Summary
DNA replication is executed only when cells have sufficient metabolic resources and undamaged DNA. Nutrient limitation and DNA damage cause a metabolic checkpoint and DNA damage checkpoint, respectively. Although SIRT1 activity is regulated by metabolic stress and DNA damage, its function in these stress-mediated checkpoints remains elusive. Here we report the SIRT1-TopBP1 axis functions as a switch for both checkpoints. Under glucose deprivation, SIRT1 is activated and deacetylates TopBP1, resulting in TopBP1-Treslin disassociation and DNA replication inhibition. Conversely, SIRT1 activity is inhibited under genotoxic stress, resulting in increased TopBP1 acetylation that is important for the TopBP1-Rad9 interaction and activation of the ATR-Chk1 pathway. Mechanistically, we showed that acetylation of TopBP1 changes the conformation of TopBP1, thereby facilitating its interaction with distinct partners in DNA replication and checkpoint activation. Taken together, our studies identify SIRT1-TopBP1 axis as a key signaling mode in the regulation of metabolic checkpoint and DNA damage checkpoint.
Introduction
To maintain genomic integrity, cells have evolved distinct cell-cycle checkpoints to arrest the cell cycle when conditions are not suitable for cell proliferation and division. Checkpoint disruption results in cell death and genomic instability, and contributes to human cancers (Bartek and Lukas, 2003; Hartwell and Weinert, 1989). Metabolic and DNA damage checkpoints are two major signaling pathways for cells to cope with unfavorable changes in cellular microenvironments. A metabolic checkpoint represents a molecular mechanism that senses metabolic stress induced by nutrient deprivation, and transduces these stress signals into a proper cellular response, including cell cycle control (Holley and Kiernan, 1974; Jones et al., 2005). Understanding the interconnection between metabolic stress and cellular signaling has emerged as a focus in the study of metabolic disorders and cancer. The DNA damage checkpoint is initiated in response to genotoxic stress to ensure the fidelity of genetic information. This checkpoint triggers cell cycle arrest, allowing the cell time to repair the damage before continuing to divide. In response to replication stress, ATR is activated and initiates signal transduction cascades that employ Chk1 kinase and Cdc25 phosphatases to block the firing of replication origins, arrest cell cycle progression, and facilitate DNA damage repair (Lopes et al., 2001; Santocanale and Diffley, 1998).
SIRT1 is widely regarded as an important regulator of energy homeostasis and is involved in a wide variety of cellular processes including metabolic diseases, cancer, and aging (Bordone and Guarente, 2005; Brooks and Gu, 2009; Haigis and Sinclair, 2010). The dependence of SIRT1 on nicotinamide adenosine dinucleotide (NAD+) links its activity with cellular metabolism and energy levels (Canto et al., 2009; Cohen et al., 2004). For instance, nutrient deficiency, including limited glucose, increases SIRT1 activity in HepG2 cells (Escande et al., 2010). On the other hand, DNA damage and oxidative stress cause a tighter association between SIRT1 and DBC1 and result in inhibition of SIRT1 activity (Yuan et al., 2012; Zannini et al., 2012). Although both nutrient limitation and DNA damage regulate SIRT1 activity, the function of SIRT1 in metabolic checkpoint and DNA damage checkpoint remains elusive. Here we report a divergent role of SIRT1 in regulating these checkpoints. SIRT1 positively regulates a metabolic checkpoint induced by glucose deprivation, while negatively regulating the DNA damage checkpoint. Importantly, we identified TopBP1 as a major SIRT1 downstream target to mediate these checkpoint processes. Furthermore, we showed that the acetylation of TopBP1 regulated by SIRT1 changes the conformation of TopBP1, thereby facilitating its interaction with distinct partners under distinct stress conditions. Thus, our studies identify an acetylation-dependent regulatory mechanism of the SIRT1-TopBP1 axis in several aspects of genome maintenance events.
Results
SIRT1 regulates metabolic checkpoint
Metabolic checkpoints are cellular mechanisms that sense metabolic stress induced by nutrient deprivation and transduce these stress signals into a proper cellular response, such as cell-cycle control (Holley and Kiernan, 1974; Jones et al., 2005; Wang and Green, 2012). SIRT1 is known as a major metabolic regulator activated by energy stresses such as glucose starvation or caloric restriction (Bordone and Guarente, 2005; Brooks and Gu, 2009; Chen et al., 2005). However, neither its involvement in metabolic checkpoint nor the molecular mechanism that mediates such an effect is clear. Here we used glucose deprivation as a means to put cultured cells into a nutrient-deprived environment, thereby inducing metabolic stress. We challenged Sirt1+/+ and Sirt1−/− mouse embryonic fibroblasts (MEFs) with glucose deprivation and examined cell cycle progression. As shown in Figure S1A, low glucose treatment dramatically reduced the S phase population in Sirt1+/+ MEFs. In contrast, no apparent decrease was detected in Sirt1−/− MEFs. BrdU incorporation was also dramatically decreased in Sirt1+/+ but not Sirt1−/− MEFs in response to glucose deprivation (Figure 1A–C). On the other hand, other nutrient deficiencies, such as amino acid or serum depletion, induced a metabolic checkpoint in both Sirt1+/+ and Sirt1−/− MEFs (Figure S1B). These results suggested that SIRT1 is specifically involved in a glucose-deprivation-induced metabolic checkpoint. Metabolic checkpoints have been reported to maintain cell viability until the metabolic stress is resolved (Gurumurthy et al., 2010; Jones et al., 2005; Wang et al., 2013a). Consistent with these previous reports, glucose deprivation induced a mild increase in apoptosis in Sirt1+/+ MEFs, while apoptosis in Sirt1−/− MEFs was dramatically more severe (Figure 1D–E). These results suggest a critical role of SIRT1 in glucose-deprivation-induced metabolic stress response.
Figure 1. SIRT1 induces cell cycle arrest in response to glucose deprivation.
(A–B) Sirt1+/+ and Sirt1−/− MEFs synchronized in G0/G1 were released and treated with various concentrations of glucose. Cells that had progressed into S phase after 24 hours were determined by BrdU incorporation. The representative image is shown in (A) and the quantification of BrdU staining cells is shown in (B). (C) Sirt1+/+ and Sirt1−/− MEFs were treated with 1 mM glucose for various times. Cells were fixed and stained with anti-BrdU antibody. (D) Sirt1+/+ and Sirt1−/− MEFs were incubated with media containing various concentrations of glucose for 48 hours and the apoptosis cell ratio was measured using FACS. (A–D) The data presented are mean ± SD for three independent experiments. **P < 0.01 (one-way ANOVA test). (E) Cells as (D) were lysed and cell lysates were blotted with the indicated antibodies. (F) List of SIRT1-associated proteins identified by mass spectrometric analysis. (G–H) HEK293T cell lysates were subjected to immunoprecipitation with control IgG, anti-SIRT1 (G), or anti-TopBP1 (H) antibodies. The immunoprecipitates were then blotted with the indicated antibodies. See also Figure S1.
SIRT1 specifically interacts with and deacetylates TopBP1
To elucidate the potential molecular mechanism that mediates the effect of SIRT1 on metabolic checkpoint, we used cells stably expressing FLAG-SIRT1 to perform tandem affinity purification and mass spectrometry analysis. In addition to known SIRT1 interacting proteins such as DBC1 and USP22 (Kim et al., 2008; Lin et al., 2012; Zhao et al., 2008), we identified TopBP1 as a major SIRT1-associated protein (Figure 1F and S1C). We confirmed the SIRT1-TopBP1 interaction by co-immunoprecipitation (Co-IP). As shown in Figure 1G, SIRT1 Co-IPed with TopBP1. Reciprocal IP with TopBP1 antibodies also brought down SIRT1 (Figure 1H). Among the Sirtuin family members, only SIRT1 specifically interacted with TopBP1 (Figure S1D). Given the known role of TopBP1 in DNA replication, we hypothesized that SIRT1 might regulate metabolic checkpoint through TopBP1.
The interaction of TopBP1 and SIRT1 prompted us to examine a potential role for SIRT1 in the regulation of TopBP1 acetylation. We found that nicotinamide (NAM, a pan-Sirtuin family inhibitor) and SIRT1 depletion led to increased TopBP1 acetylation in cells expressing FLAG-TopBP1 (Figure 2A–B). Depletion of SIRT1 also led to an increase in acetylation of endogenous TopBP1 and p53, a known SIRT1 substrate (Figure 2C). Furthermore, WT SIRT1, but not a catalytic-inactive SIRT1 mutant (H363Y), led to a significant decrease in TopBP1 acetylation in vivo and in vitro (Figure 2D and S2A). Collectively, these results suggest that SIRT1 is a bona fide TopBP1-interacting protein and mediates deacetylation of TopBP1.
