Abstract
Optimization is reported for biaryltriazoles as inhibitors of the tautomerase activity of human macrophage migration inhibitory factor (MIF), a proinflammatory cytokine associated with numerous inflammatory diseases and cancer. A combined approach was taken featuring organic synthesis, enzymatic assaying, crystallography, and modeling including free-energy perturbation (FEP) calculations. X-ray crystal structures for 3a and 3b bound to MIF are reported and provided a basis for the modeling efforts. The accommodation of the inhibitors in the binding site is striking with multiple hydrogen bonds and aryl–aryl interactions. Additional modeling encouraged pursuit of 5-phenoxyquinolinyl analogues, which led to the very potent compound 3s. Activity was further enhanced by addition of a fluorine atom adjacent to the phenolic hydroxyl group as in 3w, 3z, 3aa, and 3bb to strengthen a key hydrogen bond. It is also shown that physical properties of the compounds can be modulated by variation of solvent-exposed substituents. Several of the compounds are likely the most potent known MIF tautomerase inhibitors; the most active ones are more than 1000-fold more active than the well-studied (R)-ISO-1 and more than 200-fold more active than the chromen-4-one Orita-13.
INTRODUCTION
Macrophage migration inhibitory factor (MIF) is a cytokine that plays a central role in numerous inflammatory diseases.1–3 MIF is widely expressed in both immune and nonimmune cells including macrophages, endothelial cells, and T-cells. Upon activation, the cells release MIF, which promotes the release of other inflammatory cytokines such as TNF-α and IL-1. Excessive or chronic inflammatory response is associated with tissue damage and autoimmune diseases such as rheumatoid arthritis, Crohn’s disease, and lupus erythematosus. The connection between inflammatory disease and cancer is also well-established, and MIF has been shown to enhance cell proliferation by inhibiting accumulation of the tumor suppressor p53 and by promotion of angiogenesis.4 MIF is overexpressed in many cancer cells and can serve as a marker for disease progression. Furthermore, MIF in cancer cells is protected from degradation by Hsp90, which has led to proposed targeting of Hsp90 as an indirect way of inhibiting MIF function.5 Disruption of the inflammatory cascade and restoration of normal p53 levels have clear implications for the potential therapeutic value of inhibitors of MIF signaling. Indeed, immunoneutralization of MIF or deletion of the MIF gene is known to suppress inflammatory response, tumor growth, and angiogenesis.1–4 At the molecular level, what is needed is interference with the interaction between MIF and its cell-surface receptor CD74.6
MIF is a toroid-shaped, trimeric protein with a total of 342 amino acid residues. Besides its role as a cytokine, MIF is a keto–enol tautomerase. Though the enzymatic activity appears to be vestigial in humans, there are three tautomerase active sites at the interfaces of the monomer units opening to the outside of the toroid. Inhibition of protein–protein interactions is often challenging; however, the presence of the tautomerase sites presents an opportunity for complexation of a tautomerase inhibitor that may also interfere with MIF/CD74 binding. This notion has been supported by many studies that show correlation between the inhibition of the enzymatic and biological activities of MIF.7 For example, this has been demonstrated through assay results for tautomerase activity, MIF/CD74 binding, and MIF-induced phosphorylation of ERK1/2 in inflamed cells, production of interleukins, glucocorticoid overriding ability, and macrophage chemotactic migration.7–12 Nevertheless, the discovery of potent MIF tautomerase inhibitors is not well advanced as most inhibitors have arisen from screening exercises with no lead optimization.8,11,12 In our efforts,8–10 lead optimization has been limited by the lack of crystal structures for our tautomerase inhibitors bound to MIF, associated inconsistency between modeling results and activity data, and sensitivity of assay results to the substrate and protein source. As described here, these issues have been overcome for the series of biaryltriazoles, 1–3. In particular, we report crystal structures for two complexes of derivatives of 3 with MIF, extensive structure–activity data that is consistent with both crystallography and modeling, and reliable assay protocols. The most active compounds are ca. 1000-fold more potent as MIF tautomerase inhibitors than the known MIF inhibitor (R)-ISO-1,13,14 which has shown efficacy in rodent models as an anti-inflammatory and anticancer agent.2,4
EXPERIMENTAL SECTION
Chemistry
In a previous report, we described the synthesis of 27 1,2,3-triazole derivatives in four motifs with a 4-hydroxyphenyl group at the 1-position and a substituted benzyl or aryl group at the 4-position or vice versa.10 These constructs arose from de novo design using the program BOMB, which builds and scores libraries of compounds that it grows in a binding site.15 Only three compounds were synthesized with a an aryl group at the 4-position, namely, the 3-pyridinyl analogue 1 (X = H, Z = OH) and the corresponding 1-naphthyl and 4-isoquinolinyl alternatives. The modeling indicated that the nitrogen atom in the 3-pyridinyl and 4-isoquinolinyl compounds would coordinate with the ammonium group of a lysine residue (Lys32A) and that this interaction would likely be lost and replaced by a repulsive interaction with the oxygen of Ile64A in 2-pyridinyl and 2-quinolinyl analogues, that is, 2 and 3. However, 1 (X = H, Z = OH) and the isoquinolinyl analogue exhibited unexpected behavior as agonists in an assay for binding of MIF with the ectodomain of CD74.10 They also showed inconclusive behavior in a MIF tautomerase assay; they appeared to be inactive or weak agonists lacking dose-dependent character. This eventually raised questions: (a) would the 2-pyridinyl and 2-quinolinyl isomers of the agonists be agonists, antagonists, or inactive, and (b) if an appendage were added in the 6-position of the pyridine ring in 1, could this convert the agonist into an antagonist? As a crystal structure was not available for any of our compounds bound to MIF, the structural analyses and expectations for activity did not have firm footing. Thus, a scouting mission was initiated for 1 (X = methoxyethoxy, Z = OH), 2 (X =H, Z = OH), and 3 (X =H, Z = OH). As reported here, the surprising results led to lead optimization of 2 and 3 for inhibition of MIF tautomerase activity. The new compounds are listed as 1b–4b in Table 1; 4 is the 1,8-naphthryidine analogue of 3, while (R)-ISO-1 and 3-(3,4-dihydroxyphenyl)-7-hydroxy-4H-chromen-4-one (Orita-13) are reference compounds.13,16
Table 1.
