test for Nicotine Dependence in European-American (EA) and African-American (AA) populations.
METHODS
Our sample, from the one used in our previous GWAS, included only subjects who had smoked >100 cigarettes lifetime (2114 EA and 2602 AA subjects) and an additional 927 AA and 2003 EA subjects from the Study of Addiction: Genetics and Environment project [via the database of Genotypes and Phenotypes (dbGAP)]. GWAS analysis considered Fagerström Test for Nicotine Dependence score as an ordinal trait, separately in each population and sample and by combining the results in meta-analysis. We also conducted analyses that were adjusted for other substance use disorder criteria in a single nucleotide polymorphism (SNP) subset.
RESULTS
In EAs, one chromosome 7 intergenic region was genome-wide significant (GWS): rs13225753, p = 3.48 × 10−8 (adjusted). In AAs, GWS associations were observed at numerous SNPs mapped to a region on chromosome 14 of >305,000 base pairs (minimal p = 4.74 × 10−10). Two chromosome 8 regions were associated: p = 4.45 × 10−8 at DLC1 SNP rs289519 (unadjusted) and p = 1.10 × 10−9 at rs6996964 (adjusted for other substances), located between CSGALNACT1 and INTS10. No GWS associations were observed at the chromosome 15 nicotinic receptor gene cluster (CHRNA5-CHRNA3-CHRNB4) previously associated with nicotine dependence and smoking quantity traits. TSNAX-DISC1 SNP rs821722 (p = 1.46 × 10−7) was the most significant result with substantial contributions from both populations; we previously identified DISC1 associations with opioid dependence. Pathway analysis identified association with nitric oxide synthase and adenosine monophosphate-activated protein kinase pathways in EAs.
CONCLUSIONS
The key risk loci identified, which require replication, offer novel insights into nicotine dependence biology.
Keywords: AMPK pathway, DISC1, DLC1, eNOS pathway, FTND, GWAS, Nicotine dependence, Population differences
Genome-wide association study (GWAS), an important step in the identification of risk genes for complex traits, has only recently been applied to gene mapping for substance dependence (SD) traits. We previously reported risk genes identified by GWAS for cocaine, alcohol, and opioid dependence (1–4). By far, the most studied SD trait from a genetic perspective is nicotine dependence (ND), which is moderately heritable (h2 = .48–.72 based on twin studies) (5,6). The heritability of scores on the Fagerström Test for Nicotine Dependence (FTND), a quantitative measure frequently used to measure ND (7), was estimated to be .40 to .75 (8–10). Many GWAS studies and several meta-analyses of ND-related traits have been published. The most consistent signals identified via GWAS emerge from a set of closely mapped nicotinic receptor genes on chromosome 15 (11–13). In a meta-analysis of smoking behavior GWAS in African-Americans (AAs), the only genome-wide significant (GWS) association mapped to the same cluster (14).
We used GWAS to identify genetic variants that influence risk of ND as measured by the FTND. We included European-American (EA) and AA subjects who reported having smoked at least 100 cigarettes lifetime, derived from our substance dependence GWAS sample of 4716 subjects (1–3) (Yale-Penn sample), combined with a sample of 2930 subjects from the Study of Addiction: Genetics and Environment (SAGE), available to researchers through dbGAP (Database of Genotypes and Phenotypes) application.
METHODS AND MATERIALS
Subjects and Diagnostic Procedures
Our GWAS discovery sample included 2114 EA and 2602 AA subjects (after exclusion of those not meeting the exposure criterion: 308 AAs and 98 EAs reported never having smoked ≥100 cigarettes). All subjects were recruited for studies of the genetics of drug (opioid or cocaine) or alcohol dependence (1–3). The sample consisted of small nuclear families originally collected for linkage studies (primarily full sibs, half sibs, and parents, generally no more than one parent per family) and unrelated individuals. Subjects (Table S1 in Supplement 1) gave written informed consent as approved by the institutional review board at each site, and certificates of confidentiality were obtained from the National Institute on Drug Abuse and National Institute on Alcohol Abuse and Alcoholism. Subjects were administered the Semi-Structured Assessment for Drug Dependence and Alcoholism (15), in which the FTND is embedded. The FTND domains assessed by this instrument are how soon the subject smokes his first cigarette after awakening; whether the subject finds it difficult to refrain from smoking in places where it is forbidden; which cigarette the subject would least like to give up (e.g., the first cigarette in the morning); how many cigarettes the subject smokes per day; and whether the subject smokes even if ill enough to be confined to his bed [paraphrased from reference (7)].
Discovery phase analyses also included publicly available (via application) GWAS data from SAGE (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000092. v1.p1), containing 927 AA and 2003 EA unrelated exposed individuals (Table S1 in Supplement 1). SAGE includes individuals from the Collaborative Study on the Genetics of Alcoholism (COGA) (16), the Family Study of Cocaine Dependence (FSCD) (17), and the Collaborative Genetic Study of Nicotine Dependence (COGEND) (18). The COGA sample is a set of unrelated individuals recruited in Indiana, New York, St. Louis, Connecticut, Iowa, and San Diego selected for genotyping from a larger set of 8000 subjects. COGA cases met criteria for DSM-IV alcohol dependence. FSCD contained subjects from the greater St. Louis metropolitan area; most cases met criteria for DSM-IV alcohol dependence and cocaine dependence. Control subjects were from the same communities and had consumed alcohol but had no lifetime history of dependence on any substance. A subgroup of FSCD subjects was not alcohol dependent but had a lifetime DSM-IV diagnosis of dependence on cannabis or another illicit drug. COGEND subjects were recruited in Missouri and Michigan. COGEND cases met criteria for DSM-IV alcohol and/or nicotine dependence. Control subjects were selected from the nondependent population and did not meet criteria for alcohol, nicotine, or illicit drug dependence.
Genotyping and Quality Control
Yale-Penn GWAS samples were genotyped on the Illumina HumanOmni1-Quad v1.0 microarray, including 988,306 autosomal single nucleotide polymorphisms (SNPs; Illumina, San Diego, California), at the Center for Inherited Disease Research and the Yale Center for Genome Analysis. Genotypes were called using GenomeStudio software V2011.1 and genotyping module V1.8.4 (Illumina, San Diego, California). SAGE samples were genotyped on the Illumina Human 1M array containing 1,069,796 total SNPs (Illumina). In the Yale-Penn GWAS dataset, 44,644 SNPs on the microarray and 135 individuals with call rates <98% were excluded; 62,076 additional SNPs were removed due to minor allele frequencies (MAF) <1%. After data cleaning and quality control, 5697 individuals and 889,659 SNPs remained for imputation. Additional quality control information has been reported previously (1). After applying the same quality control procedures to the SAGE sample, 39 subjects with call rates <98% were excluded and 726,191 SNPs remained for analysis.
To verify and correct the misclassification of self-reported race, we compared the GWAS data from all subjects with genotypes from the HapMap 3 (http://hapmap.ncbi.nlm.nih.gov/) reference CEU (CEPH collection), YRI (Yoruba in Ibadan, Nigeria), and CHB (Han Chinese in Beijing, China) populations. Principal components (PC) analysis was conducted in the entire GWAS sample using Eigensoft (19,20) and 145,472 SNPs that were common to the GWAS dataset and HapMap panel (after pruning the GWAS SNPs for linkage disequilibrium [R2] >80%) to characterize the underlying genetic architecture of the samples. The first 10 PC scores were used in a k-means cluster analysis to distinguish AAs and EAs; these groups were subsequently analyzed separately. We then conducted PC analyses within the two groups and the first three PCs were used in all subsequent analyses to correct for residual population stratification.
