Abstract
Insulator-based dielectrophoresis (iDEP) has been explored as a powerful analytical technique in recent years. Unlike with larger entities such as cells, bacteria or organelles, the mechanism of iDEP transport of proteins remains little explored. In this work, we extended the pool of proteins investigated with iDEP in nanostructured devices with β-galactosidase. Our work indicates that β-galactosidase shows concentration due to negative DEP which we compare to DEP response of immunoglobulin G (IgG) encapsulated in micelles also showing negative DEP. Experimental observations are further compared with numerical simulations to elucidate the influence of electrokinetic transport and the magnitude of DEP mobility. Numerical simulations suggest that the DEP mobility calculated using the classical model underestimates the actual contribution of DEP on the experimentally monitored concentration effect of proteins. Moreover, we observed a unique voltage dependent β-galactosidase concentration which we attribute to an additional factor influencing the protein concentration at the nanoconstrictions, namely ion concentration polarization. Our work aids in understanding factors influencing protein iDEP transport which is required for the future development of protein preconcentration or separation methods based on iDEP.
Keywords: insulator-based, negative dielectrophoresis, protein, ion concentration polarization
Introduction
Manipulation of biomolecules poses serious analytical challenges in the area of biomedical and pharmaceutical research. Reliable and rapid separation techniques are in demand especially for proteins within extremely complex mixtures such as cell lysates or body fluids. Moreover, low abundance proteins such as disease biomarkers need to be identified and detected with high sensitivity for further therapeutic purposes. Another analytical challenge arises for sample available only in limited amount. Therefore, powerful methods which require only low sample volumes with the ability to concentrate analytes are demanded.
Dielectrophoresis (DEP) is a powerful analytical technique occurring in an inhomogeneous electric field with the potential to facilitate many processing steps such as preconcentration, purification, fractionation, and separation. Such a versatile applicability makes DEP an attractive analytical method for biological species, including biomolecules. For instance, a variety of applications has been demonstrated in the past including cell separation1,2, fractionation3,4, cytometry5, and patterning6. Moreover, DEP can be used to precisely manipulate and position cells7 and even single molecules8, which makes it a very attractive candidate for nanotechnological applications9. This transport phenomenon occurs in an inhomogeneous electric field when particles suspended in an aqueous solution acquire an induced dipole. Since the DEP response is based on intrinsic bioparticle properties, DEP can serve as a label-free technique which is important when further processing and/or characterization steps are necessary. In addition, since DEP relies on electric field gradients, it has the potential to serve as a preconcentration tool with potential to improve existing protein separation techniques especially in combination or series with other analytical techniques.
DEP has been exploited using two main strategies to generate inhomogeneous electric fields in the past: microelectrode and insulating topological structures. In the former case, microelectrodes are patterned onto a substrate to create electric field gradients employing microfabrication techniques. This electrode-based DEP (eDEP) has most commonly been used in the field of protein DEP such as the first examples demonstrated by Washizu et al. with interdigitated electrodes10, quadruple electrodes geometries11,12, and pairs of electrodes in close distance8. Another relatively new approach is to integrate insulating obstacles inside of the channel to create inhomogeneous electric fields, termed insulator-based DEP (iDEP). A variety of designs have been proposed for iDEP including sawtooth devices13, insulating post arrays with various geometries14,15, and nanosized constrictions16–18. With iDEP devices, particles migrate via both electrokinesis as well as DEP upon application of a DC voltage, eliminating the need of a hydrodynamic pump for sample handling. Additionally, iDEP devices can reduce issues prevalent to eDEP approaches including electrode fouling and undesirable electrode reactions, which interfere with DEP 19. Even though the iDEP device requires larger applied potentials to achieve high electric fields within the device, it establishes homogeneous electric fields throughout the entire depth of the microfluidic channel. On the other hand, high electric field gradient regions are restricted to the vicinity of the electrodes with eDEP devices, which might become disadvantageous for separation applications. Advantages and disadvantages of these DEP methods have been summarized in previous review articles 19,20.
The selectivity of DEP stems from the polarizability of biomolecules in the presence of electric field gradients. An excellent theoretical framework to describe polarizability mechanisms exists for large colloidal particles21 and biological particles such as cells, viruses, and organelles. For example, DEP response of cells is described using a shell model which assigns different permittivities to each compartment of the cell in the form of layers of shells to calculate an overall effective cell permittivity7,21–23. However, the models developed for these large cellular structures and viruses are not directly applicable to submicron-sized biomolecules such as DNA and proteins. In case of DNA, the theoretical DEP models are less developed and still under debate especially on the subject of DNA length and frequency dependence24,25. It is generally assumed that DNA polarization is mainly caused by the ion cloud surrounding the negatively charged DNA backbone. Nevertheless, a number of DNA DEP applications have been demonstrated including concentration26, fractionation10, and separation27–29 with sizes ranging from Mbp down to ~40 bp.
For proteins, the mechanism of polarization responsible for DEP transport is not well understood with much less experimental data available. Theoretically, DEP manipulation of nm- sized proteins is challenging since extremely high electric field gradients are required in order to generate DEP forces large enough to compete with molecular diffusion, electrokinetic and electrothermal forces. Regardless, nearly 20 groups have investigated protein DEP experimentally employing metal electrodes8,10,11,30, nanopipettes31, carbon nanotubes32, and in droplets33. For instance, Hölzel demonstrated single molecule DEP trapping 3 with eDEP8. Moreover, protein DEP has been applied for patterning10,34, bioprobes32, and biosensor applications35. Recently several experimental studies have reported iDEP for proteins including the first work by Lapizco-Encinas14 and the first protein DEP streaming presented by our group 15. Using nanofabrication, extremely high electric field gradients can be created and used for protein DEP, as for example demonstrated with nm-sized constriction devices16–18,36.
To achieve such high electric field gradients for manipulation of proteins, we improved our pre-existing device with triangular microposts37 creating nm-sized features using focused ion beam milling (FIBM). This nano-constriction device allowed the transition from streaming DEP to trapping DEP for λ-DNA with more than 103 fold concentration enrichment18. Here, we investigate protein DEP in this nanoconstriction device with β-galactosidase and IgG encapsulated in block-co-polymer micelles. β-galactosidase was chosen since it is an important enzyme involved in lactose hydrolysis and other catalysis reactions in animals, plants and bacteria38. With a molecular weight of 465 kDa, β-galactosidase is also employed in microbiology, such as in cloning, as a marker of cellular senescence and as an indicator of aging39, but also in food processing38 and as a molecular weight marker protein in biological assays. The experimental results obtained from β-galactosidase and IgG micelles are compared with numerical simulations in order to elucidate the influence of electrokinetic and DEP transport. Finally, we discuss additional factors influencing protein DEP concentration using this nanoconstriction DC-iDEP device.
