SUMMARY
The nucleus is a critical subcellular compartment for the pathogenesis of polyglutamine disorders, including Huntington’s disease (HD). Recent studies suggest the first 17-amino-acid domain (N17) of mutant Huntingtin (mHTT) mediates its nuclear exclusion in cultured cells. Here, we test whether N17 could be a molecular determinant of nuclear mHTT pathogenesis in vivo. BAC transgenic mice expressing mHTT lacking the N17 domain (BACHD-ΔN17) show dramatically accelerated mHTT pathology exclusively in the nucleus, which is associated with HD-like transcriptionopathy. Interestingly, BACHD-ΔN17 mice manifest more overt disease-like phenotypes than the original BACHD mice, including body weight loss, movement deficits, robust striatal neuronal loss, and neuroinflammation. Mechanistically, N17 is necessary for nuclear exclusion of small mHTT fragments that are part of nuclear pathology in HD. Together, our study suggests that N17 modifies nuclear pathogenesis and disease severity in HD mice by regulating subcellular localization of known nuclear pathogenic mHTT species.
INTRODUCTION
HD is the most common inherited neurodegenerative disease, characterized by progressive movement disorders, psychiatric symptoms, and cognitive impairment (Ross et al., 2014). Disease symptoms in HD are relentlessly progressive and patients usually die from disease complications about 20 years after onset. HD neuropathology is characterized by brain atrophy, massive loss of striatal medium spiny neurons (MSNs) and, to a lesser extent, loss of cortical pyramidal neurons (CPNs; Vonsattel and DiFiglia, 1998). Moreover, modest atrophy in other brain regions (e.g. cerebellum) has also been detected. Importantly, neurodegeneration in HD is accompanied by neuroinflammation (Moller, 2010) and signs of systemic inflammation (Björkqvist et al., 2008). The precise mechanisms underlying the pathogenesis of neurodegeneration in HD remain poorly understood.
HD is caused by a CAG repeat expansion encoding an elongated polyglutamine (polyQ) repeat in the huntingtin protein (HTT; Huntington’s Disease Collaborative Research Group, 1993). HD belongs to a group of neurodegenerative disorders caused by polyQ expansion (Orr and Zoghbi, 2007). These disorders exhibit an inverse relationship between CAG repeat length and age of disease onset, and for most of these disorders, the mutant polyQ protein can misfold and form aggregates in the brain. In HD, mutant huntingtin (mHTT) accumulates as aggregates in both the cytosol and nucleus (DiFiglia et al., 1997). The roles of polyQ protein aggregation in disease pathogenesis appear complex, and there is evidence suggesting certain aggregates may be neuroprotective (Orr and Zoghbi, 2007).
Studies of several polyQ diseases suggest mutant proteins may primarily elicit disease pathogenesis through toxicities in the nucleus. In spinocerebellar ataxia 1 (SCA1) and spinal and bulbar muscular atrophy (SBMA), blocking nuclear translocation of mutant polyQ proteins abolishes disease pathogenesis in mouse models (Klement et al., 1998; Katsuno et al., 2002). Although HTT is predominantly located in the cytoplasm, it can enter the nucleus under certain conditions (Atwal et al., 2011). In HD, available evidence suggests that mHTT can elicit both nuclear and cytoplasmic pathogenesis (Ross and Tabrizi, 2011) and form aggregates that consist of small mHTT polyQ fragments (DiFiglia et al., 1997; Lunkes et al., 2002). Studies suggest artificially enhanced nuclear entry of mHTT polyQ fragments exacerbate disease phenotypes in vivo (Benn et al., 2005; Gunawardena et al., 2003), but the endogenous mechanisms regulating nuclear cytoplasmic trafficking of mHTT or its fragments in diseased brains remain unclear.
The N-terminal 17-amino-acid domain (N17) of HTT, located immediately preceding the polyQ domain, is an evolutionarily conserved domain among vertebrates (Tartari et al., 2008). In cultured cells, N17 can mediate both cytoplasmic membrane association (Rockabrand et al., 2006; Atwal et al., 2007) and Crm1-dependent nuclear export of HTT N-terminal fragments (Maiuri et al., 2013; Zhang et al., 2013), leading to predominantly cytoplasmic localization of these HTT species (Steffan et al., 2004). Interestingly, the N17 domain can promote formation of oligomers and aggregation of mHTT exon1 fragments in vitro (Thakur et al., 2009; Tam et al., 2009). Consistent with a role for this domain in mHTT proteostasis, deletion of the N17 domain or overexpression of its binding protein Tcp1 suppresses mHTT aggregation in vitro and in cells (Tam et al., 2009; Omi et al., 2008; Jayaraman et al., 2012). The N17 domain appears to be a regulatory hub for HTT, with more than 10 known post-translational modifications (PTMs) including phosphorylation, ubiquitination, sumoylation, and acetylation (Lee et al., 2013). Among these, the phosphorylation of serines 13 and 16 has been shown to reduce oligomerization and aggregation of mHTT fragments in vitro (Mishra et al., 2012), and phosphomimetic mutation of these residues suppresses mHTT toxicity in cells, brain slices, and in BAC HD mice (Gu et al., 2009; Thompson et al., 2009; Atwal et al., 2011).
Although N17 phosphorylation has been implicated in modulating mHTT toxicities in vivo (Gu et al., 2009), two important N17 functions (i.e. influencing aggregation and nucleocytoplasmic trafficking of mHTT) have not been evaluated in vivo. In this study, we created novel BAC transgenic mouse models expressing human mutant (97Q) or WT (31Q) HTT with deletion of residues 2–16 in the N17 domain to evaluate the role of the N17 domain in modifying mHTT-induced disease pathogenesis and nuclear toxicities in vivo.
RESULTS
Generation of BAC Transgenic Mice Expressing ΔN17 Forms of Human Huntingtin
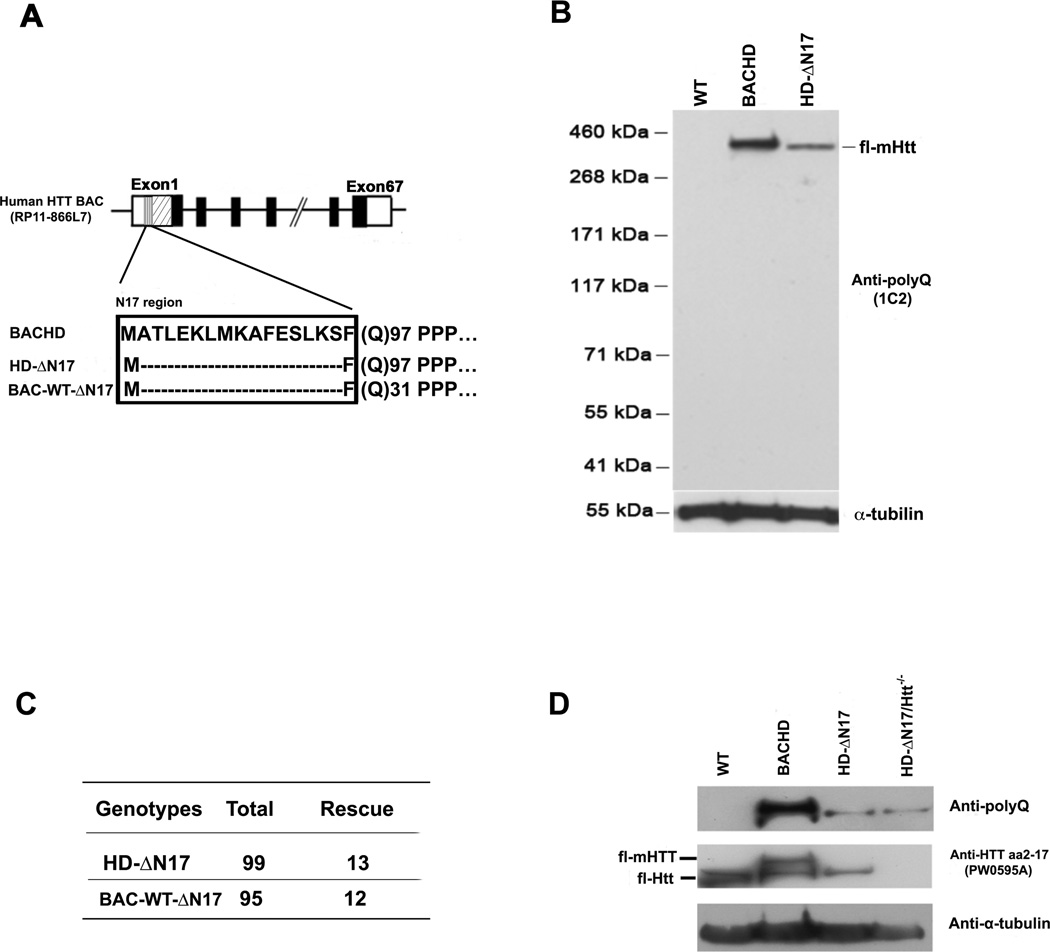
To create BAC transgenic mice expressing mutant and wildtype (WT) human HTT lacking the N17 domain, we first confirmed that deletion of residues 2–16 in N17 (ΔN17) in the context of WT or mutant HTT exon1 abolished the N17-mediated cytoplasmic localization in transfected cells (Figure S1A and S1B). We then engineered the human genomic BAC originally used to create the prior BACHD mouse models (Gray et al., 2008; Gu et al., 2009) to express ΔN17 forms of mutant (ΔN17-mHTT-97Q) or WT HTT (ΔN17-HTT-31Q; Figure 1A). These BACs were microinjected into inbred FvB/NJ embryos to generate transgenic mouse lines (N, L, and A lines) expressing ΔN17 forms of mHTT (Figures 1B, S1C and S1D). We also generated a control mouse line expressing ΔN17-HTT-31Q. Two of the three BACHD-ΔN17 lines (N and L) and the BAC-WT-ΔN17 line express comparable levels of human HTT, as assessed by Western blots using a human-HTT specific antibody (Dehay et al., 2007; Figure S1C) as well as an expanded polyQ specific antibody (1C2; Figures 1B and S1C). The level of human mHTT in BACHD-ΔN17 N line is about 25% that of the original BACHD mouse line (Figures 1B) and estimated to be about half the endogenous murine Htt level (Gray et al., 2008; Gu et al., 2009). The BACHD-ΔN17 models are suitable to assess, given the lower levels of mHTT expression, whether they show accelerated or exacerbated disease pathogenesis compared to BACHD. We focused our initial phenotypic study on the BACHD-ΔN17-N line (referred to hereafter as BACHD-ΔN17), with key phenotypes found in this line then evaluated in the other mouse lines.
Figure 1. Generation and Characterization of BAC Mice Expressing ΔN17 Forms of Huntingtin.
(A) Schematic of transgene construction depicting the constructs used in generating BACHD-ΔN17 and BAC-WT-ΔN17 mice. (B) Brain lysates of WT, BACHD and BACHD-ΔN17 mice were probed with anti-polyQ antibody to determine mHTT expression levels. The anti-α-tubulin antibody was used for loading control. (1C2). (C) BACHD-ΔN17 and BAC-WT-AN17 transgene can rescue embryonic lethality of homozygous Htt null mice. The panel shows the number of rescue mice among the total number of mice born. (D) An anti-HTT aa2-17 (PW0595A) antibody detects the N17 domain both human HTT and murine Htt. The anti-polyQ (1C2) antibody detects only human mHTT. Brain lysates of WT, BACHD, BACHD-ΔN17 and BACHD-ΔN17/Htt−/− mice were probed with 1C2 and anti-N17 (PW0595A) antibodies, with anti-α-tubulin as loading control. See also Figures S1.
