Abstract
The efficacy of radiotherapy in many tumor types is limited by normal tissue toxicity and by intrinsic or acquired radioresistance. Therefore, it is essential to understand the molecular network responsible for regulating radiosensitivity/resistance. Here, an unbiased functional screen identified four microRNAs (miR-1, miR-125a, miR-150, and miR-425) that induce radioresistance. Considering the clinical importance of radiotherapy for glioblastoma (GBM) patients, the impact of these miRNAs on GBM radioresistance was investigated. Overexpression of miR-1, miR-125a, miR-150 and/or miR-425 in GBM promotes radioresistance through upregulation of the cell cycle checkpoint response. Conversely, antagonizing with antagomiRs sensitizes GBM cells to irradiation, suggesting their potential as targets for inhibiting therapeutic resistance. Analysis of GBM datasets from TCGA revealed that these miRNAs are expressed in GBM patient specimens and correlate with Transforming Growth Factor Beta (TGF-β) signaling. Finally, it is demonstrated that expression of miR-1 and miR-125a can be induced by TGF-β and antagonized by a TGF-β receptor inhibitor. Together, these results identify and characterize a new role for miR-425, miR-1, miR-125, and miR-150 in promoting radioresistance in GBMs and provide insight into the therapeutic application of TGF-β inhibitors in radiotherapy. Systematic identification of miRs that cause radioresistance in gliomas is important for uncovering predictive markers for radiotherapy or targets for overcoming radioresistance.
INTRODUCTION
Radiation therapy is one of the most common options for cancer treatment and used for curative or adjuvant means. Systemic therapy is becoming increasingly more effective in controlling the metastatic spread of cancer, and local therapies are important in the management of disease. Local therapies typically consist of a combination of surgical resection and radiation therapy (with or without concurrent chemotherapy as a radiosensitizer). However, the efficacy of radiotherapy is limited by adjacent normal tissue toxicity and also by radioresistance, a term used in cancer biology for tumors which do not respond well to radiotherapy, i.e. tolerate doses of radiation that typically induce regression of tumors of the same lineage or across different cancers. In some cancers radiation can serve as the primary modality for local control. This includes prostate cancer, where radiation therapy can be used for curative treatment (1), as well as primary brain tumors, such as glioblastoma (GBM)(2), where many patients are only able to undergo a biopsy or subtotal resection due to the anatomical location and highly invasive nature of the tumor. Unfortunately GBMs are notoriously resistant to radiotherapy, and molecular details of radioresistance remain largely unknown.
MicroRNAs (miRNAs) are abundant small (~20–22 nts) non-coding RNAs that typically dampen gene expression at the post-transcriptional level (3, 4) and are mis-expressed in a variety of cancer cells (5),(6). A number of recent studies have shown an effect of radiation on miRNA expression patterns in an assortment of tumor lines (7–12). These changes in miRNA expression vary considerably, depending on the dose of radiation, time after exposure, cell lineage and profiling methods. High doses of radiation and later measurements (>6 hours after exposure) are very likely to have secondary effects on miRNA expression that may have limited physiological relevance. Moreover it's not clear whether any of these miRNAs influence radioresistance. To directly investigate the role of miRNAs in radioresistance we adopted a functional approach to identify miRNAs that induce radioresistance. We screened a miRNA expression library to identify four miRNAs (miR-125a, miR-150, miR-1 and miR-425) that induce radioresistance. Considering the clinical relevance of radioresistance in GBMs we characterized the impact of these miRNAs in multiple GBM lines. We observed that these miRNAs induce radioresistance in GBMs by altering the DNA damage induced cell cycle checkpoint response, and by preventing apoptosis. Investigating factors/pathways that regulate the expression of these miRNAs we observe a statistically significant correlation of these miRNAs with the transformation growth factor (TGF)-β pathway in GBMs, and manipulating TGF-β signaling influences their expression. Thus, we have defined a novel function for a set of four miRs in therapeutic resistance and determined a new mechanism regulating their expression.
MATERIALS AND METHODS
MiRNA Radioresistance Screen
The retroviral miRNA expression library was cloned into the pSCMV vector as previously described (13). Briefly, library cloning was performed by PCR-amplifying human miRNA precursors and ~150bp flanking regions on 5' and 3' sides from genomic DNA. The library of miRNA expression vectors was pooled and used to generate pooled viral mixture for infection. Two independent viral pools were generated to introduce biological noise. Each viral pool was used twice to independently infect U2OS cells, resulting in a total of four independently infected cell pools (with two independent infected cell pools for each virus preparation). The amount of virus was titrated to achieve ~20–30% infection rate. Each infected U2OS cell population was selected with puromycin and subjected to duplicate and parallel functional screens, resulting in a total of six replicates for the screen. For each replicate, genomic DNA was harvested at an initial time point after puromycin selection (untreated), and after irradiation (2 –3 weeks) from surviving clones. Genomic DNA samples were PCR-amplified using primers present in the vector but not in genomic sequence, and amplified fragments cloned into pGEM-T Easy vector and sequenced.
Glioma Cell Lines
The cell lines LN15, LN18, LN229, U138, U87, and U251 were obtained from the ATCC cell repository. U343 and NHA cells were obtained from the Lonza cell repository. A1207 was originally established by Dr. S Aaronson, National Cancer Institute, NIH, Bethesda, MD. LN444, LN340 and LN215 cells were originally established by Dr. Monika E Hegi and Dr. Nicolas de Tribolet at the University of Lausanne. These cell lines were obtained collaboratively by Dr. Clark Chen (co-author in this study).
Clonogenic Assay
Indicated cell lines (0.2–0.5 × 103 cells/well) were seeded on 6 well plate one day before transfection with 10 nM of miRNA mimics or antagomirs (Applied Biosystems). Two days later, cells were detached and 1000 cells in 4 ml suitable media (10% FBS v/v) were seeded on 6 cm dishes in four replicates and allowed to attach overnight before γ-IR treatment. Control and irradiated were incubate for 14 days to form colonies. For evaluation, formed colonies were stained with crystal violet and surviving colonies (>50 cells) were counted. Plating efficiency accounted 30% to 50%. Cells stably overexpressing miRNAs were detached and processed as above.