Results for Inhibition of the Tautomerase Activity of Human MIFa
| compd | X | Y | Z | Ki (µM)b |
|---|---|---|---|---|
| 1a | H | OH | 37 | |
| 1b | MOEO | OH | 22 | |
| 2a | H | OH | 8.8 | |
| 2b | MOEO | Cl | ND (0%) | |
| 3a | H | H | OH | 0.59 |
| 3b | MOEO | H | OH | 0.65 |
| 3c | AEOEO | H | OH | 0.77 |
| 3d | MOEO | H | F | 8.9 |
| 3e | MOEO | H | Cl | ND (3%) |
| 3f | MOEO | H | NH2 | ND (13%) |
| 3g | MOEO | H | OMe | ND (0%) |
| 3h | MOEO | H | CN | ND (8%) |
| 3i | MOEO | H | CONH2 | ND (0%) |
| 3j | MOEO | H | 3-Me, 4-OH | ND (15%) |
| 3k | MOEO | H | 3-OMe, 4-OH | ND (8%) |
| 3l | H | 3-Me | OH | 7.3 |
| 3m | H | 4-Me | OH | 2.3 |
| 3n | H | 8-Cl | OH | ND (16%) |
| 3o | H | 8-OMe | F | 56 |
| 3p | H | 8-MOEO | F | 64 |
| 3q | H | 8-p-MeOPh | OH | ND (21%) |
| 3r | H | 8-OPh | OH | 2.95 |
| 3s | H | 5-OPh | OH | 0.37 |
| 3t | Mr(CH2)3O | H | OH | 1.95 |
| 3u | Mr(CH2)3O | 3-Me | OH | 3.12 |
| 3v | MrEOEO | H | OH | 0.41 |
| 3w | MrEOEO | H | 3-F, 4-OH | 0.15 |
| 3x | MrEOEO | H | F | 29.6 |
| 3y | H | 5-p-MOEO-OPh | OH | 0.36 |
| 3z | H | 5-m-MOEO-OPh | 3-F, 4-OH | 0.082 |
| 3aa | H | 5-p-COOH-OPh | 3-F, 4-OH | 0.11 |
| 3bb | H | 5-m-COOH-OPh | 3-F, 4-OH | 0.057 |
| 4a | H | H | OH | 1.48 |
| 4b | H | H | Cl | ND (9%) |
| Orita-13 | 17 | |||
| (R)-ISO-1 | 120 |
MOEO = methoxyethoxy; AEOEO = aminoethoxyethoxy; Mr = N-morpholinyl.
ND = Ki not determined; % inhibition at 10 µM in parentheses.
The synthetic approach was conceptually straightforward featuring a 1,3-dipolar cycloaddition between an azide and a substituted 2-ethynylpyridine, quinoline, or naphthyridine; however, access to the appropriately substituted heterocycles was not always easy. Though full details are provided in the Supporting Information, Schemes 1 and 2 illustrate the synthetic routes for two cases, 3s and 3q. Typically, 2-chloroquinolinols are available commercially at low cost; however, this is not the situation for 2-chloroquinolin-5-ol, which would have facilitated the synthesis of 3s. Thus, starting with quinolin-5-ol, arylation was followed by oxidation to the N-oxide, which was chlorinated17 to yield 2-chloro-5-phenoxyquinoline. A standard sequence10 featuring Sonogashira coupling with TMS-protected acetylene followed by one-pot, Cu(I)-catalyzed reaction of the azide formed from an aryl bromide or iodide then yielded 3s.18
Scheme 1.
Synthesis of 3s
Scheme 2.
Synthesis of 3q
With 3q (Scheme 2), a substituted phenyl group was desired in the 8-position of the quinoline. This turned out to be challenging as numerous attempts at Suzuki couplings failed. For example, couplings with the triflate of 2-chloro-quinolin-8-ol gave at best 1:1 mixtures of 2-and 8-arylation, and no desired product arose from attempted couplings of either the 8-triflate or 8-pinacol boronic ester of 2-trimethylsilylethynylquinolin-8-ol. Instead, starting with commercially available 4′-methoxy-[1,1′-biphenyl]-2-amine, the desired 2-chloroquinoline was built by acylation to the acrylamide, followed by cyclization to the quinolone, and treatment with POCl3 at reflux (Scheme 2). 3q was then completed via the Sonogashira-1,3-dipolar cyclization sequence.
The identity of assayed compounds was confirmed by 1H and 13C NMR and high-resolution mass spectrometry; HPLC analyses established purity as >95%. Measurements of aqueous solubilities were carried out using a shake-flask procedure.19 Saturated solutions were made by stirring excesses of the compounds in Britton-Robinson buffer for 48 h at 30 °C. The pH of the buffer solutions was 6.5 as measured by using a Corning General Purpose pH Combination probe (4136L21). The supernatant was collected using a Pall Life Sciences Acrodisc syringe filter with a 0.2 µm pore size, and analyzed by UV–vis spectrophotometry (Agilent 8453). Piroxicam was used as a control; our values of 7–9 µg/mL are consistent with the literature value of 5.9 µg/ mL.19
Computer Modeling
All structure building was carried out using the BOMB program starting from a previously reported crystal structure of human MIF with 4-hydroxyphenylpyruvate (PDB code: 1CA7)20 or from our structure of the complex with 3b. Subsequent calculations included energy minimizations and free-energy perturbation (FEP) calculations with the MCPRO program.21 Details of the calculations are described elsewhere.22 Briefly, the OPLS-AA force field is used for the protein, OPLS/CM1A for the ligands, and TIP4P for water molecules.23 For the FEP calculations, the unbound ligands and complexes were solvated in water caps with a 25 Å radius, amounting to ca. 2000 and 1250 water molecules, respectively. The 218 amino acid residues nearest to the ligand were included in the model for the complexes. A residue-based cutoff for nonbonded interactions was invoked at 10 Å. After short conjugate-gradient optimizations, the backbone atoms of the protein were fixed. The ligand and side chains with any atom within ca. 15 Å of the ligand were fully sampled. All water molecules were sampled using translations and rigid rotations. The FEP calculations used 11 or 21 windows of simple overlap sampling. Each window covered at least 10 million configurations of equilibration and 10 million configurations of averaging for the complexes and 30 million configurations of averaging for the unbound inhibitors.
