Abstract
Bioengineered heparin is being investigated as a potential substitute for the animal-sourced anticoagulant drug. One step in the current process to prepare bioengineered heparin involves the conversion of N-sulfo heparosan, rich in →4)GlcNS(1→4) GlcA(1→ sequences (where S is sulfo, GlcN is α-D-glucosamine, and GlcA is β-D-glucuronic acid), to a critical intermediate, rich in →4)GlcNS(1→4) IdoA2S(1→ sequences (where S is sulfo and IdoA is α-L-iduronic acid), using 2-O-sulfotransferase (2-OST) and C5 epimerase (C5-epi). Until now, these heparan sulfate biosynthetic enzymes have been expressed in Escherichia coli grown in shake flask culture as fusion proteins. The current study is focused on the high-cell density fed-batch cultivation of recombinant E. coli strains expressing both enzymes. We report the high productivity expression of active 2-OST and C5-epi enzymes of 6.0 and 2.2 mg/gm dry cell weight, respectively.
Keywords: fermentation, heparin, sulfotransferase, epimerase, recombinant enzymes
Introduction
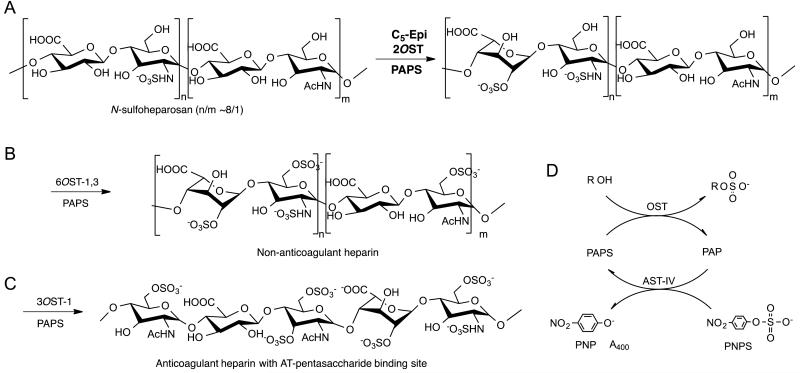
Heparin is a critically important anticoagulant drug that is derived from animal tissue [1]. A recent contamination crisis [2] has led our laboratory to undertake the preparation of a bioengineered heparin to replace this animal-sourced drug [3]. Bioengineered heparin is prepared from heparosan, →4) N-acetyl-α-D-glucosamine (1→4) β-D glucuronic acid (1→ (→4) GlcNAc (1→4) GlcA(1→), the capsular polysaccharide of E. coli K5 [4] that is chemically converted to N-sulfoheparosan and then enzymatically modified in a three-step process to obtain anticoagulant heparin [3] (Fig. 1). The first enzymatic step in this process uses heparan sulfate biosynthetic enzymes expressed in E. coli as recombinant fusion proteins with maltose binding protein (MBP), 2-O-sulfotransferase (2-OST) and C5-epimerase (C5-epi), to convert N-sulfo heparosan into an intermediate polysaccharide rich in →4)GlcNS(1→4) IdoA2S(1→ sequences (where S is sulfo and IdoA is α-L-iduronic acid). This critical step in bioengineered heparin preparation relies on the use of recombinant arylsulfotransferase IV (AST-IV) to regenerate 3′-phospho adenosine-5′-phosphosulfate (PAPS) using p-nitrophenylsulfate as a sacrificial sulfo donor [5-7].
Fig. 1.
Three-step enzymatic synthesis of heparin from N-sulfoheparosan. A. In the first step N-sulfoheparosan is treated with 2-OST and C5-epi. B. In the second step 6-O-sulfotransferase isoforms 1 and 3 are used to prepare non-anticoagulant heparin. C. In the third enzymatic step 3-O-sulfotransferase isoform 1 is used to introduce the antithrombin (AT) pentasaccharide-binding site affording anticoagulant heparin. D. PAPS, used as the sulfo donor in each step, forms PAP, which is recycled using aryl sulfotransferase IV (AST-IV) and p-nitrophenyl sulfate (PNPS) as a sacrificial sulfo donor, forming p-nitrophenol (PNP) which can be detected by its absorbance at 400 nm. The R at the top of panel D corresponds to the polysaccharide substrate.