RNA Isolation and Quantitative PCR
Total RNA was extracted with Trizol (Invitrogen) as recommended in the manufacturer's manual. Briefly, 0.5–1 × 106 cells were lyzed 1 ml Trizol. Extracted RNA was treated with 10 U DNase I for 30 min at 37°C and subsequently purified with phenolcholoroform (Ambion). Total RNA (1 μg) was reversed transcribed with MLVV (Epicentre). MiRNAs and mRNA detection was carried out with TaqMan miRNA Assay Kits (Applied Biosystems) and SYBR Green master mix (Applied Biosystems), respectively. The expression of miRNAs and mRNA was normalized to the U1 RNA. PAI F: CTCTCTCTGCCCTCACCAAC R: GTGGAGAGGCTCTTGGTCTG; U1 F: CCATGATCACGAAGGTGGTT R: ATCCGGAGTGCAATGGATAA.
Apoptosis Assay
LN229 cells stably overexpressing miR-1, miR-125a, miR-150 and miR-425 or control miR-scr were seeded at low density (0.2 × 106/plate) on 6 cm plates and next day exposed to 0, 1, 5 and 10 Gy of γ-IR. At day 0 through 4 cells were collected and analyzed for activation of Caspase 3 and 7 with Caspase-Glo 3/7 Assay System (Promega) and with cleaved anti-Capase 3 antibodies (Cell Signaling). For Caspase-Glo assay cells were lyzed in 0.2 ml lysis buffer and 10 μl were pipetted onto 96 well plate in 3 replicates. Subsequently to each well 100 μl of Caspase-Glo reagent was added and luminescence measured overtime (details see manufacturer's protocol). For Western blot analysis cells were lyzed initial in 0.1 ml lysis buffer and relative protein concentration determined by NanoDrop 2000. Samples were diluted to equal concentration and 20 μl loaded on 12% gels (details see Western blots).
Immunoblots
Cells were collected in 100 to 200 μl of lysis buffer. Relative protein concentration was determined with NanoDrop and samples adjusted to the same concentration with the lysis buffer. Between 20 to 40 μl of cell lysate supplemented with 5 to 10 μl of 5x concentrated reducing loading buffer were loaded on 8 to 12 % gels (SDS page). Proteins were transferred to nitrocellulose membrane using semi-dry apparatus. Membranes were blotted with 5 % milk in PBS buffer. Primer antibodies were diluted 500 to 2000 times in PBS buffer plus 0.1 % Tween. Fluorescent secondary antibodies were applied for detection. The following antibodies were used for immunoblots: Caspase-3 (Cell Signaling, #9665), cleaved caspase-3 (Cell Signaling, #9661), p-Chk2 T68 (Cell Signaling, #2661), p-Chk2 S33/35 (Cell Signaling, #2665), Chk2 (Cell Signaling, #2662), p-Chk1 S345 (Cell Signaling, #2341), p-Chk1 S317 (Bethyl, #A300-163A), Chk1 (Cell Signaling, #2345), p-RPA2 S33 (Bethyl, #A300-246A), RPA2 (Cell Signaling, #2208), p-H2AX (Cell Signaling, #9718), H2AX (Cell Signaling, #2595), alpha-tubulin (Sigma, #T6074-200UL).
Cloning of miRNA constructs
MiR-1, miR-125a, miR-150 and miR-425 or scrambled control miR-scr were cloned either individually or in tandem into pcDNA3.1(-)Neo vector. mCherry fluorescent protein was cloned with NheI upstream of the miRNA tandem. All cloning primers were ordered from IDT. miR-1 F: CGCGGATCCGCAAAAAGAATCAAACCAGGAC R: CCGGAATTC TCACTGGATCTTCTTTTCCTTCA; miR-125a F: CGGAATTCTGCTGTGTCTCTGTGGCTTC R:GC TCTAGAAGGCCTCCTCACACTCTTCA; miR-150 F: CGCGGATCCGAATTCGAGGAGGGGTTTTGCAGAG; miR-425 F: CGCGGATCCTCAGGCTGGAGGTGATGG R: CCGGAATTCATAGGCCTCAGTTGGAGTATCTTG
Cloning of miR-Sponges
MiRNA binding sites (RBS) were designed as published in Ebert et al (16) und Kluiver et al (17). For each miRNA 4 corresponding RBS in tandem spaced by short sequence were ordered as oligonucleotids from IDT. These were aligned to form duplexes and directly cloned into pcDNA3.1(-)Neo vector downstream of mCherry or GFP.
MiR-1-Sponge:
F:CTAGATACATACTTTCCACATTCCAGTTTATACATACTTTCCACATTCCAGTTGATACATACTTTCCACATTCCAGTTTATACATACTTTCCACATTCCA
R:AATTTGGAATGTGGAAAGTATGTATAAACTGGAATGTGGAAAGTATGTATCAACTGGAATGTGGAAAGTATGTATAAACTGGAATGTGGAAAGTATGTAT;
MiR-125a-Sponge:
F:CTAGTCACAGGTTAAAATTCTCAGGGAGTTGGCTCCCAAGGCATCACCTGTGTGTCACAGGTTAAAATTCTCAGGGA GT GGCTCCCAAG GCA TCACCTGT
R:AATTACAGGTGATGCCTTGGGAGCCACTCCCTGAGAATTTTAACCTGTGACACACAGGTGATGCCTTGGGAGCCAACTCCCTGAGAATTTTAACCTGTGA
MiR-150-Sponge:
F:CTAG CACTGGTACA CTT TTGGGAGA GTTT CACTGGTACA CTT TTGGGAGA GTTG CACTGGTACA CTT TTGGGAGA GTTT CACTGGTACA CTT TTGGGAGA
R:AATTTCTCCCAAAAGTGTACCAGTGAAACTCTCCCAAAAGTGTACCAGTGCAACTCTCCCAAAAGTGTACCAGTGAAACTCTCCCAAAAGTGTACCAGTG
MiR-425-Sponge:
F:CTAG TCAACGGGAGT TCG GTGTCATT GTT TCAACGGGAGT TCG GTGTCATT GTG TCAACGGGAGT TCG GTGTCATT GT TCAACGGGAGT TCG GTGTCATT
R:AATTAATGACACCGAACTCCCGTTGAACAATGACACCGAACTCCCGTTGACACAATGACACCGAACTCCCGTTGAAACAATGACACCGAACTCCCGTTGA
Transfection of Cells and Stable Cell Lines
LN229, U251 and U2OS cells were seeded at 70–90 % confluence on 6 well plate. Next day the cells were transfected with 10 nM mimics or antagomirs or 1 μg plasmid DNA using Lipofectamine reagent (Invitrogen). Stable cells were selected with G418 for 1 week and single colonies were picked. Expression of miRNAs was confirmed with qPCR.