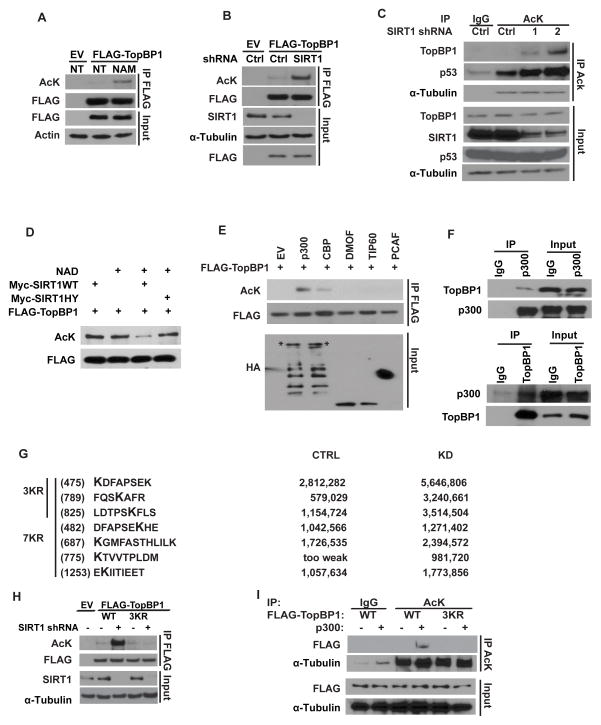
Figure 2. SIRT1 deacetylates TopBP1.
(A) HEK293T cells transfected with empty vector or FLAG-TopBP1 were either treated with vehicle or Nicotinamide (10 mM) for 16 hours. TopBP1 acetylation was determined by IP and Western blot. (B) HEK293T cells stably expressing control or SIRT1 shRNA were transfected with the indicated plasmids. TopBP1 acetylation was determined by IP and Western blot. (C) Endogenous TopBP1 acetylation was examined in cells. (D) SIRT1 deacetylated TopBP1 in vitro. Acetylated FLAG-tagged TopBP1 was purified from cells cotransfected with FLAG-TopBP1 and HA-p300 and incubated with purified SIRT1 WT or HY mutant. Acetylated TopBP1 was detected by immunoblotting with anti-acetyl lysine antibody. (E) HEK293T cells were transfected with indicated acetyltransferases. TopBP1 acetylation was then examined. (F) Endogenous interaction between TopBP1 and p300 was examined by anti-p300 or anti-TopBP1 immunoprecipitation and immunoblotting. (G) List of the Acetyl-sites analyzed by mass spectrum. (H) HEK293T cells stably expressing control or SIRT1 shRNA were transfected with indicated constructs. TopBP1 acetylation was examined. (I) HEK293T cells were transfected with the indicated constructs. TopBP1 acetylation was examined. See also Figure S2.
Acetylation is a dynamic process that can be controlled by deacetylases and acetyltransferases. Next, we tried to identify the acetyltransferases that are responsible for TopBP1 acetylation. Acetylation of TopBP1 was detected after ectopic expression of p300 and to a less extent CBP, but not other acetyltransferases, which include PCAF, Tip60, and hMOF (Figure 2E and S2B). We also detected a specific interaction between TopBP1 and p300 (Figure 2F). Furthermore, deletion of the HAT domain of p300 abolished the ability of p300 to mediate TopBP1 acetylation (Figure S2C). Next, to identify the potential acetylation sites regulated by SIRT1, TopBP1 acetylation was analyzed by mass spectrometry in control or SIRT1-depleted cells. Acetylation signals increased in seven lysines of TopBP1 when SIRT1 was depleted, among which three lysine residues (475, 789, and 825) were greatly enriched (Figure 2G and S2F–G). Depletion of SIRT1 increased the acetylation of WT TopBP1; however, in cells expressing single or double TopBP1 mutants, TopBP1 acetylation levels were only partially increased following SIRT1 depletion (Figure S2H–I). No apparent acetylation was observed when all 3 sites were mutated (3KR) (Figure 2H). Similarly, expression of p300 increased the acetylation of WT TopBP1 but not the TopBP1 3KR mutant (Figure 2I). These results demonstrate that K475, K789, and K825 are the major acetylation sites regulated by SIRT1 and p300.
Acetylation regulates TopBP1 function in DNA replication
TopBP1 enhances CDC45 chromatin loading at DNA replication origins and promotes replicative helicase activation and DNA replication initiation. We thus explored the potential role of TopBP1 acetylation in DNA replication. We found that acetylation of TopBP1 in S phase was apparently higher than in G1 phase, which was associated with a decreased interaction between SIRT1 and TopBP1 in S phase (Figure 3A–B). We next tested whether TopBP1 acetylation in S phase is important for its function in DNA replication. Knockdown of TopBP1 in cells with shRNA decreased BrdU incorporation and the S phase population (Figure 3C and S3A). Reconstituting these cells with WT TopBP1 restored BrdU incorporation and the S phase population to an extent similar to that in control cells, while reconstituting cells with 3KR mutants failed to do so (Figure 3C and S3A). Similarly, CDC45 loading was significantly reduced in TopBP1 depleted cells, which was restored by the expression of WT TopBP1 or the acetylation-mimetic 3KQ mutant, but not the 3KR mutant (Figure 3D). Because SIRT1 deacetylates TopBP1, the impact of SIRT1 on DNA replication was also examined. The levels of chromatin-bound CDC45 were significantly elevated in Sirt1−/− MEFs (Figure S3B). On the other hand, overexpression of SIRT1 decreased BrdU incorporation, CDC45 loading, and S phase populations, which could be reversed by expressing the TopBP1 3KQ mutant (Figure 3E–F and S3C). These results suggest that TopBP1 acetylation, regulated by SIRT1, is important for DNA replication. We next investigated how TopBP1 acetylation regulates DNA replication. CDK phosphorylated Treslin was demonstrated to interact with the N terminal BRCT1 and 2 domains of TopBP1, and the TopBP1-Treslin interaction is required for the recruitment of Cdc45 and for DNA replication activation in human cells (Kumagai et al., 2010, 2011; Mueller et al., 2011). We investigated whether TopBP1 acetylation regulates the interaction between TopBP1 and Treslin. As shown in Figure 3G, mutation of TopBP1 acetylation sites greatly decreased the TopBP1-Treslin interaction. To exclude the possibility of a disruption of protein conformation induced by mutating the acetylation sites, we examined the interaction between TopBP1 and other reported TopBP1 binding partners, such as EDD (Honda et al., 2002). EDD binds the center region of TopBP1 containing BRCT5-6 but not the N terminal BRCT1-2 required for TopBP1 binding. We found that TopBP1 acetylation mutants and WT TopBP1 bound EDD equally well (Figure S3D). TopBP1, when deacetylated by SIRT1 in vitro, no longer effectively pulled down Treslin in cells (Figure S3E). Furthermore, we found that Treslin-bound TopBP1 was preferentially acetylated on K825 compared to TopBP1 that did not bind Treslin (Figure S3F). To further confirm that acetylation of TopBP1 regulates TopBP1-Treslin binding, we used phosphorylated Treslin peptides corresponding to reported TopBP1 binding sites to perform a pull-down assay (Kumagai et al., 2011). The phospho-Ser1000 Treslin peptide pulled-down more WT TopBP1 than the 3KR mutant (Figure 3H), demonstrating that the acetylation of TopBP1 facilitates its binding to Treslin. Furthermore, overexpression of SIRT1 decreased the TopBP1-Treslin interaction, which can be reversed by 3KQ mutation (Figure 3I), suggesting that SIRT1 regulates the TopBP1-Treslin interacton through deacetylating TopBP1. To further clarify which TopBP1 acetylation site is essential for DNA replication, we generated single or double acetylation site mutants (K–R or K–Q) and examined TopBP1-Treslin interaction or CDC45 chromatin loading. As shown in Figure S3G–I, single or double acetylation mutants showed partial effects on TopBP1-Treslin interaction or CDC45 chromatin loading compared with the triple mutant, suggesting that all three sites are important for TopBP1 function in DNA replication. Taken together, our results demonstrate that acetylation of TopBP1 is important for its function in DNA replication by enhancing the TopBP1-Treslin interaction, CDC45 loading, and cell-cycle progression. Moreover, SIRT1 may regulate DNA replication through deacetylating TopBP1.