Biology
Protein Expression and Purification
Recombinant human MIF (rhMIF) was expressed as described previously.24 Escherichia coli cells were pelleted by centrifugation and stored at −80 °C. The purification followed published protocols24,25 with slight modifications. Cell pellets were resuspended in a lysis buffer containing 20 mM Tris-HCl pH 7.5, 20 mM sodium chloride, 10% glycerol, 2 mM magnesium chloride, and 0.2× cOmplete EDTA-free protease inhibitor cocktail (Roche), lysed by sonication and centrifuged at 27 000g for 30 min. The supernatant was filtered through a 0.22 µm syringe filter and applied to Hi-Trap SP HP and Hi-TrapQSP columns (GE Healthcare) in tandem. As rhMIF did not bind to either ion-exchange resin, the flow-through was collected, being sufficiently pure (~90%) for crystallography. Higher purity was achieved by size-exclusion chromatography on a Superdex 200 16/60 column (GE Healthcare). The resulting rhMIF was assessed by SDS gel electrophoresis to be of sufficiently high purity (>95%) for tautomerase assays. Pure protein was concentrated to 30.6 mg/mL in 20% glycerol and stored at −80 °C.
Tautomerase Assay, Ki Determination
Inhibition of the tautomerase activity of MIF was measured using 4-hydroxyphenyl pyruvic acid (HPP) as substrate, largely following previously reported protocols.26 HPP was dissolved in 0.5 M acetate buffer, pH 6.0 to a final concentration of 10 mM and incubated overnight at room temperature to allow equilibration of the keto and enol forms. MIF (6 µL) was premixed in 500 mM boric acid, pH 6.2 (142 µL) and transferred to a transparent U bottom 96-well plate to a final concentration of 200 nM MIF. It was important to optimize the protein concentration; this was performed by analysis of progress curves for enol production at protein concentrations of 50–800 nM. High signal-to-noise and linearity were observed for 200 and 400 nM MIF; below these levels, weaker signal limited accuracy of the results. Inhibitors were dissolved in DMSO to 10 mM and an initial screen was performed. For compounds that showed ca. 25% or greater inhibition at 10 µM, an inhibition constant, Ki, was measured. Compounds were placed into wells (2 µL) at six different concentrations and incubated for 30 min until the assay was started by addition of HPP (50 µL) at two concentrations (1.0 and 2.5 mM). The negative control was MIF incubated with DMSO vehicle, which in all assays was 1% and did not influence tautomerase activity. MIF activity was monitored at 305 nm for formation of the borate–enol complex using an Infinite F500 plate reader (TECAN, Morrisville, NC) for 175 s. Calculation of initial velocities and the nonlinear regression analyses for the enzyme kinetics were repeated three times with the program Prism6 (GraphPad, La Jolla, CA). The results always fit better to the competitive inhibition mode rather than the noncompetitive one, which is consistent with the crystallographic results.
Previously, we had attempted assays using rhMIF purchased from external vendors. This proved unsatisfactory as different vials of protein showed great variation in activity, including totally inactive. Even different vials received at the same time from the same vendor showed different activities. The present assay results were all obtained with protein prepared on only two occasions; when more protein was needed, another aliquot was thawed, and never refrozen. Also, Orita-13 and/or 3b were used as control compounds when new rounds of assays were conducted. The Ki results all fell in the range 13–22 µM for Orita-13, and nine independent measurements for 3b yielded results of 0.55–0.85 µM. Samples of Orita-13 and (R)-ISO-1 were purchased both from Alfa Aesar and Santa Cruz Biotechnology; consistent spectra and Ki results were obtained.
We also investigated use of l-dopachrome methyl ester (DOPA) rather than HPP as the substrate in the manner of Orita et al.16 An advantage in principle is that the absorbance is evaluated at 492 nm, which may experience less interference from the inhibitors than the 305 nm detection with HPP. However, DOPA is photosensitive, which limits the data collection to 25 s versus 175 s with HPP. The shorter linear range for calculation of the initial velocities results in poorer fits for the results at different concentrations and much less reliable Ki values.
Protein Crystallography
To obtain cocrystals of MIF in complex with 3a, 100 µM 3a in DMSO was added to rhMIF (24 µg/mL) to achieve a 3:1 molar ratio and incubated for 1 h at 5 °C. The solution was centrifuged at 13 000g to remove precipitated compound and used to set up hanging-drop crystallization experiments. A reservoir of 2.0 M ammonium sulfate, 0.1 M Tris pH 7, and 3% isopropanol was added to the protein solution in a 1:1 ratio and stored at 20 °C. Diffraction-quality crystals with a rod morphology grew within 2 weeks. The crystals were cryo-protected in 25% glycerol, 2.0 M ammonium sulfate, 0.1MTris pH 7, and 3% isopropanol and shipped to the Advanced Photon Source for remote data collection on the NE-CAT 24-ID-E beamline.