Genomic and Ingenuity Pathway Analysis
Affymetrix level 1 mRNA and Agilent level 2 microRNA data were downloaded from the public TCGA data portal (http://cancergenome.nih.gov/). Affymetrix CEL file analysis was performed in R project, a free statistical computing platform (http://www.r-project.org/) using the Bioconductor platform (http://www.bioconductor.org/). The Robust Multi-Array (RMA) algorithm was used for normalization(14). Significance and correlations of were computed using the GenePattern tool Comparative Marker Selection (CMS) (Broad Institute, MIT, Cambridge, MA). CMS is a statistical method that uses permutation testing to identify differentially regulated genomic events in one versus another predefined patient group. For miRNA target prediction we used the miRWalk(15) public database. The common gene targets of miR-1, miR-125a, miR-150, and miR-425 were analyzed using Ingenuity Pathway Analysis (IPA) (www.ingenuity.com). To interrogate glioblastoma-specific miR-1 and miR-125a dependent radioresistance pathways, we analyzed the negatively correlated TCGA target genes of the latter miRNAs by IPA.
TGF-β signaling in glioma lines
The indicated lines were treated for 12 hours with TGF-β (R&D Systems) at a concentration of 100 pM and TGF-β inhibitor (LY2157299, Selleck Chemicals) at a concentration of 10μM. The expression of each miRNA was determined by TaqMan miRNA assay. As a control for TGF-β signaling we monitored expression of PAI-1 using the following primers,
qPAI-1-fw CTCTCTCTGCCCTCACCAAC
qPAI-1-rev GTGGAGAGGCTCTTGGTCTG
Statistical analysis
We used the student's t-test for pairwise comparisons of continuous data and analysis of variance for multi-group comparisons. Significance of differences in radiosensitivity was tested separately at all four doses of radiation (0, 1, 3 and 5 Gy). When significance was shown in ANOVA we performed between-group comparisons using Tukey's post-hoc tests to determine overexpression of which miRNAs provided significant advantage over any other miRNA or the scrambled miRNA control group. Additionally, in order to determine the global impact of miRNA overexpression on cell survival we have also calculated areas under the curves (AUCs) using trapezoid approximation. This provided estimates for the overall impact of miRNA on dose-dependent cell survival. ANOVA with Tukey's post-hoc tests were used to determine significance of those comparisons. P values <0.05 were considered as statistically significant.
RESULTS
Performance of a Functional Screen for miRNAs Promoting Radioresistance
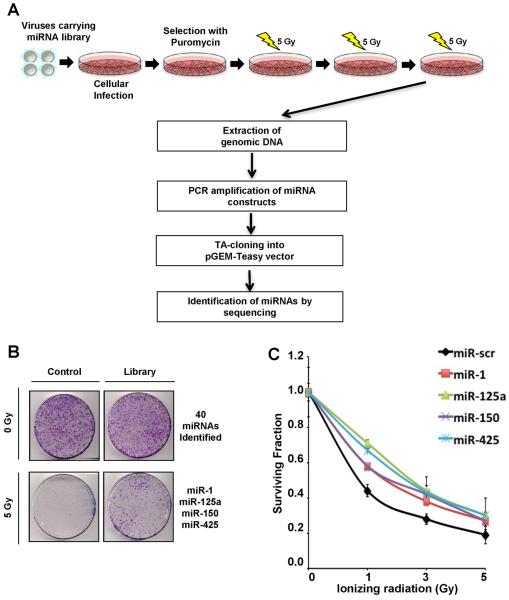
To identify miRNAs that induce radioresistance we utilized a retroviral miRNA expression library, where specific miRNA-encoding and flanking regions from 273 miRNAs were expressed (see SI Methods)(13). p53-proficient U2OS cells were transduced with the miRNA expression library to generate clonal populations over-expressing a single miRNA. To maintain clinical relevance, we treated cells with fractionated doses of radiation (5 Gy over 5 days). After multiple rounds of collecting radioresistant clones, and sequencing the over-expressed miRNA, we obtained a list of 40 miRNAs that confer a growth advantage independent of radiation (Fig. 1B and Supplemental Figure 1A). Four miRNAs (miR-1; miR-125a; miR-150; miR-425) were selectively enriched (>4 fold) in cells exposed to radiation. Importantly, these miRNAs did not confer any selective growth advantage independent of radiation indicating that the phenotype we observed was due to radioresistance. Interestingly, miR-125a has been reported to target p53, which is consistent with the radioresistant phenotype induced by its over-expression(16). We validated these results by transfecting the same cells in which the screen was performed with individual miRNA mimics (chemically modified double-stranded RNA molecules designed to mimic endogenous mature miRNAs) and assessing radiosensitivity using the clonogenic survival assays (Fig. 1C).
Figure 1. miRNA screen for radioresistance.