Theory
Dielectrophoresis (DEP) is defined as motion of a polarizable particle in the presence of a non-uniform electric field. A net electrostatic force is exerted on a particle with an induced dipole, resulting in its migration along an electric field gradient21,40. The DEP force of an ellipsoidal particle under DC condition is expressed as31:
| (1) |
where E denotes the local electric field, a, b, and c are the radii of the ellipsoid along the three major axes, and εm and ε0 refer to the medium and vacuum permittivity, respectively. Re(fMCM) is the real part of the Clausius-Mossotti (CM) factor modified for the ellipsoidal shape whose sign governs the mode of DEP and, under low frequency or DC conditions is expressed as:
| (2) |
Here, σp and σm denote the particle (p) and medium (m) conductivity, respectively and z the depolarization factor. For σp > σm, positive DEP (pDEP) occurs indicated by an attraction of the particle to high electric field regions, whereas negative DEP (nDEP) prevails for σp < σm.
Under DC conditions, DEP interplays with electrokinesis as well as diffusion. Electrokinesis is comprised of electrophoresis (EP) and electroosmosis (EO) whose velocity (uEk) is in linear relation to the electric field expressed as follows:
| (3) |
where µEK, µEP, and µEO denote the overall electrokinetic, electrophoretic, and electroosmotic mobility, respectively.
The DEP velocity (uDEP) is expressed as40:
| (4) |
where µDEP is the DEP mobility which can be calculated by balancing the DEP force with the particle’s drag force. In the case of an ellipsoid particle µDEP results in:
| (5) |
where η is the buffer viscosity and R̄ the mean translation coefficient.
Previously, we showed that a convection-diffusion model is suitable to represent protein migration considering the influence of electrokinesis, DEP, and diffusion. The total particle flux (J) is given as:
| (6) |
where D denotes the diffusion coefficient and c the concentration of the particles. Using equation (6), concentration distributions can be modeled by solving the convection-diffusion equation under steady state condition15. In addition, this model was successfully employed to explain the change in concentration distributions under varying conditions (i.e. pH, conductivity etc.) by adapting the parameters of µDEP and µEK 37. In this current work, we will use the same approach to explain the observed protein migration behavior due to DEP and electrokinesis, however, in iDEP nanoconstriction devices.
Experimental
Chemicals and Materials
Si wafers (4 in) were obtained from University Wafer. The negative photoresist SU-8 2007 and developer were purchased from Microchem, USA. (Tridecafluoro-1,1,2,2-tetrahydrooctyl)dimethylchlorosilane (TDTS) for wafer silanization was purchased from Gelest Corp., USA. Sylgard184, composed of the silicon elastomer base and the curing agent for poly(dimethylsiloxane) (PDMS) was obtained from Dow Corning Corporation, USA. For h-PDMS, vinyl PDMS prepolymer, Pt calalyst (platinum divinyltetramethyldisiloxane), and hydrosilane prepolymer were purchased from Gelest Corp, USA and a modulator (2,4,6,8-Tetramethyl-2,4,6,8-tetravinylcyclotetrasiloxane) from Sigma-Aldrich, USA. Deionized water was supplied from a Synergy purification system (Millipore, USA).
Device Fabrication
A combination of photolithography and focused ion beam milling (FIBM) was employed to fabricate an inverted Si master as previously demonstrated18. The resulting wafer was used as a master for the subsequent soft lithography to mold a PDMS replica. In detail, a master relief of SU-8 photoresist was first patterned on a Si wafer using standard photolithography. This wafer was coated with 20 nm Cr layer using a Cressington 308R Evaporator (Ted Pella Inc. USA). Subsequently FIBM was used to mill nanoposts with Nova 200 (FEI Company, USA) instrument between the tips of the triangular microposts. From this master wafer, PDMS was replica molded resulting in a structure as schematically shown in Fig 1 where both micro- and nanoposts are integrated in the channel. For β-galactosidase DEP experiments, a composite of thin toluene-diluted h-PDMS layer supported by a thick Sylgard184 PDMS layer was used as described previously41 since mere Sylgard184 PDMS structure tends to cause roof and/or lateral collapse for shallow features42. Moreover, h-PDMS prevents deformation of the relief surface, resulting in sharper edges42,43. Reservoir holes with 2 mm diameter were manually punched through the PDMS piece at both ends of the 0.8 cm channel. The resultant PDMS piece and glass slide (150µm thick) were sonicated in isopropanol and DI water baths and blow dried with nitrogen. To form a tight seal between PDMS and glass slides, both pieces were exposed to the oxygen plasma (PDC-001 Harrick Plasma, Harrick, USA) for 1 min at the highest RF setting. A 5 mm thick PDMS slab with 5 mm diameter reservoir holes was pressed above the microchip reservoirs to enlarge the reservoirs and to hold the Pt wire electrode (Alfa Aesar, USA) in position.
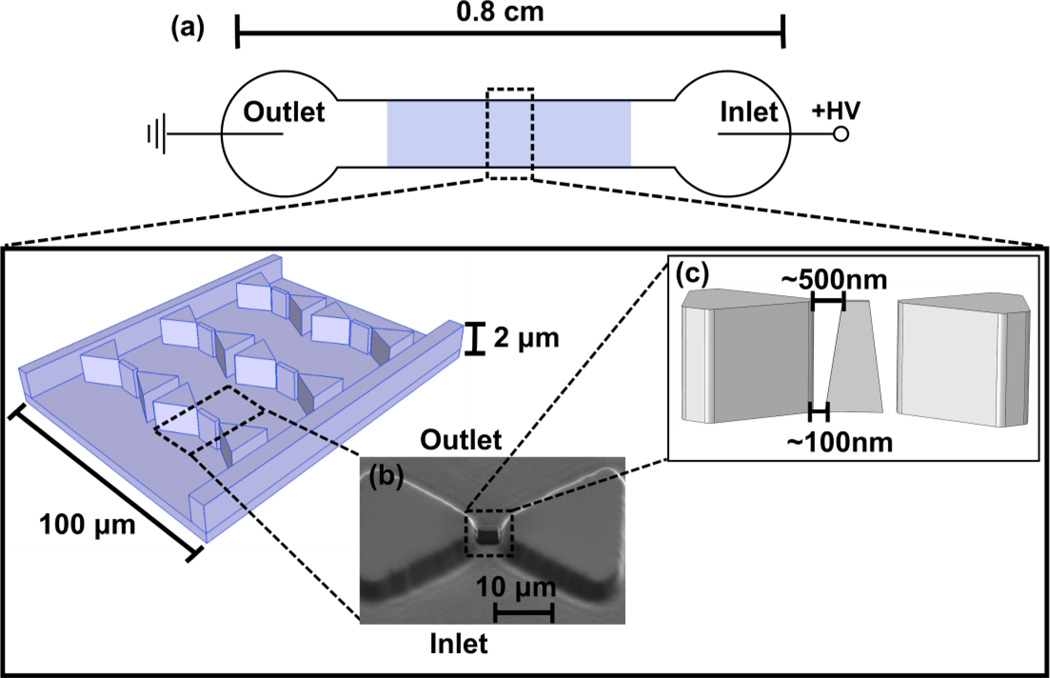
Figure 1.