Genotype Imputation
SNP genotype imputation was performed in the Yale-Penn and the SAGE GWAS datasets with IMPUTE2 (21) using genotyped SNPs with a minor allele frequency of >1% and the June 2011 1000 Genomes reference panel (22), which contains phased haplotypes for 1094 individuals of various ancestries: 379 of European descent (CEU, FIN (Finnish in Finland), GBR (British from England and Scotland), IBS (Iberian populations in Spain), and TSI (Toscani in Italia)), 286 of Asian descent CHB, JPT (Japanese in Tokyo, Japan), and CHS (Han Chinese South, China), 181 admixed American samples (PUR (Puerto Rican in Puerto Rico), CLM (Colombian in Medellin, Colombia), and MXL (Mexican ancestry in Los Angeles, California)), and 246 samples of African descent (ASW (African ancestry in southwest USA), LWK (Luhya in Webuye, Kenya), YRI) (22). All samples were imputed using every available sample in the reference panel, then split into AA and EA datasets based on the clustering techniques described above. We retained 18,564,419 SNPs with derived information content >.8 in at least one of the population groups. After excluding SNPs with MAF < 3% in both AAs and EAs, 11,995,908 SNPs common to both discovery datasets (11,106,284 in AAs, 7,535,791 in EAs) were included in association analyses.
Statistical Analysis Methods
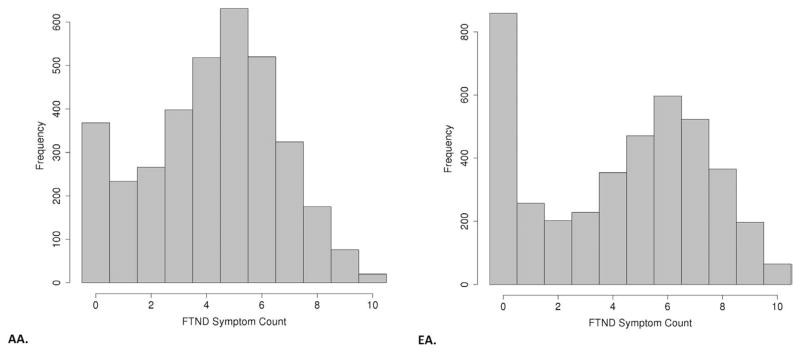
Association tests were performed for SNPs with MAF >3% using linear association models embedded in generalized estimating equations to correct for correlations among related individuals (23). We modeled the FTND score as a continuous variable that was analyzed in a standard linear regression and adjusted for age, sex, and three PCs of ancestry. Although the FTND is an ordinal variable ranging from 0 to 10, we did not use ordinal logistic regression models, which assume the same beta across each ordered transition, which was not the case for the FTND data. The distributions of FTND scores are shown in Figure 1. To investigate the possibility that true association signals may have been obscured by or confounded with comorbid dependence on other substances, we also tested models for moderately associated SNPs (p < 1 × 10−4) that contained ordinal measures for dependence on cocaine, opioids, and alcohol. Details regarding the derivation of these measures are provided elsewhere (1–3).
Figure 1.
Trait distribution of Fagerström Test for Nicotine Dependence (FTND) scores. AA, African American; EA, European American.
Analyses were performed separately within each dataset and population group, corrected for the subgroup-specific genomic inflation factor (λ), and the results were combined by meta-analysis using the inverse variance method. As described above, we then tested SNPs with a p value <1 × 10−4 in either population group or the full meta-analysis (n = 10,390) in a model adjusted for the DSM-IV criterion counts for cocaine, opioid, and alcohol dependence. We also tested 20,336 genotyped SNPs on the X chromosome and 226 on the Y chromosome for FTND association. Y chromosome SNPs were tested as binary variables in male subjects only and X chromosome SNPs were coded as homozygous in male subjects. A p value of 5.0 × 10−8 was the threshold for GWS in the GWAS; this applies a Bonferroni correction covering all independent haplotype blocks (regardless of the number of SNPs tested). Results were not adjusted for testing in two populations because we tested three distinct a priori hypotheses: SNPs are associated with FTND in AAs, SNPs are associated with FTND in EAs, and associations are evident with the same SNPs in meta-analysis in AAs and EAs. In EAs, we had 80% power to detect SNPs explaining 1% of the total variance in FTND score and the same power to detect SNPs explaining 2% of the trait variance in AAs (24).
Pathway Analysis
Meta-analyzed GWAS results from the Yale-Penn and SAGE datasets were used to identify biological pathways related to FTND. First, the number of independent SNP association tests for each gene in the genome were computed according to the method of Li and Ji (25). Next, the smallest p value for an individual SNP within each gene was multiplied by the number of independent tests in that gene to create a list of genes significantly associated with FTND after correcting for the number of tests within that gene (padj < .05). The significant genes were evaluated by pathway analysis, performed using the Ingenuity Pathway Analysis software suite (QIAGEN, Redwood City, California; http://www.ingenuity.com) to identify overrepresentation of selected genes within canonical pathways that were defined using information culled from multiple sources (Kyoto Encyclopedia of Genes and Genomes, interactome studies, manual curation, etc). Pathway analysis was done separately in AAs and EAs and a Benjamini-Hochberg false discovery rate (26) was calculated for the Fisher’s exact p value associated with each pathway.
RESULTS
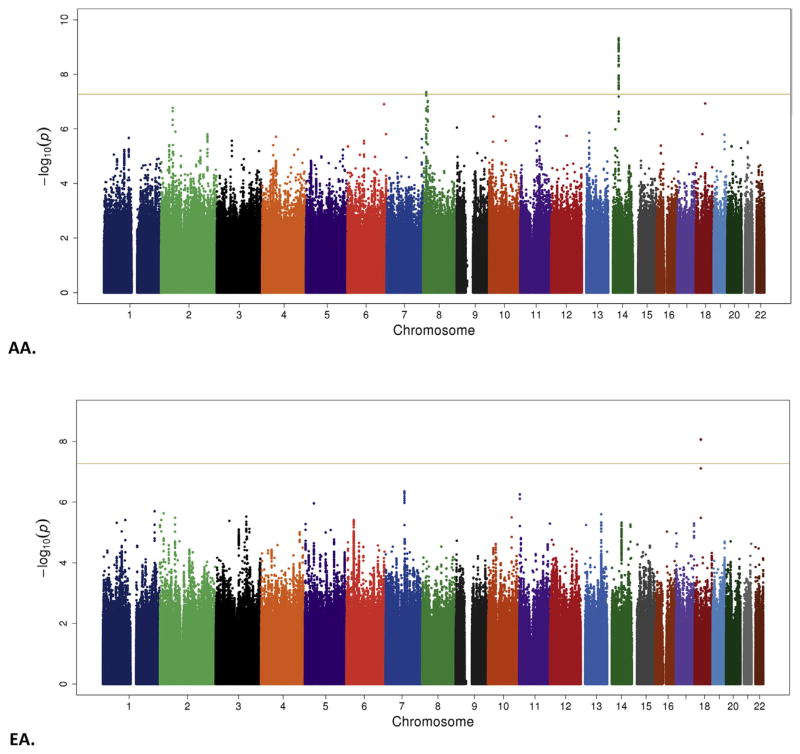
The mean FTND score in both samples is shown in Table S1 in Supplement 1, and the distribution is shown in Figure 1. In the Yale-Penn sample, EAs had higher scores than AAs among both cases and control subjects. In SAGE, the FTND scores were similar across populations (although lower than in the Yale-Penn sample). Results of the GWAS are summarized in Manhattan plots (Figure 2) and Tables 1 and 2; There was modest evidence for inflation of p values in both EAs and AAs (quantile-quantile plots, Figure S1 in Supplement 1).