(A) Schematic of radioresistant screen. U2OS cells were infected with retroviruses carrying the miRNA library(13). Cells expressing miRNAs were selected with puromycin and subjected to the fractionated doses of 5 Gy γ-irradiation (IR) over 5 days. After a round of IR exposure, 30 best-growing colonies were selected and pooled together before undergoing continuous rounds of IR. After 3 rounds of collecting radioresistant clones, genomic DNA was purified and amplified to clone into pGEM-T Easy vector and to identify the over-expressing miRNAs by sequencing. (B) Representative image of clonogenic survival assay. U2OS cells carrying control or the miRNA library were stained for colony quantification after 3 rounds of the fractionated doses of IR. Forty miRNAs that exhibited a growth advantage independent of radiation were excluded from selection and 4 miRNAs, miR-1, miR-125a, miR-150 and miR-425 that were selectively enriched in irradiated cells were chosen for further studies. (C) Clonogenic survival assay to validate the impact of selected miRNAs on radioresistance. U2OS cells were transfected with indicated miRNA mimics and subjected to indicated doses of IR before colony quantification. Curves were generated with Mean ± S.D. of 3 independent experiments. Significant differences were confirmed between the negative controls and the four experimental lines at all doses of radiation and in the area under the curve (AUC) comparison (ANOVA (p < 0.05) and Tukey's post hoc (p < 0.05) test).
Identification of Radioresistant and Radiosensitive GBM Cell Lines with Varying miRNA Levels
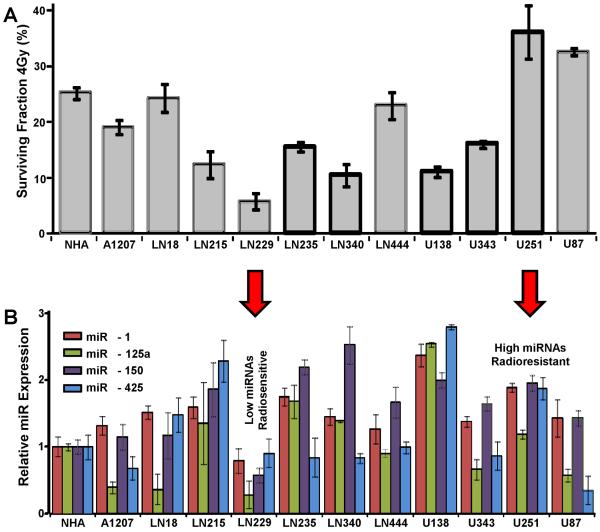
GBM is the most lethal primary brain tumor with a median survival of 9 to 12 months and the dismal prognosis of GBM patients is largely caused by the striking radioresistance of these tumors (17). We therefore sought to determine the impact of these miRNAs on GBM radiosensitivity. To achieve this goal, we first evaluated the radiosensitivity of a panel of 11 glioma lines in comparison to a normal human astrocyte control. Radiosensitivity was assessed at 2 different doses of irradiation (Fig. 2A and Supplementary Fig. 1B). In parallel, we assessed the expression of miR-1; miR-125a; miR-150 and miR-425 in these cell lines using Taqman-based RT-PCR (Fig. 2B). Following that, we ranked the cell lines by overall expression of the four studied miRNAs and cross-matched that with radiosensitivity measured by survival. In order to conduct functional studies we selected the two cell lines with the lowest (U251) and the highest radiosensitivity (LN229). In both cases, the average miRNA level corresponded to survivability – LN229 showed the lowest overall expression of the four studied miRNAs, whereas U251 showed the second highest expression levels (after U138). For both cell lines, survivability and miRNA expression differed significantly from those observed in the NHA (p<0.05). These results provide a comprehensive assessment of radioresistance and miRNA expression in GBM cell lines and allowed us to identify appropriate cells for gain or loss of function studies.
Figure 2. Radioresistance and miRNA levels in GBM cell lines.
Clonogenic assay and miRNA expression profile in a panel of GBM cell lines. (A) Clonogenic survival assay was performed with 4 Gy IR in 12 different GBM lines. (B) In the same cell lines, miRNA expression was determined by qRT-PCR using TaqMan miRNA assay. Bar graphs were generated with Mean ± S.D. of 3 independent experiments.
Elevation of miR-1, miR-125a, miR-150, and/or miR-425 promote radioresistance
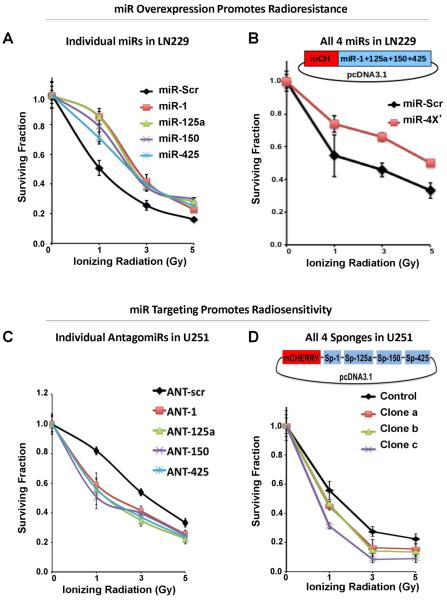
To investigate the impact of miR-1; miR-125a; miR-150 and miR-425 on radioresistance in GBM, we increased the expression of miRNAs in the relatively radiosensitive LN229 cells. Transient overexpression of miR-1; miR-125a; miR-150 and miR-425 in LN229 cells caused a significant increase in survival in clonogenic assays (Supplemental Fig. 2). Since our screen (Fig. 1) was conducted with stable expression of miRNAs, we also stably expressed miR-1, miR-125a, miR-150, or miR-425, individually (Fig. 3A) and found that these miRNAs were associated with significantly higher (ANOVA p<0.05, followed by Tukey's post-hoc test p<0.05) radioresistance relative to the scrambled miRNA control and did not show significant (Tukey's post-hoc tests p levels > 0.05) differences between one another. This was confirmed at all three doses of radiation and in AUC comparison. Furthermore, stable expression of miR-1, miR-125a, miR-150, and miR-425, in tandem using a chimeric construct (Fig. 3B) also significantly promoted radioresistance (t-tests at all 3 doses and in AUC comparison, p<0.05). These data strongly support the notion that miR-1, miR-125a, miR-150, and miR-425 can work in isolation or together to promote radioresistance in GBM.
Figure 3. MiR-1, miR-125a, miR-150, and miR-425 regulate radioresistance.