The generation of small quantities of bioengineered heparin, closely resembling animal-sourced heparin has been demonstrated [4]. In order to scale-up this process, however, substantial quantities of E. coli expressed recombinant 2-OST and C5-epi are required. It is essential to produce the enzymes in high yield in stirred tank fermentor to scale up our chemoenzymatic processes for the production of bioengineered heparin. This study describes the scale-up of the production of these two enzymes from shake-flask to stirred tank fermenter.
1.Materials and methods
2.1 Materials
All media components, fermentation reagents, chemicals used for preparation of biomass extraction buffer, and chemicals and standards used in HPLC analysis, were from Sigma-Aldrich (St. Louis, MO, USA). LB broth powder was purchased from BD Biosciences (San Jose, CA, USA). Kanamycin, Ampicillin, tetracycline, chloramphenicol, and 1-thiogalactopyranoside (IPTG) were from Gold Biotechnology (St. Louis, MO, USA).
2.2 Bacterial strain and media
The recombinant E. coli Rosetta-gami B (DE3) cells (Novagen, Cambridge MA, USA) strain with the plasmid pMalc2x and the catalytic domain of Chinese hamster ovary 2-O-sulfotransferase (Arg 51—Asn356) was used to express 2-OST-MBP fusion product [8]. The same strain and plasmid with human C5-epimerase (E53-N609) was used to express C5-epi-MBP fusion product [9]. Shake flask experiments were performed in a new formulated glucose and M9 minimal salts (64 g Na2HPO4-7H2O, 15 g KH2PO4, 2.5 g NaCl, 5.0 g NH4Cl)-based medium (200 ml/l of M9 salts, 2 ml/l of 1M MgSO4, 20 ml/l of 20% glucose, 100 μl/l of 1 M CaCl2, 20 g/l yeast extract and 1 ml/l of a trace metal solution. Trace metal solution was made according to Matsui [10] with the following modification in composition (g/l): FeSO4·7H2O, 10.0; CaCl2, 2.0; ZnSO4·7H2O, 2.2; MnSO4·4H2O, 0.5; CuSO4·5H2O, 1.0; (NH4)6MoO24·4H2O, 0.1 and Na2B4O7·10H2O dissolved in 5 M HCl, 0.02. ampicillin (50 mg/l), tetracycline (12.5 mg/l), and kanamycin (50 mg/l) were always added to the media for both growth and propagation of the strain after filtered through 0.22 μm membrane with syringe. For C5-epi expression, 50 mg/ml chloramphenicol was added with all other three antibiotics.
2.3 Shake flask experiments
Transformed cells were incubated at 37°C on shaker with 220 rpm overnight to prepare cultures from stock bacteria. A 1-l media flask was inoculated with 2.5% (v/v) of stock culture. When the fermentation reached an optical density at 600 nm (OD600) of 0.6-0.8, the temperature was decreased to 22°C for 30 min. A 200-μl aliquot of 1 M IPTG was added to each 1 liter of fermentation media for induction. For C5-epi expression, 200 mg/ml arabinose was used along with 0.2 mM IPTG. The culture was shaken for 16-18 h at 220 rpm at the reduced temperature of 22°C.
2.4 Fed-batch experiments
A 20-l bioreactor (Bio-Flo 4500, New Brunswick Scientific, Enfield, CT, USA), in situ sterilizable and equipped with pH and pO2 probes (Mettler, Toledo, Switzerland) was used for fed-batch experiments. During growth, the fermentation parameters were controlled and data were collected by Biocommand A4 software (Eppendorf, Inc., Enfield, CT, USA).