N17 Domain Is Not Required for the Essential Function of Huntingtin in Murine Development
Htt is essential for murine embryonic development (Zeitlin et al., 1995), and BAC mice expressing full-length human mHTT or phosphomimetic N17 mutants can rescue the lethality phenotype in Htt null mice (Gray et al., 2008; Gu et al., 2009). To assess whether ΔN17 forms of HTT still retain essential HTT function during development, we crossed either BACHD-ΔN17 or BAC-WT-ΔN17 transgenes onto the murine Htt null background (Zeitlin et al., 1995) and found a Mendelian ratio of rescued mice (Figure 1C, 1D) that are indistinguishable from their WT littermates up to 2 months (i.e. 2m) of age. This study demonstrates that the N17 domain is not required for the essential function of Htt during murine embryonic development, and N17 deletion mutation does not appear to affect normal HTT function in vivo.
BACHD-ΔN17 Mice Exhibit Motor and Psychiatric-like Behavioral Deficits, Weight Loss and General Health Decline
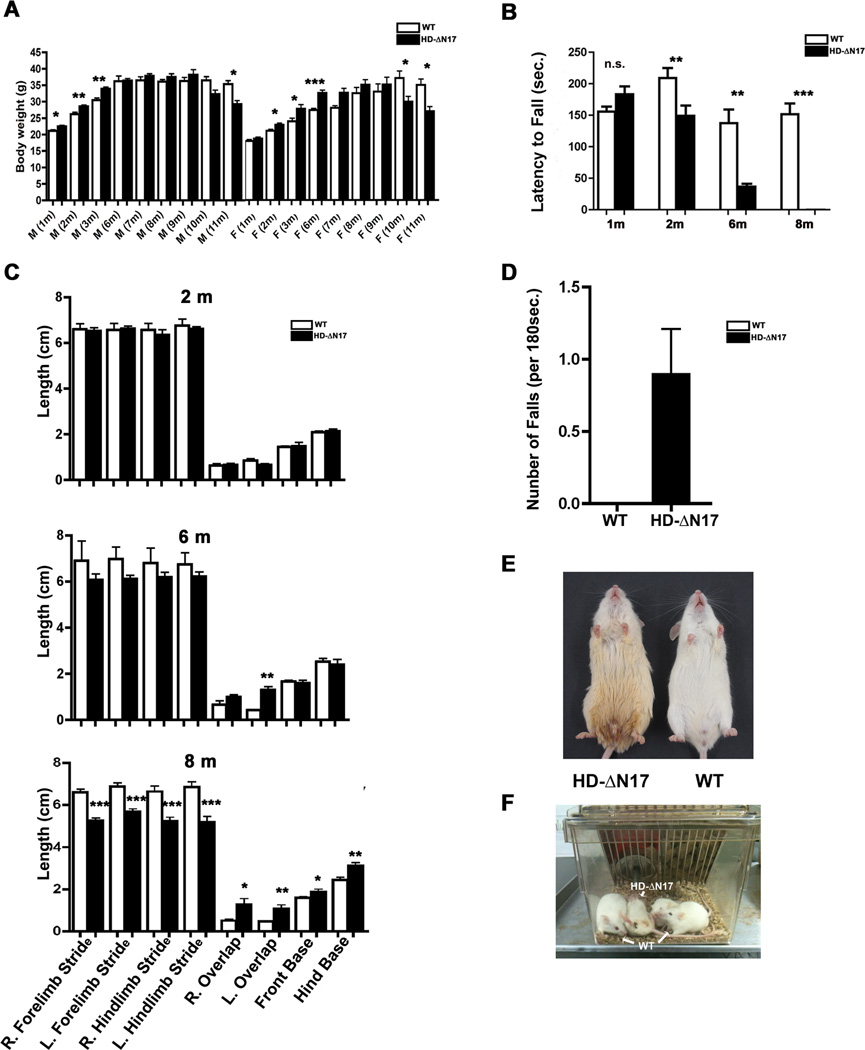
We next evaluated a cohort of BACHD-ΔN17 and WT littermates up to 11m of age for motor phenotypes and health status. We first evaluated body weight in these mice. Interestingly, both male and female transgenic mice are slightly heavier than WT at 2 and 6m of age, similar to other HD transgenic mice overexpressing human HTT (Gray et al., 2008). However, unlike BACHD mice, but similar to HD patients (Ross et al., 2014), BACHD-ΔN17 mice exhibit significant weight loss at 10m and 11m of age compared to WT (Figure 2A).
Figure 2. Age-Dependent Motor Deficits and General Health Decline in BACHD-ΔN17 Mice.
(A) Body weights of both male (M) and female (F) BACHD-ΔN17 mice and WT mice from 1m to 11m of age. Significant interaction between genotypes and age (F=3.32, p<0.01) for females and age (F=31.62, p<0.001) for males was detected (two-way ANOVA). Further Student’s t-tests at each age showed that both females and males transgenic mice gained weight before 6 months of age, before losing significant weight around 10 months of age (*p<0.05, **p<0.01, ***p<0.001). (B) Motor performance of BACHD-ΔN17 mice and WT littermates was measured using an accelerating rotarod test. (C) Gait analyses were performed at 2m, 6m and 8m of age (*p<0.05, **p<0.01, Student’s t-tests). (D) Spontaneous falls were counted in BACHD-ΔN17 mice and WT littermates at 10m of age over a period of 180 seconds. (E and F) BACHD-ΔN17 mice show unkempt fur and urine scalding at 10m of age (E) and lay upside-down in the home cage (F). Data are shown as mean ± SEM. Student’s t-tests were performed to compare the results of BACHD-ΔN17 and WT mice (*p<0.05; **p<0.01; ***p<0.001). See also Figures S2, S6, S7.
We next evaluated BACHD-ΔN17 mice for motor coordination using the accelerating rotarod test. The transgenic mice have normal rotarod performance at 1m of age (p = 0.625, Student’s t test), but exhibit progressive impairment at 2m and 6m of age (Figure 2B; two-way ANOVA, age and genotype interaction, p<0.0001; age, p<0.0001; genotype, p=0.005), similar to BACHD (Gray et al., 2008; Wang et al., 2014). However, unlike BACHD, which can still run rotarod at a compromised level at 12m of age, BACHD-ΔN17 mice can no longer run rotarod by 8m of age (Figure 2B), suggesting that BACHD-ΔN17 mice exhibit more severe motor deficits than BACHD.
We next evaluated spontaneous locomotion using the open field test (Wang et al., 2014). BACHD-ΔN17 mice showed normal locomotion at 2m of age but exhibit significant impairment in horizontal distance traveled (Two-way ANOVA, genotype and age interaction, p=0.0028; genotype, p=0.0229), horizontal speed, and vertical plane entry (rearing) at 8m of age (Figures S2A–S2C; Two-way ANOVA, genotype and age interaction, p<0.0001; genotype, p<0.0001).
HD patients exhibit psychiatric symptoms (Ross et al., 2014), and BACHD mice also show psychiatric-like behavioral deficits (Wang et al., 2014). Similar to BACHD, the BACHD-ΔN17 mice show significantly increased immobility in the forced swim test compared to WT mice at 5m of age (Figures S2D, p < 0.001, Student’s t test, n=10 per genotype), suggesting the presence of depression-like behaviors in these mice.
Gait abnormalities are common in HD patients and can lead to substantial morbidity (Ross et al., 2014). Impressively, BACHD-ΔN17 mice showed normal gait parameters at 2m and minimal deficits at 6m, but significant gait abnormalities at 8m of age (Figure 2C). Severe gait impairment in HD patients often results in spontaneous falls (Grimbergen et al., 2008), a phenotype not reported in previous HD models (Crook and Housman, 2011). To our surprise, starting around 10m of age, BACHD-ΔN17 but not WT mice show frequent spontaneous falls (Supplemental Video #1; Figure 2D). Consistent with progressive neurological decline in this model, BACHD-ΔN17 but not WT mice at 10 to 11m of age are frequently found lying upside down in their home cage, poorly groomed and with discoloration of fur due to urine scalding (Figure 2E, 2F; Supplemental Video #1).
Since BACHD-ΔN17 mice exhibit HD-like progressive motor impairment and health decline, which are either more severe than or absent in BACHD mice, we conclude that BACHD-ΔN17 mice exhibit more severe disease-like phenotypes than BACHD.
BACHD-ΔN17 Mice Exhibit Adult-onset Overt Movement Deficits and Aberrant Striatal Neuronal Activities
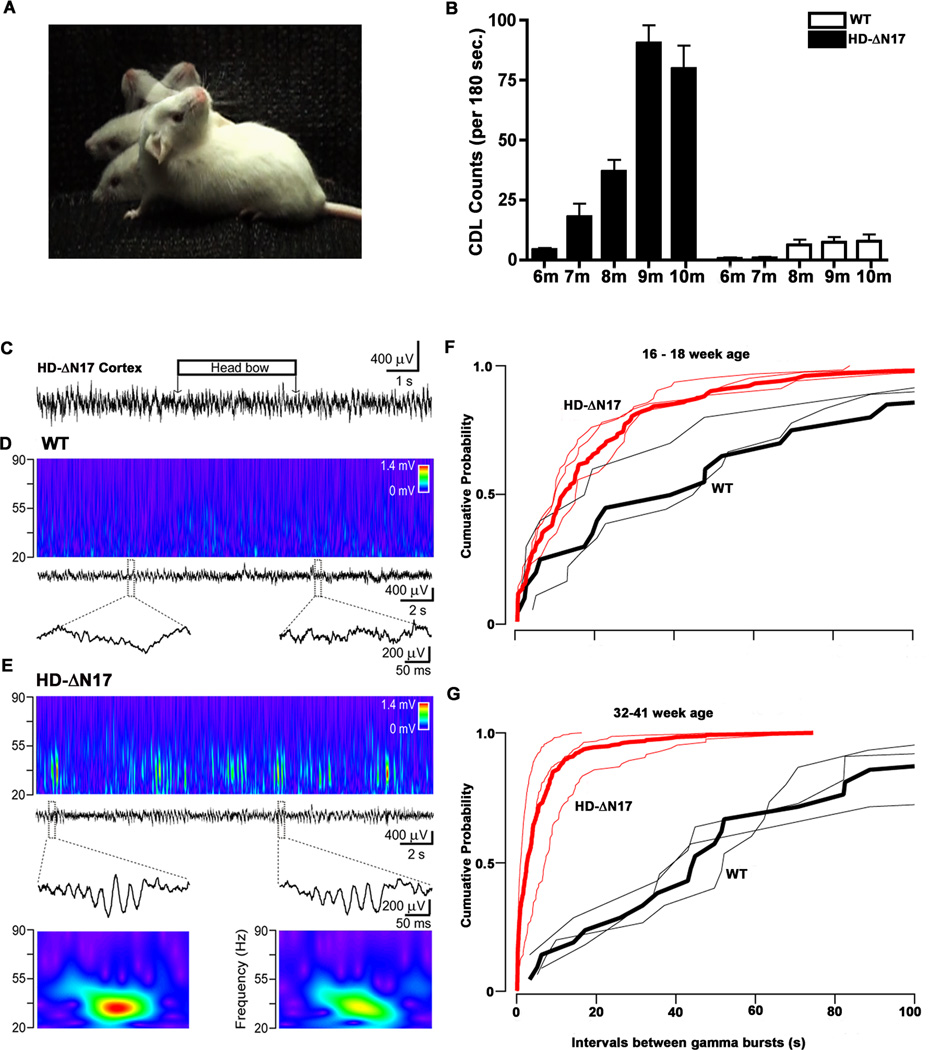
A clinical hallmark in HD is adult-onset movement disorder, including chorea and dystonia (Ross et al., 2014), a phenotype that has largely been absent in previous HD mouse models (Crook and Housman, 2011). When we visually observed 11m old mice, we were surprised to find that BACHD-ΔN17 but not WT mice show frequent abnormal head movements consisting of smooth but non-rhythmic side-to-side swinging of the head and upper body, reminiscent of choreiform movement, and also sustained retroflex of head and neck lasting up to a few seconds, reminiscent of cervical dystonia (Figure 3A; Supplemental Videos #1 and #2). The onset of abnormal head movements in BACHD-Δ17 mice is age-dependent and progressive. These movements are absent in transgenic mice at 5m of age (Supplemental Videos #3), have onset around 7m of age, and progressively increase in frequency by roughly 10-fold between 7 and 10m (Figure 3B; Supplemental Videos #4). These phenotypic characteristics appear to have some face validity for the overt movement disorder manifested in HD, and hence we refer to the abnormal movement in BACHD-Δ17 mice as Chorea/Dystonia-Like (CDL) movement deficits.