Figure 3. Acetylation regulates TopBP1 function in DNA replication.
(A–B) U2OS cells synchronized in G0/G1 were released to G1 or S phase. TopBP1 acetylation (A) or the SIRT1-TopBP1 interaction (B) was then examined. (C) U2OS cells stably expressing control or TopBP1 shRNA were reconstituted with indicated constructs. One portion of the cells was lysed and blotted with the indicated antibodies (lower panel), the other portion was stained with anti-BrdU antibody. The data presented are mean ± SD for three independent experiments. **P < 0.01. (D) U2OS cells stably expressing control or TopBP1 shRNA were transfected with indicated constructs. Cells synchronized in G0/G1 were released to S phase. Cells were collected at the indicated time and chromatin fractions were prepared. Immunoblotting was performed with the indicated antibodies. (E) U2OS cells transfected with the indicated constructs were stained with anti-BrdU antibody. The data presented are mean ± SD for three independent experiments. **P < 0.01. (F) Cells as in (E) synchronized in G0/G1 were released to S phase and chromatin fractions were prepared. Western blotting was performed. (G) Cells treated as in (D) were collected and TopBP1-Treslin interaction was examined. (H) Cells were treated as in (D) and whole cell lysates were incubated with nonphosphorylated (Pep) or phosphorylated (p-Pep) Ser1000 Treslin peptide covalently linked to beads. Immunoblotting was performed with anti-FLAG antibody. (I) Cells treated as in (F) were collected and TopBP1-Treslin interaction was examined. See also Figure S3.
TopBP1 acetylation regulates metabolic checkpoint
SIRT1 activity is regulated under conditions of metabolic stress (Canto et al., 2009; Cohen et al., 2004; Escande et al., 2010). Therefore, one mechanism by which SIRT1 regulates metabolic checkpoint might be through decreasing TopBP1 acetylation, resulting in decreased TopBP1-Treslin interaction, and blocking DNA replication initiation. We found that TopBP1 acetylation decreased in response to low glucose treatment (Figure 4A). However, in SIRT1-depleted cells, TopBP1 acetylation remained constant, suggesting that SIRT1 regulates TopBP1 acetylation during metabolic stress. To assess whether TopBP1 acetylation is important for metabolic checkpoint, we expressed WT TopBP1, 3KR, or 3KQ in cells depleted of endogenous TopBP1 and examined their effect on glucose deprivation induced metabolic checkpoint. In TopBP1-depleted cells reconstituted with WT TopBP1, the ability to enter S phase became progressively impaired as extracellular glucose was reduced in a time and glucose concentration dependent manner (Figure 4B–C). In contrast, cells reconstituted with 3KQ demonstrated a defect in metabolic checkpoint as a high percentage of cells failed to arrest and continued to enter S phase under limited glucose conditions (Figure 4B–C). This phenocopied what we observed in Sirt1−/− cells (Figure 1A–C). Moreover, knocking-down SIRT1 in cells with WT TopBP1 led to a defect in glucose deprivation induced metabolic checkpoint; conversely, in TopBP1 3KQ reconstituted cells, SIRT1 knockdown did not cause a further defect in metabolic checkpoint (Fig 4F and S4A). In addition, under both normal and glucose-limited conditions, depletion of p300 in cells decreased BrdU incorporation, which could be reversed by 3KQ mutation (Figure S4B). Furthermore, the effect of WT, 3KR, or 3KQ TopBP1 on glucose-dependent checkpoint correlated with their distinct interactions with Treslin and their effect on CDC45 chromatin loading (Figure 4D–E). Next, we examined apoptosis in cells reconstituted with TopBP1 WT or acetylation mutants. As shown in Figure 4G and S4C, glucose deprivation dramatically induced apoptosis in cells expressing 3KQ mutant but not WT TopBP1. Taken together, these results suggest that the acetylation of TopBP1, regulated by SIRT1 and p300, is critical for glucose deprivation dependent metabolic checkpoint activation.
Figure 4. TopBP1 acetylation regulates metabolic checkpoint.
(A) Cells stably expressing control or SIRT1 shRNA were transfected with indicated constructs. Cells were treated with various concentrations of glucose for 24 hours. TopBP1 acetylation was examined. (B) Cells stably expressing control or TopBP1 shRNA were transfected with indicated constructs. Cells were cultured at various concentrations of glucose. Cells that had progressed into S phase after 24 hours were determined by BrdU incorporation. (C) Cells as in (B) were treated with 1mM glucose. Cells that had progressed into S phase were determined by BrdU incorporation at the indicated times. (B–C) The data presented are mean ± SD for three independent experiments. (D) Cells as in (B) left untreated or treated with 1mM glucose were collected after 24 hours and the TopBP1-Treslin interaction was examined. (E) Cells were treated as in (D). Whole cell lysates and chromatin fractions were prepared and blotted with the indicated antibodies. (F) Cells transfected with the indicated constructs were cultured at the indicated concentrations of glucose for 24 hours. Cells were stained with anti-BrdU antibody. (G) Cells as in (B) were examined for apoptosis using FACS. (F–G) The data presented are mean ± SD for three independent experiments. See also Figure S4.
TopBP1 acetylation regulates the DNA damage response
In addition to its function in DNA replication, TopBP1 is required for ATR and Chk1 activation after genotoxic stress (Kumagai et al., 2006). First, we investigated whether TopBP1 acetylation can be regulated by genotoxic stress. Hydroxyurea (HU) dramatically elevated TopBP1 acetylation (Figure 5A–B). The increase in TopBP1 acetylation after DNA damage stress correlated with the decreased interaction between SIRT1 and TopBP1 (Figure 5C). Furthermore, the elevated interaction between TopBP1 and p300 after HU treatment might contribute to the increase in TopBP1 acetylation as well (Figure S5A). Genotoxic stress-induced TopBP1 acetylation was blocked by 3KR mutation (Figure 5D), indicating that the same lysine residues identified above may also be the major acetylation sites for TopBP1 in response to genotoxic stress.
Figure 5. TopBP1 acetylation regulates DNA damage response.
(A) U2OS Cells transfected with FLAG-TopBP1 were left untreated, or treated with 10 mM HU, and TopBP1 acetylation was examined. (B) Cells were left untreated or treated with HU, endogenous TopBP1 acetylation was examined. (C) Cells transfected with indicated constructs were left untreated or treated with HU and the SIRT1-TopBP1 interaction was examined. (D) Cells were transfected with indicated constructs and then treated with HU or left untreated. TopBP1 acetylation was then examined. (E) Cells stably expressing TopBP1 shRNA were transfected with indicated constructs. Thirty hours later, cells were left untreated or treated with UV (40 J/m2) or HU(10 mM). Immunostaining was performed with the indicated antibodies. The quantification of TopBP1 Foci was shown in the lower panel. The data presented are mean ± SD for three independent experiments. **P < 0.01 (F) Cells transfected as in (E) were left untreated (NT) or treated with HU and Chk1 phosphorylation was examined. (G) Cells as (E) were treated with HU and the colony formation assay was performed. The data presented are mean ± SD for three independent experiments. (H) Sirt1+/+ and Sirt1−/− MEFs were treated with UV, HU, or left untreated. p-Chk1 and Chk1 were detected by western blotting. (I) U2OS cells transfected with the indicated constructs. were left untreated or treated with HU and Chk1 phosphorylation was examined, (J) Cells transfected with the indicated constructs were treated with UV for different time points. Mitotic index was measure by staining cells with p-H3 S10 antibody. The data presented are mean ± SD for three independent experiments. See also Figure S5.
In response to replication stress, TopBP1 is recruited to DNA damage sites to promote the ATR-Chk1 pathway activation (Kumagai et al., 2006). We next explored whether TopBP1 acetylation regulates its chromatin recruitment at DNA damage sites following genotoxic stress. Compared to WT TopBP1, replication stress-induced focus formation was much weaker in the 3KR mutant (Figure 5E). Similarly, WT TopBP1 was efficiently loaded on chromatin, whereas the 3KR mutant exhibited a defect in chromatin accumulation (Figure S6A). Furthermore, reconstituting cells with WT TopBP1 rescued defective Chk1 phosphorylation caused by TopBP1 depletion, while reconstituting with the 3KR mutant failed to do so (Figure 5F). Consistently, cells reconstituted with the 3KR mutant were more sensitive to replication stress compared with cells reconstituted with WT TopBP1 (Figure 5G). These results suggest that TopBP1 acetylation is important for its recruitment to DNA damage sites and the activation of the ATR-Chk1 pathway.