For the 12-l fed-batch fermentation, 200 μl of glycerol stock bacteria were inoculated into a 250 ml shake flask containing 50 ml LB medium. After 12 h of cultivation in a shaker incubator (220 rpm) at 37°C, the entire pre-culture was aseptically transferred into a 2.5-l shake flask containing 500 ml LB medium and incubated under the same condition for 10 h. This was used to inoculate the 10-l fed-batch fermentation that took place in the 20-L bioreactor. Fed-batch experiments were performed at 37°C and pH of 7.0 in 10-l LB containing 4 g/l glycerol. The dissolved oxygen (DO) was set to 20% and cascade controlled by inlet airflow rate and agitation speed. The pH was set to 7.0 and cascade controlled by acid pump for hydrochloric acid and base pump for ammonia water. After 6 h of batch phase, the culture was fed with a concentrated solution (450 g/l glucose, 45 g/l yeast extract, 4.5 g/l (NH4)2 SO4 and 4.5 g/l MgSO4). In 2-OST fed-batch experiment, the culture was fed from 6 h to 10h at a constant feeding rate of 5 ml/ min, from 12 h to 22 h at a constant feeding rate of 3 ml/min, from 24 h to 26 h at a constantly feeding rate of 1 ml/min. IPTG was added to a final concentration of 0.2 mM as soon as the OD600 value reached 22.5. In the C5-epi fed-batch experiment, the culture was also constantly fed after 5 h with a feeding rate of 3 ml/min until 15 h and then from 19 h to 25.5 h at a rate of 1.5 ml/min. C5-epi fed-batch experiment was induced at an OD600 value of 22.4 at final concentrations of 0.2 mM IPTG and 1 mg/ml arabinose.
Bacterial growth was monitored at different time points by measuring the optical density at 600 nm (OD600) with a UVmini-1240 spectrophotometer (Shimadzu, Japan). The culture was diluted to the linear range. The cell pellet was collected by centrifugation at 3501 × g for 20 min at 4°C and frozen.
2.5 Biomass extraction and enzyme purification
The pellet (20 g) was suspended in 100 ml of ice-cold loading buffer A (25 mM Tris-HCl, pH 7.4, containing 500 mM NaCl) by vortexing, and cells were lysed on ice using a Q700 sonicator (Qsonica, Newtown, CT, USA) at power level 4.5 for 1 min (30 strokes, 1s on and 1s off). Sonication of the re-suspended cells was performed 3-times according to the program, taking 30 sec break between each cycle. Cell debris was spun down at 20 000 × g at 4°C for 30 min, and the supernatant was filtered through a 0.22 μm filter by using vacuum system into 50 ml tube cooled in an ice bath. The sample containing the expressed 2-OST or C5-epi was maintained on ice briefly prior to FPLC purification.
The FPLC system was manually washed, without an attached column, for 5 min at a flow rate 5 ml/min with eluting buffer B (25 mM Tris-HCl, pH 7.5, containing 500 mM NaCl and 40 mM maltose monohydrate) and then washed for 5 min at flow rate 5 ml/min with loading buffer A. The amylose column was connected to the FPLC and the column, the sample was injected and the column was washed with buffer A for 10-15 min at a flow rate 2 ml/min and then washed with elution buffer B at a flow rate 2 ml/min for 5 min by manual operation. Purified enzyme was collected on a fraction collector with UV280 detection and fractions containing the 6-OST peak were pooled together in a 50 ml tube. Glycerol 10 vol% was added to the enzyme before it was stored at −80°C.