Figure 3. Progressive Chorea/Dystonia-Like Movement Deficits and Aberrant Striatal Neuronal Activities in BACHD-ΔN17 Mice.
(A) Composited image depicting chorea/dystonia-like (CDL) head movement in BACHD-ΔN17 mouse. (B) Quantification of CDLs in 3 minutes of video recording. Two-way ANOVA reveals significant difference between BACHD-ΔN17 and WT littermates in age and genotype interaction: F=16.55, p<0.0001; phenotype: F=117.2, p<0.0001. Data are shown as mean ± SEM. (C) Cortical field potential recording showing no epileptic activity during chorea-like movement (horizontal box over the recording) in an 8-month-old BACHD-ΔN17 mouse. (D) There is no significant gamma activity in the dorsal striatum of WT littermates. Middle: local field potential recording in the dorsal striatum of a WT littermate. Top: corresponding wavelet transform shows minor activity in the low gamma band (20–50 Hz). Bottom: enlarged segments of the field potential recordings indicated by dashed boxes. (E) Frequent gamma events in BACHD-ΔN17 mice. From top second row: local field potential recording in the dorsal striatum. Top: corresponding wavelet transform shows transient increments in the gamma range. Third row: sample large gamma events, enlarged segments of the local field potential recording indicated by dashed boxes with the corresponding wavelet transform (bottom). (F and G) Progression of the large gamma events in the striatum with aging in BACHD-ΔN17 mice. Cumulative probability plot of inter-event interval in younger (F) and older (G) animals. Red: transgenic, black: wild type, thicker lines indicate the corresponding pooled cumulative probability. See also Video clips 1 to 5, Figures S6, S7.
Abnormal head-tossing movements, which are associated with cortical spike-wave epilepsy, have been shown in mice with mutated voltage-dependent calcium channels (Burgess and Noebels, 1999). However, using synchronized video monitoring and cortical local field potential recording in 5-hour-long sessions with 7–9 month old BACHD-Δ17 mice, we did not observe any epileptic activity during the entire recording period, despite frequent CDLs seen in the mutant mice (Figure 3C). Interestingly, local field potential recordings in the cortex and dorsal striatum of BACHD-Δ17 mice revealed synchronous, short duration (100–200ms), large-amplitude events in the low gamma range (25–55 Hz; Figure 3E; data not shown), while WT littermates showed only minor gamma activity (Figure 3D). This finding is consistent with prior observation of increased low gamma frequency events in the striatum of other HD mouse models (e.g. Cayzac et al., 2011). To investigate whether the abnormal striatal gamma frequency activity in the BACHD-Δ17 mice is progressive, we recorded younger transgenic mice (16–18 weeks of age) and found that the mutant mice already show significant differences in gamma frequency events from WT littermates (Kolmogorov-Smirnov test, p = 0.0078), but importantly such differences progressively worsen with age (Kolmogorov-Smirnov test, p = 5.8 × 10−11; Figure 3F and 3G). Our study shows that CDLs are not due to cortical epileptic activity and are associated with an age-dependent increase in the abnormal striatal gamma frequency activities in BACHD-ΔN17 mice.
Selective Cortical and Striatal Atrophy, Striatal Neuronal Loss and Reactive Gliosis in BACHD-ΔN17 Mice
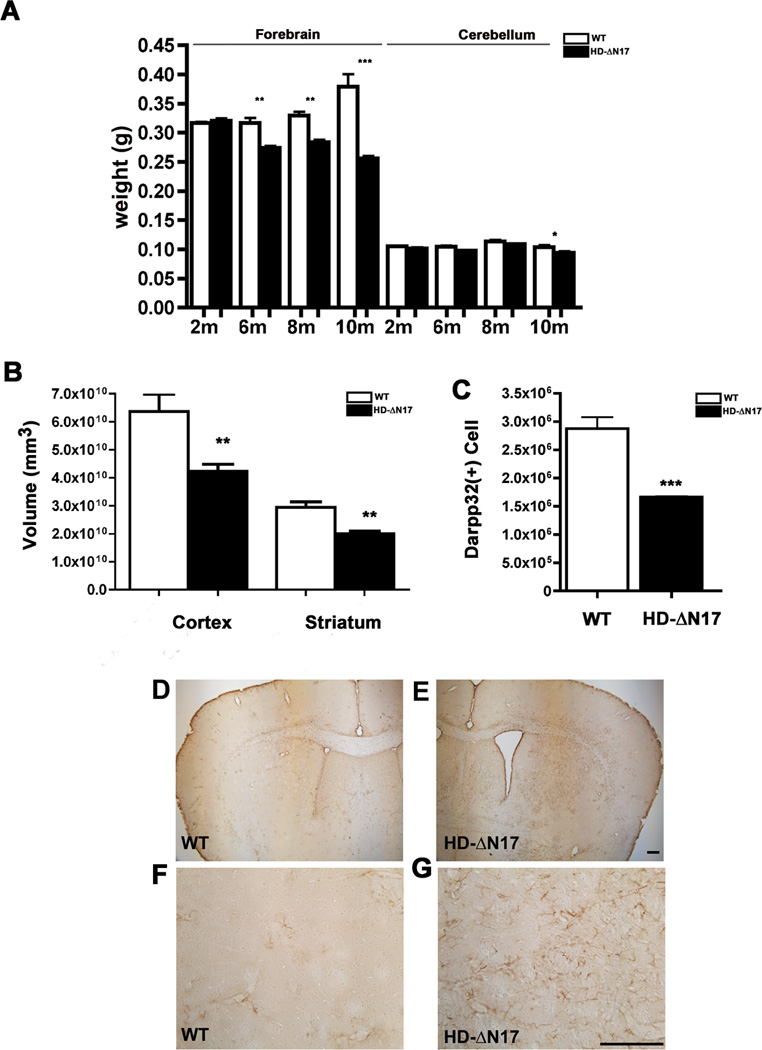
HD neuropathology is characterized by selective cortical and striatal atrophy with massive striatal neuronal loss, but modest atrophy of other brain regions, such as the cerebellum (Vonsattel and Difiglia, 1998). Existing HD mouse models often recapitulate the brain atrophy but lack robust striatal neuronal loss (Crook and Housman, 2011). We evaluated BACHD-ΔN17 mice for HD-like neurodegeneration phenotypes. The transgenic mice have similar forebrain weight compared to WT littermates at 2m, but at 6–10m of age BACHD-ΔN17 mice show progressive and significant forebrain weight loss (Figure 4A; two-way ANOVA, genotype and age interaction, p < 0.0001; genotype, p = 0.0039; age, p < 0.0001). Impressively, at 10m of age, BACHD-ΔN17 mice have 34% forebrain weight loss compared to WT. Moreover, the brain atrophy in BACHD-ΔN17 is relatively selective since the cerebellum shows only modestly significant weight loss at 10m of age and not younger (Student’s t test, p<0.05; Figure 4A). We next performed unbiased stereology and found significant cortical (p < 0.005, Student’s t test) and striatal (p < 0.001, Student’s t test) volume loss in 10m old BACHD-ΔN17 mice compared to WT littermates (Figure 4B). Importantly, using stereological counting of DARPP-32 immunoreactive MSNs, we found that BACHD-ΔN17 mice exhibit robust loss of striatal MSNs (about 40%) at 10m of age (Figure 4C; N = 5 per genotype, p < 0.001, Student’s t test). Finally, since neurodegeneration in HD caudate/putamen is accompanied by reactive gliosis (Vonsattel and DiFiglia, 1998), we examined BACHD-ΔN17 striatum and cortex at 10m of age and found reactive astrocytes with elevated GFAP expression and hypertrophic processes (Figure 4D–4G). We conclude that BACHD-ΔN17 mice exhibit progressive HD-like neurodegenerative pathology, including relatively selective cortical and striatal atrophy, striatal MSN loss, and reactive gliosis.
Figure 4. Progressive and Selective Brain Atrophy, Striatal Neuronal Cell Loss, and Gliosis in BACHD-ΔN17 Mice.
(A) Weight of the forebrain and cerebellum of BACHD-ΔN17 and WT littermates was measured at 2, 6, 8 and 10 months of age. Progressive forebrain weight loss is detected in BACHD-ΔN17 mice starting at 6 months of age and progresses between 6m and 10m of age Two-way ANOVA reveals significant differences between HD-AN17 and WT littermates in age and genotype interaction (F=49.94, p<0.0001; genotype: F=173.2, p<0.0001). No weight-loss is detected in the cerebellum until 10m of age. (B) Unbiased stereological measurement of brain regions was performed to measure the cortical and striatal volume in BACHD-ΔN17 mice and WT littermates. (C). Unbiased stereological counting revealed robust loss of Darpp32+ neurons in 10m old BACHD-ΔN17 mice compared to WT controls. (D – G) Reactive astrogliosis is detected in striatum and deep cortical layers of BACHD-ΔN17 mouse brain with GFAP immunohistochemical staining (E, G). No such gliosis is seen in control mouse brain (D, F). Data are shown as mean ± SEM. Student’s t-tests were performed to compare BACHD-ΔN17 and WT samples (*p<0.05; **p<0.01; ***p<0.001). Scale bar = 100 µm. See also Figures S3, S6, S7.
BACHD-ΔN17 Mice Exhibit HD-like Peripheral Pathology
HD is characterized by certain peripheral pathology, such as muscle atrophy and testicular degeneration (van der Burg et al., 2009). To assess peripheral pathology, we evaluated the weights of multiple peripheral organs and tissues in BACHD-ΔN17 and WT mice at 10m of age (Figure S3A–S3C). Importantly, the weights of multiple peripheral organs (i.e. heart, spleen, kidney, and liver) in the transgenic mice are similar to WT mice at this relatively advanced disease stage (Figure S3A), suggesting the toxicity elicited by ΔN17-mHTT is not ubiquitous. We next evaluated the BACHD-ΔN17 mice for the testicular degeneration phenotype of HD (Van Raamsdonk et al., 2007), finding marked testicular atrophy in the mutant mice (Fig. S3B) associated with age-dependent infertility starting at around 5m. Finally, BACHD-ΔN17 mice also show quadriceps muscle pathology (Figure S3D–S3G), including significant atrophy, reduction of muscle fiber caliber, and aberrant central localization of nuclei within the fiber (Sopher et al., 2004). There results show that BACHD-ΔN17 mice recapitulate several aspects of peripheral pathology found in HD.