Figure 2.
Manhattan plots. AA, African American; EA, European American.
Table 1.
Findings in African-Americans
| chr | chr.pos | Marker | Effect Allele | Ref Allele | MAF | Type | Gene | Yale-Penn p | SAGE p | Meta p | Meta Adjusted p |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 202346327 | rs2714493 | T | C | .28 | NA | NA | 1.96E-05 | 3.74E-02 | 1.57E-06 | 3.95E-07 |
| 2 | 202348303 | rs2080326 | C | A | .31 | NA | NA | 3.93E-05 | 2.30E-02 | 2.08E-06 | 3.98E-07 |
| 2 | 202352867 | rs2540436 | C | A | .32 | INT | ALS2CR11 | 3.57E-05 | 2.35E-02 | 1.91E-06 | 4.92E-07 |
| 2 | 202373160 | rs2714491 | T | C | .31 | INT | ALS2CR11 | 3.12E-05 | 2.67E-02 | 1.86E-06 | 4.59E-07 |
| 6 | 159476628 | rs114561596 | A | G | .04 | NA | NA | 1.58E-04 | 1.41E-04 | 1.25E-07 | 2.04E-07 |
| 6 | 168397398 | rs181977128 | A | G | .03 | U5 | KIF25-AS1 | 6.58E-05 | 7.94E-03 | 1.56E-06 | 3.41E-07 |
| 8 | 13234600 | rs1729163 | G | A | .23 | INT | DLC1 | 4.42E-07 | 6.15E-02 | 6.03E-08 | 1.98E-07 |
| 8 | 13235841 | rs1799660 | T | C | .24 | INT | DLC1 | 3.82E-07 | 5.79E-02 | 4.89E-08 | 1.66E-07 |
| 8 | 13236486 | rs289520 | T | A | .24 | INT | DLC1 | 3.97E-07 | 5.70E-02 | 4.99E-08 | 1.60E-07 |
| 8 | 13237048 | rs289519 | T | C | .24 | INT | DLC1 | 3.83E-07 | 5.28E-02 | 4.45E-08 | 1.37E-07 |
| 8 | 19623695 | rs13263337 | T | C | .31 | NA | NA | 2.80E-05 | 6.64E-04 | 1.05E-07 | 1.30E-09 |
| 8 | 19623911 | rs6996964 | T | C | .31 | NA | NA | 2.47E-05 | 6.99E-04 | 9.47E-08 | 1.10E-09 |
| 8 | 19624046 | rs6997137 | T | C | .28 | NA | NA | 9.43E-05 | 2.98E-03 | 1.18E-06 | 3.03E-08 |
| 8 | 19624063 | rs6995952 | T | G | .31 | NA | NA | 2.95E-05 | 1.04E-03 | 1.50E-07 | 2.31E-09 |
| 8 | 19624119 | rs6997291 | A | C | .31 | NA | NA | 3.76E-05 | 1.26E-03 | 2.24E-07 | 3.16E-09 |
| 8 | 19624402 | rs6996589 | A | G | .41 | NA | NA | 1.17E-05 | 2.49E-03 | 1.02E-07 | 1.40E-08 |
| 8 | 19624520 | rs6586864 | T | C | .31 | NA | NA | 4.66E-05 | 9.18E-04 | 2.32E-07 | 3.27E-09 |
| 8 | 19625027 | rs11337452 | G | GT | .31 | NA | NA | 4.66E-05 | 8.95E-04 | 2.28E-07 | 3.27E-09 |
| 8 | 19625232 | rs7819691 | G | T | .31 | NA | NA | 4.66E-05 | 7.53E-04 | 2.04E-07 | 2.57E-09 |
| 8 | 19625464 | rs13282247 | T | C | .44 | NA | NA | 2.37E-06 | 3.04E-02 | 1.53E-07 | 1.95E-08 |
| 8 | 19625510 | rs1492637 | T | A | .40 | NA | NA | 3.25E-06 | 6.99E-02 | 4.73E-07 | 1.79E-08 |
| 8 | 19625619 | rs13280698 | T | G | .32 | NA | NA | 8.50E-05 | 1.04E-03 | 5.10E-07 | 8.61E-09 |
| 8 | 19625762 | rs13248141 | A | C | .32 | NA | NA | 8.21E-05 | 1.18E-03 | 5.40E-07 | 9.51E-09 |
| 8 | 19626356 | rs13254708 | T | G | .32 | NA | NA | 7.08E-04 | 4.47E-04 | 3.69E-06 | 3.48E-08 |
| 14 | 45301142 | rs75063231 | T | C | .07 | NA | NA | 1.12E-04 | 8.91E-04 | 5.11E-07 | 1.89E-07 |
| 14 | 45307535 | rs146754986 | A | T | .04 | NA | NA | 2.40E-06 | 7.96E-05 | 1.06E-09 | 4.37E-07 |
| 14 | 45315320 | rs201547800 | C | CT | .04 | NA | NA | 2.32E-06 | 6.52E-05 | 8.54E-10 | 4.94E-07 |
| 14 | 45322802 | rs118042324 | T | C | .04 | NA | NA | 2.05E-06 | 4.17E-05 | 5.08E-10 | 4.25E-07 |
| 14 | 45323581 | rs144667340 | T | G | .04 | NA | NA | 1.94E-06 | 4.17E-05 | 4.74E-10 | 4.07E-07 |
| 14 | 45324092 | rs201864750 | T | TAAAC | .04 | NA | NA | 1.94E-06 | 4.17E-05 | 4.74E-10 | 4.07E-07 |
| 14 | 45324218 | rs139503483 | G | A | .04 | NA | NA | 3.10E-06 | 2.34E-05 | 5.57E-10 | 3.92E-07 |
| 14 | 45326352 | rs141086819 | A | T | .04 | NA | NA | 1.94E-06 | 4.17E-05 | 4.74E-10 | 4.07E-07 |
| 14 | 45333376 | rs141232514 | T | G | .04 | NA | NA | 1.94E-06 | 4.17E-05 | 4.74E-10 | 4.12E-07 |
| 14 | 45334768 | rs117517701 | T | C | .04 | NA | NA | 1.94E-06 | 4.17E-05 | 4.74E-10 | 4.12E-07 |
| 14 | 45335572 | rs141064100 | G | C | .04 | NA | NA | 1.94E-06 | 4.17E-05 | 4.74E-10 | 4.12E-07 |
| 14 | 45337321 | rs117018253 | T | A | .04 | NA | NA | 3.35E-05 | 4.57E-07 | 4.73E-10 | 1.07E-07 |
| 14 | 45344383 | rs147624171 | T | C | .04 | NA | NA | 2.05E-06 | 4.84E-05 | 5.66E-10 | 4.86E-07 |
| 14 | 45376406 | rs45590239 | C | T | .04 | U3 | C14orf28 | 2.96E-06 | 6.32E-05 | 1.08E-09 | 2.35E-07 |
| 14 | 45413169 | rs114630737 | C | T | .04 | INT | KLHL28 | 3.05E-06 | 3.87E-03 | 3.46E-08 | 2.49E-07 |
| 14 | 45418368 | rs115843672 | C | T | .04 | INT | KLHL28 | 2.21E-06 | 2.87E-03 | 2.00E-08 | 2.20E-07 |
| 14 | 45428887 | rs115555158 | C | T | .04 | INT | KLHL28 | 2.22E-06 | 1.96E-03 | 1.52E-08 | 1.23E-07 |
| 14 | 45434239 | rs114318796 | A | T | .04 | INT | FAM179B | 2.29E-06 | 1.88E-03 | 1.52E-08 | 1.23E-07 |