2.6 SDS PAGE and protein concentration analysis
Protein concentration was determined with Nanodrop 3300 (Thermo Fisher Scientific Inc., Wilmington, DE, USA). SDS-PAGE protein analysis was performed in a Mini-Protean Tetra system (BIORAD, Hercules, CA, USA), by loading 30 μl of 1:1 (v:v) boiled protein solution and dye buffer on a 4–20% precast gel (Mini protean TGX gels, BIORAD) and running in a Tris, glycine and SDS buffer (10× Tris/glycine/SDS buffer, BIORAD) at 100 V for 45 min. A 250 kDa to 10 kDa protein standard (Precision Plus Protein Kaleidoscope, BIORAD) was used as ladder. Gels were washed with tap water for 30 min and then stained by soaking for 5 h in Coomassie blue solution (Gel Code Blue Safe Protein Stain, Thermo Fisher Scientific).
2.7 Activity assay
The 2-OST activity assay was performed as previously described with some modification [6,7,9]. The activity was analyzed by incubating 50 μg of purified 2-OST with 25 μg of N-sulfoheparosan, 25 μg of AST-IV, 0.5 μM para-nitrophenylsulfate (PNPS) and 2 μg 3′-phosphoadenosine-5′-phosphosulfate (PAPS) in 250 μl system with supplement of 50 mM MES buffer pH 7.0. 10 mM MnCl2, 5 mM MgCl2, and 1% Triton X-100 (pH 7). For assaying the activity of C5-epi, in addition to 50 μg purified C5-epi, 50 μg purified 2-OST was also added to the assay system mentioned above [11]. The assays were conducted in transparent, 96-well plates purchased from Greiner Bio-One (Monroe, NC, USA). The plates were incubated at 37°C in SpectraMax plate reader (Molecular Devices, Sunnyvale, CA, USA). A kinetic model was used to measure the absorbance at 400 nm/min over 1 h. The enzyme activity was calculated using the following equation:
Abst2 and Abst1 are the absorbance at 400 nm at t2 min and t1 min, respectively. The time range was chosen between a linear segment of the plot. ε0 is the extinction coefficient (10.5 × 10 −3 ), RT is the reaction time (min), is the concentration of the assayed enzyme divided by ten. One unit of the activity equals to per mg enzyme produce 1 nmol PNP/min.
2.8 Mass analysis
The disaccharide analysis was carried out as reported previously [12]. Heparin lyase 1, 2, and 3 (10 mU each) in 5 μl of 25 mM Tris, 500 mM NaCl, 300 mM imidazole buffer (pH 7.4) were added to 10 μg of 2-OST or C5-epi treated sample in 100 μl of distilled water and incubated at 35 °C for 10 h to degraded the hydrolyzed sample completely. The products were recovered by centrifugal filtration using an YM-10 micro-concentrator, and the disaccharides were recovered in the flow-through and freeze-dried. The digested disaccharides were dissolved in water to concentration of 50-100 ng/2 μl for liquid chromatography (LC)-mass spectrometric (MS) analysis.
LC-MS analyses were performed on an Agilent 1200 LC/MSD instrument (Agilent Technologies, Inc. Wilmington, DE) equipped with a 6300 ion trap and a binary pump followed by a UV detector equipped with a high-pressure cell. The column used was a Poroshell 120 C18 column (2.1 × 100 mm, 2.7 μm, Agilent, USA). Eluent A was water/acetonitrile (85:15) v/v, and eluent B was water/acetonitrile (35:65) v/v. Both eluents contained 12 mM tributylamine (TrBA) and 38 mM ammonium acetate with pH adjusted to 6.5 with acetic acid. The gradient of solution A for 5 min followed by a linear gradient from 5 to 15 min (0-40% solution B) was used at flow rate of 150 μl/min.
2.9 HPLC analysis
Concentrations of glucose and acetic acid in the medium during the fermentation were measured by HPLC as previously described [13].