BACHD-ΔN17 Mice Show Accelerated and Exclusively Nuclear mHTT Pathology
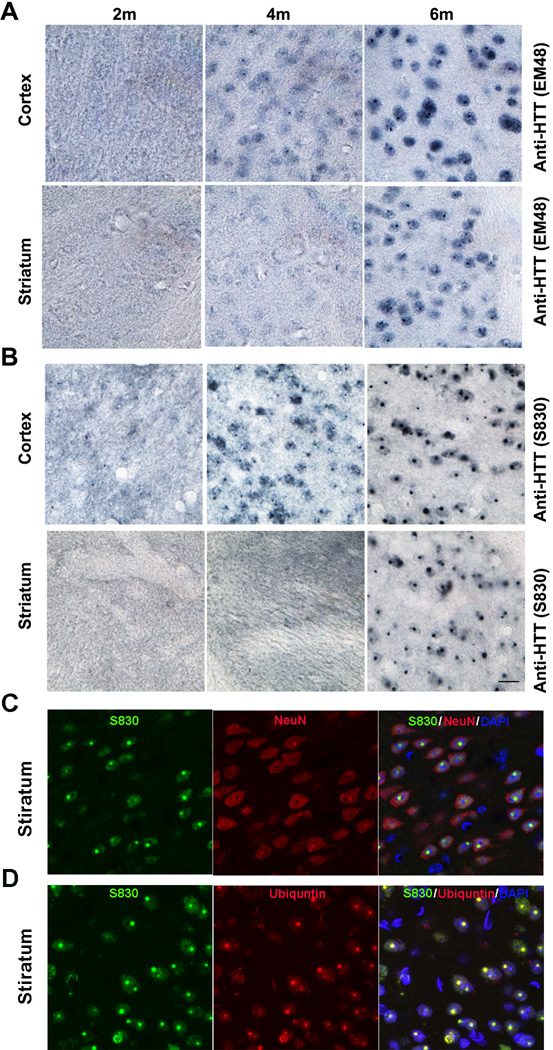
A marker of HD neuropathology is the presence of aggregated mHTT polyQ fragments in the nucleus and cytoplasm (DiFiglia et al., 1997). Using two human mHTT-exon1-specific antibodies that are sensitive to aggregated mHTT (S830 and EM48; Sathasivam et al., 2001; Gutekunst et al., 1999), we characterized patterns of mHTT aggregation in BACHD-ΔN17 mice (Figure 5). We did not detect any mHTT aggregates in 2m old transgenic brain sections. At 4m of age, BACHD-ΔN17 mice show diffuse mHTT nuclear staining, as well as small nuclear inclusions (NIs), in the cortex but rarely in the striatum (Figure 5A, 5B). At 6m and older, BACHD-ΔN17 mice show both diffuse nuclear staining and prominent nuclear inclusions (NIs) in both cortex and striatum (Figure 5A, 5B). Interestingly, the mHTT aggregates are exclusively found in the nucleus, with cytoplasmic aggregates not found in the transgenic mice (Figure 5C, 5D). Moreover, the nuclear mHTT aggregate pathology can also be found in other brain regions at 6m and 10m of age, including the hippocampus, thalamus, cerebellum, and brain stem (Figure S4). Double immunofluorescent staining showed that mHTT aggregates are almost exclusively in NeuN+ neurons in the striatum (Figure 5C) and that the NIs are also immunoreactive for ubiquitin (Figure 5D), a pathological feature similar to HD (DiFiglia et al., 1997) but absent in BACHD mice (Gray et al., 2008; data not shown). Our result demonstrates that N17 is a critical determinant of the rate and subcellular localization of mHTT aggregate pathology in vivo.
Figure 5. Deletion of N17 Selectively Accelerates mHTT Nuclear Accumulation and Aggregation in BACHD-ΔN17 Mouse Brains.
(A and B) Immunohistochemistry with EM48 (A) and S830 (B) antibodies to detect mHTT nuclear accumulation and aggregation in 2m, 4m and 6M old BACHD-ΔN17 mouse cortex and striatum. (C). Immunofluorescent staining to simultaneously detect S830+ mHTT aggregates (green), NeuN+ neurons (red), and merged signals with DAPI nuclear staining (Yellow and Purple). (D). Immunofluorescent staining to simultaneously detect S830+ mHTT aggregates (green), Ubiquitin+ nuclear inclusions (red), and merged signals with DAPI nuclear staining (Yellow and Purple). Scale bar = 50 µm. See also Figures S4, S6, S7.
Evidence Supports Selective Requirement of N17 to Mitigate Nuclear Translocation and Aggregation of Small mHTT PolyQ Fragments
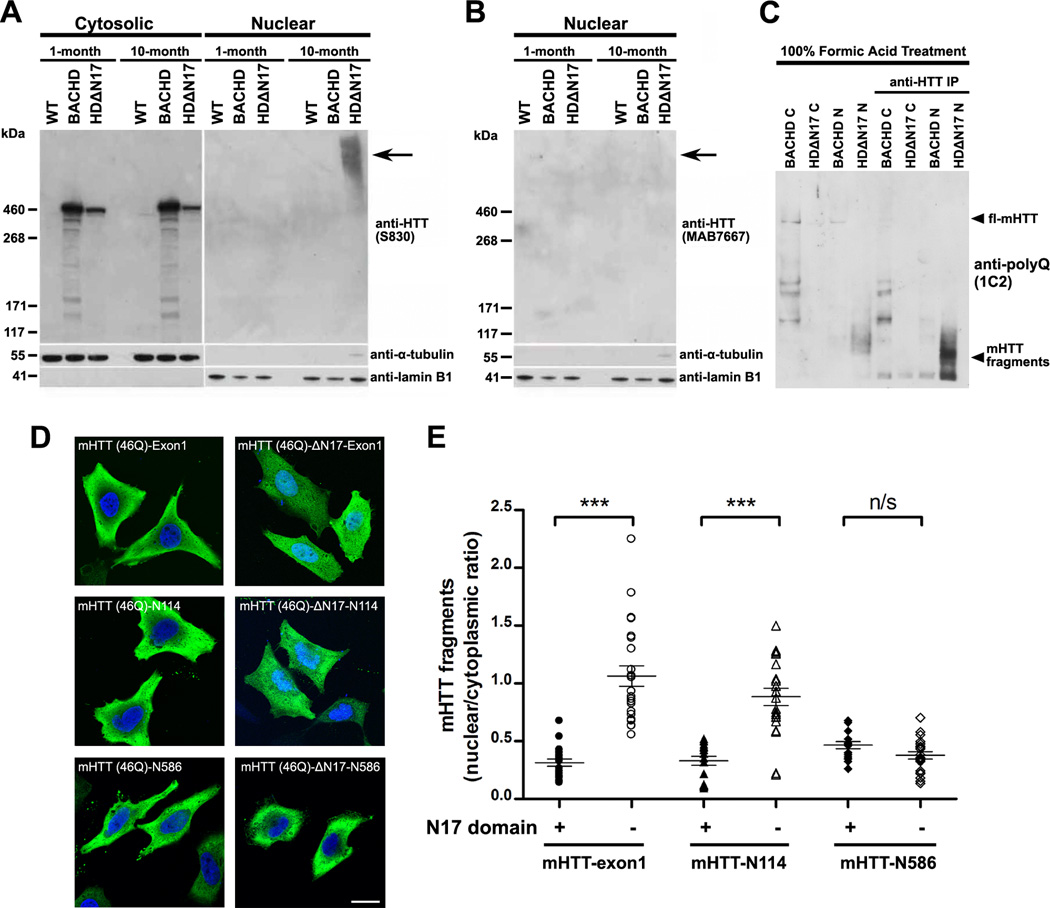
To obtain biochemical evidence that N17 is crucial for nuclear exclusion of mHTT in vivo, we performed subcellular fractionation of nuclear and cytoplasmic protein extracts from 1m and 10m–old BACHD, BACHD-ΔN17, and WT control mice (Figure 6A–6C). Prior studies in transfected cells suggest that certain N17 mutations (e.g. M8P) can lead to increased nuclear translocation of fragment or full-length HTT (Atwal et al., 2007), suggesting N17 may be required for cytoplasmic targeting of all HTT species. To our surprise, we found that in both 1m and 10m old mouse forebrains, the soluble ΔN17-mHTT (full-length mHTT lacking only 2–16 residues) is distributed almost exclusively in the cytosol, a pattern similar to full length mHTT in BACHD (Figure 6A). Interestingly, at 10m but not at 1m of age, high molecular weight (HMW) mHTT species accumulate exclusively in the nucleus of BACHD-ΔN17 mice but not in BACHD or WT controls (Figure 6A). Similar mHTT HMW species have been found in brains of HD patients and mouse models (DiFiglia et al., 1997; Landles et al., 2010; Lunkes et al., 2002). Consistent with the notion that such HMW mHTT species are aggregated mHTT N-terminal fragments, we found that they can be detected with antibodies against mHTT-exon1 epitopes (Figure 6A), but cannot be stained with an antibody against a C-terminal HTT epitope (Figure 6B; Shirasaki et al., 2012). Moreover, by using formic acid to dissociate the nuclear mHTT aggregates (Lunkes et al., 2002), we found that nuclear aggregates in BACHD-ΔN17 mice appear to consist predominantly of two small mHTT polyQ fragments of around 55kD (Figure 6C). Thus, our biochemical analyses provide strong evidence that the absence of N17 in the context of otherwise full-length mHTT results in a dramatic acceleration of nuclear accumulation and aggregation of small mHTT N-terminal fragments in vivo.
Figure 6. N17-Dependent Nuclear Accumulation of mHTT In Vivo and In Vitro.
(A and B) Western blot analysis of cytosolic and nuclear fractions from the forebrain extracts from 1m and 10m old WT, BACHD, and BACHD-ΔN17 mice. The blot was probed with antibodies against (A) the N-terminal human mHTT exon-1 (S830), and (B) the C-terminal HTT (MAB7667; Shirasaki et al., 2012). Lamin B1 and α-actin served as loading controls for nuclear and cytosolic fractions, respectively. (C) Nuclear (N) and cytosolic (C) extracts of forebrains from 1 and 10 month old WT, BACHD, and BACHD-ΔN17 were treated with 100% formic acid to dissolve the SDS-insoluble mHTT aggregates (Lunkes et al, 2002), followed by Western analysis. For better detecting the presence of mHTT fragments shown in lanes 5–8, mHTT were immunoprecipitated and detected by an anti-polyQ antibody (1C2). (D) HEK-293 cells were transfected with plasmids carrying mHTT fragments of different sizes, i.e. Exon1, CpA (N114), Caspase 6 (N586), and either with (left) or without N17 (right, deletion of amino acid residues 2–16). All constructs of fragments were fused at C-terminal with HA-tag and are as labeled on the images. The subcellular localization of mHTT was revealed by immunostaining of the HA-tag and imaged by confocal microscopy. (E) The nuclear and cytosolic distribution of mHTT fragments in (D) was quantified based on an established protocol (Ch'ng et al., 2012). The ratios of mean intensity of nuclear and cytosolic fluorescent signals of individual cells were plotted and analyzed using Student’s t test. *** p < 0.001; Scale bar = 20 µm. See also Figure S8.
To further explore the discrepancy between the in vitro (Thakur et al., 2009; Tam et al., 2009) and our in vivo findings on the impact of N17 on the rate of mHTT aggregation, we used a cell-based FRET assay in transfected primary neurons to study the difference between intact N17 and ΔN17 in modifying the aggregation of mHTT-exon1 (Zhang et al., 2013). Consistent with prior in vitro studies, we found that overall levels of aggregation were significantly reduced in the ΔN17 mutants compared to mHTT-exon1 with intact N17 (Figure S5A–S5B). However, similar to our in vivo finding, deletion of N17 leads to significantly increased nuclear aggregation (Figure S5C).