SIRT1 regulates TopBP1 function in the DNA damage response
Since SIRT1 regulates TopBP1 deacetylation, its impact on DNA damage induced ATR-Chk1 activation was also examined. Depletion of SIRT1 dramatically enhanced Chk1 phosphorylation induced by HU or UV (Figure S6B). Similarly, Chk1 phosphorylation is significantly increased in Sirt1−/− MEFs (Figure 5H). This is consistent with the increased TopBP1 acetylation and its role in Chk1 activation. Next, we explored whether SIRT1 regulates the focus formation and chromatin recruitment of TopBP1 after replication stress. As shown in Figure S6C, TopBP1 focus formation was greatly reduced when SIRT1 was cotransfected in U2OS cells. This is consistent with the role of TopBP1 acetylation in its focus formation. Conversely, SIRT1-depleted cells displayed an apparent increase in TopBP1 chromatin loading following HU treatment (Figure S6D). We further investigated whether SIRT1 regulates Chk1 activation through deacetylating TopBP1. SIRT1 knockdown increased Chk1 phosphorylation after HU treatment compared to control shRNA (Figure S6E, lane 2 versus lane 1). However, in TopBP1 depleted cells, no apparent Chk1 phosphorylation was detected even when SIRT1 was depleted. In addition, SIRT1 downregulation could only increase Chk1 phosphorylation in cells expressing WT TopBP1, but not the 3KR mutant (Figure S6E, lane 5–8). Conversely, SIRT1 overexpression decreased Chk1 phosphorylation in cells expressing WT TopBP1 but not the 3KQ mutant (Figure 5I). Collectively, these data demonstrate that SIRT1 negatively regulates the ATR-Chk1 pathway through deacetylating TopBP1. Activation of ATR-Chk1 signaling is essential for the DNA damage checkpoint. Next, we examined whether the SIRT1-TopBP1 axis regulates DNA damage checkpoint. As shown in Figure 5J, depleted of SIRT1 resulted in a more sustained cell cycle checkpoint activation in cells expressing WT TopBP1, while in cells expressing the 3KQ mutant, SIRT1 depletion could not induce further effects. These data suggest SIRT1 is able to regulate DNA damage induced cell-cycle checkpoint through deacetylating TopBP1.
Acetylation regulates TopBP1 interaction with Rad9
Since TopBP1 acetylation regulates its chromatin loading in response to DNA damage, we next investigated the underlying mechanism. TopBP1 was shown to interact with Rad9 of the 9-1-1 complex via its N terminal tandem BRCT domain and phosphorylate Ser387 of Rad9; this interaction is important for TopBP1 chromatin loading and the ATR-Chk1 activation (Delacroix et al., 2007; Lee and Dunphy, 2010; Lee et al., 2007). First, we determined whether Rad9 is essential for TopBP1 loading in response to replication stress in mammals. As shown in Figure 6A–C, TopBP1 chromatin loading and focus formation after HU treatment were severely compromised upon Rad9 depletion. Given that TopBP1 focus formation requires Rad9 and TopBP1 acetylation, we next determined whether TopBP1 acetylation regulates the TopBP1-Rad9 interaction. We found that replication stress increased the interaction between WT TopBP1 and Rad9; however, the increased interaction was abolished in the case of the acetylation-defective TopBP1 mutant (Figure 6D). In contrast, the acetylation-mimetic TopBP1 showed enhanced and constitutive interaction with Rad9 (Figure 6D). Furthermore, we performed a pull-down assay using Flag-TopBP1 that was deacetylated by SIRT1 in vitro. As shown in Figure S7A, deacetylated TopBP1 ineffectively pulled down Rad9 in cells. These results suggest that TopBP1 acetylation is a major mechanism of stress-induced TopBP1-Rad9 interaction. To further clarify which TopBP1 acetylation site is essential for the DNA damage response, we examined the TopBP1-Rad9 interaction and Chk1 phosphorylation using single or double acetylation site mutants. As shown in Figure S7B–D, single or double acetylation site mutants showed partial effects on TopBP1-Rad9 interaction and Chk1 phosphorylation compared with the triple mutant, suggesting all three sites are important for TopBP1 function in the DNA damage response. Furthermore, phospho-Ser387 Rad9 peptide was able to pull down WT TopBP1, but not 3KR or 7KR mutant, in cells treated with HU (Figure 6E). Overall, these results suggest that TopBP1 acetylation facilitates its binding to Rad9 and its subsequent chromatin loading.
Figure 6. Acetylation regulates TopBP1 interaction with Rad9.
(A) U2OS cells were transfected with control or Rad9 shRNA. Rad9 levels were examined by western blot. (B) U2OS cells stably expressing control or Rad9 shRNA were transfected with WT TopBP1. Cells were left untreated or treated with UV or HU. Immunostaining was performed with the indicated antibodies. The quantification of TopBP1 Foci was shown in the lower panel. The data presented are mean ± SD for three independent experiments. **P < 0.01(C) U2OS cells stably expressing control or Rad9 shRNA were left untreated or treated with HU and harvested at the indicated time. Soluble and chromatin fractions were prepared and immunoblotted with indicated antibodies. (D) Cells stably expressing TopBP1 shRNA were transfected with the indicated constructs and left untreated or treated with HU. The TopBP1-Rad9 interaction was examined. (E) Cells stably expressing TopBP1 shRNA were transfected with the indicated constructs. and left untreated or treated with HU. Whole-cell lysates were incubated with nonphosphorylated (P) or phosphorylated (p-P) Ser387 Rad9 peptide covalently linked to beads. Immunoblotting was performed. See also Figure S6 and S7.
Acetylation regulates intramolecular interactions in TopBP1
It is interesting that TopBP1 acetylation is important for its BRCT domain to bind both phosphorylated Treslin and Rad9. We speculated that the TopBP1 N-terminal BRCT domains that are responsible for binding phosphorylated Treslin and Rad9 are not available to recognize these phospho-motifs when TopBP1 is not acetylated. TopBP1 acetylation might induce a conformational change which exposes the N-terminal BRCT domains to bind Treslin and Rad9. To test this possibility, we first examined whether the N-terminal BRCT domains of TopBP1 interact with other regions of TopBP1. GST pull-down results indicated that the N-terminal BRCT1 and 2 domain was able to interact with the C-terminal region of TopBP1 (Figure 7A). This raised the possibility of an intramolecular interaction between the N-terminus and C-terminus of TopBP1. To test whether this intracellular interaction can be regulated by TopBP1 acetylation, we performed a Duo-link in situ assay using WT TopBP1 and its mutants. Duo-link in situ assay is an antibody-based method in which two antigens (HA-tagged N-terminal TopBP1 and FLAG-tagged C-terminal TopBP1) are immunolabeled first with two primary antibodies and then with different species-specific secondary antibodies conjugated to complementary oligonucleotides. When two antibody molecules are in close proximity, the complementary DNA strands can be ligated, amplified, and visualized with a fluorescent probe (Figure 7B and S7E). Under unstressed conditions, the N-terminal region of WT TopBP1 interacts with its C-terminal region, as demonstrated by positive Duo-Link signals in untreated cells (Figure 7C). HU treatment resulted in a loss of Duo-Link signals, suggesting a disassociation between the N-terminus and C-terminus of TopBP1. Conversely, glucose deprivation led to enhanced Duo-Link signals, suggesting increased intracellular interaction of TopBP1. In cells expressing the 3KR mutant, Duo-Link signals were not affected by HU stress or glucose starvation, suggesting a lack of conformational change when TopBP1 acetylation is abolished. In cells expressing the 3KQ mutant, there was no Duo-Link signal even in the absence of stress, suggesting TopBP1 is always in an open conformation. We also examined the possibility that TopBP1 acetylation affects intermolecular interactions. We found that TopBP1 could form oligomers; however, the oligomerization was not affected by metabolic stress or replication stress (Figure S7F). In addition, mutation of TopBP1 acetylation sites did not affect TopBP1 oligomerization. These results suggest that TopBP1 acetylation does not affect intermolecular interactions, and TopBP1 intermolecular interactions cannot explain the phenotypes we observed. Collectively, these results suggest that TopBP1 acetylation disrupts its intramolecular interactions, causes a conformational change, and exposes its N-terminal BRCT domains to allow for its interaction with Rad9 or Treslin.
Figure 7. Acetylation regulates the intramolecular interaction in TopBP1.