3. Results
3.1 Culture conditions of shake flask experiments
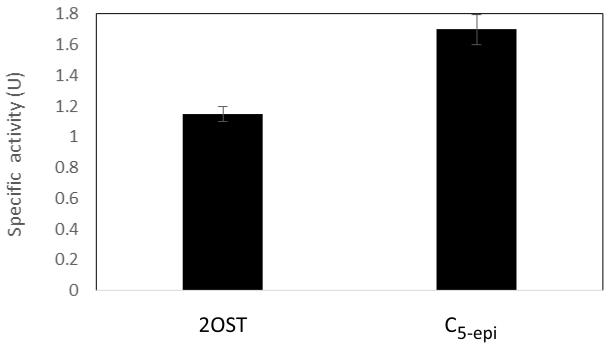
Shake flask experiments were performed to investigate the medium and culture and induction conditions that are suitable for fed-batch expression. The batch cultures of recombinant E. coli harboring pMAL-c2X -2-OST and pMAL-c2X-C5-epi usually were carried out in LB medium [8,11]. In order to identify medium conditions suitable for batch fed fermentation, we investigated the effect of different media components on the expression of 2-OST and C5-epi. A new M9 salt and glucose based medium was identified that can be used to express both of these enzymes. The cultures grown until OD600 = 0.6 were induced once using IPTG at 0.2 mM for 2-OST, and IPTG plus 200 mg/ml of arabinose for C5-epi. The results showed that the specific activity of 2-OST and C5-epi were 1.7 ± 0.1 U and 1.15 ± 0.05 U, respectively (Fig. 2).
Fig. 2.
Specific activity of 2-OST and C5-epi expressed in shake flask fermentation.
3.2 High cell density cultivation
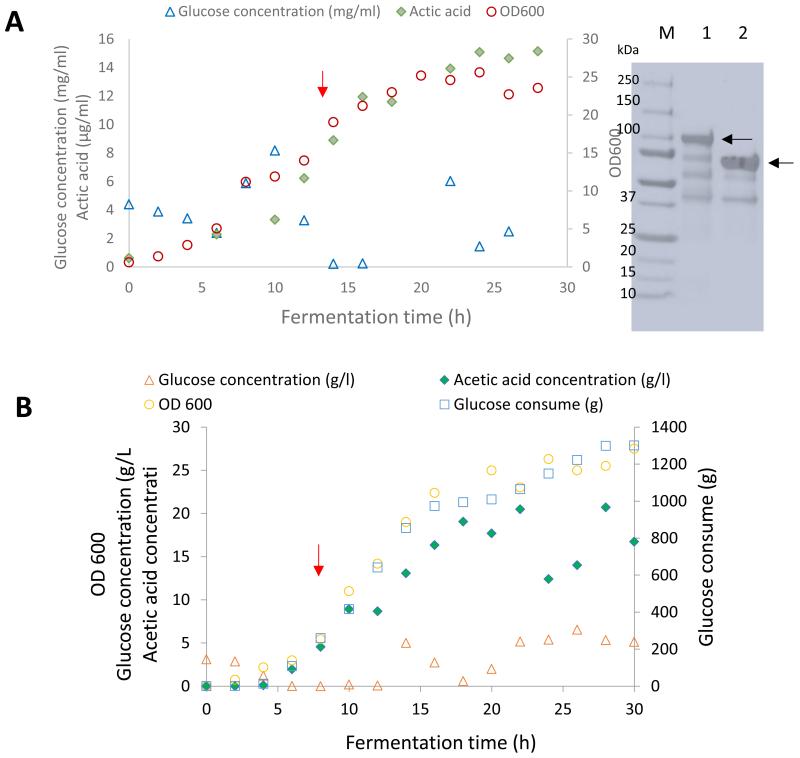
In most processes the aim is to reach highest possible cell densities in a short time under aerobic conditions to meet the demand of large-scale bioengineering heparin biosynthesis. For the fed-batch fermentation of 2-OST, the maximum cell density was 5.5 gcdw/l (Table 1). The concentration of purified enzyme was 167 mg/l. The productivity was 6.0 mg/l-h, and the final specific activity was 0.54 ± 0.01 nmol/min mg. After 20 h, the percent conversion of NS2S was 63.4%. Acetic acid, which was produced as a by-product during batch mode, is generally produced at a high specific growth rate when there is excess carbon source or oxygen-limited conditions [14]. In this experiment, acetic acid accumulated up to 15.1 g/l in the cell-free culture medium; therefore, a constant feeding rate did not meet the demand of exponential cell growth, and the glucose concentration was changed over a wide range (Fig. 3).