Cocrystals of MIF in complex with 3b were obtained by soaking crystals of apo MIF. Crystals were obtained by the hanging-drop method at 20 °C. A reservoir of 2.2 M ammonium sulfate, 0.1 M Tris pH 7, and 3% isopropanol mixed in a 1:1 ratio with rhMIF (16 mg/mL) was used to produce 2 µL drops. Once crystals formed, 0.5 µL of a suspension of 10 mM 3b in 10% DMSO, 2.0Mammonium sulfate, 90 mM Tris pH 7, and 2.7% isopropanol was added to the drop and allowed to incubate for 14 days. Crystals were cryo-protected with 25% glycerol, 2.2 M ammonium sulfate, 0.1 M Tris pH 7, and 3% isopropanol and diffracted on a Rigaku 007 HF+ X-ray source equipped with a Saturn 944+ CCD detector at Yale. Full details of the data collection and refinement for 3a and 3b are provided in the Supporting Information.
RESULTS AND DISCUSSION
In presenting the results, a sense of the progression of events will be given. The previous report was for work through mid-2010 and included 1a.10 The triazole series were not pursued again until early 2012, when 1b, 2a, and 3a were synthesized. However, the assay issues and protein access discussed above, which were needed to allow report of the carefully controlled results in Table 1, were not fully worked out until 2013.
With the present assay protocols, the parent compounds 1a, 2a, and 3a are all MIF tautomerase inhibitors. Progression from the 3-pyridinyl (1a) to 2-pyridinyl (2a) to 2-quinolinyl (3a) derivatives shows strong enhancement in activity from 37 to 8.8 to 0.59 µM. For comparison, (R)-ISO-1, which is reported14 to have an IC50 of 7 µM in a MIF tautomerase assay with DOPA as the substrate, has a Ki of 120 µM in our assay. Issues with IC50 measurements are well-known; they depend on the concentration and Michaelis constant (Km) of the substrate, while Ki is an intrinsic measure of the binding of the protein and inhibitor.27 In an independent assay of ISO-1 using DOPA, an IC50 of >100 µM was reported; the discrepancy with the 7 µM value was suggested to arise from use of different concentrations of rhMIF.28 We also find Orita-13 with a Ki averaging 17 µM to be much less active than from the previously reported Ki of 0.038 µM, again in a dopachrome assay.16 Orita-13 would seem to be the most potent MIF inhibitor in the journal literature.11 It arose from a screening study that reported Ki values for 14 compounds with the next most potent compound at 0.28 µM.16 There have been no follow-up reports with the compound except for a crystal structure29 that found it rotated 180° in the binding site from the original X-ray study,16 and it appears not to have been assayed again until now. Though we find Orita-13 to be 7-fold more active than ISO-1, it is 30-fold less active than 3a.
Crystallography
To progress from this point, a recurrent conformational issue complicated modeling efforts.10 For 3a, as an example, four principal geometries could be constructed in the binding site, which can be labeled 3a-ud, 3a-dd, 3a-du, and 3a-uu. These represent two conformers that can both be rotated 180° about the long axis in the binding site. The structure building and energy minimizations with BOMB and MCPRO could rule out 3a-ud and 3a-uu, but the preference between 3add and 3a-du was uncertain. 3a-dd is the higher-energy conformer in the gas-phase than 3a-du, by 6 kcal/mol according to OPLS/CM1A calculations, owing to the added quinoline N– triazole N3 repulsion; however, 3a-dd appeared to make better hydrogen-bonding interactions in the binding site.
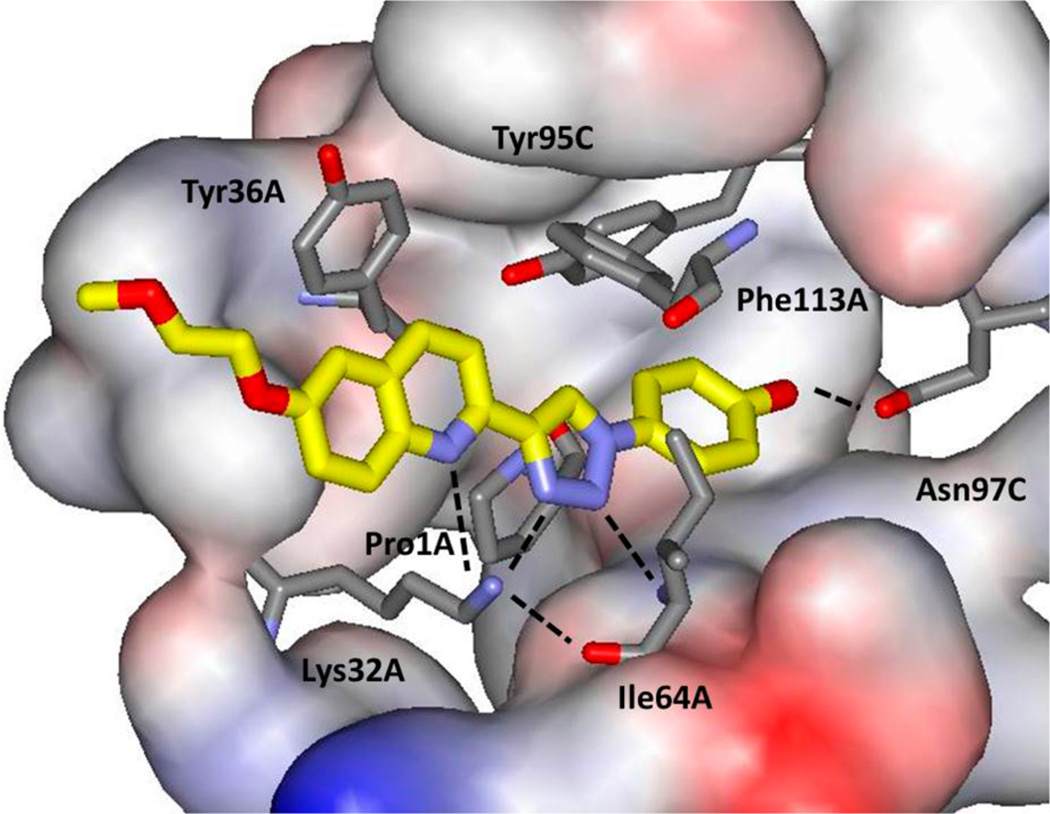
This issue was resolved by obtaining the crystal structures for 3a and 3b in complex with MIF at resolutions of 2.60 and 1.81 Å, respectively. As illustrated in Figure 1 for 3b, the crystal structures of both complexes showed that 3a-dd is the preferred geometry. The inhibitor is inserted such that the phenolic hydroxyl group is hydrogen-bonded with Asn97C (r(OO) = 2.52 Å); there is also a hydrogen bond between N2 of the triazole and the backbone NH of Ile64A (r(NN) = 2.90 Å), and there is the striking complexation of the ammonium group of Lys32A by the quinoline N, triazole N3, and O of Ile64A (r(NN) = 3.33, 2.95; r(NO) = 2.81 Å), which requires the higher-energy 3a-dd geometry. There are also aryl–aryl interactions between the phenolic and quinolinyl fragments of the inhibitor and Tyr95C and Tyr36A, respectively, and Phe113A has contacts at the junction of the quinoline