(A) HEK293T cells transfected with indicated constructs were pulled-down with GST or GST-BRCT1-2 and analyzed by western blotting. (B) A diagram of Duo-link in situ assay. (C) WT, 3KR, or 3KQ mutants were expressed in U2OS cells stably expressing TopBP1 shRNA. Cells were left untreated or treated with HU or low glucose. Duo-link in situ assay was performed. The quantification of Duo-link positive staining cells is shown in the right panel. The data presented are mean ± SD for three independent experiments. *P<0.05; **P<0.01 See also Figure S7.
Discussion
Here, our study demonstrates that SIRT1 differentially regulates metabolic checkpoint and DNA damage checkpoint through deacetylating TopBP1. Our studies identify an acetylation-dependent regulatory mechanism of the SIRT1-TopBP1 axis in several aspects of genomic maintenance events. TopBP1 and Treslin cooperate in the loading of Cdc45 onto replication origins (Kumagai et al., 2010, 2011; Zegerman and Diffley, 2007, 2010). Here we found that TopBP1 acetylation regulated by SIRT1 is essential for the TopBP1-Treslin interaction and DNA replication (Figure 3). The acetylation of TopBP1 provides a new mechanism to regulate DNA replication under both normal and stress conditions. Under normal conditions, TopBP1 is acetylated in S phase, which allows TopBP1 binding to Treslin and initiation of DNA replication. Under nutrient deprivation conditions, SIRT1 activity is upregulated (Canto et al., 2009; Cohen et al., 2004; Escande et al., 2010) and TopBP1 acetylation is decreased, leading to a decrease in TopBP1-Treslin interaction and the activation of metabolic checkpoint. To our knowledge, this is the first report showing the critical role of the SIRT1-TopBP1 pathway in the control of cellular metabolic checkpoint by nutrient availability. Other regulatory mechanisms of metabolic checkpoint may also be involved during limited glucose conditions as cells expressing an acetylation-mimetic mutant (3KQ) cannot completely block this metabolic checkpoint. A previous report showed SIRT1 deacetylates Mcm10 and regulates DNA replication (Fatoba et al., 2013). It is possible that SIRT1 may also regulate metabolic checkpoint through Mcm10, and further studies are required to confirm this. Recent evidence also suggests that the LKB1-AMPK-Raptor cascade mediates metabolic checkpoints in response to various energy stresses (Gwinn et al., 2008; Inoki et al., 2003; Jones et al., 2005). It has been noted that there is a similarity between AMPK and SIRT1 in sensing energy stress and nutrient status, regulating mitochondria function, modulating glucose/lipid homeostasis, and controlling the activity of many common target molecules such as PGC-1α, FOXOs, and p53 (Canto and Auwerx, 2009; Finley and Haigis, 2009). Furthermore, the activities of SIRT1 and AMPK are reciprocally modulated (Canto et al., 2009; Canto et al., 2010; Lan et al., 2008). Therefore, additional studies are required to examine the potential crosstalk between AMPK and the SIRT1-TopBP1 pathway in controlling metabolic checkpoint.
While SIRT1-mediated TopBP1 deacetylation is important for metabolic checkpoint activation, this reaction is blocked following replication stress. Replication stress results in dissociation of TopBP1 and SIRT1 and increases TopBP1 acetylation (Figure 5). Meanwhile, decreased SIRT1 activity in response to DNA damage may also contribute to the increased TopBP1 acetylation (Yuan et al., 2012; Zannini et al., 2012). Increased TopBP1 acetylation in turn facilitates Rad9 binding and checkpoint activation. The interaction between the 9-1-1 complex and TopBP1 involves Ser-387 of Rad9 and the N-terminal region of TopBP1 (Delacroix et al., 2007; Lee and Dunphy, 2010; Lee et al., 2007). Interestingly, Ser-387 of Rad9 in human cells is constitutively phosphorylated by casein kinase 2 (St Onge et al., 2003), which apparently does not require the presence of DNA damage or stalled DNA replication forks. Despite this, the TopBP1-Rad9 interaction is enhanced by replication stress. These results indicate that an additional DNA-damage-dependent regulatory mechanism must exist to coordinate the TopBP1-Rad9 interaction and ATR-Chk1 activation in response to genotoxic stress. Our findings of genotoxic-induced acetylation of TopBP1 could explain the inducible interaction between TopBP1 and Rad9 and this represents a new regulatory mechanism for checkpoint activation. Although SIRT1 has previously been shown to maintain genomic stability (Oberdoerffer et al., 2008; Wang et al., 2008), our studies also suggest that SIRT1, when overexpressed or hyperactivated, might compromise genomic stability, at least in the context of replication stress. A recent study also linked SIRT1 with higher acquisition of genetic mutations in cancers (Wang et al., 2013b).
Based on our results, we propose that acetylation of TopBP1 changes its conformation, exposing its N-terminal BRCT domains that are required for Treslin and Rad9 interactions. However, further structural studies and conformational analyses of TopBP1 are necessary to better understand the precise mechanism. An intriguing question is how TopBP1 determines its distinct binding partners under different conditions. A possible scenario is that during normal replication, TopBP1 is acetylated and interacts with Treslin to facilitate DNA replication initiation. Although acetylated TopBP1 could also interact with Rad9, unlike the Treslin-TopBP1 complex, the Rad9-TopBP1 complex is not recruited to chromatin in the absence of replication stress. Under nutrient limitation conditions, SIRT1 activity is switched on, which in turn abolishes the TopBP1-Treslin interaction and arrests the cell cycle. On the other hand, SIRT1 activity is switched off during DNA damage, which facilitates the TopBP1-Rad9 interaction, activates the ATR-Chk1 pathway, and causes cell-cycle arrest. Activated Chk1 inactivates S-phase CDK, thereby reducing Treslin phosphorylation that is required for TopBP1 binding (Sanchez et al., 1997). Consistent with this, the TopBP1-Treslin interaction greatly decreases in a checkpoint-dependent manner (Boos et al., 2011). Once the DNA damage checkpoint response is terminated, acetylated TopBP1 may associate with Treslin again and promote the initiation of DNA replication. The on and off function of the SIRT1-TopBP1 axis in response to different cellular stresses makes it a key switch that enables cells to regulate biological events more efficiently and specifically. The SIRT1-TopBP1 pathway must therefore be tightly regulated and dysfunction of this pathway might compromise many aspects of cell metabolism and proliferation.
Experimental Procedures
Cell Culture, plasmids and antibodies
293T cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS). HCT116 and U2OS cells were cultured in McCoy’s 5A supplemented with 10% FBS. Mouse embryonic fibroblasts (MEFs) were cultured in DMEM containing 15% FBS. TopBP1 was cloned into pIRES-EGFP and p3XFLAG-CMV™-13 vector. All site mutants were generated by site-directed mutagenesis (Stratagene) and verified by sequencing. WT SIRT1 and its mutants have been previously described (Kim et al., 2008). Acetyltransferases p300, CBP, PCAF, Tip60 and hMOF constructs were kindly provided by Dr. Jianyuan Luo (University of Maryland).
Antibodies against phosphoChk1 (345), CDC45, and α-Tubulin were purchased from Cell Signaling. TopBP1 and Treslin antibody were obtained from Bethyl laboratories. Anti-p53 and Chk1 antibody were purchased from Santa Cruz. Rad9 antibodies have been previously described (Delacroix et al., 2007). Anti-FLAG (m2), anti-HA, and anti-β-actin antibodies were purchased from Sigma. Anti-acetylated lysine antibodies were purchased from Rockland and Millipore. Acetyl lysine antibody agarose was purchased from Immunechem.
RNA Interference
SIRT1 shRNAs and TopBP1 shRNA (NM_007027.3-4768s21c1) targeting 3′UTR region were purchased from Sigma. The sequences for SIRT1 shRNA are GCAAAGCCTTTCTGAATCTAT and GCGGGAATCCAAAGGATAATT. The TopBP1 3′UTR shRNA sequence is GATAGATTTGGGTAGTAATTT. Rad9 shRNA was purchased from Open Biosystems.