Table 1.
Fed-batch fermentation experiments of 2-OST and C5-epi
| 2-OST | C5-epi | |
|---|---|---|
| Induction time (h) | 17 | 16 |
| Total hours of induction (h) | 11 | 14 |
| Feeding start time (h) | 6 | 5 |
| Fermentation time (h) | 28 | 30 |
| Maximum cell density (gcdw/l) | 5.5 | 5.9 |
| Purified enzyme concentration (mg/l) | 167 | 65 |
| Enzyme yield /biomass (mg/gcdw) | 30.3 | 11.1 |
| Productivity (mg/l h) | 6.0 | 2.2 |
| Final specific activity (nmol/min mg) | 0.54 ± 0.01 | 1.78 ± 0.05 |
| Heparin disaccharide (ΔUA2S – GlcNS)(%) | 63 ± 0.3 | 70 ± 1.1 |
Fig. 3.
E. coli 2-OST and C5-epi fed batch fermentation. A. 2-OST fermentation and B. C5-epi fermentation: bacterial growth (open circles), glucose concentration (open triangles), Acetic acid concentration (filled diamonds), and glucose consume (open squares). Induction times for 2-OST and C5-epi were indicated by red arrows. C. SDS PAGE analysis of purified enzyme; lanes 1, 250–10 kDa ladder; 2, purified C5-epi; 3, purified 2-OST. The specific enzyme bands are indicated by black arrows.
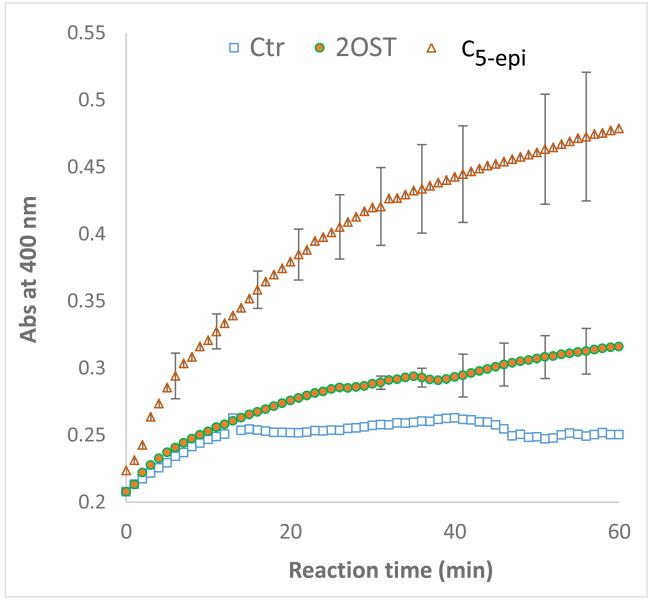
For the fed-batch fermentation of C5-epi, feeding was initiated after acetic acid was consumed. The maximum cell density achieved was 5.9 gcdw/l. The concentration of purified enzyme was 65.3 mg/l. The productivity was 2.2 mg/l-h, and the final specific activity was 1.78 ± 0.05 nmol/ min mg (Table 1, Fig. 4). After 20 h, NS2S conversion was 70%. As mentioned above, the acetic acid concentration was high, which suggested that the feeding model should be optimized for higher cell density and/or higher enzyme production.
Fig. 4.
Measurement of specific activity of the enzyme during a 1-h reaction catalyzed by 2-OST and C5-epi, both the enzymes were purified enzymes from fed batch fermentation. Control without substrate (open squares), 2-OST (filled circles), C5-epi (open triangle).