| 14 | 45449377 | rs117098369 | A | G | .04 | INT | FAM179B | 6.33E-06 | 1.15E-03 | 3.24E-08 | 3.14E-07 |
| 14 | 45454771 | rs146530309 | T | G | .04 | INT | FAM179B | 6.33E-06 | 1.15E-03 | 3.24E-08 | 3.14E-07 |
| 14 | 45456187 | rs185900235 | A | G | .04 | INT | FAM179B | 3.81E-06 | 9.23E-04 | 1.58E-08 | 1.30E-07 |
| 14 | 45457326 | rs186282840 | T | G | .04 | INT | FAM179B | 6.33E-06 | 1.15E-03 | 3.24E-08 | 3.14E-07 |
| 14 | 45457465 | rs114962601 | T | C | .05 | INT | FAM179B | 2.18E-06 | 6.13E-05 | 9.84E-10 | 3.61E-08 |
| 14 | 45461748 | rs114821783 | G | T | .05 | INT | FAM179B | 2.53E-06 | 1.29E-03 | 1.36E-08 | 1.27E-07 |
| 14 | 45464042 | rs149421422 | C | T | .05 | INT | FAM179B | 3.10E-06 | 1.15E-03 | 1.58E-08 | 1.52E-07 |
| 14 | 45465895 | rs139535864 | C | G | .05 | INT | FAM179B | 2.53E-06 | 1.29E-03 | 1.36E-08 | 1.27E-07 |
| 14 | 45470860 | rs115446694 | G | A | .05 | INT | FAM179B | 2.44E-06 | 1.29E-03 | 1.31E-08 | 1.22E-07 |
| 14 | 45473087 | rs116189259 | C | T | .05 | INT | FAM179B | 3.10E-06 | 1.15E-03 | 1.58E-08 | 1.58E-07 |
| 14 | 45484261 | rs138311939 | G | C | .04 | INT | FAM179B | 4.86E-06 | 4.33E-03 | 6.56E-08 | 4.55E-07 |
| 14 | 45497025 | rs148752305 | T | C | .05 | INT | FAM179B | 2.12E-06 | 1.29E-03 | 1.10E-08 | 1.00E-07 |
| 14 | 45500080 | rs143082021 | C | T | .05 | INT | FAM179B | 2.12E-06 | 1.29E-03 | 1.10E-08 | 1.00E-07 |
| 14 | 45502277 | rs146751744 | G | A | .05 | INT | FAM179B | 2.12E-06 | 1.29E-03 | 1.10E-08 | 1.00E-07 |
| 14 | 45523954 | rs115778832 | A | G | .05 | INT | FAM179B | 2.12E-06 | 1.29E-03 | 1.10E-08 | 1.02E-07 |
| 14 | 45525202 | rs114669325 | G | C | .05 | INT | FAM179B | 2.12E-06 | 1.29E-03 | 1.10E-08 | 1.02E-07 |
| 14 | 45528945 | rs115409819 | T | G | .05 | INT | FAM179B | 2.12E-06 | 4.28E-03 | 2.77E-08 | 1.73E-07 |
| 14 | 45530189 | rs114956580 | T | C | .05 | INT | FAM179B | 2.12E-06 | 1.29E-03 | 1.10E-08 | 1.02E-07 |
| 14 | 45534085 | rs139470339 | T | TA | .04 | INT | FAM179B | 5.02E-06 | 1.15E-03 | 2.47E-08 | 2.51E-07 |
| 14 | 45547337 | rs115837469 | A | G | .04 | NA | NA | 4.91E-06 | 1.17E-05 | 5.80E-10 | 6.98E-08 |
| 14 | 45557602 | rs115289722 | C | A | .04 | INT | PRPF39 | 1.04E-05 | 5.79E-05 | 4.42E-09 | 1.71E-07 |
| 14 | 45561675 | rs145792754 | A | G | .04 | INT | PRPF39 | 1.07E-05 | 5.79E-05 | 4.58E-09 | 1.74E-07 |
| 14 | 45568644 | rs200997489 | G | GATT | .04 | INT | NA | 9.48E-06 | 1.01E-05 | 1.10E-09 | 8.68E-08 |
| 14 | 45581037 | rs201994106 | TA | T | .04 | INT | NA | 9.48E-06 | 1.01E-05 | 1.10E-09 | 9.18E-08 |
| 14 | 45591744 | rs149743773 | A | G | .04 | INT | LOC100652866 | 1.62E-05 | 2.94E-06 | 8.55E-10 | 1.65E-07 |
| 14 | 45592328 | rs140331847 | A | G | .04 | INT | LOC100652866 | 1.67E-05 | 2.94E-06 | 8.87E-10 | 1.62E-07 |
| 14 | 45605463 | rs61746895 | G | A | .04 | NSM | FANCM | 1.90E-05 | 2.43E-06 | 8.90E-10 | 1.61E-07 |
| 14 | 45612664 | rs117577361 | T | A | .04 | INT | FANCM | 3.21E-05 | 4.49E-06 | 2.60E-09 | 1.71E-07 |
| 14 | 45613093 | rs145624594 | T | C | .04 | INT | FANCM | 3.31E-05 | 4.49E-06 | 2.69E-09 | 1.72E-07 |
| 14 | 91329707 | rs114643642 | T | A | .05 | NA | NA | 5.13E-05 | 2.57E-01 | 2.86E-05 | 3.04E-07 |
| 14 | 91342649 | rs145411601 | C | CATGTAACT | .05 | INT | RPS6KA5 | 3.24E-05 | 2.96E-01 | 2.28E-05 | 2.91E-07 |
| 14 | 91343673 | rs111534394 | T | C | .05 | INT | RPS6KA5 | 3.33E-05 | 2.94E-01 | 2.33E-05 | 2.94E-07 |
| 14 | 91346004 | rs80060912 | T | C | .05 | INT | RPS6KA5 | 3.31E-05 | 2.88E-01 | 2.23E-05 | 3.17E-07 |
| 14 | 91347824 | rs112462778 | A | G | .05 | INT | RPS6KA5 | 3.62E-05 | 2.83E-01 | 2.37E-05 | 3.48E-07 |
| 14 | 91349561 | rs112440489 | C | T | .05 | INT | RPS6KA5 | 3.96E-05 | 2.80E-01 | 2.55E-05 | 3.71E-07 |
| 16 | 19459690 | rs7184765 | C | A | .10 | INT | TMC5 | 6.62E-07 | 8.86E-01 | 4.09E-06 | 2.85E-07 |
| 16 | 19460842 | rs6497373 | A | G | .10 | INT | TMC5 | 6.62E-07 | 8.91E-01 | 4.14E-06 | 2.82E-07 |
Summary of significant results and also results up to p = 5 × 10−7. All study and population specific results were corrected for their respective genomic inflation factors. Only those rows are shown where the marker allele frequency was >.02. Top GWAS results, those that meet genome-wide significance are in bold.
chr, chromosome; chr.pos, chromosome position base pairs; GWAS, genome-wide association study; INT, intron; MAF, minor allele frequencies; NA, not applicable; NSM, non-synonymous mutation; Ref, reference; SAGE, Study of Addiction: Genetics and Environment.
Table 2.