BrdU incorporation, phospho-histone-H3 staining, and fluorescent microscopy
Cells growing on coverslips were incubated with 20 μM bromodeoxyuridine (BrdU) for 30 min. Cells were washed with PBS for two times and fixed by 3% paraformaldehyde for 15 min. Then cells were permeabilized with 0.5% Triton for 10 min and 4M HCl for 30 min. Cells were then washed three times with 1×PBS containing 0.5% Tween-20 and incubated with the mouse anti-BrdU primary antibody (Roche Pharmaceuticals) at 37°C for 25 min. The cells were washed and labeled with FITC-conjugated goat anti-mouse IgG. The nuclei were stained with 4′ and 6-diamidino-2-phenylindole (DAPI).
phospho-histone-H3 staining was performed as pervious described (Luo et al., 2009). Cells were fixed by 3% paraformaldehyde and permeabilized with 0.5% Triton. Then cells were incubated with mouse anti-phospho-histone-H3 (anti-PH3) monoclonal antibody (Cell Signaling Technology, Beverly, MA, USA) at 37°C for 25 min. The cells were washed and labeled with FITC-conjugated goat anti-mouse IgG. The nuclei were stained with DAPI.
BrdU-incorporated cells or pH3 staining cells were examined with a Fluorescence microscope (ECLIPSE 80i; Nikon) using a 4× NA 0.10 objective lens (Nikon). For each condition, randomly selected 600 cells were counted. Images were captured with a camera (SPOT 2 Megasample; Diagnostic Instruments, Inc.) and processed using SPOT software (version 4.6; Diagnostic Instruments, Inc.).
Immunofluorescence staining
Cells grown on coverslips were fixed in Acetone-Methanol (1:1) at −20 °C for 20 min. Slides were washed in phosphate-buffered saline and blocked with 5% goat serum for 1 hour at room temperature, then incubated with primary antibodies at 37 °C for 30 min. After washing with PBS twice, cells were incubated with FITC or rhodamine-conjugated secondary antibodies at 37°C for 30 min. Nuclei were counterstained with 4′6-diamidino-2-phenylindole (DAPI). After a final wash with PBS, coverslips were mounted with glycerin containing paraphenylenediamine.
Duo-link in situ assay
U2OS with stable TopBP1 depletion were transfected with WT, 3KR, and 3KQ TopBP1. At 24 hour after transfection, cells were treated with vehicle or HU (10mM) for 3 hours and fixed with 3% paraformaldehyde solution in PBS containing 50mM sucrose at room temperature for 15 min. After permeabilization with 0.5% Triton X-100 buffer containing 20mM HEPES pH 7.4, 50mM NaCl, 3mM MgCl2 and 300mM sucrose at room temperature for 5 min, cells were incubated for 1 hour in blocking solution. The slides were incubated with the mixture of primary antibodies containing anti-HA mouse monoclonal antibody (targeting N-terminal region of TopBP1) and anti-FLAG rabbit polyclonal sera (targeting C-terminal region of TopBP1). Control experiments used no primary antibody. Intramolecular interactions were detected and visualized by proximity ligation assay and rolling circle amplification using duo-link in situ kit according to the manufacturer’s instructions (Olink Bioscience).
Supplementary Material
Figure S1. SIRT1 is involved in glucose deprivation dependent metabolic checkpoint.
(A) Cell as in 1C were stained with PI and cell-cycle profiles were analyzed by FACS. (B) Sirt1+/+ and Sirt1−/− MEFs were cultured with normal medium or medium without amino acids or FBS for 24 hours. Cells were incubated with 20 μM BrdU; 30 min later, cells were fixed and stained with anti-BrdU antibody. The data presented are mean ± SD for three independent experiments. (C) List of the TopBP1 peptides identified by mass spectrum analysis. (D) HEK293T cells were transfected with empty vector or plasmids encoding full-length FLAG-SIRT1-7. Whole-cell lysates were subjected to immunoprecipitation with anti-FLAG beads. Immunoblotting was performed with anti-TopBP1 and anti-FLAG antibodies. Figure S1, related to Figure 1.
Figure S2. SIRT1 and p300 regulate TopBP1 acetylation
(A) Cells stably expressing SIRT1 shRNA were transfected with the indicated constructs. 48 hours later, TopBP1 acetylation was examined. LE: longer exposure; SE: shorter exposure. (B) HEK293T cells were transfected with the indicated constructs and histone acetylation was examined. (C) HEK293T cells were transfected with empty vector, WT, or HAT-deleted HA-p300. TopBP1 or tubulin was then examined. (D) HEK293T cells were co-transfected with FLAG-TopBP1 and different HA-tagged acetyltransferases, CBP, p300, PCAF, Tip60, or hMOF. TopBP1 acetylation was detected with anti-AcK825 antibodies. (E) SIRT1 deacetylated TopBP1 in vitro. Acetylated FLAG-tagged TopBP1 was purified from cells contransfected with FLAG-TopBP1 and HA-p300 and incubated with NAD, purified WT SIRT1, or HY mutant SIRT1, or in the absence of the indicated reagents. Acetylated TopBP1 was detected by immunoblot with anti-AcK825. (F–G) Raw data from TopBP1 acetyl-specific mass spectrum analysis. (H–I) Cells were transfected with the indicated constructs. 48 hours later, TopBP1 acetylation was examined. Figure S2, related to Figure 2.
Figure S3. SIRT1 regulates DNA replication through deacetylating TopBP1
(A) Cells as in 3C were stained with PI and cell-cycle profiles were examined by FACS. (B) Sirt1+/+ and Sirt1−/− MEFs were serum starved for 48 hours (0.1% serum) and stimulated to enter cell-cycle progression by serum addition. Cells were collected at the indicated times and chromatin fractions were prepared and immunoblotting was performed with the indicated antibodies. (C) Cells as in 3E were stained with PI and cell-cycle profiles were examined by FACS. Protein expression was analyzed by Western blot (upper panel). (D) Cells were transfected with the indicated constructs and the TopBP1-EDD interaction was examined. (E) Acetylated FLAG-tagged TopBP1 was purified from cells cotransfected with FLAG-TopBP1 and HA-p300 using FLAG beads and then incubated with NAD and purified SIRT1 (WT or HY mutant) in vitro. The TopBP1-Treslin interaction was then examined. (F) Cells transfected with Myc-Treslin and FLAG-TopBP1 were lysed and immunoprecipitated 4 times with Myc-Beads (containing FLAG-TopBP1 interacted with Myc-Treslin). Myc-Treslin depleted cell lysates were immuprecipated with anti-FLAG antibody (containing FLAG-TopBP1 not bound to Myc-Treslin). Acetylation of TopBP1 (AcK825) was examined in both Myc and FLAG immunoprecipitates. (G) Cells stably expressing TopBP1 shRNA were transfected with the indicated constructs. After 48 hours, the TopBP1-Treslin interaction was examined. (H) Cells stably expressing TopBP1 shRNA were transfected with the indicated constructs. After 24 hours, cells were incubated with media containing 1mM glucose for 24 hours, and the TopBP1-Treslin interaction was examined. (I) Cells stably expressing TopBP1 shRNA were transfected with the indicated constructs and were serum starved for 48 hours (0.1% serum) before being induced with media containing 10% serum for 18 hours to enter S phase. Whole cell lysates and chromatin fractions were prepared. Western blotting was performed with the indicated antibodies. Figure S3, related to Figure 3.
Figure S4. TopBP1 acetylation regulated by p300 and SIRT1 plays an important role in glucose limitation dependent metabolic checkpoint
(A) Cells as in 4F were stained with PI and cell-cycle profiles were examined by FACS. Protein expression was analyzed by western blot (lower panel). (B) Cells expressing the indicated constructs were treated with various concentrations of glucose (25mM or 1mM) for 24 hours. Cells were incubated with 20 μM BrdU; 30 min later, cells were fixed and stained with anti-BrdU antibody. The data presented are mean ± SD for three independent experiments. *P<0.05; **P<0.01 (one-way ANOVA test). Protein expression was analyzed by western blot (lower panel). (C) Cells stably expressing TopBP1 shRNA were transfected with the indicated constructs treated with various concentrations of glucose (25mM or 0.1mM) for 48 hours. PARP1 cleavage was then examined by western blot. Figure S4, related to Figure 4.
Figure S5. p300-TopBP1 interaction is regulated by DNA damage
(A) Cells were transfected with the indicated plasmids and were treated with HU (10mM) or left untreated. The p300-TopBP1 interaction was then examined. Figure S5, related to Figure 5.