Validation of miRNA impact on radioresistance in GBM. In the radiosensitive GBM cell line LN229, transient transfection of either miRNA mimics alone (A) or stable incorporation of miRNAs in combination (B) increases the survival of cells exposed to irradiation in the clonogenic assay. The construct of 4x miR carrying vector is illustrated in B. In the radioresistant GBM cell line U251, transient transfection of either antagomiRs alone (C) or stable inhibition of all the miRs via sponge (D) decreases the survival of cells exposed to irradiation in the clonogenic assay. The construct of sponge carrying vector is illustrated in D. In all panels curves were generated with mean ± S.D. of 3 independent experiments. Significant differences were confirmed between the negative controls and the remaining groups at all doses of radiation and in the AUC comparison (ANOVA (p < 0.05) and Tukey's post hoc (p < 0.05) test).
Targeting miR-1, miR-125a, miR-150, and miR-425 Increases Radiosensitivity
In converse experiments, we sought to determine whether miR-1, miR-125a, miR-150, or miR-425 could be targeted to increase sensitivity to irradiation. Using the relatively radioresistant cell line U251, we transiently transfected antisense oligonucleotides (termed antagomirs, ANT) to miR-1, miR-125a, miR-150, or miR-425 (Fig. 3C). We found that expression of each of the antagomirs sensitized cells to irradiation by significantly reducing survival at all three doses and in AUC comparison (Fig. 3C), as in the previous experiment, survivability did not differ between the cells transfected with different ANTs. Although these results are encouraging, it is important to determine from the therapeutic perspective whether a more `stable' inhibition of these miRNAs produces the same effect on radioresistance of GBM. Therefore the miRNAs were next inhibited using a sponge based method that has been previously described (18, 19). A combination-sponge was constructed to impede the expression of all four miRNAs (19), and we ascertained using a reporter system that the sponges were indeed inhibiting the miRNAs (Supplementary Fig. 3). Stable expression of sponges for each of the miRNAs together also increased the sensitivity of these cells to irradiation in the colony formation assay (Fig 3D). Together these data strongly suggest that miR-1, miR-125a, miR-150, and miR-425 may be important targets for prevention of therapeutic resistance.
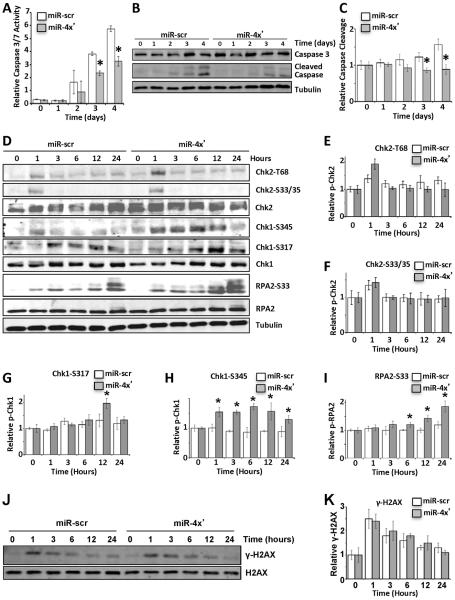
miR-1, miR-125a, miR-150, and miR-425 Decrease Radiation Induced Apoptosis in Association with Increased Checkpoint Activation
The radioresistance induced by the miRNAs may be due to accelerated cell growth or diminished apoptosis. To investigate the impact of these miRNAs on apoptosis we assessed caspase 3/7 enzymatic activity after IR in LN229 cells stably expressing miR-1, miR-125a, miR-150, and miR-425. Caspase activity was determined using a luciferase-based assay (Fig. 4A) and also monitored via the cleavage of caspase 3 in immunoblots (Fig. 4B, 4C). Both approaches clearly indicate that radiation-induced apoptosis is diminished in cells expressing miR-1, miR-125a, miR-150, and miR-425 individually (Supplemental Fig. 4A) or in combination (Fig. 4A–C). As aberrant checkpoint responses can correlate with radioresistance (20–23), we next determined the impact of the miRNAs on checkpoint activation. We evaluated the phosphorylation of the checkpoint proteins, Chk2 and Chk1 at different time points after irradiation (Fig. 4D–H). Although there is moderate increase in phosphorylation of Thr68 on Chk2 (Fig. 4D, 4E) in cells expressing the miRNAs this is not statistically significant. Phosphorylation of Ser33/35 on Chk2 (Fig. 4D, 4F) is comparable to control cells. The overexpression of miR-1, miR-125a, miR-150, and miR-425 also has a strong impact on Chk1 phosphorylation (Fig. 4D, 4G–H). Phosphorylation of Chk1 at Ser317 was significantly increased at 12 hrs post IR (Fig. 4D, 4G), but phosphorylation of Chk1 at Ser345 was significantly increased at all time points in cells over expressing these miRNAs (p<0.05; Fig. 4D, 4H). Ser345 is a consensus site (SQ/TQ motif) for the PI3-like kinases and is typically phosphorylated by ATR in response to DNA damage (24, 25). RPA2, another ATR substrate involved in the intra-S and G/M checkpoint is also significantly hyper-phosphorylated at Ser33 in cells overexpressing miR-1, miR-125a, miR-150, and miR-425 (Fig. 4D, 4I). Enhanced phosphorylation of Chk1 at Ser345 and RPA2 at Ser33 in the presence of the miRNAs may be caused by impaired DNA repair and persistence of double-strand DNA break (DSB). However, cell lysates probed for γ-H2AX (markers for DSBs) by immunoblot show that irradiation-induced formation and dissipation of γ-H2AX was comparable in control cells and cells expressing the miRNAs (Fig. 4J, 4K). The underlying cause of the impact of the miRNAs on Chk1 at S345 at all time points post IR over S317 only at 12 hrs, or lack of impact of the miRNAs on Chk2 phosphorylation remains unclear. However it is noteworthy that phosphorylation of Ser345 has been previously correlated with radioresistance in gliomas (22, 23). From the mechanistic standpoint, Ser345 is critical for Chk1 activity and is necessary for implementing the intra-S and G2/M checkpoint (26). Considering the importance of Ser345 phosphorylation in radioresistance and the combinatorial impact of the miRNAs on Ser345 phosphorylation we also confirmed that individual expression of miR-1, miR-125a, miR-150, and miR-425 significantly increases Ser345 phosphorylation (Supplemental Fig. 4B). Together, these results indicate that expression of miR-1, miR-125a, miR-150, and miR-425 enhances cellular checkpoint response after IR and does not impair DSB repair.
