Abstract
Humans, animals, and plants are constantly under attack from pathogens and pests, resulting in severe consequences on global human health and crop production. Small RNA (sRNA)-mediated RNA interference (RNAi) is a conserved regulatory mechanism that is involved in almost all eukaryotic cellular processes, including host immunity and pathogen virulence. Recent evidence supports the significant contribution of sRNAs and RNAi to the communication between hosts and some eukaryotic pathogens, pests, parasites, or symbiotic microorganisms. Mobile silencing signals—most likely sRNAs—are capable of translocating from the host to its interacting organism, and vise versa. In this review, we will provide an overview of sRNA communications between different kingdoms, with a primary focus on the advances in plant-pathogen interaction systems.
Introduction
Cell-to-cell communication occurs between organisms that form pathogenic, parasitic, or symbiotic relationships. Such communication involves transportation of regulatory molecules across the cellular boundaries between the host and its interacting pathogens/pests/parasites or symbionts. Recently, mobile small RNAs (sRNAs) have been indicated to function in communication between hosts and advanced pathogens/pests/parasites. sRNAs are non-coding regulatory RNAs that are loaded into Argonaute (AGO) proteins to silence genes with complementary sequences in a mechanism called RNA interference (RNAi). The RNAi machinery is conserved in most eukaryotes and mediated by non-coding small interfering RNAs (siRNAs), microRNAs (miRNAs) and piwi-associated RNAs (piRNAs). RNAi functions not only as a defense mechanism to silence foreign DNA and RNA species such as those from viruses, transposons, and transgenes, but also plays an important role in regulating and fine-tuning the expression of genes in a plethora of diverse physiological and cellular processes, including host immune responses [1–4]. Mobile, cell non-autonomous sRNAs that translocate within an organism have been observed in various plant [5–9] and animal systems [10–13]. Some sRNAs can even move across the boundaries between hosts and their interacting pathogenic, parasitic, or symbiotic organisms and trigger gene silencing in trans in the non-related species, a mechanism termed cross-kingdom or cross-organism RNAi [4,14,15]. Here, we review the latest discoveries on cross-kingdom regulatory sRNAs with an emphasis on plant-microbial interactions and potential applications for mobile sRNAs in the future of plant biotechnology. sRNA-directed RNAi enriches the toolbox for plant researchers to manipulate gene expression to bolster plant resistance and, furthermore, to modulate the outcome of plant interactions with other organisms.
Mobile Small RNAs
Mobile sRNAs, or possibly their precursor RNAs in certain conditions, which spread gene silencing into adjacent cells and tissues or even spread systemically, have fascinated scientists for the last two decades. From the time the phenomenon was first discovered [16–18], genetic determinants, pathways, and mechanisms have been revealed in a variety of organisms [5,7,19,20]. Diverse functions and useful applications for extracellular sRNAs have been established, encompassing cell-to-cell signaling and communication in multi-cellular organisms [13], trans-generational RNAi [21,22] and memorization [23–26], cell fate differentiation and vascular formation [27–31], systemic antiviral immunity [32], environmental RNAi [11,33], cancer prevention and diagnosis [34], and intercellular immune activation [13,35–37].
Cross-kingdom RNAi is a form of communication between two, often unrelated, interacting organisms such as a host and its pathogen, pest, parasite, or symbiont. This phenomenon has been overlooked in the past due to technical limitations. In respect to extracellular interacting organisms, such as bacteria, fungi, oomycetes, protozoa, nematodes, parasites, or herbivores, cross-kingdom RNAi implies that a translocation of gene silencing signals occurs between hosts and these organisms. These silencing signals may utilize conserved cell-to-cell as well as systemic RNAi pathways present in plants and animals, and may also use organism-specific pathways. The language of RNAi-based inter-species cross talk could be termed ‘social RNAs’ [38].