and triazole. The structure appears very well packed with a large number of favorable intermolecular features given the size of the inhibitor. The crystal structures for 3a and 3b are essentially the same, and it was pleasing that the methoxyethoxy substituent of 3b was well-resolved (Figure S2). Though the substituent would be solvent-exposed in dilute solution, in the crystal it is in contact with an adjacent MIF trimer. The fact that 3a and 3b show the same activity (Table 1) is fully consistent with the crystal structures. It may also be noted that both crystal structures only have one of the three tautomerase sites occupied by 3a or 3b, the remaining sites being occupied by solvent and/or glycerol.
Figure 1.
Rendering of 3b bound to human MIF from the 1.81 Å crystal structure (PDP ID: 4WRB). Carbon atoms of 3b are shown in yellow; some residues have been removed for clarity. Hydrogen bonds are highlighted with dashed lines.
In previously reported crystal structures, hydrogen bonding interactions for an inhibitor with the side chains of Asn97 and Lys32 and with the backbone NH of Ile64 have frequently been observed along with aryl–aryl contacts involving Tyr95 and Phe113.16,20,29 The interactions with Lys32 have typically come from a carbonyl or carboxylate group of the inhibitor. For example, in the 3L5R structure for Orita-13, the hydroxyl group of the chromen-4-one is hydrogen bonded with Asn97, and the carbonyl oxygen atom is hydrogen-bonded to both Lys32 and the NH of Ile64.29 The present structures are particularly striking because the triazolylquinoline substructure provides three separate hydrogen bonds with Lys32A and Ile64A as well as an additional aryl–aryl interaction with Tyr36A (Figure 1). It may also be noted that, in the present structures Pro1A, the catalytic base for the tautomerase reaction and nucleophile for formation of covalent inhibitors11 is probably protonated. One hydrogen atom on the nitrogen is likely pointing toward the carbonyl oxygen atom of Tyr36A with an N–O distance of 3.22 Å, while there is a clear hydrogen bond between this carbonyl oxygen atom and the hydroxyl group of Tyr95C at an O–O distance of 2.51 Å. If there is a second hydrogen atom on the Pro1A nitrogen atom, it would be pointing toward N1 of the triazole (r(NN) = 3.25 Å), and there would be favorable cation–π interactions with the electron-rich ring.
Phenyl and Quinolinyl Substituents
Having resolved the structural issue, FEP calculations were performed to seek substituent modifications in the phenyl and quinolinyl fragments that could improve potency. The results are summarized in Table 2. It should be noted that Clx and Brx refer to modeling the halogens with an extra partial positive charge, which allows representation of halogen bonding.30 There is little empty space in the vicinity of the phenolic hydroxyl group. The FEP results indicated that all replacements of the hydroxyl group would be unfavorable, though the penalty for switching to fluorine should not be severe. There is a trade-off between loss of hydrogen-bonding with Asn97C and a diminished dehydration penalty. An amino group is not favorable since only one of the two amino hydrogens can form a hydrogen bond to Asn97C. The activity data for 3b and 3d–3i in Table 1 are consistent with the predictions. Only the fluoro analogue 3d showed significant activity with a Ki of 8.9 µM, ca. 15-fold higher than that for 3b. For 3j and 3k, a methyl or methoxy substituent adjacent to the hydroxyl group was also considered, but as expected from the tight packing, these additions were unfavorable.
Table 2.
MC/FEP Results for Changes in Free Energy of Binding to Human MIFa
 | |||||
|---|---|---|---|---|---|
| H to X | ΔΔGb | H to Yb | ΔΔGb | H to Yb | ΔΔGb |
| F | −2.27 | 3-F | 0.15 | 4-OMe | 0.02 |
| Cl | −1.77 | 3-Cl | −0.82 | 4-OH | 1.64 |
| Clx | −1.06 | 3-Clx | −0.32 | 8-F | −0.79 |
| Br | −1.16 | 3-Me | −2.69 | 8-Cl | −1.49 |
| Brx | −0.62 | 3-Et | −1.56 | 8-Clx | −2.06 |
| Me | 2.73 | 3-OMe | 0.23 | 8-Me | 0.61 |
| Et | 6.26 | 4-F | 0.46 | 8-Et | −0.11 |
| NH2 | 1.89 | 4-Cl | 0.32 | 8-OMe | −0.51 |
| OMe | 0.14 | 4-Clx | 0.18 | 8-OEt | −0.16 |
| OH | −2.70 | 4-Me | −1.00 | 8-MOM | 0.27 |
| 4-Et | −0.34 | 8-CH2F | 0.40 | ||
ΔΔGb is the computed change in free energy of binding (kcal/mol); Clx and Brx include X-sites to allow halogen bonding; the statistical uncertainty in the results (±1σ) is 0.2 kcal/mol.