Findings in European-Americans
| chr | chr.pos | Marker | Effect Allele | Ref Allele | MAF | Type | Gene | Yale-Penn p | SAGE p | Meta p | Meta Adjusted p |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 7 | 82157289 | rs35653049 | C | T | .05 | NA | NA | 1.30E-01 | 1.39E-08 | 4.44E-07 | 4.56E-08 |
| 7 | 82159767 | rs12532806 | G | A | .05 | NA | NA | 1.47E-01 | 1.17E-08 | 4.91E-07 | 3.96E-08 |
| 7 | 82158523 | rs13225753 | G | A | .05 | NA | NA | 1.37E-01 | 1.78E-08 | 5.28E-07 | 3.48E-08 |
| 7 | 82163223 | rs12535073 | G | A | .05 | NA | NA | 1.60E-01 | 1.17E-08 | 6.38E-07 | 5.08E-08 |
| 7 | 82154194 | rs12532927 | C | T | .05 | NA | NA | 1.23E-01 | 3.47E-08 | 7.67E-07 | 9.95E-08 |
| 7 | 82151812 | rs10486966 | G | A | .05 | NA | NA | 1.22E-01 | 4.62E-08 | 8.97E-07 | 1.54E-07 |
| 7 | 82149544 | rs35521884 | T | C | .05 | NA | NA | 1.22E-01 | 6.12E-08 | 1.07E-06 | 1.72E-07 |
| 7 | 82165099 | rs35763698 | A | G | .05 | NA | NA | 2.15E-01 | 1.07E-07 | 5.70E-06 | 4.48E-07 |
| 15 | 78961282 | rs199970818 | A | AAAAAT | .40 | NA | NA | 4.74E-04 | 2.57E-02 | 2.73E-05 | 2.72E-07 |
Summary of significant results and also results up to p = 5 × 10−7. All study and population specific results were corrected for their respective genomic inflation factors. Only those rows are shown where the marker allele frequency was >.02. Top GWAS results, those that meet genome-wide significance are in bold.
chr, chromosome; chr.pos, chromosome position base pairs; GWAS, genome-wide association study; MAF, minor allele frequencies; NA, not applicable; Ref, reference; SAGE, Study of Addiction: Genetics and Environment.
Most of the top-ranked findings were population specific (Tables 1 and 2; Table S3 in Supplement 2). The only GWS association that was specific to EAs was for the chromosome 7 SNP rs13225753. This SNP (at p = 3.48 × 10−8) and two other SNPs nearby with similar p values, were tested in the adjusted (for substance use disorder criteria) ordinal model (regional Manhattan plot; Figure 3A). The gene mapped closest to this region is CACNA2D1, calcium channel, voltage-dependent, alpha 2/delta subunit 1, which encodes a component of a voltage-dependent calcium channel.
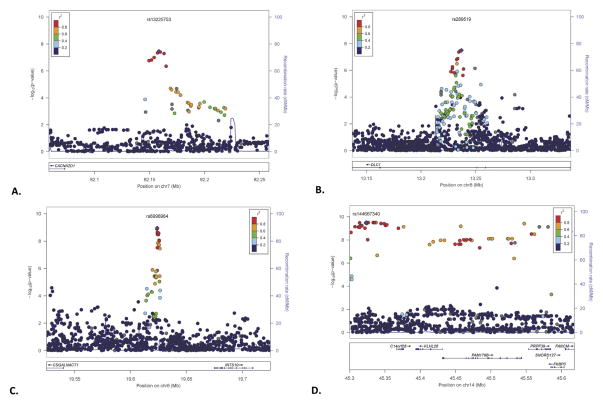
Figure 3.
Regional Manhattan plots. (A) Meta-analysis of the association results from single nucleotide polymorphisms (SNPs) in the 82.1 to 82.25 mega-base pair (MBP) region on chromosome 7 in Yale-UPenn + Study of Addiction: Genetics and Environment (SAGE) European American (EAs) with Fagerström Test for Nicotine Dependence (FTND) score adjusted for co-occurring substance dependence severity. The SNPs are color coded according to R2, with the most significant SNP (rs13225753) shown in purple. The light blue line and right y axis show the observed recombination rate in the HapMap CEU samples. (B) Meta-analysis of the association results from SNPs in the 13.15 to 13.3 MBP region on chromosome 8 in Yale-UPenn + SAGE African Americans (AAs) with FTND score. The SNPs are color coded according to R2, with the most significant SNP (rs289519) shown in purple. The light blue line and right y axis show the observed recombination rate in the HapMap Yoruba in Ibadan, Nigeria (YRI) samples. (C) Meta-analysis of the association results from SNPs in the 19.55 to 19.7 MBP region on chromosome 8 in Yale-UPenn + SAGE AAs with FTND score adjusted for co-dependent substance dependence severity. The SNPs are color coded according to R2, with the most significant SNP (rs6996964) shown in purple. The light blue line and right y axis show the observed recombination rate in the HapMap YRI samples. (D) Meta-analysis of the association results from SNPs in the 45.3 to 45.6 MBP region on chromosome 14 in Yale-UPenn + SAGE AAs with FTND score. The SNPs are color coded according to R2, with the most significant SNP (rs144667340) shown in purple. The light blue line and right y axis show the observed recombination rate in the HapMap YRI samples.
All other GWS results were observed in the AA part of the sample. This included two distinct chromosome 8 regions, one region under the ordinal model and one under the adjusted ordinal model. Several closely mapped SNPs at the DLC1 (deleted in liver cancer 1) locus yielded p values just under 5 × 10−8, the most significant being rs289519 (MAF = .23) under the ordinal model (regional Manhattan plot; Figure 3B). More than six megabases distal to DLC1, numerous tightly mapped markers in an intergenic region showed GWS under the adjusted ordinal model, with rs6996964 being the most significant at p = 1.1 × 10−9. This region is flanked by CSGALNACT1 (chondroitin sulfate N-acetylgalactosaminyltransferase 1) and INTS10 (integrator complex subunit 10) (regional Manhattan plot; Figure 3C).
There were numerous associations at a region on chromosome 14 from 45,307,535 to 45,613,093 base pairs (rs146754986 to rs145624594), spanning >300,000 base pairs (regional Manhattan plot; Figure 3D). The region includes seven named loci: C14orf28, KLHL28 (kelch-like family member 28), FAM179B (family with sequence similarity 179, member B), PRPF39 (pre-mRNA processing factor 39), SNORD127 (small nucleolar RNA, C/D box 127), FKBP3 (FK506 binding protein 3, 25kDa), and FANCM (Fanconi anemia, complementation group M). Although support came from both the Yale-Penn and SAGE samples, it was generally greater in the former. GWS p values were as low as 4.73 × 10−10.
Another noteworthy finding that did not reach GWS was an association with TSNAX-DISC1, SNP rs821722. TSNAX-DISC1 is a read-through transcription between TSNAX (translin-associated factor X) and DISC1 (disrupted in schizophrenia 1). This is the most significant result in the GWAS (meta p = 1.46 × 10−7) of loci with substantial contributions from both the AA (p = 2.12 × 10−5) and EA (p = 3.28 × 10−3) samples (Table S3 in Supplement 2).
Although many previous studies have demonstrated significant associations with SNPs that map to the chromosome 15 nicotinic receptor cluster (CHRNA5/CHRNA3/CHRNB4), we found no GWS results for this region (Table S2 in Supplement 1). The most significant p value that we observed for this gene cluster was 6.78 × 10−7 for rs11633958, which is intronic at CHRNA5. We observed p values of <10−5 at each of the three loci. Both the Yale-Penn and the SAGE samples contributed to these findings, and the EA contribution was in most cases greater than the AA contribution.