Since our studies showed that fl-mHTT does not require N17 for its predominant cytoplasmic localization in vivo, we next asked what sizes of mHTT N-terminal polyQ fragments are dependent on N17 for their nuclear exclusion, at least in cultured cells. We focused on three mHTT fragments that have been implicated in HD pathogenesis: the caspase 6 (C6) fragment, consisting of the N-terminal 586 amino acid residues of mHTT (Graham et al., 2006); CpA fragment, with N-terminal 114 residues (Lunkes et al, 2002); and Exon-1 (Sathasivam et al., 2013). Importantly, both CpA and Exon1 fragments have been shown to be part of mHTT nuclear aggregate pathology in HD (Lunkes et al, 2002; Landles et al., 2010). We generated plasmid constructs overexpressing these mHTT-46Q fragments either with or without N17 and transiently transfected them into HEK-293 cells (Figure 6D). By quantifying the ratio of nuclear vs. cytoplasmic mHTT fragment signals in the transfected cells (Ch'ng et al., 2012; Figure 6E), we found that the C6 fragments either with or without N17 are located predominantly in the cytoplasm, but both the mHTT-exon1 and CpA fragments appear to depend on N17 for nuclear exclusion, since deletion of N17 in these fragments leads to significant increase in their nuclear localization (Fig. 6D-6E). Together, our study provides the initial evidence that N17 appears to be selectively required for nuclear exclusion of certain small mHTT fragments that are known to be nuclear pathogenic in HD.
Phenotypic Characterization of Additional BACHD-ΔN17 and BAC-WT-ΔN17 Mouse Lines
We next studied additional BACHD-ΔN17 mouse lines to ensure key phenotypes in BACHD-ΔN17 mice are reproducible in independent lines. The BACHD-ΔN17-L line, with comparable ΔN17-mHTT expression levels as BACHD-ΔN17 (Figure S1C–S1D), exhibits significant body weight loss at 5m of age (Figure S6A). These mice show more aggressive disease, with premature lethality at around 5m of age (Figure S6B). Importantly, BACHD-ΔN17-L mice also show CDLs starting at 4–5m of age (Figure S6C; Supplemental Video #5) and forebrain weight loss with modest cerebellar weight loss (Figure S6D). Finally, immunostaining of cortical and striatal sections reveals robust mHTT nuclear aggregation in transgenic but not WT mice (Figure S6E; data not shown). Thus, multiple disease features of BACHD-ΔN17 are reproduced in the L line mice.
A third BACHD-ΔN17 mouse line that we have analyzed is the A line. These mice express very little ΔN17-mHTT, which is not detectable in the soluble fraction but can be detected as a faint band in the insoluble fraction (Figure S6F). Interestingly, the A line mice appear indistinguishable from WT littermates until 18m of age, when they show significant body weight loss (Figure S6G), abnormal and progressive CDLs (Figure S6H), and significant forebrain and cerebellar atrophy (Figure S6I). Moreover, mHTT aggregate pathology is absent at 6m but progressively increases between 18m and 24m in the A line mice (Figure S6J). Together, our study suggests that a very small amount of ΔN17-mHTT is sufficient to elicit multiple disease-like features in vivo, albeit with much later age of disease onset.
Finally, we analyzed BAC-WT-ΔN17 mice that express ΔN17-HTT-31Q at a level comparable to BACHD-ΔN17 (Figure S1C–S1D). The transgenic mice do not show body weight loss by 12m of age (Figure S7A), nor any brain or organ atrophy up to 19m of age (Figure S7B–S7C). The transgenic mice are comparable to WT littermates in rotarod test (Figure S7D) and do not exhibit CDLs (Supplemental Video #6) up to 12m of age. Finally, the transgenic mice do not show nuclear HTT pathology by 12m (Figure S7E–S7F). Thus, we conclude that HD-like phenotypes elicited by ΔN17 forms of mHTT are polyQ-length dependent.
Gene Expression Profiling and Network Analyses Reveal HD-like Transcriptional Profiles and Progressive Neuroinflammation in BACHD-ΔN17 Striatum
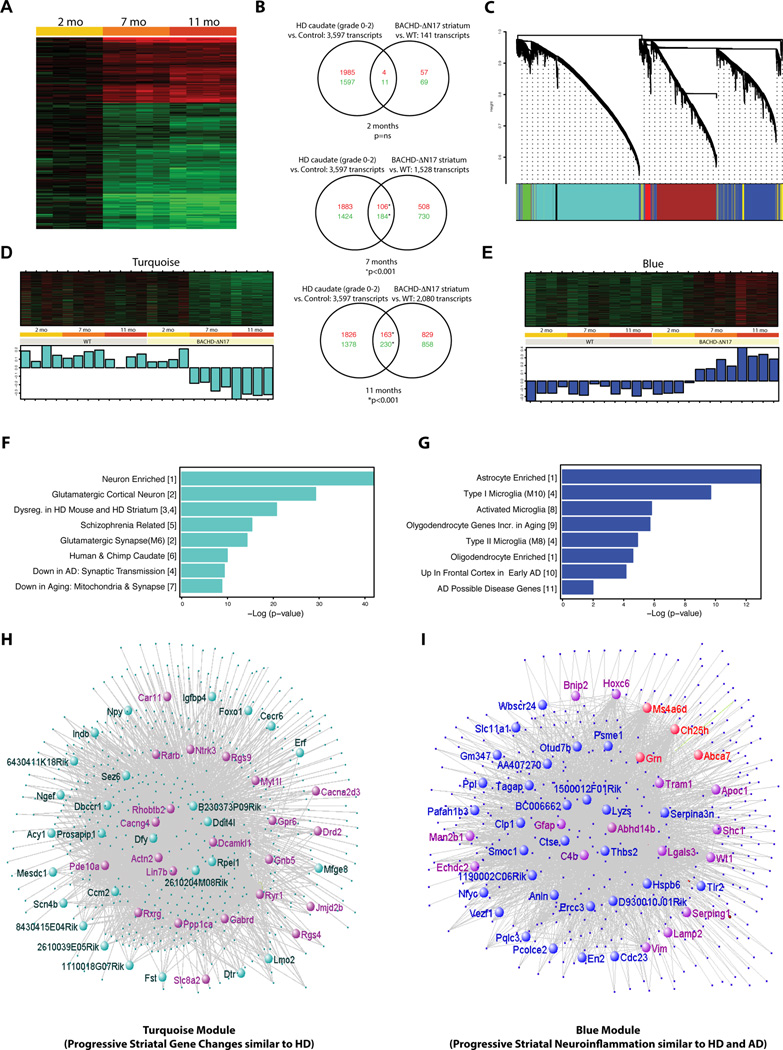
A molecular signature of mHTT nuclear pathogenesis is transcriptional dysregulation (Hodges et al., 2006; Ross and Tabrizi, 2011). To assess whether accelerated nuclear mHTT accumulation in BACHD-ΔN17 mice could also induce transcriptional signatures similar to HD, we used microarrays to profile progressive striatal gene expression changes in BACHD-ΔN17 and WT control mice. We identified a set of time-dependent gene expression changes when comparing BACHD-ΔN17 and WT mice at 2m, 7m and 11m (Supplemental Table 1). We then compared the gene expression profile in BACHD-ΔN17 striatum with that observed in the postmortem HD caudate/putamen (Hodges et al., 2006; Kuhn et al., 2007) and found a progressive overlap for both down- and up-regulated genes (Figure 7A). This overlap was significant at 7m and 11m, but not 2m (Figure 7B; hypergeometric test), suggesting BACHD-ΔN17 mice exhibit progressive HD-like transcriptionopathy.
Figure 7. Progressive HD-like Transcriptional Dysregulation and WGCNA Analyses of Gene Expression Profiles in BACHD-ΔN17 Striatum.
(A) Heat map representing differentially expressed genes at different ages in the striatum of BACHD-ΔN17 compared to WT mice. Up-regulated genes are shown in red and down-regulated genes in green; color intensity corresponds to fold change in expression. (B) Venn diagrams representing the overlap between genes previously reported as dysregulated in HD patients (Kuhn et al., 2007, left circles) and genes dysregulated in the BACHD-ΔN17 model (this study, right circles). An increasing degree of overlap between the two datasets is observed in both up-regulated (red numbers) and down-regulated (green numbers) genes as the BACHD-ΔN17 animals age (p = hypergeometric test). (C) Cluster dendrogram generated by hierarchical clustering of probes on the basis of topological overlap. Seven modules of highly co-expressed transcripts were identified using a dynamic cutting algorithm, and color-coded modules (Blue, Red, Green, Turquoise, Brown, and Pink respectively). (D) Heatmap and first principal component of expression data for genes in the Turquoise and Blue modules identified age and genotype-dependent, coordinated changes in gene expression within these two modules. (E) Enrichment analyses indicate overrepresentation of neuronal genes and genes implicated in other neurodegenerative disorders (e.g. schizophrenia, AD) in the Turquoise module, and of gene transcripts related to neuroinflammation, including microglia and astrocyte markers, in the Blue module. (F) Network visualization of the Turquoise and Blue modules with top hubs highlighted (inner circle). Genes already shown to have transcriptional changes in HD caudate (Hodges et al., 2006) are depicted in purple. Genes involved in AD and frontal temporal dementia (e.g. Grn) are depicted in red.
To further illuminate the molecular networks dysregulated in the longitudinal transcriptional profiles from the BACHD-ΔN17 and WT brains, we analyzed our entire gene expression dataset using Weighted Gene Coexpression Network Analysis (WGCNA; Zhang and Horvath, 2005). This analysis allows the identification of highly co-expressed gene groups (modules) and their central hubs (the most connected genes in the module), and its application to both normal and diseased brain transcriptomes is highly informative in identifying gene networks and in helping generate novel hypotheses (Geschwind and Konopka, 2009). WGCNA identified seven main modules, ranging between 63 and 3,247 probes in size (Figure 7C; Supplemental Table 1). We correlated the consensus gene expression pattern within each module (“module eigengene” or ME, Oldham et al., 2008), with genotype and age, and identified two modules with consistent genotype- and age-dependent changes in gene expression. The Turquoise module (Figure 7D, 7F, 7H) revealed a cluster of 3,247 genes that are unchanged in WT striatum at 2m, 7m, and 11m of age, but are progressively reduced in expression at 7 and 11 months in BACHD-ΔN17 striatum. The 2,141 genes in the Blue module (Figure 7E, 7G, 7I) showed the opposite pattern, with WT unchanged over time and age-dependent progressive increase in the BACHD-ΔN17 striatum.
Gene set enrichment analysis (Miller et al., 2011) of the Turquoise module (Figure 7F) shows significant enrichment of neuron-specific genes, glutamatergic synapse transcripts, genes enriched in the human/chimp caudate, and genes changed in R6/2 HD mice and HD patients (Figure 7F; Woodman et al., 2007). This module contains many of the well-known MSN marker genes that are down-regulated in HD caudate/putamen, including Drd2, Drd1, Gpr6, Actn2, Pde10a, Adora2a, Ppp1r1b (Darpp-32), Cnr1, and Rgs9 (Figure 7H; Supplemental Table 2). The down-regulation of multiple Turquoise module genes was confirmed by quantitative RT-PCR (Figure S8A) and by quantifying immunostained brain sections to assess protein levels (i.e. Drd2 and Drd1; Figure S8B–S8D). This latter finding suggests that Turquoise module genes do not simply reflect the loss of striatal MSNs in the BACHD-ΔN17 mice, but also neuronal dysfunction, since the surviving MSNs show reduction of dopamine receptor levels.