Figure 4. Impact of miRNAs on DNA damage reponse.
LN229 cells stably expressing scramble (scr) or miR-1, miR125a, miR-150 and miR-425 (miR-4X') were exposed to 10 Gy IR. (A–C) Impact of miRNAs on apoptosis. Induction of caspase3/7 enzymatic activities was measured using the Caspase Glo kit at indicated days after IR (A). Cleavage of caspase 3 was monitored by immunoblotting (B), which was quantified by Image J (C). (D–I) Impact of miRNAs on checkpoint response. Levels of total and phosphorylated Chk2, Chk1 and RPA2 were detected at indicated time points after IR via immunobot with tubulin as a loading control (D). Quantification with normalization of phospho-form to total protein was graphically demonstrated for Chk2 pT68 (E), Chk2 S33/35 (F), Chk1 S317 (G), Chk1 S345 (H), and RPA2 S33 (I). (J–K) The kinetics of H2AX phosphorylation was demonstrated at indicated time points after IR via immunoblot (J) and phospho-form relative to total H2AX was quantified (K). In all panel but B, D and J the mean ± S.D. of 3 independent experiments is demonstrated. Comparison was done with one-way ANOVA (p < 0.05) and * represents p < 0.05 calculated with Tukey's post hoc test.
miRNAs Expression in Human GBMs and Regulation by Transforming Growth Factor Beta
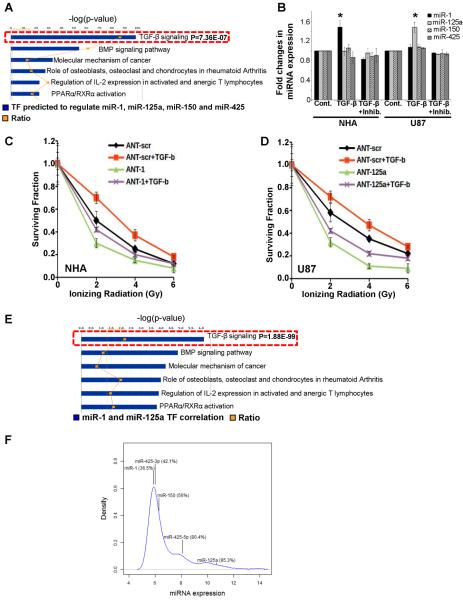
A key question is how expression of these miRNAs is regulated in GBM. To address this issue we used two algorithms, MotifScanner and CisGenome, to search for binding sites of all listed transcription factors in TRANSFAC in the promoter regions of the 4 miRNAs (Supplementary Table 1). We focused on common factors and these were integrated into known signaling pathways using Ingenuity Pathway Analysis (IPA). Based on this prediction based-pathway analysis which is not restricted to any cell lineage, the transformation growth factor (TGF)-β signaling pathway is significantly correlated with expression of miR-1, miR-125a, miR-150, and miR-425 (Fig. 5A). Interestingly, TGF-β signaling has been correlated with radioresistance in gliomas and TGF-β inhibitors are being considered as radio-sensitizing agents (27, 28). However, the tremendous heterogeneity of GBM complicates this issue, as it is clear that TGF-β signaling is differentially active in GBM. We investigated TGF-β signaling in a panel of GBM lines using the downstream target gene PAI-1 as a marker for TGF-β response (Supplementary Fig. 5). Similar to previously published studies, we identified multiple GBM lines which responded to TGF-β and a receptor antagonist. The impact of TGF-β signaling on the expression of miR-1, miR-125a, miR-150, and miR-425 was evaluated in these cell lines. In U87, there is a statistically significant increase of miR-125a that was induced by TGF-β and abrogated by the addition of a TGF-β signaling inhibitor. In NHA we obtained similar results for miR-1 (Fig. 5B). There was no impact of TGF-β on miR-150 and miR-425 in NHA or U87 cells (Fig. 5B). The other two GBM lines, U251 and U138, which had active TGF-β signaling, showed no significant impact on the miRNAs indicating other pathways are likely important for regulating miRNA expression in these cells (data not shown). Based on our previous data we hypothesized that TGF–β induced radioresistance in these cell lines maybe mediated by miR-125a and miR-1. Therefore antagonizing these miRNAs should `rescue' the radioresistance induced by TGF-β. Consistent with our hypothesis antagonizing miR-1 in NHA (Fig. 4C) and miR-125a in U87 (Fig. 4D) sensitizes these cell lines to radiation, and neutralizes any radioresistance caused by TGF-β signaling. It is noteworthy that inhibition of these miRNAs prior to exogenous induction of TGF-β signaling sensitizes NHA and U87 cells to radiation, and this effect is further manifested as TGF-β causes radioresistance. To further interrogate the physiological relevance of the link between TGF-β signaling and miR-1/miR-125a expression, we analyzed the Cancer Genome Atlas (TCGA) database. There is a statistically significant correlation of TGF-β signaling and expression of miR-1 and miR-125a in primary GBMs (Fig. 5E). Together, these results indicate that the TGF-β signaling pathway may be involved in regulating the expression of miR-1 and miR-125a and suggest the involvement of these miRNAs in TGF-β mediated radioresistance. Finally, the most important issue is whether these miRNAs are expressed in primary GBMs. Based on analysis of TCGA database, miR-125a (~95 percentile), miR-425-5p (~80 percentile)/miR-425-3p (~42 percentile) and miR-150 (–56 percentile) are expressed in majority of human GBMs, and miR-1 (34 percentile) is also expressed in a significant proportion of primary GBMs. These data strongly suggest that the miRNAs we have identified have the potential to be important for human disease (Fig. 5F).