Cross-kingdom gene silencing in animal systems
There are few instances that point to the existence of cross-kingdom gene silencing in animal systems. One example is environmental RNAi in Caenorhabditis elegans, in which the worms uptake environmental RNA signals that have gene suppressive effects [11]. RNAi can be induced by soaking the worms in RNA solutions or by feeding them antisense RNA-expressing bacteria, such as Escherichia coli. A number of genes that are required for the uptake of environmental long dsRNAs as well as systemic silencing have been discovered in C. elegans, such as systemic RNAi defective-1 (sid-1) and sid-2, two transmembrane RNA transporters [19,20] (Figure 1). However, many identified transporter-like proteins are specific to worms or invertebrates. In a recent feeding experiment, two natural non-coding RNAs from E. coli, OxyS and DsrA, could suppress protein-coding genes in C. elegans [39]. Gene suppression of the che-2 mRNA (a WD-40 protein involved in chemosensory) by OxyS relied heavily on distinct RNAi genes, such as the AGO protein ALG-1, the dsRNA-binding protein RDE-4, and the ABC transporter HAF-2. The sid-1 and sid-2 mutants did not show any alteration in gene suppression, probably due to the redundant function of OxyS and DsrA-mediated gene silencing. OxyS is induced by oxidative stress, while its primary role is translational repression of E. coli mRNAs rpoS (sigma subunit of RNA polymerase) and fhlA (transcription activator). “Why” and “how” E. coli regulatory RNAs evolved to target genes from an unrelated species like C. elegans in trans, remains to be illustrated.
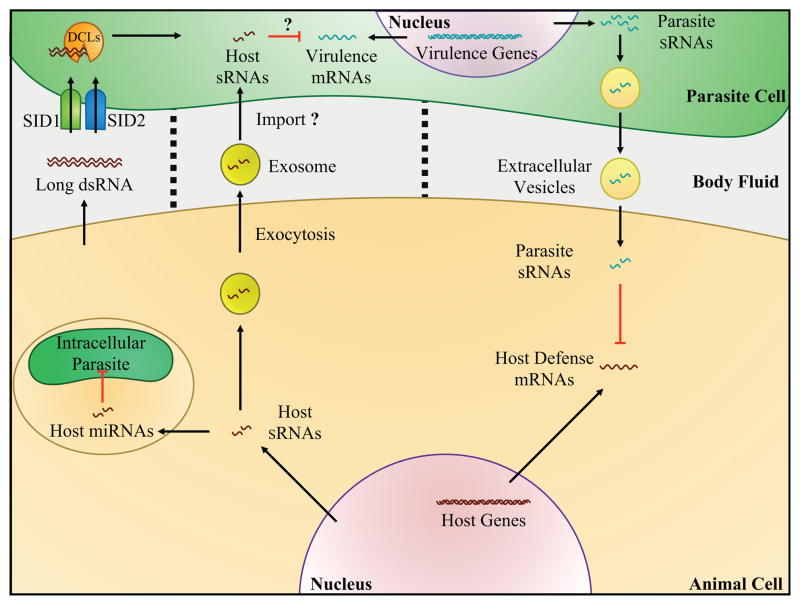
Figure 1.
Cross-kingdom gene silencing between animal cell and parasites. Animal cells produce host sRNAs, and selected host miRNAs translocate into intracellularly phagocytized parasites and target parasitic mRNAs. Some mammalian sRNAs are selectively sorted into vesicles for secretion via exocytosis (exosomes). Extracellular parasite cells likely internalize exosomal sRNAs and, in addition, take up extracellular long dsRNAs via cell membrane-associated RNA transporters, SID-1 and SID-2. Long dsRNAs are further processed into mature sRNAs by DCLs; both pathways may trigger gene silencing in the parasite. Parasites encapsulate and secrete parasitic sRNAs that circulate in body fluids of infected individual and are internalized by host cells, triggering parasitic-induced host gene silencing.
Cross-kingdom RNAi has also been observed in host-parasite interactions (Figure 1). The protozoan malaria parasite Plasmodium falciparum infects humans by entering the blood stream and multiplies intracellularly. It has long been known that individuals with sickle cell disease resist infection by P. falciparum, however the underlying mechanism was not fully understood. The dysregulated miRNA composition in these cells was recently found to contribute to this resistance. The erythrocytes infected by P. falciparum produce miRNAs that are translocated into the parasitic cells in high concentrations [40]. Two highly enriched human miRNAs in erythrocytes of sickle cell individuals, miR451 and let-7i, were demonstrated to bind to Plasmodium mRNAs. One target gene of miR451 is the cAMP-dependent protein kinase PKA-R. Overexpression of miR451 and let-7i led to reduced parasitemia, suggesting that translocated human miRNAs suppress virulence-associated mRNAs in the parasite (Figure 1). It is worth noting that Plasmodium lacks essential RNAi components, such as AGOs and Dicer or Dicer-like (DCL) proteins that process the double-stranded RNA precursors into sRNAs, which suggests that the mode of action of sRNA-mediated gene suppression in this interaction may be independent of the canonical RNAi pathway. Binding of miR451 to PKA-R mRNA is likely to block ribosomal loading and causes translation inhibition [40].