Importantly, the hub (i.e. most connected) genes in the Turquoise module highlight novel candidates that may be relevant to striatal MSN dysfunction and degeneration in the BACHD-ΔN17 model (Figure 7H). The top hub gene in Turquoise model is Actinin-2, a post-synaptic density (PSD) protein enriched in the striatum and cortex (Dunah et al., 2000) and that complexes with NMDA receptors (Wyszynski et al., 1997). Our recent study revealed that striatal Actn2 reduction in BACHD striatum is due to both non-cell-autonomous toxicities of cortical mHTT as well as cell-autonomous toxicities of striatal mHTT (Wang et al., 2014). Examples of other Turquoise module hub genes that could be candidates to study their roles in HD pathogenesis include, but are not limited to, Pde10a, Pde2a, Dcamkl1, Gabrd, Cacng4, Ddit4l, Ryr1, Igfbp4, Ntrk3, Gpr6, Rxrg1, Rarb (Figure 7H).
Blue module genes are progressively up-regulated in BACHD-ΔN17, but not WT striatum. Gene set enrichment analysis reveals a significant over-representation of genes specific for astrocytes, microglia, and oligodendrocytes (Figure 7G). We also confirmed the up-regulation of several Blue module genes in aged BACHD-ΔN17 striatum using quantitative RT-PCR (Figure S8A). Consistent with the interpretation that the Blue module consists of genes involved in progressive neuroinflammation, the module contains well-known marker genes involved in reactive astrocytosis (e.g. GFAP and S100a6; Figure 4D-4G) and microgliosis (e.g. Lgals3/Mac-2, Tspo, and CD68; Figure 7I; Supplemental Table 2; Moller, 2010). To confirm the presence of progressive microgliosis in the BACHD-ΔN17 mice between 6 and 10m of age, we performed immunostaining for Lgals3 (Mac-2/Galectin-3), a marker protein that is selectively up-regulated in activated microglia during brain injury and neurodegenreative disease (e.g. AD; Rotshenker, 2009). We found that Lgals3 staining is apparent only in BACHD-ΔN17 but not WT striatum at 11m of age and is restricted to Iba1+ microglia with hypertrophic (reactive) morphology (Figure S8E–S8K), and they are progressively increase in BACHD-ΔN17 striatum between 6m and 11m of age.
Since the Blue module is enriched in genes involved in neuroinflammation, it is interesting to note that multiple genes in the module are also up-regulated in the cortex of early AD, with some being AD risk genes (e.g. ApoE, Trem2, Abca7, Ms4a6d, and Grn; Figure 7G, 7I; Karch et al., 2014). In particular, we found a marked overlap of Blue module hub genes with the recently defined hub genes for the Bayesian Brain Immune and Microglia gene modules for late-onset AD (Figure 7I; Supplemental Table 2; Zhang et al., 2013). These shared hub genes include complement cascade genes (e.g. C1qa, C1qb, C3, Tyrobp, CD33), toll-like receptor signaling genes (e.g. Tlr2, Myd88, CD86), chemokine/cytokine genes (e.g. Cx3cr1, Il17ra), and Fc-receptor system genes (e.g. Trem2, S100a11, Apoc1, Serpina1). The intriguing overlap of neuroinflammatory transcription signatures between BACHD-ΔN17 striatum and AD suggests that this model could be useful to study disease-associated neuroinflammatory genes that may be shared between HD and other neurodegenerative disorders.
DISCUSSION
In summary, we have generated and characterized novel BAC transgenic mice expressing mutant or WT human HTT lacking the N17 domain to evaluate the impact of this domain in regulating mHTT nucleocytoplasmic trafficking, aggregation, and disease pathogenesis in vivo. We showed that BACHD-ΔN17 mice exhibit earlier onset and more severe disease phenotypes compared to BACHD mice. Importantly, BACHD-ΔN17 mice show multiple HD-like disease phenotypes (e.g. CDLs, robust striatal MSN loss and neuroinflammation) that are not observed in previous HD mouse models. Mechanistically, our study reveals the important function of the N17 domain in mitigating nuclear pathogenesis elicited by small mHTT polyQ fragments in vivo.
Our study provides novel insight into N17 domain function in vivo. The N17 domain is evolutionarily conserved in the vertebrate lineage (Tartari et al., 2008), suggesting it may confer conserved functions for HTT amongst vertebrates. Our study shows that BAC transgenes expressing mutant or WT HTT lacking N17 domain can rescue the embryonic lethality phenotype of murine Htt null mice, suggesting the N17 domain is not required for the essential HTT function during murine development. Our study, together with prior genetic evidence (Neveklovska et al., 2012; Zeng et al., 2010) suggests that essential embryonic developmental functions of HTT in vertebrates do not appear to require the three domains near the HTT N-terminus (i.e. the N17, polyQ, and polyproline domains). It would be tempting to speculate that these HTT exon1 domains, which co-evolved within the vertebrate lineage (Tartari et al., 2008), could be sub-serving a function of HTT that is specific to mature organisms, possibly within the nervous system. One hint of this function may come from a study where mice expressing Htt lacking the polyQ domain show enhanced neuronal autophagy and longevity (Zeng et al., 2010).
Our study reveals N17 as a critical molecular determinant of mHTT-induced nuclear pathogenesis in vivo. Although prior cell-based studies have demonstrated that the N17 domain confers cytoplasmic membrane association (Rockabrand et al., 2006; Atwal et al., 2007) and nuclear export of mutant and WT HTT (Maiuri et al., 2013; Zhang et al., 2013), these studies were done in the context of small HTT fragments (i.e. exon1) and in transfected cells, hence the impact of N17 in regulating mHTT subcellular trafficking in the context of full-length mHTT and disease pathogenesis in vivo remained yet unexplored. Our study provides crucial evidence that BAC mice expressing mHTT that lacks the N17 domain, despite lower level expression of mHTT than the BACHD model, show dramatic acceleration of mHTT aggregation (4m in BACHD-ΔN17 vs. 11m-12m in BACHD). Moreover, the pattern of mHTT pathology is also drastically different between the two models, with BACHD showing predominantly cytoplasmic mHTT aggregates (Gray et al., 2008) while BACHD-ΔN17 show exclusively nuclear mHTT aggregates. These results demonstrate a pivotal in vivo function of N17 domain in regulating mHTT proteostasis in the nuclear and cytoplasmic compartments, suggesting that compromising N17 function can dramatically accelerate nuclear pathogenesis in HD mouse brains.
Extensive prior studies suggest that generation of mHTT N-terminal fragments by proteolysis (DiFiglia et al., 1997) or alternative splicing (Sathasivam et al., 2011) may be an important mechanism in eliciting nuclear pathogenesis in HD (Ross and Tabrizi, 2011). Perhaps the best evidence supporting this idea is that aggregated mHTT species isolated from neuronal nuclei in HD patients or mice are small mHTT fragments, including exon1 (Landles et al., 2010) and CpA fragments (Lunkes et al., 2002). Moreover, transgenic mice expressing mHTT polyQ fragments exhibit exacerbated disease and increased mHTT nuclear pathology compared to those models expressing full-length mHTT (Crook et al., 2011; Li et al, 1999). However, an intriguing set of studies shows that the soluble mHTT-exon1 in transgenic mouse brains is predominantly cytoplasmic, even when the fragment is fused with an exogenous nuclear localization signal (Benn et al., 2005), suggesting that additional molecular element(s) may be involved in regulating the nuclear pathogenesis of mHTT fragments. Our current study suggests that N17 could function in the context of small mHTT fragments to mitigate their potential in eliciting nuclear pathogenesis in vivo. Our in vivo biochemical analyses reveal that the aggregated nuclear mHTT species in BACHD-ΔN17 brains consists predominantly of two small mHTT polyQ fragments, while the intact soluble full-length ΔN17-mHTT was always cytoplasmic (Figure 6). This latter finding is somewhat surprising, since point mutations in N17 (i.e. M8P), in the context of full-length HTT, have been shown to increase nuclear translocation in transfected cells (Atwal et al., 2007). Moreover, our cell-based study provides initial support for a selective requirement of N17 to mediate nuclear exclusion of small mHTT fragments (e.g. exon1 and CpA) that are known to be nuclear pathogenic in HD (Lunkes et al., 2002; Landles et al., 2010), but not for larger mHTT fragments (e.g. C6). Given the limitation of our current study using a transfected non-neuronal cell line to study N17-dependecy in nucleocytoplasmic trafficking of various mHTT fragments, future studies should validate and extend such observations into primary neuronal cells and into WT and HD mouse brains.
Based on the phenotypic findings of BACHD-N17 mice and our mechanistic insights that N17 appears to preferentially act on smaller mHTT fragments to mediate their nuclear exclusion, we would like to propose the following hypothesis on how N17 may regulate nuclear pathogenesis in HD (Figure 8). In HD neurons, full-length or fragmented mHTT species are predominantly located in the cytoplasm and may contribute to cytoplasmic pathogenesis. For nuclear pathogenesis to occur, two events are likely needed. First, the generation of small mHTT fragments (e.g. exon1 or CpA), which are at baseline protected from nuclear translocation or accumulation by N17. Moreover, N17 may also promote their cytoplasmic clearance and suppress their toxicities through protective PTMs (Steffan et al., 2004, Thompson et al., 2009, Gu et al., 2009). Thus, for nuclear pathogenesis to occur, we speculate that a second event would be required to impair the N17 function in mediating nuclear export and cytoplasmic membrane association of the mHTT fragments. This may involve either functional impairment (i.e. intact, but non-functional N17, such as N17-mediated oligomerization or aggregation; Jayaraman et al., 2012) or physical impairment of this domain (e.g. deleterious PTMs; N17 truncation). One important possibility in our model is that the accumulated nuclear mHTT species may be a mixture of those with intact N17 and those with physically impaired N17, which may help to explain the presence of N17 epitope in at least some of the nuclear mHTT species in HD brains (DiFiglia et al., 1997). We acknowledge that future studies are needed to rigorously test this hypothetical model. This may require detailed proteomic analyses of the mHTT species accumulating in the nuclei of HD patients or mouse models and elucidation of the mechanisms that regulate N17-mediated HTT nucleocytoplasmic trafficking in both normal and diseased brains.
Figure 8. A Working Model of N17 Domain Role in Regulating Nuclear Pathogenesis in HD.

In HD neurons, full-length or fragmented mHTT species are predominantly located in the cytoplasm due to N17 function in Crm1-dependent nuclear export and cytoplasmic membrane association. For nuclear pathogenesis to occur, two events are likely needed to occur. First, the generation of small mHTT N-terminal fragments (e.g. mHTT-exon1 or CpA), which are known to be part of nuclear mHTT pathology in HD brains. Since these small mHTT fragments have functional N17, they are located predominantly in the cytoplasm, where N17 can facilitate their clearance (Thompson et al., 2009). The various mHTT species can also elicit cytoplasmic pathogenesis. For nuclear pathogenesis to occur, N17 domain on the small mHTT fragments has to be impaired either functionally (e.g. oligomerization or aggregation of mHTT fragments in the nucleus) or physically (e.g. deleterious N17 PTMs or truncation/proteolysis of N17 itself). Future studies are needed to determine whether N17 impairment can be demonstrated among the nuclear accumulated mHTT species in HD patients or mouse models. Moreover, the molecular pathways that may regulate N17 function in nucleocytoplasmic trafficking and whether manipulating these pathways could modify nuclear mHTT pathogenesis and overall disease phenotypes in HD models or HD remain to be elucidated.