The 2-OST and C5-epi were purified using an amylose column. There were no more than four bands observed in SDS-PAGE analysis with the most intense band corresponding to each of the target proteins (Fig. 3). These results demonstrate that that the MBP tag of these recombinant proteins represents and efficient means for their purification.
4. Discussion
The chemoenzymatic preparation of a bioengineered heparin relies on three enzymatic steps, the first and most difficult step requires the extensive epimerization of GlcA to IdoA by C5-epi and its subsequent sulfation with 2-OST to afford IdA2S. Since the epimerization reaction catalyzed by C5epi is reversible 2-OST must be present to quickly trap IdoA as IdoA2S in order to obtain a polysaccharide product, such as heparin, which contains up to 80-90% IdoA2S residues. Thus, there is a need for larger quantities of active C5-epi and 2-OST than can be produced in shake flask culture. As part of our efforts to scale-up the production of these enzymes we turned to fed-batch stirred tank fermenters. Unfortunately, initial attempts to prepare these critical enzymes resulted in either low levels of these enzymes or inactive enzymes.
Preventing the accumulation of toxic levels of acetic acid is a major task to achieve high cell concentrations in the bioreactor. The accumulation of acetic acid in the media may have toxicity effects on the cells in the culture. The primary factor contributing to the toxicity of acetic acid is its ability to diffuse across E. coli cellular membranes. The diffusing acetic acid or other organic acids entering into the cytoplasm will disrupt the pH and anion pool of the cytoplasm. The resulting increase in internal acidity can affect the integrity of purine bases and result in denaturing of essential enzymes inside the cell, both of which negatively affect cell viability and enzyme expression. The specific growth rate of E. coli K12 batch cultured in the fermenter decreased from 15 to 5g/l as the acetic acid concentration increased [14]. Growth-inhibiting acidic by-products of incomplete substrate oxidation, such as acetic acid, are produced in response to oxygen limitation or excess carbon. The formation of acetic acid and subsequent growth inhibition can be avoided by limiting the amount of carbon source in the culture media via an exponential feeding strategy [15] or by controlling the dissolved oxygen level [16]. Although active 2-OST and C5-epi were obtained, the fed-batch fermentations were not optimized, cell densities achieved were not sufficiently high, and the relationship between the growth rate and enzyme expression is unclear. Additional work remains to be performed before this process step can be taken to pilot-scale production.
5. Conclusions
This study demonstrates the high-density cultivation of recombinant E. coli strains for the high productivity expression of recombinant 2-OST and C5-epi. These enzymes are critically important in the first enzymatic step in the chemoenzymatic preparation of bioengineered heparins.
Acknowledgments
The authors gratefully acknowledge funding from the NIH in the form of grant HL096972, from the National Natural Science Foundation of China under Grant No. 31171737 and the PRC grant 210208310507 supporting the visit of Jianhua Zhang, and from the Bioengineered Heparin Consortium.