With X = OH.
Turning to the quinoline ring, the 3-, 4-, and 8-positions were first considered for possible additions of small groups. Structure-building with BOMB suggested that at least a small alkyl group might be accommodated at C3 or C4 in the space near Tyr36A. However, the FEP results indicated that about the only hope would be for a methyl group at C3 (Table 2), since we have generally observed that the computed enhancement needs to be beyond −2 kcal/mol to have good confidence in an observed activity increase.15 Indeed, the assay results for 3l and 3m (7.3 and 2.3 µM) showed weaker inhibition than for the parent 3a (0.59 µM), while 3t and its 3-methyl analogue 3u showed essentially the same activity. In view of the FEP results for the other options, further exploration at these sites was not pursued.
Structure building at C8 then suggested that addition of an alkoxy group or substituted phenyl might provide additional coordination with Lys32A (Figure 1). Enticing images could be generated as in Figure 2 showing a possible cation–π interaction in the latter case. Such interactions require representation of explicit polarization effects in the force fields,23 so a confident FEP result could not be obtained with OPLS/CM1A. The FEP calculations were executed for nine options at C8, as shown in Table 2 with the result that chlorine might be most promising. However, the predicted ΔΔGb values of −1.49 and −2.06 kcal/ mol are on the fringe of the −2 kcal/mol threshold. In fact, the 8-Cl analogue 3n was found to have diminished activity. The ΔΔGb for an 8-methoxy group of −0.51 kcal/mol made it likely that this modification would also be unproductive. This was tested with 3o that has a fluorine replacing the phenolic hydroxyl group; its Ki of 56 µM suggests that the phenol would be 4 or 0.8 µM, if the 3d/3b or 3x/3v ratio is transferable. Though the oxygen of the methoxy group of 3o can form a hydrogen bond with the ammonium group of Lys32, it is replacing a water molecule that is fulfilling this role and it requires orientation of the oxygen toward the quinoline N, which promotes lone pair–lone pair repulsion. In the balance, consistent with the FEP prediction, the 8-methoxy group in 3o and the methoxyethoxy (MOEO) group in 3p did not provide a notable activity boost.
Figure 2.
Computed structure for an analogue of 3a with a phenyl substituent on C8 of the quinoline highlighting a possible cation–π interaction with Lys32A. Carbon atoms of the ligand are in yellow; the ammonium group of Lys32A is shown as a blue sphere. Some residues have been removed for clarity.
Returning to the possibility of a phenyl substituent at C8, as noted in Scheme 2, there were challenges in the synthesis of such compounds. Nevertheless, there was success with the p-methoxy analogue 3q. In spite of the attractive graphics for 8-phenyl analogues (Figure 2), 3q only showed 21% inhibition of MIF’s tautomerase activity at 10 µM (Table 1). Lys32 is on the surface of MIF, and it appears difficult to substitute favorably for the water molecules on its solvent-exposed side. The possibility of additional coordination from the ligand with Lys32A via the 1,8-naphthyridine 4a was also considered; the result was a respectable Ki of 1.48 µM, but not an improvement over 3a.
Still not relenting, structures were built with BOMB with a phenoxy group at C5, C6, C7, and C8 of the quinoline ring. A conformational search on the 8-phenoxy analogue indicated that it would be better preorganized for binding and interaction with Lys32A than the 8-methoxy alternative, and perhaps the phenoxy groups might interact with other surface residues. In fact, energy minimizations showed that this could be the case for the C5 (Figure 3) and C8 phenoxy compounds. Thus, this notion was pursued yielding 3r and 3s. The 8-phenoxy analogue 3r is a 3.0 µM inhibitor, while the 5-phenoxy 3s at 0.37 µM did provide an advance over 3a and 3b (Table 1). The presumption is that the added intermolecular contacts illustrated in Figure 3 are beneficial for binding.
Figure 3.
Computed structure for 5-phenoxy-containing 3s bound to human MIF illustrating surface contact and a possible aryl–aryl interaction with Tyr36A. Carbon atoms of the ligand are in yellow. Some residues have been removed for clarity.
Aqueous Solubility
From the crystal structures and modeling, it was expected that physical properties of the compounds could be modulated by variation of substituents that would be solvent exposed. Attachments at the 6- or 7-positions in the quinoline ring seemed likely to be appropriate (Figures 1–3). For illustration, aqueous solubility was considered. Thus, polyether-containing groups were appended at C6 with the 6-hydroxy compounds as precursors. Aqueous solubilities were measured for a series of four compounds as summarized in Scheme 3. The solubilities of 3a (2.2 µg/mL or 7.6 µM) and 3b (3.6 µg/mL or 9.9 µM) are surprisingly low given the small size, percentage of heteroatoms, and the computed octanol/water partition coefficients of 3.3 to 4.0 from ChemDraw and QikProp.31,32 When experimental data for log S and log Po/w are analyzed,33 a rule-of-thumb is log S = −log Po/w − 0.2 with r2 = 0.70 and rms = 1.0 for 271 compounds with log Po/w > 0. Thus, log S ≈ 10−4 or S ≈ 100 µM for 3a and 3b could be expected within a factor of 10. The observed results are at the very bottom of this range. Addition of the amino group in 3c has the expected qualitative effect, raising the solubility to 13.9 µg/mL. A further 3–4-fold boost is obtained upon replacement of the amino group with N-morpholinyl, a well-known solubilizing group.34,35 3v is potent with a Ki of 0.41 µM, and its solubility of 48.5 µg/mL (1.05 × 10−4 M) is well inside the range normally observed for oral drugs, 4–4000 µg/mL.33 The ClogP appears to be high in this case; the pattern for the four compounds from QikProp seems more reasonable. The predicted solubility from QikProp of 0.2 × 10−4 M for 3v is also within normal error bounds.32
Scheme 3.