In mammals, cell-to-cell communication is mediated by exosomal vesicles that contain miRNAs (Figure 1) [13]. Exosomal miRNAs have specific functions such as immune response activation [37]. The helminth nematode Heligmosomoides polygyrus also utilizes exosomal vesicles to increase virulence in a fashion similar to that of the mammalian miRNA transport mechanism [41]. H. polygyrus secretes miRNA-loaded vesicles that are accompanied by a nematode AGO protein, most likely to stabilize the miRNAs. Remarkably, H. polygyrus vesicles are internalized by mice cells, which results in suppression of host immunity. Some H. polygyrus miRNAs were shown to target in vitro host mRNAs that are related to host immunity [41]. However, it needs to be determined through which AGO protein—nematode AGO or host AGO—these nematode-derived miRNAs silence host genes. Taken together, nematode vesicles resemble their mammalian exosomal miRNA transport counterparts [42,43].
Evidence has shown that parasites can secrete sRNAs into their host during infection. sRNAs originating from parasites, such as protozoa Trypanosomas cruzi [44] and Schistosoma japonicum [45] and the nematode Litomosoides sigmodontis [41], have been found in the body fluids of infected individuals, indicating that an invasion of circulating sRNAs in host systems may be a common event (Figure 1).
Plants communicate with their interacting organisms using mobile sRNAs
Due to their sessile nature, it is of vital importance that plants are in constant communication with their interacting organisms and environment. Host defense responses induced by pathogens/pests/parasites, or signal transduction triggered by the communication between hosts and symbionts, or communities of endophytic organisms, are all initiated by molecular signals. sRNAs, and possibly their precursor RNAs in certain situations, function as mobile signals that spread silencing information to influence the interacting organisms.
RNAi in fungi has been best studied in the model systems Neurospora crassa and the fission yeast Schizosaccharomyces pombe [46,47]; yet, neither species are natural pathogens or symbionts of plants. Fungal sRNA biogenesis pathways are diverse, and include both DCL-dependent and DCL-independent pathways [48,49]. Most eukaryotic microbes that come into intimate contact with plants, including pathogenic fungi and oomycetes, possess functional RNAi pathways and produce regulatory sRNAs. Remarkably, scientists have developed an effective disease control strategy, called host-induced gene silencing (HIGS) [50,51], by generating transgenic plants that express exogenous RNAi triggers to successfully silence essential genes in pathogens and pests. In addition to its successful use in model plants such as Arabidopsis thaliana and tobacco Nicotiana benthamiana, HIGS has been also successfully applied in important crops, including wheat, barley, Medicago, and banana, to efficiently work against a variety of fungal and oomycete pathogens, such as Blumeria graminis, Puccinia tritici, Fusarium spp., and Phytophthora capsici (Figure 2A) [51,52]. The use of HIGS to combat fungal pathogens caused alteration in fungal morphology, growth inhibition in planta, and most importantly, reduced virulence. In addition, HIGS is a powerful tool to study gene function in non-transformable species [50,53]. A HIGS approach was applied to study gene function of the Monosaccharide Transporter 2 from Glomus sp. [54], demonstrating that HIGS is functional also on arbuscular mycorrhiza, which forms symbiotic relationship with hosts. The artificial sRNAs generated from host plants could be transported into arbuscular mycorrhiza and to be functional. Most surprisingly, silencing effects were also observed after external treatment of fungal mycelium with corresponding duplex sRNAs, indicating that a sufficient RNA uptake system must exist in fungi [55].
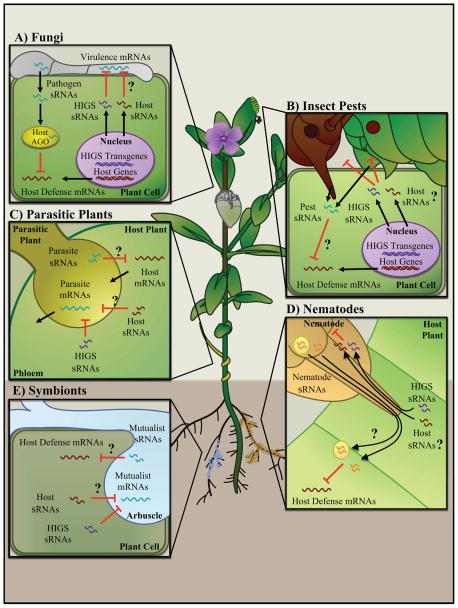
Figure 2. Cross-kingdom gene silencing between plants and interacting organisms. Host-induced gene silencing (HIGS) sRNAs are produced via transgenes in plants.