Two pathways in EAs with several overlapping genes were significantly associated with FTND (false discovery rate <.05): endothelial nitric oxide synthase (eNOS) and adenosine monophosphate-activated protein kinase (AMPK) signaling. Both pathways contain the genes CHRNA5 and CHRNA3, each of which had an SNP with a gene-wide independent test-corrected p value <.05. No pathways were significant in AAs.
DISCUSSION
We present herein results from a GWAS study of the ordinal trait, FTND score. Our results differ substantially from those of most other studies of nicotine-related traits. In many prior studies, an association peak was observed over the chromosome 15 nicotinic receptor cluster that was much larger than other association peaks. We also found little overlap with risk genes identified from our previous GWAS of cocaine, opioid, and alcohol dependence (1–3), the one exception being DISC1, discussed below. Some risk genes that we identified appear to be of biological relevance. More GWS results were identified in the AA than the EA part of the sample, which is similar to what we observed for the SD traits studied in this sample previously.
In EAs, we identified one GWS signal, mapped near the CACNA2D1 locus. This gene encodes a protein that is part of a calcium channel (27)—we previously identified calcium signaling genes as important for risk for opioid (2) and cocaine (1) dependence. Variation at this locus, the protein product of which interacts indirectly with the μ-opioid receptor (28), has previously been associated with opioid sensitivity in a small human sample (29).
We identified three GWS regions in AAs, perhaps the most interesting and compelling of which was the span on chromosome 14 delimited by the C14ORF28 and FANCM (Fanconi anemia, complementation group M) loci. The regional Manhattan plot shows an extensive region characterized by numerous GWS association findings, including KLHL28 (kelch-like family member 28), FAM179B (family with sequence similarity 179, member B), SNORD127 (small nucleolar RNA, C/D box 127), PRPF39 (PRP39 pre-mRNA processing factor 39 homolog [S. cerevisiae]), and FKBP3 (FK506 binding protein 3, 25kDa). This is suggestive of more than one risk locus. Of these, the most immediately appealing candidate is C14ORF28, which encodes a protein that, although of unknown function, interacts with D1 dopamine receptors and is differentially expressed in both bipolar disorder and schizophrenia, compared with control subjects (30). We also observed an association with SNPs mapped to an intergenic region on chromosome 8 and at DLC1 at a different chromosome 8 region.
Results in the ordinal model compared with the ordinal model adjusted for dependence on other substances (which was evaluated only on the SNPs with p < 1.0 × 10−4 in the unadjusted model) were somewhat different. The only GWS region in EAs was observed under the adjusted model, with findings about an order of magnitude less significant under the nonadjusted model. In AAs, DLC1 was significant only under the ordinal model and the more distal intergenic region, only under the adjusted model. The extended chromosome 14 region was GWS in AAs under the ordinal model, with only one SNP in the region meeting significance criteria under the adjusted model as well. Regions significant under the adjusted model should be considered more specific to ND than those significant under the unadjusted ordinal model, and the use of an adjusted model could account for some of our novel association findings. Although ND frequently co-occurs with dependence on other substances, especially the three SD disorders on which our sample was ascertained, this is rarely adjusted analytically and sometimes not even measured.
Finally, we observed evidence for association with TSNAX-DISC1, a finding that at p = 1.46 × 10−7 is less compelling statistically than the others described here but which gains interest in the context of our previous observations of association of common (2) and rare (31) variants at this locus with opioid dependence. We also previously observed an association of an SNP near CCDC88A, a protein that interacts with DISC1, with alcohol dependence (3). The DISC1 protein product plays a role in cognitive function (32) and several psychiatric traits and is emerging as an important contributor to SD risk.
In the present study, we observed numerous GWS signals that have not been reported previously but none in the chromosome 15 cluster of genes encoding nicotinic receptors. This may be attributable to two features of our sample and the nature of the reported association. First, we had excellent representation of AA subjects, and it was from this part of the sample that the most interesting findings were derived. Second, our subjects were recruited for the purpose of studying other types of SD (opioid, cocaine, and alcohol dependence) without reference to cigarette smoking or other tobacco use. Thus, our sample had high affection with and comorbidity for other SD traits. Although comorbidity is often not reported in ND GWAS, we surmise that our sample was more severely affected with drug phenotypes than other, previously reported samples. We controlled for this comorbidity analytically.
Also, our trait of interest was FTND as an ordinal trait, whereas the strongest reported chromosome 15 cluster associations have been for smoking quantity or closely related traits. For example, in a study of >8000 Finnish subjects, each risk allele at CHRNA5*rs16969968 corresponded to about one additional cigarette smoked per day (33). Similar smoking quantity-based findings have emerged (34), including from large meta-analyses (11–13,35). Although one item in the FTND assesses smoking quantity (7) and contributes to the overall score, it is not a distinctive or predominant component of the assessment. Although CHRNA5-CHRNA3-CHRNB4 candidate SNPs have also been associated with ND based on dichotomized FTND scores (36), the results fell just short of the GWAS threshold of 5 × 10−8. Thus, the explanation for our lack of observation of significant associations to this cluster could also be our phenotype, i.e., FTND, as an ordinal trait.
As noted above, the majority of our interesting findings were seen in the AA part of the sample, despite our having slightly lower power in the AA part of the sample. This has been the case for all other SD traits that we studied in this sample [alcohol, cocaine, and opioid dependence (1–3)] but not for posttraumatic stress disorder, where the most interesting results were in the EA part of the sample (37). We believe that this is most likely an artifact of population history and differing selection pressures on populations, but this hypothesis remains to be tested.
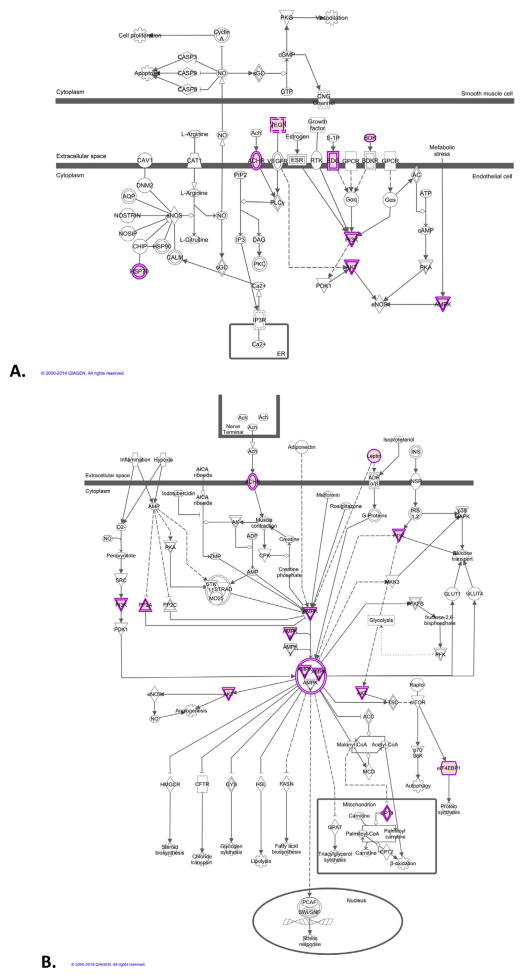
Our pathway analysis identified two associated pathways in the EA part of the sample: eNOS and AMPK signaling (Figure 4). The eNOS pathway is important for blood pressure regulation and vascular disorders. AMPK has been tied directly to some nicotine effects, including effects on energy balance that might contribute to the effects of nicotine on weight loss (38) and its anti-inflammatory effects (39). Somewhat surprisingly, there were no significant pathway associations identified in the AA part of the sample, despite the greater number of GWS SNPs in this population.
Figure 4.