Experimental Solubilites (S, µg/mL) and Computed log P
Final Compounds
Given the good results for 3s and 3v, additional analogues were synthesized to again test the impact of replacing the phenolic hydroxyl group with fluorine (3x) and for addition of a fluorine adjacent to the hydroxyl group (3w, 3z, 3aa, 3bb). Though methyl and methoxy groups could not be accommodated at the meta position (3j and 3k), fluorine is smaller and it is expected to enhance the hydrogen-bond donor character of the hydroxyl group and strengthen the hydrogen bond with Asn97C. This idea was fruitful, as 3w with a Ki of 0.15 µM is 3-fold more potent than 3v. The added fluorine makes 3w somewhat less soluble with S = 27.2 µg/mL. The same idea was then applied to the 5-phenoxy compounds. Addition of a para-methoxyethoxy group in 3y did not change the potency from that of 3s, since the added group is expected to be largely solvent-exposed (Figure 3). However, again several-fold enhancements of the potency accompanied introduction of a meta solubilizing group along with a fluorine adjacent to the hydroxyl group yielding the most potent compounds, 3z and 3bb, with Ki values of 0.082 and 0.057 µM. The solubility of 3bb was also measured; the result of 47.2 µg/mL further supports the potential value of this compound.
CONCLUSION
The purpose of this work was to optimize biaryltriazoles as inhibitors of the tautomerase activity of human MIF. A combined approach was taken featuring organic synthesis, enzymatic assaying, crystallography, and modeling including FEP calculations. The acquisition of the crystal structures for 3a and 3b bound to MIF provided important proof that the inhibitors were bound in the tautomerase active site and a firmer structural foundation for the modeling. Exploration of the structure–activity relationships using a carefully optimized and controlled tautomerase assay showed that it was challenging to improve on the activity of 3a and 3b owing to the limited size of the binding site and the exquisite accommodation of the inhibitors through multiple hydrogen bonds and aryl–aryl interactions (Figure 1). Some enticing possibilities such as introduction of an alkoxy or phenyl substituent at the 8-position in the quinoline ring (Figure 2) did not produce more potent compounds. Additional modeling with the BOMB program encouraged pursuit of 5-phenoxy analogues (Figure 3), which did deliver a gain in potency with 3s. Consideration of appendages at the 6-position of the quinoline ring led to 3v, which is both highly soluble and potent. Finally, addition of a meta-fluorine to enhance the hydrogen bonding with Asn97C yielded additional gains in potency with 3w, 3z, 3aa, and 3bb. With Ki values below 100 nM, 3z and 3bb are likely the most potent known inhibitors of the tautomerase activity of human MIF; they are more than 1000-fold more active than the well-studied (R)-ISO-1 and more than 200-fold more active than the chromen-4-one Orita-13.
Supplementary Material
ACKNOWLEDGMENTS
Gratitude is expressed to the National Institutes of Health (GM32136), Yale University, and Debiopharm SA for research support, to the National Science Foundation (DGE-1122492) for a fellowship to M.J.R., to Drs. Richard Bucala and Lin Leng for the MIF expression vector and interactions on the tautomerase assay, and to Dr. Binh Le for assistance with crystallizations and X-ray data collection. The data collection for the structure of 3a with human MIF was conducted at the Advanced Photon Source (APS) on the Northeastern Collaborative Access Team beamlines which are supported by the National Institutes of Health (P41 GM103403); the APS is operated for the U.S. Department of Energy Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Synthetic details, NMR and HRMS spectral data for compounds in Tables 1, and crystallographic details. The crystal structure data for the complexes of 3a and 3b with MIF have been deposited in the RCSB Protein Data Bank with the codes 4WR8 and 4WRB. These materials are available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Morand EF, Leech M, Bernhagen J. Nat. Rev. Drug. Discovery. 2006;5:399–410. doi: 10.1038/nrd2029. [DOI] [PubMed] [Google Scholar]
- 2.Greven D, Leng L, Bucala R. Expert Opin. Ther. Targets. 2010;14:253–264. doi: 10.1517/14728220903551304. [DOI] [PubMed] [Google Scholar]
- 3.Asare Y, Schmitt M, Bernhagen J. Thromb. Haemost. 2013;109:391–398. doi: 10.1160/TH12-11-0831. [DOI] [PubMed] [Google Scholar]
- 4.Conroy H, Mawhinney L, Donnelly SC. Q. J. Med. 2010;103:831–836. doi: 10.1093/qjmed/hcq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schulz R, Moll UM. Curr. Opin. Oncol. 2014;26:108–113. doi: 10.1097/CCO.0000000000000036. [DOI] [PubMed] [Google Scholar]
- 6.Leng L, Metz C, Fang Y, Xu J, Donnelly S, Baugh J, Delonery T, Chen Y, Mitchell RA, Bucala R. J. Exp. Med. 2003;197:1467–1476. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Senter PD, Al-Abed Y, Metz CN, Benigni F, Mitchell RA, Chesney J, Han J, Gartner CG, Nelson SD, Todaro GJ, Bucala R. Proc. Natl. Acad. Sci. U. S. A. 2002;99:144–149. doi: 10.1073/pnas.011569399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cournia Z, Leng L, Gandavadi S, Du X, Bucala R, Jorgensen WL. J. Med. Chem. 2009;52:416–424. doi: 10.1021/jm801100v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hare AA, Leng L, Gandavadi S, Du X, Cournia Z, Bucala R, Jorgensen WL. Bioorg. Med. Chem. Lett. 2010;20:5811–5814. doi: 10.1016/j.bmcl.2010.07.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jorgensen WL, Gandavadi S, Du X, Hare AA, Trofimov A, Leng L, Bucala R. Bioorg. Med. Chem. Lett. 2010;20:7033–7036. doi: 10.1016/j.bmcl.2010.09.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.For reviews, see: Orita M, Yamamoto S, Katayama N, Fujita S. Curr. Pharm. Des. 2002;8:1297–1317. doi: 10.2174/1381612023394674. Garai J, Lóránd T. Curr. Med. Chem. 2009;16:1091–1114. doi: 10.2174/092986709787581842. Xu L, Li Y, Sun H, Zhen X, Qiao C, Tian S, Hou T. Drug Discovery Today. 2013;18:592–600. doi: 10.1016/j.drudis.2012.12.013.