Schematic of the iDEP device set-up (not to scale). a) A potential difference is applied to a microchannel exhibiting an insulator post array. b) Nanoconstrictions are created between the tips of triangular microposts by FIBM to achieve high electric field gradients for manipulation of nm-sized proteins. The location of these posts may vary up to 200 nm in between the tips of the larger triangular posts due to instrument limitations in positioning the focused ion beam. c) A scanning electron microscopy image of the triangular post with nanoconstrictions in the PDMS mold is also shown. d) A schematics illustrating the vertical positioning of the nanopost in between the microposts and the variations of distance to the microposts due to FIBM.
Sample Preparation and Experimental Set-up
Prior to the experiment, the channel was coated overnight with 500 µM tri-block-copolymer F108 to reduce undesirable protein adsorption onto the PDMS surface. After overnight incubation, F108 solution was washed away and exchanged with the buffer used for the subsequent DEP experiment in case of F108 static coating condition. Channels employed under F108 dynamic coating condition for IgG micelle DEP experiments were filled with pH 8 phosphate buffer containing 3 mM F108 while no buffer exchange was required prior to experiments. The buffers were prepared with different conductivities including 32 µS/cm HEPES buffer at pH 6.4, 100 µS/cm phosphate buffer at pH 8, and 1 mS/cm phosphate buffer at pH 8. The pH and conductivity of all the buffers were assessed with a pH meter (SB70P sympHony, VWR, USA) and a conductivity meter (ORION 3 STAR, Thermo scientific, USA). For the DEP experiments, the reservoirs were filled with 80 µL buffer containing 3 mg/mL CHAPS. For IgG micelle experiments the buffer also contained 3mM F108. The inlet reservoir was filled with a sample buffer containing the analyte. Pt electrodes attached to both reservoirs were connected to the high voltage power supply (HVS448 6000V, LabSmith, Livermore, CA) to apply DC voltages.
Two different proteins were employed in DEP experiments including β-galactosidase (Sigma-Aldrich, USA) and immunoglobulin G (IgG) (Invitrogen, USA) with the concentration of 21 µg/mL and 20 µg/mL, respectively. Prior to the experiments, proteins required labeling with fluorescence for visual detection. Alexa Fluor 488 labeled IgG was used as received and β-galactosidase was labeled using an Alexa Fluor 488 protein labeling kit (Invitrogen, USA) following the basic protocol. Labeled proteins were purified using a suitable molecular weight cutoff centrifugal filter (EMD Millipore Corp., USA) after which the purity was tested using thin layer chromatography. Recovered protein concentration was determined using the Bradford protein assay with a plate reader spectrophotometer (Synergy HT, BioTek Instruments, VT).
Detection and Data Analysis
For fluorescence microscopy imaging, an inverted microscope (IX 71, Olympus, USA), with a 100x objective (LUCPlan FL N, Olympus, USA), a mercury burner (U-RFL-T, Olympus, USA) and fluorescent filter set (exciter ET470/40x, dichroic T495LP, emitter ET525/50m, Olympus, USA) was used. Images were acquired at 150 ms/frame using a CCD camera (Quantum 512 SC, Photometrics, USA) and Micro-Manager software (University of California, USA) and analyzed with Image J software (version 1.43).
Results and Discussion
β-galactosidase DC iDEP
We investigated the DEP behavior of β-galactosidase using the nanoconstriction DC-iDEP device shown schematically in Fig 1. For iDEP experiments, protein was dissolved in a low conductivity buffer (100 µS/cm) at pH 8 with the zwitterionic additive CHAPS to reduce protein aggregation15. β-galactosidase is known to form a tetramer in native state with a molecular weight of 465 kDa 44.
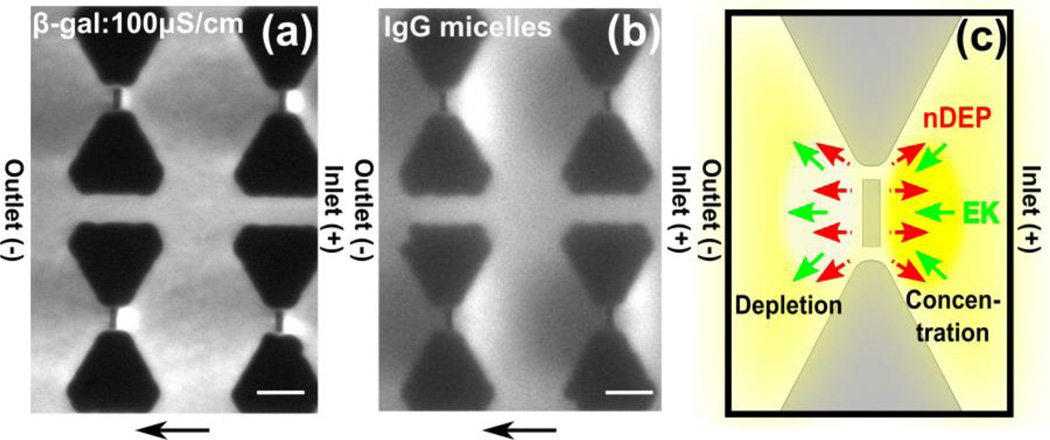
After the channel was filled with the protein solution and a steady state was established, β-galactosidase started to concentrate at the inlet side of a nanoconstriction (as shown in Figure 1) upon application of 100 V across the 0.8 cm channel as shown in Fig 2a. As indicated by arrows in Fig 2a, β-galactosidase was transported by cathodic electrokinetic flow, which was verified by EOF measurements with the current monitoring method45 (data not shown). Since the isoelectric point of β-galactosidase is ~4.646, the protein is negatively charged in the pH 8 buffer used for the iDEP experiments. The cathodic flow direction indicates an overall stronger EOF component counteracting the electrophoretic transport. Fig 2a also indicates β-galactosidase depletion at the outlet. We can attribute this unique protein concentration/depletion to nDEP based on the interplay of electrokinetic and dielectrophoretic forces at the nanoposts as schematically depicted in Fig 2c. Protein concentration occurs at the inlet side of the nanopost constriction since the nDEP force directing away from the nanopost counteracts electrokinetic flow. On the other hand, protein depletion occurs at the outlet side due to a similar overlay of nDEP with electrokinesis. Fig 2c summarizes the observed concentration and depletion characteristic for nDEP of β-galactosidase.