Figure S6. SIRT1 regulates the DNA damage response through deacetylating TopBP1
(A) Cells stably expressing TopBP1 shRNA were transfected with WT TopBP1 or 3KR mutant. Thirty hours later, cells were mock-treated or treated with HU (10 mM). Cells were harvested at the indicated time points. The soluble and chromatin fractions were prepared and immunoblotted with antibodies as indicated. (B) HCT116 (left panel) or U2OS (right panel) cells stably expressing control shRNA or SIRT1 shRNA were treated with UV (40 J/m2), HU (5 mM) or left untreated. Chk1 phosphorylation was then examined. (C) U2OS cells transfected with plasmids encoding WT TopBP1, myc-SIRT1, or empty vector were treated with 5 mM HU or UV (40 J/m2). TopBP1 focus formation was examined by IF. Quantification of TopBP1 foci was shown in the right panel. The data presented are mean ± SD for three independent experiments. (D) Cells stably expressing control or SIRT1 shRNA were mock-treated or treated with HU (10 mM) for the indicated times. The soluble and chromatin fractions were prepared and immunoblotted with antibodies as indicated. (E) Cells expressing control, SIRT1, or TopBP1 shRNA were transfected with empty vector, WT TopBP1, or the 3KR mutant and, then treated with HU (10mM) or left untreated. Chk1 phosphorylation was then examined. Figure S6, related to Figure 6.
Figure S7. TopBP1 acetylation is important for the DNA damage response
(A) Acetylated FLAG-tagged TopBP1 was purified from cells cotransfected with FLAG-TopBP1 and HA-p300 using FLAG beads and incubated with NAD, purified WT SIRT1 or HY mutant SIRT1 for deacetylation in vitro. The TopBP1-Rad9 interaction was then examined. (B) Cells stably expressing TopBP1 shRNA were transfected with the indicated constructs. After 48 hours, cells were left untreated or treated with HU (10 mM), and the TopBP1-Rad9 interaction was examined. (C) Cells stably expressing TopBP1 shRNA were transfected with the indicated constructs. After 48 hours, the TopBP1-Rad9 interaction was examined. (D) Cells stably expressing TopBP1 shRNA were transfected with the indicated constructs. After 48 hours, cells were treated with HU (10mM) and Chk1 phosphorylation was examined. (E) U2OS cells stably expressing TopBP1 shRNA were stably expressing TopBP1 3KR mutant. Negative control experiments for the Duo-link assay (HA antibody only and FLAG antibody only) were performed. (F) Empty vector or HA-TopBP1 were cotransfected with FLAG-WT, 3KR, or 3KQ mutants in HEK293T cells stably expressing TopBP1 shRNA. Cells were left untreated or treated with HU (10 mM) or low glucose (0.1 mM). Cell lysates were subjected to immunoprecipitation with anti-HA antibodies. The immunoprecipitates were then blotted with the indicated antibodies. Figure S7, related to Figure 6 and 7.
Acknowledgments
We thank Dr. Larry Karnitz (Mayo Clinic, USA) for providing Rad9 antibody, C-tail phospho-387 and non-phospho peptide and Dr Jianyuan Luo for providing acetylatransferases p300, CBP, PCAF, Tip60 and hMOF constructs (University of Maryland, USA). This work was supported by the National Basic Research Program of China (973 Program, Grant No. 2013CB530700), National Natural Science Foundation of China (81222029, 31270806 and 81322031), Shanghai Pujiang Program (12PJ1407500) and National Institutes of Health grants (CA130996, CA189666, CA129344, CA108961 and CA148940).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–429. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- Boos D, Sanchez-Pulido L, Rappas M, Pearl LH, Oliver AW, Ponting CP, Diffley JF. Regulation of DNA replication through Sld3-Dpb11 interaction is conserved from yeast to humans. Curr Biol. 2011;21:1152–1157. doi: 10.1016/j.cub.2011.05.057. [DOI] [PubMed] [Google Scholar]
- Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat Rev Mol Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W. How does SIRT1 affect metabolism, senescence and cancer? Nat Rev Cancer. 2009;9:123–128. doi: 10.1038/nrc2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr Opin Lipidol. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Jiang LQ, Deshmukh AS, Mataki C, Coste A, Lagouge M, Zierath JR, Auwerx J. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab. 2010;11:213–219. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes & development. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escande C, Chini CC, Nin V, Dykhouse KM, Novak CM, Levine J, van Deursen J, Gores GJ, Chen J, Lou Z, et al. Deleted in breast cancer-1 regulates SIRT1 activity and contributes to high-fat diet-induced liver steatosis in mice. J Clin Invest. 2010;120:545–558. doi: 10.1172/JCI39319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatoba ST, Tognetti S, Berto M, Leo E, Mulvey CM, Godovac-Zimmermann J, Pommier Y, Okorokov AL. Human SIRT1 regulates DNA binding and stability of the Mcm10 DNA replication factor via deacetylation. Nucleic Acids Res. 2013;41:4065–4079. doi: 10.1093/nar/gkt131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley LW, Haigis MC. The coordination of nuclear and mitochondrial communication during aging and calorie restriction. Ageing Res Rev. 2009;8:173–188. doi: 10.1016/j.arr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurumurthy S, Xie SZ, Alagesan B, Kim J, Yusuf RZ, Saez B, Tzatsos A, Ozsolak F, Milos P, Ferrari F, et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 2010;468:659–663. doi: 10.1038/nature09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Holley RW, Kiernan JA. Control of the initiation of DNA synthesis in 3T3 cells: low-molecular weight nutrients. Proc Natl Acad Sci U S A. 1974;71:2942–2945. doi: 10.1073/pnas.71.8.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda Y, Tojo M, Matsuzaki K, Anan T, Matsumoto M, Ando M, Saya H, Nakao M. Cooperation of HECT-domain ubiquitin ligase hHYD and DNA topoisomerase II-binding protein for DNA damage response. J Biol Chem. 2002;277:3599–3605. doi: 10.1074/jbc.M104347200. [DOI] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Shevchenko A, Dunphy WG. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell. 2010;140:349–359. doi: 10.1016/j.cell.2009.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Shevchenko A, Dunphy WG. Direct regulation of Treslin by cyclin-dependent kinase is essential for the onset of DNA replication. J Cell Biol. 2011;193:995–1007. doi: 10.1083/jcb.201102003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Cacicedo JM, Ruderman N, Ido Y. SIRT1 modulation of the acetylation status, cytosolic localization, and activity of LKB1. Possible role in AMP-activated protein kinase activation. J Biol Chem. 2008;283:27628–27635. doi: 10.1074/jbc.M805711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Dunphy WG. Rad17 plays a central role in establishment of the interaction between TopBP1 and the Rad9-Hus1-Rad1 complex at stalled replication forks. Mol Biol Cell. 2010;21:926–935. doi: 10.1091/mbc.E09-11-0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- Lin Z, Yang H, Kong Q, Li J, Lee SM, Gao B, Dong H, Wei J, Song J, Zhang DD, et al. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol Cell. 2012;46:484–494. doi: 10.1016/j.molcel.2012.03.024. [DOI] [PubMed] [Google Scholar]
- Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- Luo K, Yuan J, Chen J, Lou Z. Topoisomerase IIalpha controls the decatenation checkpoint. Nat Cell Biol. 2009;11:204–210. doi: 10.1038/ncb1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller AC, Keaton MA, Dutta A. DNA replication: mammalian Treslin-TopBP1 interaction mirrors yeast Sld3-Dpb11. Curr Biol. 2011;21:R638–640. doi: 10.1016/j.cub.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, Elledge SJ. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- St Onge RP, Besley BD, Pelley JL, Davey S. A role for the phosphorylation of hRad9 in checkpoint signaling. J Biol Chem. 2003;278:26620–26628. doi: 10.1074/jbc.M303134200. [DOI] [PubMed] [Google Scholar]
- Wang R, Green DR. Metabolic checkpoints in activated T cells. Nat Immunol. 2012;13:907–915. doi: 10.1038/ni.2386. [DOI] [PubMed] [Google Scholar]
- Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Huang G, Zeng H, Yang K, Lamb RF, Chi H. Tuberous sclerosis 1 (Tsc1)-dependent metabolic checkpoint controls development of dendritic cells. Proc Natl Acad Sci U S A. 2013a;110:E4894–4903. doi: 10.1073/pnas.1308905110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Yuan H, Roth M, Stark JM, Bhatia R, Chen WY. SIRT1 deacetylase promotes acquisition of genetic mutations for drug resistance in CML cells. Oncogene. 2013b;32:589–598. doi: 10.1038/onc.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Luo K, Liu T, Lou Z. Regulation of SIRT1 activity by genotoxic stress. Genes Dev. 2012;26:791–796. doi: 10.1101/gad.188482.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannini L, Buscemi G, Kim JE, Fontanella E, Delia D. DBC1 phosphorylation by ATM/ATR inhibits SIRT1 deacetylase in response to DNA damage. J Mol Cell Biol. 2012;4:294–303. doi: 10.1093/jmcb/mjs035. [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF. Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature. 2010;467:474–478. doi: 10.1038/nature09373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–590. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. SIRT1 is involved in glucose deprivation dependent metabolic checkpoint.