- 12.(a) Xu L, Zhang Y, Zheng L, Qiao C, Li Y, Li D, Zhen X, Hou T. J. Med. Chem. 2014;57:3737–3745. doi: 10.1021/jm401908w. [DOI] [PubMed] [Google Scholar]; (b) Tsai L-T, Lin T-H. J. Biomol. Screening. 2014;19:1116–1123. doi: 10.1177/1087057114523317. [DOI] [PubMed] [Google Scholar]
- 13.Al-Abed Y, Dabideen D, Aljabari B, Valster A, Messmer D, Ochani M, Tanovic M, Ochani K, Bacher M, Nicoletti F, Metz C, Pavlov VA, Miller EJ, Tracey KJ. J. Biol. Chem. 2005;280:36541–36544. doi: 10.1074/jbc.C500243200. [DOI] [PubMed] [Google Scholar]
- 14.Chang KF, Al-Abed Y. Bioorg. Med. Chem. Lett. 2006;16:3376–3379. doi: 10.1016/j.bmcl.2006.04.038. [DOI] [PubMed] [Google Scholar]
- 15.Jorgensen WL. Acc. Chem. Res. 2009;42:724–733. doi: 10.1021/ar800236t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orita M, Yamamoto S, Katayama N, Aoki M, Takayama K, Yamagiwa Y, Seki N, Suzuki H, Kurihara H, Sakashita H, Takeuchi M, Fujita S, Yamada T, Tanaka A. J. Med. Chem. 2001;44:540–547. doi: 10.1021/jm000386o. [DOI] [PubMed] [Google Scholar]
- 17.Georg J. 200770078155. U.S. Patent Appl. 2007 Apr 5;
- 18.Andersen J, Bolvig S, Liang X. Synlett. 2005:2941–2947. [Google Scholar]
- 19.Baka E, Comer JEA, Takács-Novák K. J. Pharm. Biomed. Anal. 2008;46:335–341. doi: 10.1016/j.jpba.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 20.Lubetsky JB, Swope M, Dealwis C, Blake P, Lolis E. Biochemistry. 1999;38:7346–7354. doi: 10.1021/bi990306m. [DOI] [PubMed] [Google Scholar]
- 21.Jorgensen WL, Tirado-Rives J. J. Comput. Chem. 2005;26:1689–1700. doi: 10.1002/jcc.20297. [DOI] [PubMed] [Google Scholar]
- 22.Jorgensen WL, Bollini M, Thakur VV, Domaoal RA, Spasov K, Anderson KS. J. Am. Chem. Soc. 2011;133:15686–15696. doi: 10.1021/ja2058583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jorgensen WL, Tirado-Rives J. Proc. Natl. Acad. Sci. U. S. A. 2005;102:6665–6670. doi: 10.1073/pnas.0408037102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernhagen J, Mitchell RA, Calandra T, Voelter W, Cerami A, Bucala R. Biochemistry. 1994;33:14144–14155. doi: 10.1021/bi00251a025. [DOI] [PubMed] [Google Scholar]
- 25.Sun H-W, Bernhagen J, Bucala R, Lolis E. Proc. Natl. Acad. Sci. U. S. A. 1996;93:5191–5196. doi: 10.1073/pnas.93.11.5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor AB, Johnson WH, Czerwinski RM, Li HS, Hackert ML, Whitman CP. Biochemistry. 1999;38:7444–7452. doi: 10.1021/bi9904048. [DOI] [PubMed] [Google Scholar]
- 27.Cheng Y-C, Prusoff WH. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 28.Balachandran S, Rodge A, Gadekar PK, Yadav VN, Kamath D, Chetrapal-Kunwar A, Bhatt P, Srinivasan S, Sharma S, Vishwakarma RA. Bioorg. Med. Chem. Lett. 2009;19:4773–4776. doi: 10.1016/j.bmcl.2009.06.052. [DOI] [PubMed] [Google Scholar]
- 29.McLean LR, Zhang Y, Li H, Choi Y-M, Hon Z, Vaz RJ, Li Y. Bioorg. Med. Chem. Lett. 2010;20:1821–1824. doi: 10.1016/j.bmcl.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 30.(a) Jorgensen WL, Schyman P. J. Chem. Theory Comput. 2012;8:3895–3901. doi: 10.1021/ct300180w. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Yan XC, Schyman P, Jorgensen WL. J. Phys. Chem. A. 2014;118:2820–2826. doi: 10.1021/jp501553j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.ChemBioDraw, ver. 12.0.2. Cambridge, MA: CambridgeSoft Inc.; 2014. [Google Scholar]
- 32.QikProp, ver. 4.0. New York: Schrödinger Inc.; 2014. [Google Scholar]
- 33.Jorgensen WL, Duffy EM. Adv. Drug Delivery Rev. 2002;54:355–366. doi: 10.1016/s0169-409x(02)00008-x. [DOI] [PubMed] [Google Scholar]
- 34.Roughley SD, Jordan AM. J. Med. Chem. 2011;54:3451–3479. doi: 10.1021/jm200187y. [DOI] [PubMed] [Google Scholar]
- 35.Bollini M, Cisneros JA, Spasov KA, Anderson KS, Jorgensen WL. Bioorg. Med. Chem. Lett. 2013;23:5213–5216. doi: 10.1016/j.bmcl.2013.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.