References
- 1.Linhardt RJ. Heparin: Structure and activity. Journal of Medicinal Chemistry. 2003;46:2551–2554. doi: 10.1021/jm030176m. [DOI] [PubMed] [Google Scholar]
- 2.Liu H, Zhang Z, Linhardt RJ. Lessons learned from the contamination of heparin. Natural Product Reports. 2009;26:313–321. doi: 10.1039/b819896a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z, Yang B, Zhang Z, Ly M, Takieddin M, Mousa S, Liu J, Dordick JS, Linhardt RJ. Control of the heparosan N-deacetylation leads to an improve bioengineered heparin. Applied Microbiology and Biotechnology. 2011;91:91–99. doi: 10.1007/s00253-011-3231-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z, Ly M, Zhang F, Zhong W, Suen A, Dordick JS, Linhardt RJ. E. coli K5 fermentation and the preparation of heparosan, a bioengineered heparin precursor. Biotechnology and Bioengineering. 2010;107:964–973. doi: 10.1002/bit.22898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burkart MD, Izumi M, Chapman E, Lin C, Wong C. Regeneration of PAPS for enzymatic synthesis of sulfated oligosaccharides. Journal of Organic Chemistry. 2000;65:5565–5574. doi: 10.1021/jo000266o. [DOI] [PubMed] [Google Scholar]
- 6.Paul P, Liu J, Dordick JS, Linhardt RJ. Recent advances in sulfotransferase enzyme activity assays. Analytical and Bioanalytical Chemistry. 2012;403:1491–1500. doi: 10.1007/s00216-012-5944-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sterner E, Li L, Paul P, Beaudet JM, Liu J, Linhardt RJ, Dordick JS. Assays for determining heparan sulfate and heparin O-sulfotransferase activity and specificity. Analytical and Bioanalytical Chemistry. 2014;406:525–536. doi: 10.1007/s00216-013-7470-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Avci FY, Muñoz EM, McDowell LM, Chen M, Pedersen LC, Zhang L, Linhardt RJ, Liu J. Enzymatic redesigning of biologically active heparan sulfate. Journal of Biological Chemistry. 2005;280:42817–42825. doi: 10.1074/jbc.M504338200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munoz E, Xu D, Avci F, Kemp M, Liu J, Linhardt RJ. Enzymatic synthesis of heparin related polysaccharides on sensor chips: Rapid screening of heparin–protein interactions. Biochemical Biophysical Research Communications. 2006;339:597–602. doi: 10.1016/j.bbrc.2005.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsui T, Sato H, Yamamuro H, Shinzato N, Matsuda H, Misawa S, Sato S. High cell density cultivation of recombinant E. coli for hirudin variant 1 production by temperature shift controlled by pUC18-based replicative origin. Applied Microbiology and Biotechnology. 2008;80:779–783. doi: 10.1007/s00253-008-1611-2. [DOI] [PubMed] [Google Scholar]
- 11.Li K, Bethea HN, Liu J. Using engineered 2-O-sulfotransferase to determine the activity of heparan sulfate C5-epimerase and its mutants. Journal of Biological Chemistry. 2010;285:11106–11113. doi: 10.1074/jbc.M109.081059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang B, Weyers A, Baik JY, Sterner E, Sharfstein S, Mousa SA, Zhang F, Dordick JS, Linhardt RJ. Ultra-performance ion-pairing liquid chromatography with on-line electrospray ion trap mass spectrometry for heparin disaccharide analysis. Analytical Biochemistry. 2011;415:59–66. doi: 10.1016/j.ab.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Restaino OF, Bhaskar U, Paul P, Li L, De Rosa M, Dordick JS, Linhardt RJ. High cell density cultivation of a recombinant E. coli strain expressing a key enzyme in bioengineered heparin production. Applied Microbiology and Biotechnology. 2013;97:3893–3900. doi: 10.1007/s00253-012-4682-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakano K, Rischke M, Sato S, Maèrkl H. Influence of acetic acid on the growth of Escherichia coli K12 during high-cell-density cultivation in a dialysis reactor. Applied Microbiology and Biotechnology. 1997;48:597–601. doi: 10.1007/s002530051101. [DOI] [PubMed] [Google Scholar]
- 15.Yoon SK, Kang WK, Park TH. Fed-batch operation of recombinant Escherichia coli containing trp promoter with controlled specific growth rate. Biotechnology and Bioengineering. 1994;43:995–999. doi: 10.1002/bit.260431013. [DOI] [PubMed] [Google Scholar]
- 16.Konstantinov K, Kishimoto M, Seki T, Yoshida T. A balanced DO-stat and its application to the control of acetic acid excretion by recombinant Escherichia coli. Biotechnology and Bioengineering. 1990;36:750–758. doi: 10.1002/bit.260360714. [DOI] [PubMed] [Google Scholar]