Our study also provides important evidence that the N17 domain can act as a bidirectional molecular switch to modify the severity of disease elicited by full-length mHTT in HD mice. Using the same human genomic transgenic constructs expressing mHTT with the same polyQ length, differing only in the N17-coding regions, we have previously shown that BAC mice expressing mHTT with phosphomimetic mutations at serines 13 and 16 abolish all the disease phenotypes found in BACHD, suggesting that promoting these N17 PTMs may suppress disease pathogenesis in vivo (Gu et al., 2009). Our current study showed that deletion of the N17 domain in the same BAC transgenic HD model system leads to dramatic exacerbation of disease phenotypes, including worsening rotarod deficits, body weight loss, robust striatal neurodegeneration and neuroinflammation, and the emergence of new HD-like phenotypes that are not seen other HD mouse models (e.g. CDLs, spontaneous falls, and urine scalding). Importantly, BACHD-ΔN17 mice also show peripheral pathology reminiscent of HD (van der Burg et al., 2009). Together, our study reveals that deletion of N17 in the context of the BAC transgenic HD model system results in more severe, yet still HD-like disease phenotypes in vivo. Despite the improved face validity of BACHD-ΔN17 mice compared to prior HD mouse models such as BACHD, we are cognizant that this mouse model has limitations in terms of construct validity (i.e. deletion of 15 amino acid residues in the N17 domain of mHTT). We believe the BACHD-ΔN17 model may offer new opportunities to study aspects of disease elicited by mHTT (e.g. nuclear pathogenesis, mHTT proteolysis, striatal neuronal degeneration and neuroinflammation). Moreover, our in vivo insights gained from HD BAC mice with N17 mutations suggest the importance of elucidating the molecular bases for the bidirectional disease modifications conferred by N17 mutations and exploration of molecular targets interacting with this crucial domain for HD therapy.
Finally, from a translational research perspective, BACHD-ΔN17 has the potential to be a novel model system to study the interplay between age-dependent neurodegeneration and neuroinflammation in neurodegenerative disorders. Emerging evidence suggests that neuroinflammation may play a key role in many, if not all, major neurodegenerative disorders, including AD, PD, and HD (Glass et al., 2010). Previous HD mouse models exhibit limited neuroinflammation, not approaching the extent seen in HD patients (Moller, 2010). Importantly, our longitudinal transcriptome profiling and subsequent validation studies showed that BACHD-N17 mice exhibit age-dependent neuroinflammation that partially overlaps that seen in both HD and AD, and includes causal or risk allele genes involved in AD (e.g. ApoE, Trem2, Abca7, Ms4a4 and Clu) and FTD genes (e.g. Grn; Karch et al., 2014). These findings underscore potential common neuroinflammatory mechanisms between our mouse model and multiple neurodegenerative disorders and suggest that BACHD-ΔN17 mice could be an invaluable in vivo paradigm to study molecular and cellular interactions between degenerating neurons and reactive glia, as well as to test candidate therapeutics against neurodegeneration or neuroinflammation.
EXPERIMENTAL PROCEDURES
Transgene Constructs and Generation of BACHD-ΔN17 and BAC-WT-ΔN17 Mice
HTT BAC DNA modification was performed as previously reported (Yang et al. 1997; Gu et al., 2009). The modified BAC constructs were confirmed by sequencing before being microinjected into FvB/NJ fertilized oocytes at the UCLA Transgenic Core to generate BACHD-ΔN17 and BAC-WT-ΔN17 mice. Transgenic mice were bred and maintained under standard conditions consistent with NIH guidelines, and all procedures complied with regulations and approved by the UCLA Institutional Animal Care and Use Committees.
Quantitation of Transgene Expression
Three brain extracts from the transgenic mouse lines were used for mHTT quantification by Western blot with mHTT specific antibody, MAB5492 (against human HTT polyproline domain; EMD Millipore; Dehay et al., 2007), 1C2 and α-tubulin as reported previously (Gu et al., 2009).
Behavioral Tests and Quantifications
Quantitation of chorea/dystonia-like (CDL) behavior, rearing, and spontaneous falling of each mouse was manually calculated for the last 3 minutes of a 5-minute video clip (details see Supplemental Experimental Procedures). Accelerating rotarod test was performed at 1, 2, 4, 6 and 8 months of age, and automated open field tests was performed at 2, 4, and 8 months of age with established protocols (Gray et al., 2008; Wang et al., 2014). Gait impairment testing was performed at 2, 6, and 8 months of age. All tests used over 10 mice. The investigators were blinded to the genotypes when performing these behavioral tests.
Electroencephalography
Synchronous video-EEG recordings were carried out at least 7 days after surgery in freely moving mice in an 8-inch diameter recording chamber. Field potential recordings were bandpass filtered and amplified (0.3 Hz – 1 kHz, gain: 1000, Model 3500, A-M Systems, Carlsborg, WA), digitized on-line at a rate of 2048 s−1 (PCI-MIO16E-4 data acquisition board National Instruments, Austin, TX), and acquired continuously (SignalExpress, National Instruments, Austin, TX). All data analysis was performed under IGOR Pro v6.22A (Lake Oswego, OR). Morlet wavelet analysis was performed for time-frequency analysis of local field potential recordings.
Analysis of Fragment Size-dependent Subcellular Distribution of mHTT
HEK-293 cells were transfected with DNA constructs of mHTT (46Q) fragments fused with a HA-tag (see supplemental information) for 48 h. The expression and localization of mHTT fragments were probed with an antibody against the HA-tag, followed by an Alexa 488 conjugated secondary antibody. Hoechst 33258 (Sigma) was used to visualize nuclei. Confocal images were analyzed by ImageJ (NIH) using an established method (Ch'ng et al., 2012). In brief, the fluorescence intensity of the nuclear and cytosolic mHTT fragments was quantified and the ratio of the mean intensity of nuclear/cytosolic signal was calculated. Student’s t test was used to measure the statistical significance.
Microarray and Data Analysis
Microarray processing was performed by the UCLA Neuroscience Genomics Core (UNGC, www.semel.ucla.edu/ungc). The transcription profiling dataset in this study is available at GEO (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE64386). Enrichment analysis was performed using the collapseRows function as described (Miller et al. 2011b; http://www.inside-r.org/packages/cran/WGCNA/docs/userListEnrichment). For methods and data analysis in detail see Supplemental Experiment Procedures.
Statistical Analyses
Western blot quantification statistical analysis was determined by one-way ANOVA, followed by Tukey’s Multiple Comparison post hoc test. For electroencephalograph data, statistical analysis was performed using the Kolmogorov-Smirnov test. Significance was set to p < 0.05. To determine significant main effects and interactions between main factors, two-way ANOVA was used to evaluate the effects of time and genotype followed by post hoc tests using the Bonferroni method. When appropriate, differences between genotype means were evaluated with unpaired Student’s t-test. Overlap between gene sets was assessed using the hypergeometric test in R (function phyper), using the number of shared gene symbols between array platforms as background. All data are expressed as mean ± standard error of the mean (SEM). Significance level was p < 0.05. Prism software was used to conduct these analyses (Graphpad Inc., La Jolla, CA, USA). Detailed materials and methods can be found in Supplemental Experiment Procedures.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of Yang lab for helpful input and H.W. Dong (LONI, USC) for help in confocal microscopy. The research was supported by the NINDS/NIH grants (R01NS049501, R01NS074312, R01NS084298-01) and by the Hereditary Disease Foundation (HDF) to XWY. HD research in the Yang lab is also supported by CHDI Foundation, Inc., the David Weil Fund to the Semel Institute at UCLA, and The McKnight Foundation. We acknowledge the support of the NINDS Informatics Center for Neurogenetics and Neurogenomics (P30 NS062691).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Atwal RS, Desmond CR, Caron N, Maiuri T, Xia J, Sipione S, Truant R. Kinase inhibitors modulate huntingtin cell localization and toxicity. Nature Chemical Biology. 2011;7:453–452. doi: 10.1038/nchembio.582. [DOI] [PubMed] [Google Scholar]
- Atwal RS, Xia J, Pinchev D, Taylor J, Epand RM, Truant R. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Human Molecular Genetics. 2007;16:2600–2615. doi: 10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- Benn CL, Landles C, Li H, Strand AD, Woodman B, Sathasivam K, Li SH, Ghazi-Noori S, Hockly E, Faruque SM, et al. Contribution of nuclear and extranuclear polyQ to neurological phenotypes in mouse models of Huntington's disease. Hum Mol Genet. 2005;14:3065–3078. doi: 10.1093/hmg/ddi340. [DOI] [PubMed] [Google Scholar]
- Björkqvist M, Wild EJ, Thiele J, Silvestroni A, Andre R, Lahiri N, Raibon E, Lee RV, Benn CL, Soulet D, et al. A novel pathogenic pathway of immune activation detectable before clinical onset in Huntington's disease. J Exp Med. 2008;205:1869–1877. doi: 10.1084/jem.20080178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess DL, Noebels JL. Single gene defects in mice: the role of voltage-dependent calcium channels in absence models. Epilepsy Res. 1999;36:111–122. doi: 10.1016/s0920-1211(99)00045-5. [DOI] [PubMed] [Google Scholar]
- Cayzac S, Delcasso S, Paz V, Jeantet Y, Cho YH. Changes in striatal procedural memory coding correlate with learning deficits in a mouse model of Huntington disease. Proc. Natl. Acad. Sci. U.S.a. 2011;108:9280–9285. doi: 10.1073/pnas.1016190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch’ng TH, Uzgil B, Lin P, Avliyakulov NK, O’Dell TJ, Martin KC. Activity-dependent transport of the transcriptional coactivator CRTC1 from synapse to nucleus. Cell. 2012;150:207–221. doi: 10.1016/j.cell.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook ZR, Housman D. Huntington's Disease: Can Mice Lead the Way to Treatment? Neuron. 2011;69:13–13. doi: 10.1016/j.neuron.2010.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay B, Weber C, Trottier Y, Bertolotti A. Mapping of the epitope of monoclonal antibody 2B4 to the proline-rich region of human Huntingtin, a region critical for aggregation and toxicity. Biotechnol J. 2007;2:559–564. doi: 10.1002/biot.200600249. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel J, Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- Dunah A, Sheng M, Standaert D. a-Actinin-2 in rat striatum: localization and interaction with NMDA glutamate receptor subunits. Molecular Brain Research. 2000:1–11. doi: 10.1016/s0169-328x(00)00102-9. [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Konopka G. Neuroscience in the era of functional genomics and systems biology. Nature. 2009;461:908–915. doi: 10.1038/nature08537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Saijo K, Winner B, Marchetto MC, Gage FH. Mechanisms underlying inflammation in neurodegeneration. Cell. 2010;140:918–934. doi: 10.1016/j.cell.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, Pearson J, Shehadeh J, Bertram L, Murphy Z, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Gray M, Shirasaki DI, Cepeda C, Andre VM, Wilburn B, Lu XH, Tao J, Yamazaki I, Li SH, Sun YE, et al. Full-Length Human Mutant Huntingtin with a Stable Polyglutamine Repeat Can Elicit Progressive and Selective Neuropathogenesis in BACHD Mice. Journal of Neuroscience. 2008;28:6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimbergen YAM, Knol MJ, Bloem BR, Kremer BPH, Roos RAC, Munneke M. Falls and gait disturbances in Huntington's disease. Mov. Disord. 2008;23:970–976. doi: 10.1002/mds.22003. [DOI] [PubMed] [Google Scholar]