Figure 5. Identification of the regulatory pathways of miRNAs.
(A) Ingenuity pathway (Gene Ontology) analysis was applied to identify the signaling pathways most significantly represented by the transcriptional factors most strongly predicted to be regulating miRNA-1, miRNA-125a, miRNA 150, and miRNA-425. The analyzed transcriptional factors were predicted to bind within 5000 base pairs upstream of each mature miRNA. The top predicted pathways based-on their correlation to miRNA expression are shown. (B) NHA and U87 cells were treated with TGF-β (100 pM) with or without TGF-β inhibitor (LY2157299, 10μM) for 12 hours. The expression of each miRNA was determined by TaqMan miRNA assay. The mean ± S.D. of 3 independent experiments is shown and comparison was one with one-way ANOVA (p < 0.05) whereas * indicates p<0.05 calculated with Tukey's post hoc test. (C, D) represents survival assay in NHA and U87 cell lines, respectively in response to TGF-β. Curves were generated with mean ± S.D. of 3 independent experiments. Comparison was done with one-way ANOVA (p < 0.05) and Tukey's post hoc (p < 0.05) test. (E) Ingenuity pathway analysis was applied to identify the signaling pathways that regulate miRNA-1 and miRNA-125a expression. (F) The mean expression distribution of 534 miRNAs across 475 GBM samples in the TCGA database is shown. The numbers in parentheses indicate the percentile rank of the corresponding miRNA.
DISCUSSION
The cellular response to radiation involves a signaling cascade that induces the cross-talk of multiple DNA repair pathways (DSB repair, nucleotide excision repair etc), the cell cycle checkpoint response and the activation of programmed cell death. As anticipated miRNAs down-regulating DNA repair factors also influence radiosensitivity, that is, their over-expression sensitizes cells to radiation [reviewed in (7) (29)]. MiRNAs can also cause radiosensitivity by suppressing other pathways. For example, miR-125b, shares the seed-sequence with miR-125a and can be expected to induce radioresistance, but it in oral squamous cell carcinoma (OSCC) cells it down-regulates intercellular adhesion molecule-2 (ICAM2) and causes radiosensitivity (30). However, there are only a limited number of studies where miRNAs induce radioresistance. The miR-221/miR-222 confers radioresistance in glioblastoma (31) and gastric carcinoma (32); miR-95 (33), miR-210 (34) and the miR-17-92 (35) cluster have been reported to induce radioresistance in individual tumor lines. However, a systematic study of miRNA-induced radioresistance has not been conducted.
A major obstacle to the miRNA research field has been that the bioinformatic approach to predict target transcripts is hampered by the fact that the existing algorithms have a high margin of error. For many miRNAs, current algorithms predict hundreds or even thousands of potential targets, making it difficult to identify the most important targets. This is more pertinent because we are studying a cluster of miRNAs, and the cumulative list of predicted targets of the four miRNAs is even longer. We used RNA22 to predict targets of miR-1, miR-125a-5p/miR-125a-3p, miR-150, and miR-425, and have compiled a list of potential targets involved in the radiation response. RNA22 is distinct from other methods in that it obviates the use of (i) canonical `seed' regions in the MRE and (ii) cross-species sequence conservation filter (36). This is noteworthy because critical DSB repair factors (such as MDC1, 53BP1, DNA-PK, BRCA1 and BRCA2) are not found in lower eukaryotes, potentially making algorithms using evolutionary conservation as a key criterion less effective in identifying DSB repair targets. Since miRNAs typically suppress gene expression the two questions that emerge are: 1) Depletion of which genes cause radioresistance? 2) Are these gene transcripts targeted by miR-1, miR-125a, miR-150, and miR-425? A recent study utilized a genome-wide RNAi screen (37) to identify genes that influence radiosensitivity, and reported a list of genes whose loss correlates with radioresistance. We compared this list to the predicted targets of miR-1, miR-125a, miR-150 and miR-425 and a significant number of these factors impact pathways (DNA repair, cell cycle, apoptosis) that have been implicated in radioresistance (Supplemental Table 2). Although these results are preliminary, they do provide some support for our experimental data indicating a role for miR-1, miR-125a, miR-150, and miR-425 in the regulation of radiosensitivity.
TGF-β is a major mediator of cellular and tissue responses to radiation, and inhibitors of the TGFβ pathway are being explored as therapeutic agents that enhance the radiosensitivity of tumors while protecting normal tissue (28). Our results suggest that the radiosensitivity induced by inhibiting TGF-β is partially mediated by the diminished expression of miRNAs (such as miR-125a and miR-1). Increasingly effective pharmacological means to modulate miRNA activities are emerging with studies reporting efficient manipulation of miRNAs in vivo in non-human primates (38),(39). Specifically, a miR-122 antagonist, SPC3649, was administered to hepatocytes to block replication of the hepatitis C (HCV). No adverse effects associated with SPC3649 have been reported. SPC3649 is currently being evaluated in Phase 2 clinical trials and may be the first miRNA-based therapeutic that is brought to market. Therefore in the coming years it is feasible miRNAs that induce radioresistance, such as, miR-1, miR-125a, miR-150, and miR-425 may become key therapeutic targets for enhancing the efficacy of radiotherapy.
Supplementary Material
ACKNOWLEDGEMENTS
DC is supported by R01CA142698 (NCI), R01 AI101897-01 (NIAID), Basic Scholar Grant (American Cancer Society), Leukemia and Lymphoma Scholar Award, Ann-Fuller Foundation and Mary Kay Foundation. ABH is supported by R01CA151522. WF is supported by grant number 2012/05/E/NZ5/02130 of the National Science Center (Poland).
Footnotes
Conflict of Disclosure: The authors have no conflicts of interest to disclose.