Figure 2.
(a.b) Fluorescence microscopy images obtained experimentally by DC iDEP experiments with β-galactosidase and IgG encapsulated in micelles. Flow direction is from right to left. Scale bar indicates 10 µm. (a) β-galactosidase shows concentration at the inlet side of the nanoposts and depletion at the outlet side due to negative DEP with 100 µS/cm phosphate buffer at 100V applied across a 0.8 cm channel. (b) IgG micelles concentrate at the inlet side of the nanoposts and deplete at the opposite side due to nDEP with 100 µS/cm phosphate buffer at 50 V applied across a 1 cm channel. (c) Schematics showing the flow directions due to DEP and electrokinesis and the resultant species concentration and depletion around the nanoconstriction. Negative DEP counteracts electrokinesis at the inlet side of the nanopost, resulting in protein concentration at the inlet side, whereas depletion occurs at the outlet side (yellow).
In order to provide a strong evidence that the β-galactosidase concentration occurs due to nDEP, we performed iDEP experiments using a previously tested analyte showing nDEP. Similar to our previous study37, we employed IgG encapsulated in micelles of the tri-block co-polymer F108 demonstrating nDEP. As shown in Fig 2b, IgG micelle concentration occurred at the inlet side of the nanopost similar to the β-galactosidase concentration. Therefore, we conclude that β-galactosidase exhibits nDEP using our nanoconstriction DC-iDEP device.
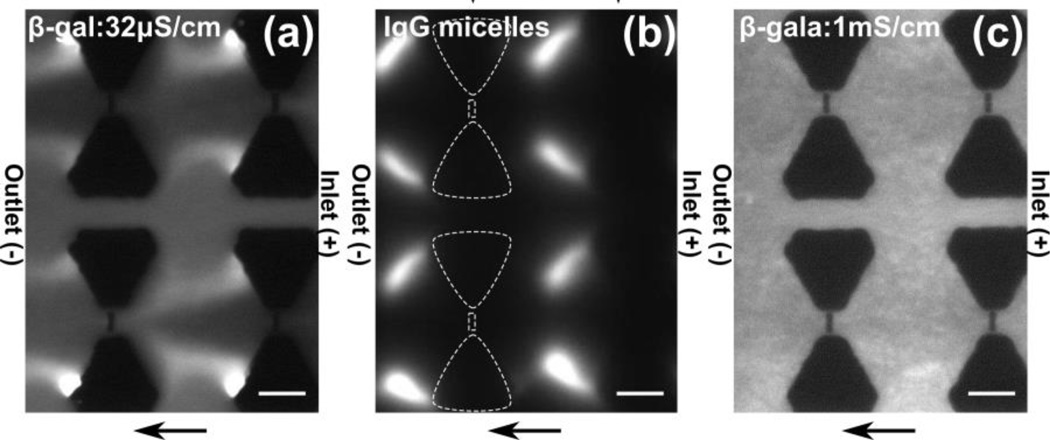
Both DEP and electrokinesis are influenced by the buffer medium conductivity. Therefore we also investigated the influence on protein DEP concentration 32 µS/cm and 1 mS/cm. Fig 3a showing the DEP behavior with 32 µS/cm buffer conductivity demonstrates a strong depletion around the nanoposts which even expands to the regions between the rows of the microposts. This β-galactosidase concentration behavior is similar to what we observed with IgG micelles for applied potential of 200 V shown in Fig 3b. In contrast to the IgG micelle concentration at 50 V (see Fig 2a), a strong depletion was observed around the nanoposts under an application of 200 V, which even expands to the regions between two rows of the microposts. In case of higher conductivity buffer of 1 mS/cm, we observed no concentration or depletion as shown in Fig 3c.
Figure 3.
(a-c) Fluorescence microscopy images obtained experimentally by DC-iDEP experiments. Flow direction is from right to left. Scale bar indicates 10 µm. (a) β-galactosidase concentration with 32 µS/cm HEPES buffer with 100 V applied across a 0.8 cm channel. Depletion at the nanoconstrictions is shown at the outlet side and concentration at the edge of the microposts. (b) IgG micelles concentrate at the outlet side of the microposts with 200 V applied across a 1 cm channel. White dashed lines indicate a row of posts with µm sized microposts and nm-sized post in between. (c) β-galactosidase shows no apparent concentration change throughout the channel with 1 mS/cm phosphate buffer at 100V applied across a 0.8 cm channel.
The observations obtained with varying medium conductivity are surprising. Based on the classical DEP theory, we would expect a higher nDEP response since the CM factor should be more negative compared to the 100µS/cm case. Conductivity dependent changes in the electrokinetic mobility would however counteract the observed protein concentration. We can speculate on the possible reason for this behavior in relation to the contribution of electrical double layer (EDL) polarization for sub-micrometer particles. It was previously shown that nanoparticles with thick EDL exhibit extraordinary large DEP response mostly due to their electrophoretic motion distorting the ion distributions within the EDL47. We estimate an EDL thickness of ~18 nm for 32 µS/cm buffer and 4 nm for 1 mS/cm, respectively. Therefore, proteins in the more dilute buffer would show increased DEP response compared to the ones in the higher conductivity buffer. Recently, Zhao and Bau demonstrated that a thick EDL accounts for a major contribution to the total dipole moment in the case of DNA48. Although this model has not yet been extended to proteins, it might hold for our experimental observations.
Comparison of experiments and numerical simulations
Numerical simulations serve as a helpful tool allowing the comparison with experimental observations. We can assess the concentration distribution by solving the convection-diffusion model as described in the theoretical section. To estimate µDEP, we approximated the shape as an oblate ellipsoid from the dimension of β-galactosidase reported via X-ray crystallography44. Using the classical model developed for this shape15 and assuming σp = 0 S/m as an extreme nDEP case, we obtained µDEP in the order of 10−24 m4/V2s. We then investigated the concentration distribution with various µDEP and µEK values attempting to match the experimentally observed distributions in protein concentration at the nanoconstriction under iDEP conditions with numerical simulations.