(A)HIGS sRNAs translocate into pathogens and suppress virulence mRNAs in pathogenic fungi and oomycetes. We speculate that there are natural host sRNAs that target virulence mRNAs for host defense via similar pathways. Pathogen-derived sRNAs that mimic host sRNAs are translocated into host cells to hijack the host AGO/RISC machinery to suppress host immunity mRNAs.
(B) HIGS sRNAs are effective against insect pests by translocating into insect pest cells and silence their virulence genes. Similarly, natural host sRNAs could also potentially target pest mRNAs for silencing. At the same time, pest sRNAs are also likely to be injected into host cells to suppress host immunity or manipulate other cellular pathways.
(C) Parasitic plants, such as Cuscuta sp., form haustoria to acquire nutrients from the phloem source. Bi-directional exchange of a large array of mRNAs has been observed. It is likely sRNAs are also transported between hosts and parasitic plants as gene regulators.
(D)HIGS sRNAs are effective against plant-parasitic nematodes. It is possible that natural host sRNAs may target nematode mRNAs for defense. On the other hand, nematode-induced gene silencing of host mRNAs is also likely, either via encapsulated or non-vesicular nematode sRNAs.
(E) HIGS sRNAs are effective against mRNAs from symbiotic organisms, such as mycorrhiza. Similarly, natural host sRNAs, as well as symbiotic sRNAs, are also likely to be exchanged during the regulation of symbiosis.
The successful application of HIGS demonstrates the ability of plants to deliver mobile gene silencing signals to communicate with and manipulate diverse interacting organisms. However, some pathogen-produced sRNAs are capable of inducing gene silencing in the plants, too. A positive role of sRNAs in fungal virulence is supported by the fact that fungal sRNAs differentially accumulate during the infection process [56,57]. Moreover, the aggressive fungal plant pathogen Botrytis cinerea produces sRNAs (Bc-sRNAs) that move into the host plant cell during early infection and hijack the host AGO, the key protein in the RNAi machinery, to silence important host immunity genes [56]. This observation points to the bi-directional nature of cross-kingdom RNAi in plant-pathogen interactions (Figure 2A). Some Bc-sRNAs structurally mimic plant sRNAs that specifically bind to Arabidopsis AGO1 (AtAGO1) and target genes involved in plant defense against B. cinerea infection. Similar results were obtained also in tomato [56]. By using stringent target prediction criteria, more than 70 Bc-siRNAs that are enriched during infection have predicted host targets in both Arabidopsis and tomato. It is worthwhile to investigate whether similar sRNA effectors that suppress host immunity also exist in other pathogens or pests.
B. cinerea possesses two DCL genes, both of which are required for the production of mobile Bc-sRNAs. Gene knockout of both DCLs in B. cinerea led to reduced virulence capacities due to the absence of plant immune-suppressing Bc-sRNAs [56]. The majority of predicted Bc-sRNA effectors (including the three experimentally confirmed Bc-sRNAs: Bc-siR3.1, Bc-siR3.2 and Bc-siR5) are mapped to clusters within long-terminal repeat (LTR) retrotransposons in the genome of B. cinerea. Retrotransposons are hot spots of sRNA production for transposon silencing, a mechanism called quelling in fungi. Interestingly, these Botrytis LTR retrotransposons, called Boty elements, are genetically associated with virulence and host preference in natural populations of B. cinerea [58], supporting the notion that these Boty elements give rise to sRNA effectors that enhance the pathogenicity of B. cinerea. Bc-sRNA effectors are physically linked to Boty elements and may facilitate fast turnover of Bc-sRNAs, which may provide an evolutionary advantage to pathogens in the arms race against host plants [4]. Similarly, fungal and oomycete protein effector genes are also enriched in the retrotransposon regions [59].
Taking advantage of environmental RNAi in invertebrates, scientists have engineered crop plants to express artificial sRNAs that can silence essential genes of plant-parasitic nematodes and herbivores (Figure 2B & 2C) [52,60–62]. Diverse host plant species have been successfully engineered to manipulate interacting pests in order to limit their virulence or to reduce their fecundity on host plants, to achieve advanced host resistance [51]. Furthermore, mobile silencing signals are not limited only to sRNAs. HIGS in the pest cotton bollworm was retained when they were fed on dcl2dcl3dcl4 triple mutant Arabidopsis plants, suggesting that long dsRNA precursor rather than mature siRNAs are translocated, and which are likely to be processed in the bollworm to be functional [63]. This observation is consistent with long dsRNA uptake by insects and nematodes.