REFERENCES
- 1.D'Amico AV, Whittington R, Malkowicz SB, Schultz D, Blank K, Broderick GA, et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. Jama. 1998;280(11):969–74. doi: 10.1001/jama.280.11.969. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–66. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–79. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 5.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–79. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 6.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10(6):389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chowdhury D, Choi YE, Brault ME. Charity begins at home: non-coding RNA functions in DNA repair. Nature reviews Molecular cell biology. 2013;14(3):181–9. doi: 10.1038/nrm3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishii H, Saito T. Radiation-induced response of micro RNA expression in murine embryonic stem cells. Med Chem. 2006;2(6):555–63. doi: 10.2174/1573406410602060555. [DOI] [PubMed] [Google Scholar]
- 9.Cao F, Li X, Hiew S, Brady H, Liu Y, Dou Y. Dicer independent small RNAs associate with telomeric heterochromatin. Rna. 2009;15(7):1274–81. doi: 10.1261/rna.1423309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simone NL, Soule BP, Ly D, Saleh AD, Savage JE, Degraff W, et al. Ionizing radiation-induced oxidative stress alters miRNA expression. PLoS One. 2009;4(7):e6377. doi: 10.1371/journal.pone.0006377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Josson S, Sung SY, Lao K, Chung LW, Johnstone PA. Radiation modulation of microRNA in prostate cancer cell lines. Prostate. 2008;68(15):1599–606. doi: 10.1002/pros.20827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weidhaas JB, Babar I, Nallur SM, Trang P, Roush S, Boehm M, et al. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer research. 2007;67(23):11111–6. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo S, Bai H, Megyola CM, Halene S, Krause DS, Scadden DT, et al. Complex oncogene dependence in microRNA-125a-induced myeloproliferative neoplasms. Proc Natl Acad Sci U S A. 2012;109(41):16636–41. doi: 10.1073/pnas.1213196109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 15.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk - Database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011 doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, et al. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23(7):862–76. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noda SE, El-Jawahri A, Patel D, Lautenschlaeger T, Siedow M, Chakravarti A. Molecular advances of brain tumors in radiation oncology. Semin Radiat Oncol. 2009;19(3):171–8. doi: 10.1016/j.semradonc.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4(9):721–6. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kluiver J, Gibcus JH, Hettinga C, Adema A, Richter MK, Halsema N, et al. Rapid generation of microRNA sponges for microRNA inhibition. PLoS One. 2012;7(1):e29275. doi: 10.1371/journal.pone.0029275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim YC, Roberts TL, Day BW, Harding A, Kozlov S, Kijas AW, et al. A role for homologous recombination and abnormal cell-cycle progression in radioresistance of glioma-initiating cells. Mol Cancer Ther. 2012;11(9):1863–72. doi: 10.1158/1535-7163.MCT-11-1044. [DOI] [PubMed] [Google Scholar]
- 21.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 22.Squatrito M, Brennan CW, Helmy K, Huse JT, Petrini JH, Holland EC. Loss of ATM/Chk2/p53 pathway components accelerates tumor development and contributes to radiation resistance in gliomas. Cancer Cell. 2010;18(6):619–29. doi: 10.1016/j.ccr.2010.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ropolo M, Daga A, Griffero F, Foresta M, Casartelli G, Zunino A, et al. Comparative analysis of DNA repair in stem and nonstem glioma cell cultures. Mol Cancer Res. 2009;7(3):383–92. doi: 10.1158/1541-7786.MCR-08-0409. [DOI] [PubMed] [Google Scholar]
- 24.Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14(12):1448–59. [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21(13):4129–39. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Sanchez Y. Chk1 in the DNA damage response: conserved roles from yeasts to mammals. DNA Repair (Amst) 2004;3(8–9):1025–32. doi: 10.1016/j.dnarep.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Barcellos-Hoff MH, Newcomb EW, Zagzag D, Narayana A. Therapeutic targets in malignant glioblastoma microenvironment. Semin Radiat Oncol. 2009;19(3):163–70. doi: 10.1016/j.semradonc.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andarawewa KL, Paupert J, Pal A, Barcellos-Hoff MH. New rationales for using TGFbeta inhibitors in radiotherapy. Int J Radiat Biol. 2007;83(11–12):803–11. doi: 10.1080/09553000701711063. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Taniguchi T. MicroRNAs and DNA damage response: implications for cancer therapy. Cell cycle. 2013;12(1):32–42. doi: 10.4161/cc.23051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shiiba M, Shinozuka K, Saito K, Fushimi K, Kasamatsu A, Ogawara K, et al. MicroRNA-125b regulates proliferation and radioresistance of oral squamous cell carcinoma. British journal of cancer. 2013;108(9):1817–21. doi: 10.1038/bjc.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W, Guo F, Wang P, Hong S, Zhang C. miR-221/222 confers radioresistance in glioblastoma cells through activating Akt independent of PTEN status. Current molecular medicine. 2014;14(1):185–95. doi: 10.2174/1566524013666131203103147. [DOI] [PubMed] [Google Scholar]
- 32.Chun-Zhi Z, Lei H, An-Ling Z, Yan-Chao F, Xiao Y, Guang-Xiu W, et al. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC cancer. 2010;10:367. doi: 10.1186/1471-2407-10-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang X, Taeb S, Jahangiri S, Emmenegger U, Tran E, Bruce J, et al. miRNA-95 mediates radioresistance in tumors by targeting the sphingolipid phosphatase SGPP1. Cancer research. 2013;73(23):6972–86. doi: 10.1158/0008-5472.CAN-13-1657. [DOI] [PubMed] [Google Scholar]
- 34.Grosso S, Doyen J, Parks SK, Bertero T, Paye A, Cardinaud B, et al. MiR-210 promotes a hypoxic phenotype and increases radioresistance in human lung cancer cell lines. Cell death & disease. 2013;4:e544. doi: 10.1038/cddis.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang P, Rao EY, Meng N, Zhao Y, Wang JJ. MicroRNA-17-92 significantly enhances radioresistance in human mantle cell lymphoma cells. Radiation oncology. 2010;5:100. doi: 10.1186/1748-717X-5-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1203–17. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 37.Hurov KE, Cotta-Ramusino C, Elledge SJ. A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev. 2010;24(17):1939–50. doi: 10.1101/gad.1934210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7189):896–9. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 39.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327(5962):198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.