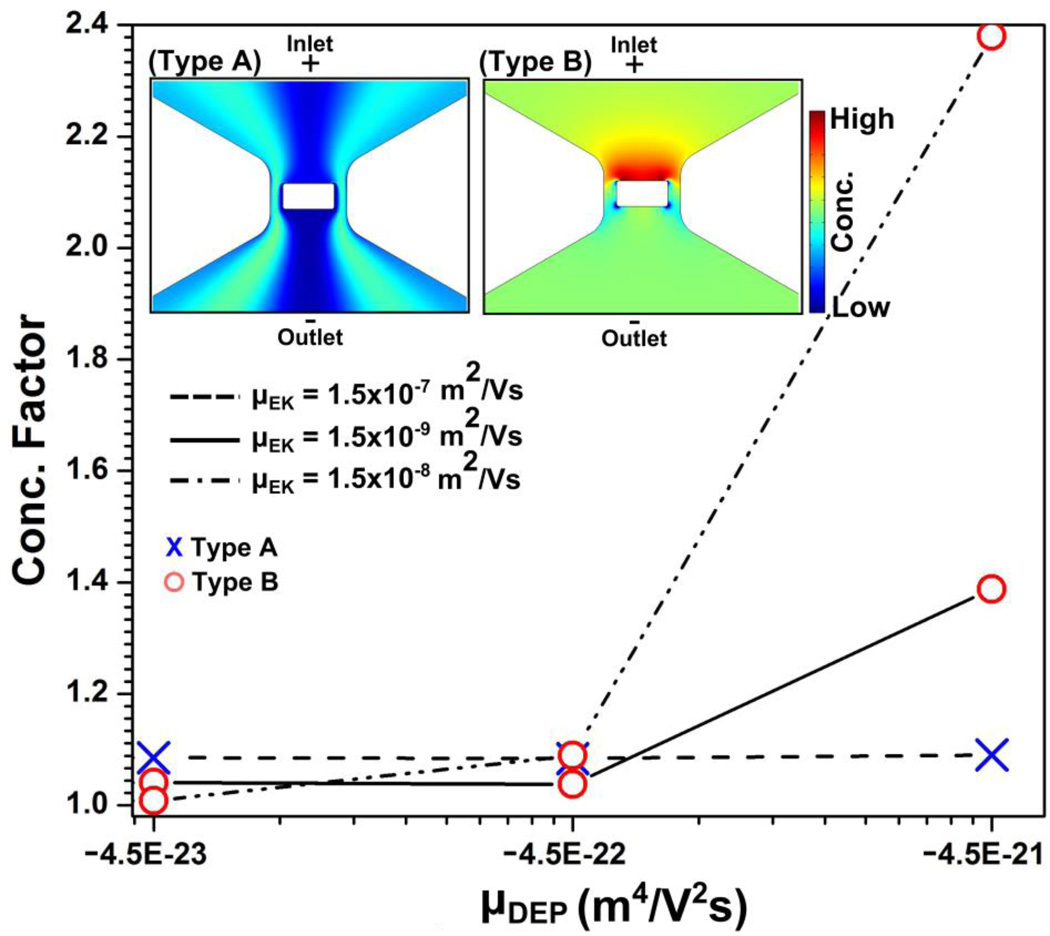
We varied µEK from 1.5×10−7 to 1.5×10−9 m2/Vs and µDEP from −4.5×10−24 to −4.5×10−21 m4/V2s. Protein concentration or depletion was not obtained with µDEP of −4.5×10−24 m4/V2s, while in the range of µDEP = −4.5×10−23 to −4.5×10−21 m4/V2s two distinctive types of concentration distributions were found: type A in which the protein concentration is depleted around the nanopost and type B in which the protein concentration is enhanced at the inlet side of the constriction as shown in the inset of Fig 4.
Figure 4.
The resultant concentration factors obtained by numerical simulations are plotted as a function of µDEP for three different µEK mobilities. Different markers are used in order to represent the different types of concentration distributions: red circle markers and blue cross markers representing the type A and B concentration distributions, respectively. The inset shows type A and B concentration distributions as obtained by numerical simulation.
These simulated concentration patterns were compared with the β-galactosidase iDEP experiment at a conductivity of 100 µS/cm as shown in Fig. 2a. We observe that the type B distribution qualitatively best represents the experimental results where the concentration enriches at the inlet side and depletes at the opposite side. By analyzing the variations of µEK and µDEP, we found that the parameter set of −4.5 × 10−23 m4/V2s ≥ µDEP and µEK ≥ 1.5×10−8 m2/Vs shows type B behavior similar to the experimentally observed concentration effect at 100 µS/cm for β-galactosidase. It is important to remark that previously a value of 1.5×10−8 m2/Vs was reported for µEK under similar buffer conditions in PDMS devices45. This leads to the conclusion that µDEP is underestimated with the classical model, since the values estimated with the oblate model were one order of magnitude smaller.
We also discuss the numerically obtained concentration patterns in relation to variations in the medium conductivity with β-galactosidase. In the case of 32 µS/cm, the simulation results indicate that the experimentally observed concentration qualitatively fest best to a type A concentration distribution. Although the type A concentration profile obtained in numerical modeling as shown in Fig. 4 does not entirely match experimentally observed location of concentrated regions (Fig. 3a), the numerical simulations show that the concentration of the protein shifts sideways from the nanoconstriction region (characteristic for type A). We can explain this transition with the increase in the zeta potential of the channel surface, thus enhanced electrokinetic mobility induced through a decreased ion concentration of the buffer medium. Note that the discrepancies between numerical simulations and experiments might be due to additional effects influencing the concentration profile such as ion concentration polarization as we will discuss in the following section.