(A) Cell as in 1C were stained with PI and cell-cycle profiles were analyzed by FACS. (B) Sirt1+/+ and Sirt1−/− MEFs were cultured with normal medium or medium without amino acids or FBS for 24 hours. Cells were incubated with 20 μM BrdU; 30 min later, cells were fixed and stained with anti-BrdU antibody. The data presented are mean ± SD for three independent experiments. (C) List of the TopBP1 peptides identified by mass spectrum analysis. (D) HEK293T cells were transfected with empty vector or plasmids encoding full-length FLAG-SIRT1-7. Whole-cell lysates were subjected to immunoprecipitation with anti-FLAG beads. Immunoblotting was performed with anti-TopBP1 and anti-FLAG antibodies. Figure S1, related to Figure 1.
Figure S2. SIRT1 and p300 regulate TopBP1 acetylation
(A) Cells stably expressing SIRT1 shRNA were transfected with the indicated constructs. 48 hours later, TopBP1 acetylation was examined. LE: longer exposure; SE: shorter exposure. (B) HEK293T cells were transfected with the indicated constructs and histone acetylation was examined. (C) HEK293T cells were transfected with empty vector, WT, or HAT-deleted HA-p300. TopBP1 or tubulin was then examined. (D) HEK293T cells were co-transfected with FLAG-TopBP1 and different HA-tagged acetyltransferases, CBP, p300, PCAF, Tip60, or hMOF. TopBP1 acetylation was detected with anti-AcK825 antibodies. (E) SIRT1 deacetylated TopBP1 in vitro. Acetylated FLAG-tagged TopBP1 was purified from cells contransfected with FLAG-TopBP1 and HA-p300 and incubated with NAD, purified WT SIRT1, or HY mutant SIRT1, or in the absence of the indicated reagents. Acetylated TopBP1 was detected by immunoblot with anti-AcK825. (F–G) Raw data from TopBP1 acetyl-specific mass spectrum analysis. (H–I) Cells were transfected with the indicated constructs. 48 hours later, TopBP1 acetylation was examined. Figure S2, related to Figure 2.
Figure S3. SIRT1 regulates DNA replication through deacetylating TopBP1
(A) Cells as in 3C were stained with PI and cell-cycle profiles were examined by FACS. (B) Sirt1+/+ and Sirt1−/− MEFs were serum starved for 48 hours (0.1% serum) and stimulated to enter cell-cycle progression by serum addition. Cells were collected at the indicated times and chromatin fractions were prepared and immunoblotting was performed with the indicated antibodies. (C) Cells as in 3E were stained with PI and cell-cycle profiles were examined by FACS. Protein expression was analyzed by Western blot (upper panel). (D) Cells were transfected with the indicated constructs and the TopBP1-EDD interaction was examined. (E) Acetylated FLAG-tagged TopBP1 was purified from cells cotransfected with FLAG-TopBP1 and HA-p300 using FLAG beads and then incubated with NAD and purified SIRT1 (WT or HY mutant) in vitro. The TopBP1-Treslin interaction was then examined. (F) Cells transfected with Myc-Treslin and FLAG-TopBP1 were lysed and immunoprecipitated 4 times with Myc-Beads (containing FLAG-TopBP1 interacted with Myc-Treslin). Myc-Treslin depleted cell lysates were immuprecipated with anti-FLAG antibody (containing FLAG-TopBP1 not bound to Myc-Treslin). Acetylation of TopBP1 (AcK825) was examined in both Myc and FLAG immunoprecipitates. (G) Cells stably expressing TopBP1 shRNA were transfected with the indicated constructs. After 48 hours, the TopBP1-Treslin interaction was examined. (H) Cells stably expressing TopBP1 shRNA were transfected with the indicated constructs. After 24 hours, cells were incubated with media containing 1mM glucose for 24 hours, and the TopBP1-Treslin interaction was examined. (I) Cells stably expressing TopBP1 shRNA were transfected with the indicated constructs and were serum starved for 48 hours (0.1% serum) before being induced with media containing 10% serum for 18 hours to enter S phase. Whole cell lysates and chromatin fractions were prepared. Western blotting was performed with the indicated antibodies. Figure S3, related to Figure 3.
Figure S4. TopBP1 acetylation regulated by p300 and SIRT1 plays an important role in glucose limitation dependent metabolic checkpoint
(A) Cells as in 4F were stained with PI and cell-cycle profiles were examined by FACS. Protein expression was analyzed by western blot (lower panel). (B) Cells expressing the indicated constructs were treated with various concentrations of glucose (25mM or 1mM) for 24 hours. Cells were incubated with 20 μM BrdU; 30 min later, cells were fixed and stained with anti-BrdU antibody. The data presented are mean ± SD for three independent experiments. *P<0.05; **P<0.01 (one-way ANOVA test). Protein expression was analyzed by western blot (lower panel). (C) Cells stably expressing TopBP1 shRNA were transfected with the indicated constructs treated with various concentrations of glucose (25mM or 0.1mM) for 48 hours. PARP1 cleavage was then examined by western blot. Figure S4, related to Figure 4.
Figure S5. p300-TopBP1 interaction is regulated by DNA damage
(A) Cells were transfected with the indicated plasmids and were treated with HU (10mM) or left untreated. The p300-TopBP1 interaction was then examined. Figure S5, related to Figure 5.
Figure S6. SIRT1 regulates the DNA damage response through deacetylating TopBP1
(A) Cells stably expressing TopBP1 shRNA were transfected with WT TopBP1 or 3KR mutant. Thirty hours later, cells were mock-treated or treated with HU (10 mM). Cells were harvested at the indicated time points. The soluble and chromatin fractions were prepared and immunoblotted with antibodies as indicated. (B) HCT116 (left panel) or U2OS (right panel) cells stably expressing control shRNA or SIRT1 shRNA were treated with UV (40 J/m2), HU (5 mM) or left untreated. Chk1 phosphorylation was then examined. (C) U2OS cells transfected with plasmids encoding WT TopBP1, myc-SIRT1, or empty vector were treated with 5 mM HU or UV (40 J/m2). TopBP1 focus formation was examined by IF. Quantification of TopBP1 foci was shown in the right panel. The data presented are mean ± SD for three independent experiments. (D) Cells stably expressing control or SIRT1 shRNA were mock-treated or treated with HU (10 mM) for the indicated times. The soluble and chromatin fractions were prepared and immunoblotted with antibodies as indicated. (E) Cells expressing control, SIRT1, or TopBP1 shRNA were transfected with empty vector, WT TopBP1, or the 3KR mutant and, then treated with HU (10mM) or left untreated. Chk1 phosphorylation was then examined. Figure S6, related to Figure 6.
Figure S7. TopBP1 acetylation is important for the DNA damage response
(A) Acetylated FLAG-tagged TopBP1 was purified from cells cotransfected with FLAG-TopBP1 and HA-p300 using FLAG beads and incubated with NAD, purified WT SIRT1 or HY mutant SIRT1 for deacetylation in vitro. The TopBP1-Rad9 interaction was then examined. (B) Cells stably expressing TopBP1 shRNA were transfected with the indicated constructs. After 48 hours, cells were left untreated or treated with HU (10 mM), and the TopBP1-Rad9 interaction was examined. (C) Cells stably expressing TopBP1 shRNA were transfected with the indicated constructs. After 48 hours, the TopBP1-Rad9 interaction was examined. (D) Cells stably expressing TopBP1 shRNA were transfected with the indicated constructs. After 48 hours, cells were treated with HU (10mM) and Chk1 phosphorylation was examined. (E) U2OS cells stably expressing TopBP1 shRNA were stably expressing TopBP1 3KR mutant. Negative control experiments for the Duo-link assay (HA antibody only and FLAG antibody only) were performed. (F) Empty vector or HA-TopBP1 were cotransfected with FLAG-WT, 3KR, or 3KQ mutants in HEK293T cells stably expressing TopBP1 shRNA. Cells were left untreated or treated with HU (10 mM) or low glucose (0.1 mM). Cell lysates were subjected to immunoprecipitation with anti-HA antibodies. The immunoprecipitates were then blotted with the indicated antibodies. Figure S7, related to Figure 6 and 7.