Pathways associated with Fagerström Test for Nicotine Dependence score in the European-American sample. (A) Endothelial nitric oxide synthase (eNOS) signaling. (B) Adenosine monophosphate-activated protein kinase (AMPK) signaling. AC, adenylate cyclase; ACC, acetyl coenzyme A carboxylase; Ach, acetylcholine; ACHR, acetylcholine receptor; ADP, adenosine diphosphate; ADR, adrenergic receptor; AICA, 5-Aminoimidazole-4-carboxamide; AK, adenylate kinase; AKT, v-akt murine thymoma viral oncogene homolog 1; AMP, adenosine 5-monophosphate; AQP, aquaporin; ATP, adenosine triphosphate; BDK, bradykinin; BDKR, bradykinin receptor; Ca2+, calcium; CALM, calmodulin; cAMP, cyclic adenosine monophosphate; CASP, caspase; CAT1, cationic amino acid transporter-1; CAV1, caveolin-1; CFTR, cystic fibrosis transmembrane conductance regulator; cGMP, cyclic guanosine monophosphate; CHIP, C-terminal Hsp70-interacting protein; CNG, cyclic nucleotide gateway; CPT, carnitine palmitoyltransferase; DAG, diacylglycerol; DNM2, dynamin 2; EDG, lysophosphatidic acid receptor; eF4EBP1, eukaryotic initiation factor 4e binding protein; ER, endoplasmic reticulum; ESR, estrogen receptor; FASN, fatty acid synthase; GLUT, glucose transporter; GPAT, glycerol-3-phosphate acyltransferase; GPCR, G protein-coupled receptor; GTP, guanosine triphosphate; GYS, glycogen synthase; HMGCR, 3-hydroxy-3-methylglutaryl-coenzyme-A reductase; HSL, hormone-sensitive lipase; HSP, heat shock protein; INS, insulin; INSR, insulin receptor; IP3, inositol 1,4,5-triphosphate; IP3R, inositol 1,4,5-triphosphate receptor; IRS, insulin receptor substrate; MAPK, mitogen-activated protein kinase; MCD, Malonyl-CoA decarboxylase; MKK3, mitogen-activated protein kinase 3; mTOR, mammalian target of rapamycin; NO, nitric oxide; NOSIP, eNOS interacting protein; NOSTRIN, nitric oxide synthase traffic inducer; O2-, superoxide; PI3K, phosphoinositide kinase-3; PCAF, p300/CBP-associated factor; PDK1, phosphoinositide-dependent kinase-1; PFK, phosphofructokinase; PIP2, phosphatidylinositol 4,5-bisphosphate; PKA, protein kinase A; PKC, protein kinase C; PKG, protein kinase G; PP2A, protein phosphatase 2A; PP2C, protein phosphatase 2C; RTK, receptor tyrosine kinase; S-1P, sphingosine 1-phosphate; sGC, soluble guanylyl cyclase; SNF, sucrose non-fermentable; SRC, sarcoma; STK, serine/threonine kinase; SWI, switch; TSC, tumor suppressor complex; VEGF, vascular endothel growth factor; VEGFR, vascular endothel growth factor receptor; ZMP, zinc-dependent protease. (The networks were generated through the use of QIAGEN’s Ingenuity Pathway Analysis (IPA[r]), QIAGEN Redwood City, California; www.qiagen.com/ingenuity).
The FTND distribution in our sample is nonnormal (Figure 1), but we do not believe this is a major limitation. Linear models assume normally distributed residuals, not traits. Skewed trait distributions may be a concern in GWAS, but the primary risk with such traits is the potential for outliers with extreme values to create spurious results (especially for rare SNPs shared by a small number of individuals with extreme trait values). This is not the case with the FTND symptom count in our sample. The large proportion of the sample with FTND = 0, in fact, limits the potential for this group to produce spurious results for low MAF SNPs. Further, the small genomic inflation factors observed (Figure S1 in Supplement 1) suggest that the trait distribution did not significantly alter the distribution of p values genome-wide. We did, however, correct for the minimal inflation that exists.
Although we had a moderately large sample available for analysis, our study is limited by the lack of a separate replication sample. Our sample was, as noted, ascertained on the basis of other SD traits or lack thereof, whereas many other studies have selected for ND subjects. It is not clear to what extent this is a limitation for identifying risk alleles, but it is a difference with respect to many other (but far from all) published ND studies. Also, we note that the SAGE sample (but not the Yale-Penn sample) has been studied previously in GWAS with respect to several ND-related traits.
In conclusion, we identified several novel loci that associate with FTND score. The adjusted model allowed us to isolate ND risk from risk of dependence on alcohol, opioids, and cocaine, important because ND frequently co-occurs with these disorders. The key risk loci that we identified participate, or may participate, in pathways known to be relevant to SD: calcium signaling, dopaminergic function, neuronal differentiation, synapse formation, and cognitive function. None of these SNPs overlap with variants identified as affecting SD risk in our previous studies. However, in some cases, similar pathways are involved (e.g., calcium signaling), and in one case, the same locus, DISC1, is implicated. These results, if replicated, should increase our understanding of the biological mechanisms of ND and may identify novel pharmacologic targets for treatment and biomarkers to identify risk for prevention efforts.
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health Grants RC2 DA028909, R01 DA12690, R01 DA12849, R01 DA18432, R01 AA11330, and R01 AA017535 and the Veterans Affairs Connecticut and Philadelphia Veterans Affairs Mental Illness Research, Educational, and Clinical Centers.
We appreciate the work in recruitment and assessment provided at Yale University School of Medicine and the APT Foundation by James Poling, Ph.D.; at McLean Hospital by Roger Weiss, M.D.; at the Medical University of South Carolina by Kathleen Brady, M.D., Ph.D., and Raymond Anton, M.D.; and at the University of Pennsylvania by David Oslin, M.D. Genotyping services for a part of our genome-wide association study were provided by the Center for Inherited Disease Research and Yale University (Center for Genome Analysis). Center for Inherited Disease Research is fully funded through a Federal contract from the National Institutes of Health to The Johns Hopkins University (contract number N01-HG-65403). We are grateful to Ann Marie Lacobelle, Catherine Aldi, and Christa Robinson for their excellent technical assistance; to the Semi-structured Assessment for Drug Dependence and Alcoholism interviewers, led by Yari Nuñez and Michelle Slivinsky, who devoted substantial time and effort to phenotype the study sample; and to John Farrell and Alexan Mardigan for database management assistance.
The publicly available datasets used for the analyses described in this manuscript were obtained from the dbGAP at http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000092.v1.p1 through dbGAP accession number phs000092.v1.p. Funding support for the Study of Addiction: Genetics and Environment was provided through the National Institutes of Health Genes, Environment and Health Initiative (U01 HG004422). Study of Addiction: Genetics and Environment is one of the genome-wide association studies funded as part of the Gene Environment Association Studies under the Genes, Environment and Health Initiative. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the Gene Environment Association Studies Coordinating Center (U01 HG004446).
Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (P01 CA089392), and the Family Study of Cocaine Dependence (R01 DA013423). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the National Institutes of Health Genes, Environment and Health Initiative (U01HG004438); the National Institute on Alcohol Abuse and Alcoholism; the National Institute on Drug Abuse; and the National Institutes of Health contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C).
Footnotes
Supplementary material cited in this article is available online at http://dx.doi.org/10.1016/j.biopsych.2014.08.025.