Applied potential dependent β-galactosidase iDEP
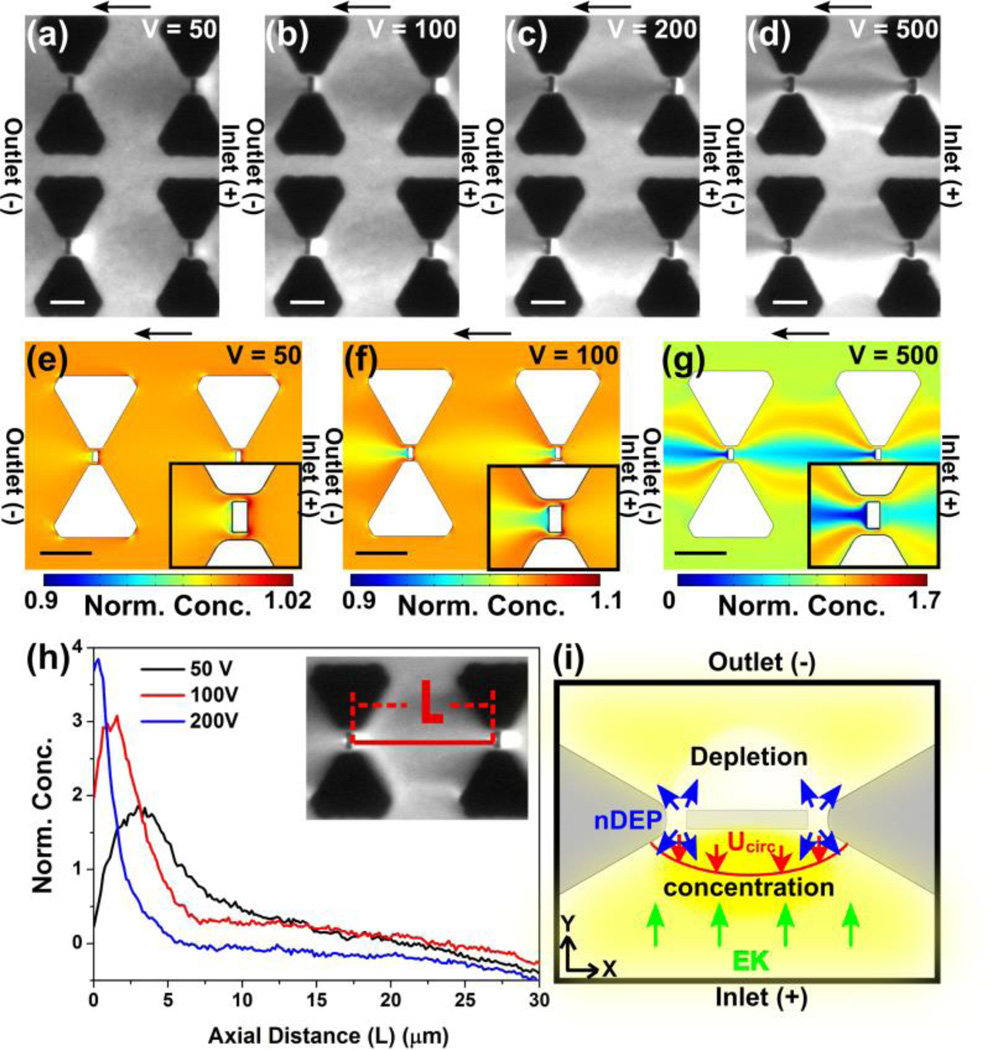
We investigated DEP behavior of β-galactosidase in dependence of the applied potentials in a range from 50V to 500V at a medium conductivity of 100 µS/cm. Fluorescence microscopy images shown in Fig 5a-d demonstrate a transition of the concentration distribution with increasing applied potential. With only 10~20V, β-galactosidase concentration was depleted at the outlet side (data not shown). Subsequently, by gradually increasing the applied potential, β-galactosidase started to concentrate on the inlet side, while depletion at the outlet side intensified (Fig 5a, at 50V). This protein enrichment at the inlet side was enhanced with increasing the voltage further (Fig 5b, at 100V) and a similar concentration trend was observed with higher applied potentials up to 200 V (Fig 5c). However, protein streaming from the inlet side started with applied potentials higher than 200 ~300V (Fig 5d, at 500V).
Figure 5.
(a-d) Experimental results and numerical simulations for the iDEP device with integrated triangular microposts and rectangular nanoposts between the tips of the triangles. Flow direction is from right to left in the cathodic direction. Scale bar indicates 10 µm. Fluorescence microscopy images obtained by DC-iDEP experiments with β-galactosidase, demonstrating voltage dependent concentration distributions due to nDEP with the following applied voltages: (a) 50V (b) 100V, (c) 200V, and (d) 500V for a 0.8 cm long channel. (e-g) Numerical simulation results obtained by solving eq. 6 with the same external electric field as the experiments: (e) 63 V/cm, (f) 125 V/cm, and (g) 625 V/cm. The insets show the close-up around the nanopost region where the highest electric field gradient is expected. (h) Protein concentration profiles extracted from the concentration distribution at the regions perpendicular to the nanopost as indicated in the inset image and plotted as a function of voltages 50, 100, and 200 V applied voltage for a 0.8 cm channel. Fluorescence intensity is normalized with the intensity at the same region at 0V. (i) The force balance around the nanoconstriction whose size scales down to ~100 nm including electrokinesis, negative DEP, and ICP and the resultant concentration distribution (yellow).
It is interesting to discuss the transition of concentration distributions as shown in Fig 5 a-d in dependence of applied potentials. To clearly visualize the concentration distribution around the nanopost regions where the higher electric field gradients are created, fluorescence intensity profiles perpendicular to the nanopost were plotted with different applied voltages (Fig 5h). For this operation, the fluorescence intensities were normalized with the intensities at 0 V and the corresponding intensity during iDEP is analyzed along the line, L, as shown in Fig. 5h. The maximum concentration was observed ~5 µm away from the nanopost at the inlet side at 50 V. By increasing the potential, the concentration maximum approached the nanopost and was closest to the nanopost at 200 V. Concomitantly, the peak maximum increased with increasing applied potential with a maximum concentration factor of 3.8 at 200 V.
To further characterize the voltage dependent protein DEP behavior, we carried out numerical simulations to reveal concentration distributions in the iDEP device solving eq. 6. Fig 5 e-g show the normalized concentration distribution around the post regions at 50 V, 100 V, and 500 V with µEK = 1.5 × 10−8 m2/Vs and µDEP = − 9.0 × 10−22 m4/V2s for β-galactosidase exhibiting nDEP. These values were chosen, since simulation results using these parameters revealed type B concentration distribution and a similar voltage dependency as observed in the experiments. Specifically, the region of protein concentration is located at the inlet side of the nanopost at 50 V (Fig. 5e) and 100 V (Fig. 5f), whereas the opposite side is depleted. With the higher applied voltages of 500 V, the concentration distribution changed its shape drastically as shown in Fig 5g where streamlines similar to the experimental observations were apparent. Although the concentration distribution obtained by simulation appears similar to our experiments, we noticed a difference. The experimentally observed concentration at 50 to 200 V is more delocalized compared to the simulation where the protein concentration only occurs at a very small region adjacent to the nanopost. Based on the comparison of the experimental results and numerical simulations, the experimentally observed protein concentration cannot solely be explained through DEP.
It is likely that multiple phenomena contributing to the protein migration play a role and change their balance in dependence of the applied potentials. For instance, numerical simulations previously showed that the change in electrophoresis, electroosmosis, and DEP influences the protein concentration profiles in DC iDEP with microposts37. However, in the nanopost device as employed in this study, additional factors may influence the concentration behavior due to the nm-sized constrictions. For example, Liao et al.16 recently showed that electrothermal effects influence concentration of streptavin at a nanoconstriction. Under high ionic strength conditions, electrothermal effects shifted the protein concentration zone away from the nanoconstriction considerably interplaying with DEP and electrokinetic effects. However, in our work, a low conductivity buffer and potential ranges were employed for which we can exclude considerable Joule heating effects as recently investigated in another study49. Therefore, we consider changes in the ionic concentration around the nanoconstriction to explain our experimental observation next.