- Gu X, Greiner ER, Mishra R, Kodali R, Osmand A, Finkbeiner S, Steffan JS, Thompson LM, Wetzel R, Yang XW. Serines 13 and 16 Are Critical Determinants of Full-Length Human Mutant Huntingtin Induced Disease Pathogenesis in HD Mice. Neuron. 2009;64:828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardena S, Her L-S, Brusch RG, Laymon RA, Niesman IR, Gordesky-Gold B, Sintasath L, Bonini NM, Goldstein LS. Disruption of Axonal Transport by Loss of Huntingtin or Expression of Pathogenic PolyQ Proteins in Drosophila. Neuron. 2003;40:25–40. doi: 10.1016/s0896-6273(03)00594-4. [DOI] [PubMed] [Google Scholar]
- Gutekunst CA, Li SH, Yi H, Mulroy JS, Kuemmerle S, Jones R, Rye D, Ferrante RJ, Hersch SM, Li XJ. Nuclear and neuropil aggregates in Huntington's disease: relationship to neuropathology. J Neurosci. 1999;19:2522–2534. doi: 10.1523/JNEUROSCI.19-07-02522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges A, Strand AD, Aragaki AKA, Sengstag T, Hughes G, Elliston LA, Hartog C, Goldstein DR, Thu D, et al. Regional and cellular gene expression changes in human Huntington's disease brain. Human Molecular Genetics. 2006;15:965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- Huntington’s Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Jayaraman M, Mishra R, Kodali R, Thakur AK, Koharudin LM, Gronenborn AM, Wetze lR. Kinetically competing huntingtin aggregation pathways control amyloid polymorphism and properties. Biochemistry. 2012;51:2706–2716. doi: 10.1021/bi3000929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch CM, Cruchaga C, Goate AM. Alzheimer's disease genetics: from the bench to the clinic. Neuron. 2014;83:11–26. doi: 10.1016/j.neuron.2014.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuno M, Adachi H, Kume A, Li M, Nakagomi Y, Niwa H, Sang C, Kobayashi Y, Doyu M, Sobue G. Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Neuron. 2002;35:843–854. doi: 10.1016/s0896-6273(02)00834-6. [DOI] [PubMed] [Google Scholar]
- Klement IA, Skinner PJ, Kaytor MD, Yi H, Hersch SM, Clark HB, Zoghbi HY, Orr HT. Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell. 1998;95:41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- Kuhn A, Goldstein DR, Hodges A, Strand AD, Sengstag T, Kooperberg C, Becanovic K, Pouladi MA, Sathasivam K, Cha JH, et al. Mutant huntingtin's effects on striatal gene expression in mice recapitulate changes observed in human Huntington's disease brain and do not differ with mutant huntingtin length or wild-type huntingtin dosage. Hum Mol Genet. 2007;16:1845–1861. doi: 10.1093/hmg/ddm133. [DOI] [PubMed] [Google Scholar]
- Landles C, Sathasivam K, Weiss A, Woodman B, Moffitt H, Finkbeiner S, Sun B, Gafni J, Ellerby LM, Trottier Y. Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington disease. J. Biol. Chem. 2010;285:8808–8823. doi: 10.1074/jbc.M109.075028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CYD, Cantle JP, Yang XW. Genetic Manipulations of Mutant Huntingtin in Mice: New Insights into HD Pathogenesis. Febs J. 2013;280:4382–4394. doi: 10.1111/febs.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li SH, Cheng AL, Mangiarini L, Bates GP, Li XJ. Ultrastructural localization and progressive formation of neuropil aggregates in Huntington's disease transgenic mice. Hum Mol Genet. 1999;8:1227–1236. doi: 10.1093/hmg/8.7.1227. [DOI] [PubMed] [Google Scholar]
- Lunkes A, Lindenberg KS, Weber C, Devys D, Landwehrmeyer GB, Mandel JL, Trottier Y. Proteases acting on mutant huntingtin generate cleaved products that differentially build up cytoplasmic and nuclear inclusions. Molecular Cell. 2002;10:259–269. doi: 10.1016/s1097-2765(02)00602-0. [DOI] [PubMed] [Google Scholar]
- Maiuri T, Woloshansky T, Xia J, Truant R. The huntingtin N17 domain is a multifunctional CRM1 and Ran-dependent nuclear and cilial export signal. Human Molecular Genetics. 2013;22:1383–1394. doi: 10.1093/hmg/dds554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Cai C, Langfelder P, Geschwind DH, Kurian SM, Salomon DR, Horvath S. Strategies for aggregating gene expression data: the collapseRows R function. BMC Bioinformatics. 2011;12:322. doi: 10.1186/1471-2105-12-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra R, Hoop CL, Kodali R, Sahoo B, van der Wel PCA, Wetzel R. Serine Phosphorylation Suppresses Huntingtin Amyloid Accumulation by Altering Protein Aggregation Properties. Journal of Molecular Biology. 2012;424:1–14. doi: 10.1016/j.jmb.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveklovska M, Clabough EB, Steffan JS, Zeitlin SO. Deletion of the huntingtin proline-rich region does not significantly affect normal huntingtin function in mice. J Huntingtons Dis. 2012;1:71–87. doi: 10.3233/JHD-2012-120016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omi K, Hachiya NS, Tanaka M, Tokunaga K, Kaneko K. 14-3-3zeta is indispensable for aggregate formation of polyglutamine-expanded huntingtin protein. Neurosci Lett. 2008;431:45–50. doi: 10.1016/j.neulet.2007.11.018. [DOI] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. Trinucleotide Repeat Disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- Rockabrand E, Slepko N, Pantalone A, Nukala VN, Kazantsev A, Marsh JL, Sullivan PG, Steffan JS, Sensi SL, Thompson LM. The first 17 amino acids of Huntingtin modulate its sub-cellular localization, aggregation and effects on calcium homeostasis. Human Molecular Genetics. 2006;16:61–77. doi: 10.1093/hmg/ddl440. [DOI] [PubMed] [Google Scholar]
- Ross CA, Aylward EH, Wild EJ, Langbehn DR, Long JD, Warner JH, Scahill RI, Leavitt BR, Stout JC, Paulsen JS, et al. Huntington disease: natural history, biomarkers and prospects for therapeutics. Nat Rev Neurol. 2014;10:204–216. doi: 10.1038/nrneurol.2014.24. [DOI] [PubMed] [Google Scholar]
- Ross CA, Tabrizi SJ. Huntington’s disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10:83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- Rotshenker S. The role of Galectin-3/MAC-2 in the activation of the innate-immune function of phagocytosis inmicroglia in injury and disease. J Mol Neurosci. 2009;39:99–103. doi: 10.1007/s12031-009-9186-7. [DOI] [PubMed] [Google Scholar]
- Sathasivam K, Woodman B, Mahal A, Bertaux F, Wanker EE, Shima DT, Bates GP. Centrosome disorganization in fibroblast cultures derived from R6/2 Huntington's disease (HD) transgenic mice and HD patients. Hum Mol Genet. 2001;10:2425–2435. doi: 10.1093/hmg/10.21.2425. [DOI] [PubMed] [Google Scholar]
- Sathasivam K, Neueder A, Gipson TA, Landles C, Benjamin AC, Bondulich MK, Smith DL, Faull RL, Roos RA, Howland D, Detloff PJ, Housman DE, Bates GP. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc Natl Acad Sci. 2013;110:2366–2370. doi: 10.1073/pnas.1221891110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasaki D, Greiner ER, Al-Ramahi I, Gray M, Boontheung P, Geschwind DH, Botas J, Coppola G, Horvath S, Loo JA, et al. Network Organization of the Huntingtin Proteomic Interactome in Mammalian Brain. Neuron. 2012;75:41–57. doi: 10.1016/j.neuron.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopher BL, Thomas PS, Jr, LaFevre-Bernt MA, Holm IE, Wilke SA, Ware CB, Jin LW, Libby RT, Ellerby LM, La Spada AR. Androgen receptor YAC transgenic mice recapitulate SBMA motor neuronopathy and implicate VEGF164 in the motor neuron degeneration. Neuron. 2004;41:687–699. doi: 10.1016/s0896-6273(04)00082-0. [DOI] [PubMed] [Google Scholar]
- Steffan JS, Agrawal N, Pallos J, Rockabrand E, Trotman LC, Slepko N, Illes K, Lukacsovich T, Zhu YZ, Cattaneo E, et al. SUMO modification of Huntingtin and Huntington’s disease pathology. Science. 2004;304:100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- Tam S, Spiess C, Auyeung W, Joachimiak L, Chen B, Poirier MA, Frydman J. The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nature Structural & Molecular Biology. 2009;16:1279–1285. doi: 10.1038/nsmb.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartari M, Gissi C, Sardo Lo V, Zuccato C, Picardi E, Pesole G, Cattaneo E. Phylogenetic comparison of huntingtin homologues reveals the appearance of a primitive polyQ in sea urchin. Mol. Biol. Evol. 2008;25:330–338. doi: 10.1093/molbev/msm258. [DOI] [PubMed] [Google Scholar]
- Thakur AK, Jayaraman M, Mishra R, Thakur M, Chellgren VM, Byeon IJL, Anjum DH, Kodali R, Creamer TP, Conway JF, et al. Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nature Structural & Molecular Biology. 2009;16:380–389. doi: 10.1038/nsmb.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LM, Aiken CT, Kaltenbach LS, Agrawal N, Illes K, Khoshnan A, Martinez-Vincente M, Arrasate M, O’Rourke JG, Khashwji H, et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. The Journal of Cell Biology. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Burg JM, Björkqvist M, Brundin P. Beyond the brain: widespread pathology in Huntington’s disease. Lancet Neurol. 2009;8:765–774. doi: 10.1016/S1474-4422(09)70178-4. [DOI] [PubMed] [Google Scholar]
- Van Raamsdonk JM, Murphy Z, Selva DM, Hamidizadeh R, Pearson J, Petersén Bjorkqvist M, Muir C, Mackenzie IR, Hammond GL. Testicular degeneration in Huntington disease. Neurobiology of Disease. 2007;26:512–520. doi: 10.1016/j.nbd.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Vonsattel JPG, Difiglia M. Huntington disease. J. Neuropathol. Exp. Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- Waldron-Roby E, Ratovitski T, Wang X, Jiang M, Watkin E, Arbez N, Graham RK, Hayden MR, Hou Z, Mori S, Swing D, Pletnikov M, Duan W, Tessarollo L, Ross CA. Transgenic mouse model expressing the caspase 6 fragment of mutant huntingtin. J Neurosci. 2012;32:183–193. doi: 10.1523/JNEUROSCI.1305-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Gray M, Lu XH, Cantle JP, Holley SM, Greiner E, Gu X, Shirasaki D, Cepeda C, Li Y, et al. Neuronal targets for reducing mutant huntingtin expression to ameliorate disease in a mouse model of Huntington's disease. Nat. Med. 2014;20:536–541. doi: 10.1038/nm.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodman B, Butler R, Landles C, Lupton MK, Tse J, Hockly E, Moffitt H, Sathasivam K, Bates GP. The HdhQ150/Q150 knock-in mouse model of HD and the R6/2 exon 1 model develop comparable and widespread molecular phenotypes. Brain Research Bulletin. 2007;72:83–97. doi: 10.1016/j.brainresbull.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Wyszynski M, Lin J, Rao A, Nigh E, Beggs AH, Craig AM, Sheng M. Competitive binding of alpha-actinin and calmodulin to the NMDA receptor. Nature. 1997;385:439–442. doi: 10.1038/385439a0. [DOI] [PubMed] [Google Scholar]
- Yang XW, Model P, Heintz N. Homologous recombination based modification in Esherichia coli and germline transmission in transgenic mice of a bacterial artificial chromsome. Nature Biotechnology. 1997;15:859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- Zeitlin S, Liu JP, Chapman DL, Papaioannou VE, Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington's disease gene homologue. Nat Genet. 1995;11:155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- Zheng S, Clabough EB, Sarkar S, Futter M, Rubinsztein DC, Zeitlin SO. Deletion of the huntingtin polyglutamine stretch enhances neuronal autophagy and longevity in mice. PLoS Genet. 2010;6:e1000838. doi: 10.1371/journal.pgen.1000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li A, Holmes BB, Marasa JC, Diamond MI. An N-terminal Nuclear Export Signal Regulates Trafficking and Aggregation of Huntingtin (HTT) Protein Exon 1. Journal of Biological Chemistry. 2013;288:6063–6071. doi: 10.1074/jbc.M112.413575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.