DISCLOSURES
Although unrelated to the current study, Dr. Kranzler has been a consultant or advisory board member for Alkermes, Lilly, Lundbeck, Otsuka, Pfizer, and Roche. He is also a member of the American Society of Clinical Psychopharmacology Alcohol Clinical Trials Initiative, which is supported by Lilly, Lundbeck, AbbVie, Pfizer, and Ethypharm and has a US patent pending entitled “Test for Predicting Response to Topiramate and Use of Topiramate.” All other authors report no biomedical financial interests or potential conflicts of interest.
References
- 1.Gelernter J, Sherva R, Koesterer R, Almasy L, Zhao H, Kranzler HR, Farrer L. Genome-wide association study of cocaine dependence and related traits: FAM53B identified as a risk gene. Mol Psychiatry. 2014;19:717–723. doi: 10.1038/mp.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gelernter J, Kranzler HR, Sherva R, Koesterer R, Almasy L, Zhao H, Farrer L. Genome-wide association study of opioid dependence: Multiple associations mapped to calcium and potassium pathways. Biol Psychiatry. 2014;76:66–74. doi: 10.1016/j.biopsych.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, et al. Genome-wide association study of alcohol dependence: Significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry. 2014;19:41–49. doi: 10.1038/mp.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quillen EE, Chen XD, Almasy L, Yang F, He H, Li X, et al. ALDH2 is associated to alcohol dependence and is the major genetic determinant of “daily maximum drinks” in a GWAS study of an isolated rural chinese sample. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:103–110. doi: 10.1002/ajmg.b.32213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maes HH, Sullivan PF, Bulik CM, Neale MC, Prescott CA, Eaves LJ, Kendler KS. A twin study of genetic and environmental influences on tobacco initiation, regular tobacco use and nicotine dependence. Psychol Med. 2004;34:1251–1261. doi: 10.1017/s0033291704002405. [DOI] [PubMed] [Google Scholar]
- 6.Kendler KS, Neale MC, Sullivan P, Corey LA, Gardner CO, Prescott CA. A population-based twin study in women of smoking initiation and nicotine dependence. Psychol Med. 1999;29:299–308. doi: 10.1017/s0033291798008022. [DOI] [PubMed] [Google Scholar]
- 7.Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO. The Fagerstrom Test for Nicotine Dependence: A revision of the Fagerstrom Tolerance Questionnaire. Br J Addict. 1991;86:1119–1127. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- 8.Broms U, Madden PA, Heath AC, Pergadia ML, Shiffman S, Kaprio J. The Nicotine Dependence Syndrome Scale in Finnish smokers. Drug Alcohol Depend. 2007;89:42–51. doi: 10.1016/j.drugalcdep.2006.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vink JM, Willemsen G, Boomsma DI. Heritability of smoking initiation and nicotine dependence. Behav Genet. 2005;35:397–406. doi: 10.1007/s10519-004-1327-8. [DOI] [PubMed] [Google Scholar]
- 10.Edwards AC, Maes HH, Pedersen NL, Kendler KS. A population-based twin study of the genetic and environmental relationship of major depression, regular tobacco use and nicotine dependence. Psychol Med. 2011;41:395–405. doi: 10.1017/S0033291710000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tobacco and Genetics Consortium . Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.David SP, Hamidovic A, Chen GK, Bergen AW, Wessel J, Kasberger JL, et al. Genome-wide meta-analyses of smoking behaviors in African Americans. Transl Psychiatry. 2012;2:e119. doi: 10.1038/tp.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, et al. Diagnostic reliability of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA) Drug Alcohol Depend. 2005;80:303–312. doi: 10.1016/j.drugalcdep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Edenberg HJ. The collaborative study on the genetics of alcoholism: An update. Alcohol Res Health. 2002;26:214–218. [PMC free article] [PubMed] [Google Scholar]
- 17.Bierut LJ, Strickland JR, Thompson JR, Afful SE, Cottler LB. Drug use and dependence in cocaine dependent subjects, community-based individuals, and their siblings. Drug Alcohol Depend. 2008;95:14–22. doi: 10.1016/j.drugalcdep.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 20.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.1000 Genomes Project Consortium. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeger SL, Liang KY. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42:121–130. [PubMed] [Google Scholar]
- 24.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: Design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinb) 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 26.Yoav Benjamin YH. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 27.Gao B, Sekido Y, Maximov A, Saad M, Forgacs E, Latif F, et al. Functional properties of a new voltage-dependent calcium channel alpha(2)delta auxiliary subunit gene (CACNA2D2) J Biol Chem. 2000;275:12237–12242. doi: 10.1074/jbc.275.16.12237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Currie KP. G protein modulation of CaV2 voltage-gated calcium channels. Channels (Austin) 2010;4:497–509. doi: 10.4161/chan.4.6.12871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhodin A, Gronbladh A, Ginya H, Nilsson KW, Rosenblad A, Zhou Q, et al. Combined analysis of circulating beta-endorphin with gene polymorphisms in OPRM1, CACNAD2 and ABCB1 reveals correlation with pain, opioid sensitivity and opioid-related side effects. Mol Brain. 2013;6:8. doi: 10.1186/1756-6606-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhan L, Kerr JR, Lafuente MJ, Maclean A, Chibalina MV, Liu B, et al. Altered expression and coregulation of dopamine signalling genes in schizophrenia and bipolar disorder. Neuropathol Appl Neurobiol. 2011;37:206–219. doi: 10.1111/j.1365-2990.2010.01128.x. [DOI] [PubMed] [Google Scholar]
- 31.Xie P, Kranzler HR, Krystal JH, Farrer LA, Zhao H, Gelernter J. Deep resequencing of 17 glutamate system genes identifies rare variants in DISC1 and GRIN2B affecting risk of opioid dependence. Addict Biol. 2013;19:955–964. doi: 10.1111/adb.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomson PA, Parla JS, McRae AF, Kramer M, Ramakrishnan K, Yao J, et al. 708 Common and 2010 rare DISC1 locus variants identified in 1542 subjects: Analysis for association with psychiatric disorder and cognitive traits. Mol Psychiatry. 2014;19:668–675. doi: 10.1038/mp.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hallfors J, Loukola A, Pitkaniemi J, Broms U, Mannisto S, Salomaa V, et al. Scrutiny of the CHRNA5-CHRNA3-CHRNB4 smoking behavior locus reveals a novel association with alcohol use in a Finnish population based study. Int J Mol Epidemiol Genet. 2013;4:109–119. [PMC free article] [PubMed] [Google Scholar]
- 34.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saccone NL, Culverhouse RC, Schwantes-An TH, Cannon DS, Chen X, Cichon S, et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: A meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 2010;6:8. doi: 10.1371/journal.pgen.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saccone NL, Wang JC, Breslau N, Johnson EO, Hatsukami D, Saccone SF, et al. The CHRNA5-CHRNA3-CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African-Americans and in European-Americans. Cancer Res. 2009;69:6848–6856. doi: 10.1158/0008-5472.CAN-09-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie P, Kranzler HR, Yang C, Zhao H, Farrer LA, Gelernter J. Genome-wide association study identifies new susceptibility loci for posttraumatic stress disorder. Biol Psychiatry. 2013;74:656–663. doi: 10.1016/j.biopsych.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez de Morentin PB, Whittle AJ, Ferno J, Nogueiras R, Dieguez C, Vidal-Puig A, López M. Nicotine induces negative energy balance through hypothalamic AMP-activated protein kinase. Diabetes. 2012;61:807–817. doi: 10.2337/db11-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng PY, Lee YM, Law KK, Lin CW, Yen MH. The involvement of AMP-activated protein kinases in the anti-inflammatory effect of nicotine in vivo and in vitro. Biochem Pharmacol. 2007;74:1758–1765. doi: 10.1016/j.bcp.2007.08.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.