It is known that nm-sized channels with critical dimension of 10 ~100 nm exhibit a unique ion permselectivity, which is termed ion concentration polarization (ICP)50–53. We observed by close inspection of the nanopost region via scanning electron microscopy (see Figure 1), that the smallest constrictions of our device scale down to ~100 nm. Such small constrictions are known to generate ICP which may dynamically change ion concentration around the nanoconstriction under the ionic strengths employed here 51. To emphasize the interplay of ICP with DC iDEP, Fig 5i schematically shows the migration directions of the various effects around the nanoconstriction regions schematically. At low voltages, protein transported through the channel by cathodic electrokinetic flow is depleted at the outlet side of the nanopost and concentrated on the opposite side due to nDEP in accordance with the simulation shown in Fig 5e-f. However, during DEP concentration, proteins finally concentrated several µm away from the nanopost which we assume is ICP triggered. Since ICP is known to create parabolic-like backflow (Ucirc) at the ion depletion zone formed in front of the nanostructure, we suggest that ICP enhances the protein concentration caused by nDEP. While increasing the voltage, the concentration zone due to ICP moves closer to the nanoposts since the forwarding electrokinetic flow increases. As the nanopost region is approached, protein concentration due to nDEP may also enhance due to the larger electric field gradient at the nanoconstriction, resulting in an overall concentration enhancement adjacent to the nanoposts.
This aforementioned scenario involving the interplay of DEP, EK, and ICP creates a unique voltage dependent concentration distribution caused by a dynamic change of the local environment (i.e. electric field distribution, ion distribution). Such dynamic changes in vicinity of nanoconstrictions should affect both electrokinetic and dielectrophoretic behavior of proteins. For instance, Kim et al. previously measured the electric field strength in the depletion zone to be as high as ~1000 V/cm with the externally applied electric field of 30 V/cm51. This largely enhanced electric field amplifies the electrokinetic transport at the inlet side of the nanostructure thereby counteracting DEP. Moreover, one would expect the increase of nDEP forces due to larger electric field gradients or increases in the negative CM-factor with increased medium conductivity at the outlet side due to ICP. Even though it is challenging to quantitatively assess the effect of ICP with our current device, we can conclude that the observed concentration distribution resulted from dynamic changes of electrokinesis and iDEP due to the change in ion concentration originating from ICP at the nanopost. Moreover, ICP can influence iDEP migration and concentration due to the amplified electric field at the anodic inlet side, whereas it enhances nDEP at the cathodic outlet side.
Conclusion
Our work successfully demonstrated β-galactosidase concentration due to nDEP under DC conditions using a nanoconstriction iDEP device. β-galactosidase concentration was observed at the inlet side of the nanoconstrictions, which can be explained by a combination of electrokinesis and DEP. Similar observations resulted from iDEP experiments with IgG micelles, which have previously been demonstrated to exhibit nDEP. Additionally, numerical simulation showed transitions in the iDEP concentration around the nanocontrictions between two distinct types, which could be correlated with experimental observations. Moreover, we observed a unique voltage dependent β-galactosidase concentration distribution at the nanoconstriction which we suggest to be caused by ion concentration polarization occurring at the nanoconstrictions influencing particle transport around the nanoconstrictions and the resultant protein concentration.
Acknowledgements
This project was supported by grants from the National Center for Research Resources (5R21RR025826-03) and the National Institute of General Medical Sciences (8R21GM103522-03) from the National Institutes of Health. We thank Prof. Daniel Buttry and Dr. Poonam Singh in the Department of Chemistry and Biochemistry at Arizona State University for assistance with EOF measurements related to this work.
References
- 1.Hu X, Bessette PH, Qian J, Meinhart CD, Daugherty PS, Soh HT. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15757–15761. doi: 10.1073/pnas.0507719102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pethig R. Crit. Rev. Biotechnol. 1996;16:331–348. [Google Scholar]
- 3.Wang X-B, Yang J, Huang Y, Vykoukal J, Becker FF, Gascoyne PRC. Anal. Chem. 2000;72:832–839. doi: 10.1021/ac990922o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang J, Huang Y, Wang X-B, Becker FF, Gascoyne PRC. Anal. Chem. 1999;71:911–918. doi: 10.1021/ac981250p. [DOI] [PubMed] [Google Scholar]
- 5.Voldman J, Gray ML, Toner M, Schmidt MA. Anal. Chem. 2002;74:3984–3990. doi: 10.1021/ac0256235. [DOI] [PubMed] [Google Scholar]
- 6.Albrecht DR, Underhill GH, Wassermann TB, Sah RL, Bhatia SN. Nat. Methods. 2006;3:369–375. doi: 10.1038/nmeth873. [DOI] [PubMed] [Google Scholar]
- 7.Gagnon ZR. Electrophoresis. 2011;32:2466–2487. doi: 10.1002/elps.201100060. [DOI] [PubMed] [Google Scholar]
- 8.Hölzel R, Calander N, Chiragwandi Z, Willander M, Bier FF. Phys. Rev. Lett. 2005;95:128102(1)–128102(4). doi: 10.1103/PhysRevLett.95.128102. [DOI] [PubMed] [Google Scholar]
- 9.Koh SJ. Nanoscale Res. Lett. 2007;2:519–545. doi: 10.1007/s11671-007-9091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Washizu M, Suzuki S, Kurosawa O, Nishizaka T, Shinohara T. IEEE Trans. Ind. Appl. 1994;30:835–843. [Google Scholar]
- 11.Bakewell DJG, Hughes MP, Milner JJ, Morgan H. Ann Int IEEE Embs. 1998;2:1079–1082. [Google Scholar]
- 12.Zhang C, Khoshmanesh K, Mitchell A, Kalantar-zadeh K. Anal. Bioanal. Chem. 2010;396:401–420. doi: 10.1007/s00216-009-2922-6. [DOI] [PubMed] [Google Scholar]
- 13.Staton SJR, Jones PV, Ku G, Gilman SD, Kheterpal I, Hayes MA. Analyst. 2012;137:3227–3229. doi: 10.1039/c2an35138b. [DOI] [PubMed] [Google Scholar]
- 14.Lapizco-Encinas BH, Ozuna-Chacón S, Rito-Palomares M. J. Chromatogr. A. 2008;1206:45–51. doi: 10.1016/j.chroma.2008.05.077. [DOI] [PubMed] [Google Scholar]
- 15.Nakano A, Chao T-C, Camacho-Alanis F, Ros A. Electrophoresis. 2011;32:2314–2322. doi: 10.1002/elps.201100037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao K-T, Tsegaye M, Chaurey V, Chou C-F, Swami NS. Electrophoresis. 2012;33:1958–1966. doi: 10.1002/elps.201100707. [DOI] [PubMed] [Google Scholar]
- 17.Liao K-T, Chou C-F. J. Am. Chem. Soc. 2012;134:8742–8745. doi: 10.1021/ja3016523. [DOI] [PubMed] [Google Scholar]
- 18.Camacho-Alanis F, Gan L, Ros A. Sens Actuators B. 2012;173:668–675. doi: 10.1016/j.snb.2012.07.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Duarte R. Electrophoresis. 2012;33:3110–3132. doi: 10.1002/elps.201200242. [DOI] [PubMed] [Google Scholar]
- 20.Li M, Li WH, Zhang J, Alici G, Wen W. J. Phys. -Appl. Phys. 2014;47:063001. [Google Scholar]
- 21.Jones TB. Electromechanics of Particles. Cambridge: Cambridge University Press: 2005. [Google Scholar]
- 22.Voldman J. Annu. Rev. Biomed. Eng. 2006;8:425–454. doi: 10.1146/annurev.bioeng.8.061505.095739. [DOI] [PubMed] [Google Scholar]
- 23.Pethig R. Biomicrofluidics. 2010;4:022811. doi: 10.1063/1.3456626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao H. Phys. Rev. E. 2011;84:021910(1)–021910(6). doi: 10.1103/PhysRevE.84.021910. [DOI] [PubMed] [Google Scholar]
- 25.Henning A, Bier FF, Hölzel R. Biomicrofluidics. 2010;4:022803(1)–022803(9). doi: 10.1063/1.3430550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swami N, Chou C-F, Ramamurthy V, Chaurey V. Lab. Chip. 2009;9:3212–3220. doi: 10.1039/b910598k. [DOI] [PubMed] [Google Scholar]
- 27.Regtmeier J, Duong TT, Eichhorn R, Anselmetti D, Ros A. Anal. Chem. 2007;79:3925–3932. doi: 10.1021/ac062431r. [DOI] [PubMed] [Google Scholar]
- 28.Regtmeier J, Eichhorn R, Bogunovic L, Ros A, Anselmetti D. Anal. Chem. 2010;82:7141–7149. doi: 10.1021/ac1005475. [DOI] [PubMed] [Google Scholar]
- 29.Huang LR, Tegenfeldt JO, Kraeft JJ, Sturm JC, Austin RH, Cox EC. Nat. Biotechnol. 2002;20:1048–1051. doi: 10.1038/nbt733. [DOI] [PubMed] [Google Scholar]
- 30.Zheng L, Brody JP, Burke PJ. Biosens. Bioelectron. 2004;20:606–619. doi: 10.1016/j.bios.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 31.Clarke RW, Piper JD, Ying L, Klenerman D. Phys. Rev. Lett. 2007;98:198102(1)–198102(4). doi: 10.1103/PhysRevLett.98.198102. [DOI] [PubMed] [Google Scholar]
- 32.Maruyama H, Nakayama Y. Appl. Phys. Express. 2008;1:124001(1)–124001(3). [Google Scholar]
- 33.Agastin S, King MR, Jones TB. Lab. Chip. 2009;9:2319–2325. doi: 10.1039/b903831k. [DOI] [PubMed] [Google Scholar]
- 34.Asokan SB, Jawerth L, Carroll RL, Cheney RE, Washburn S, Superfine R. Nano Lett. 2003;3:431–437. [Google Scholar]
- 35.Gong J-R. Small. 2010;6:967–973. doi: 10.1002/smll.200902132. [DOI] [PubMed] [Google Scholar]
- 36.Kovarik ML, Jacobson SC. Anal. Chem. 2008;80:657–664. doi: 10.1021/ac701759f. [DOI] [PubMed] [Google Scholar]
- 37.Nakano A, Camacho-Alanis F, Chao T-C, Ros A. Biomicrofluidics. 2012;6:034108. doi: 10.1063/1.4742695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sen P, Nath A, Bhattacharjee C, Chowdhury R, Bhattacharya P. Biochem. Eng. J. 2014;90:59–72. [Google Scholar]
- 39.Huang T, Rivera-Pérez JA. genesis. 2014;52:300–308. doi: 10.1002/dvg.22761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pohl HA. Dielectrophoresis. Cambridge: Cambridge University Press; 1978. [Google Scholar]
- 41.Kang H, Lee J, Park J, Lee HH. Nanotechnology. 2006;17:197. [Google Scholar]
- 42.Odom TW, Love JC, Wolfe DB, Paul KE, Whitesides GM. Langmuir. 2002;18:5314–5320. [Google Scholar]
- 43.Li H-W, Muir BVO, Fichet G, Huck WTS. Langmuir. 2003;19:1963–1965. [Google Scholar]
- 44.Jacobson RH, Zhang X-J, DuBose RF, Matthews BW. Nature. 1994;369:761–766. doi: 10.1038/369761a0. [DOI] [PubMed] [Google Scholar]
- 45.Hellmich W, Regtmeier J, Duong TT, Ros R, Anselmetti D, Ros A. Langmuir. 2005;21:7551–7557. doi: 10.1021/la0510432. [DOI] [PubMed] [Google Scholar]
- 46.Wallenfels K, Weil R. In: The Enzymes. Boyer Paul D., editor. Vol. 7. Academic Press; 1972. pp. 617–663. [Google Scholar]
- 47.Zhao H, Bau HH. J. Colloid Interface Sci. 2009;333:663–671. doi: 10.1016/j.jcis.2009.01.056. [DOI] [PubMed] [Google Scholar]
- 48.Zhao H, Bau HH. Langmuir. 2010;26:5412–5420. doi: 10.1021/la903842z. [DOI] [PubMed] [Google Scholar]
- 49.Nakano A, Luo J, Ros A. Anal. Chem. 2014;86:6516–6524. doi: 10.1021/ac501083h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Napoli M, Eijkel JCT, Pennathur S. Lab. Chip. 2010;10:957–985. doi: 10.1039/b917759k. [DOI] [PubMed] [Google Scholar]
- 51.Kim SJ, Li LD, Han J. Langmuir. 2009;25:7759–7765. doi: 10.1021/la900332v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim SM, Burns MA, Hasselbrink EF. Anal. Chem. 2006;78:4779–4785. doi: 10.1021/ac060031y. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y-C, Stevens AL, Han J. Anal. Chem. 2005;77:4293–4299. doi: 10.1021/ac050321z. [DOI] [PubMed] [Google Scholar]