The animal-parasitic nematode H. polygyrus secretes vesicular miRNAs to suppress host immunity. It is well known that plant-parasitic nematodes feed on roots of plants that cause damages of the root system leading to reduced plant health and biomass production. Whether plant parasites also generate natural sRNA silencing signals to be transloated into host cells has yet to be explored. Thus, further research on the ability of pests and plant-parasitic nematodes to generate extracellular sRNAs that target plant immunity genes using host sRNA transport systems is of particular importance to the future of crop production (Figure 2B & 2C).
Mobile sRNAs or long dsRNAs, as cross-kingdom RNAi triggers, are fascinating; yet, it is enigmatic how these RNA molecules “travel,” sometimes over long distances through diverse cellular boundaries between plants and interacting organisms. Cell-to-cell movement of plant sRNAs has previously been studied [8]. It is likely that mobile pathogen sRNAs can spread similarly from the site of infection into adjacent cells and impact the surrounding plant tissue. Importantly, sRNA transfer is not a random process through a concentration gradient, but rather a selective transport of functional sRNAs [64–67]. This is supported by the fact that profiles of mobile sRNA pools are very different from the total sRNA populations within the cells. Such selective transport mechanisms could likely be overcome when the concentration of silencing signals reaches a high level, as in the case of HIGS. RNA-protective factors such as AGOs, other RNA-binding proteins, or encapsulation into extracellular vesicles likely play important roles in protecting mobile RNAs against degradation during transport [13,14]. These RNA-binding proteins or transport machinery may also be involved in the sRNA selection process.
The parasitic plant dodder (Cuscuta pentagona) establishes a symplastic junction—via a haustorium—with their hosts to gain access to water and nutrients. Bidirectional transfer of thousands of mRNAs between Cuscuta and two hosts, Arabidopsis and tomato, has been observed [68,69]. Host mRNA transcripts were tracked back in the dodder parenchyma at a distance of up to 30 cm away from the tomato/dodder connection [70]. Because the profiles of the transferred parasite mRNAs and the total mRNAs within the invaded cells are rather different, it is likely selective transport is responsible. However, the fate and function of these transferred mRNAs remains unclear, e.g. whether these transferred transcripts are translated into functional proteins, or are simply degraded. The evidence of mRNA exchange makes it very likely that sRNAs that affect gene expression also travel bi-directionally via the haustorium (Figure 2D). This is supported by the successful application of HIGS against Cuscuta [71].
Mobile RNAs as ligands of Toll-Like Receptors in immune signaling
Circulating miRNAs have been shown to be internalized by recipient cells functioning as gene expression regulators. Recent studies revealed that miRNAs can also act as ligands of Toll-Like Receptors (TLRs). TLRs are a conserved family of receptor proteins that play a major role in immune signaling in animals and plants. Two exosomal tumor-related miRNAs bind to murine TLR7 and human TLR8 in immune cells, activating a prometastatic inflammatory response [72]. Interestingly, the mouse TLR13 recognizes a conserved 23S ribosomal RNA molecule of the bacterial pathogen Staphylococcus aureus and triggers an immune response [73]. These findings suggest that conserved nucleic acids can serve in pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI). In the model plant A. thaliana, treatment of bacterial plasmid DNA was able to elicit PTI. DNA-induced PTI was abolished when plants were pre-treated with endocytosis-inhibitory chemicals suggesting that uptake of bacterial DNA was endocytosis-dependent [74]. Whether mobile sRNAs of plant-interacting pathogens/pests/parasites or symbionts act as signals to trigger a plant immune response, perhaps by binding to TLRs or other types of receptor-like proteins, needs to be investigated.
Biotechnological use of mobile sRNAs in plants
The discovery of sRNAs as mobile gene regulators creates exciting new opportunities to further investigate plant-pathogen interactions and to develop novel strategies for plant defense against pathogens and pests [50,51]. This is supported by the fact that HIGS has effectively worked in a variety of plant species against diverse plant herbivores, nematodes, and filamentous pathogens, when targeting important virulence genes. HIGS is also a well-established tool in specific host plant cultivars against particular pathogen strains under controlled lab-scale conditions. An important step remains to test the broader applicability of HIGS under field conditions, where HIGS plants are exposed to fluctuating environmental stresses that include pathogen and pest populations containing tremendous genetic variability, rather than clonal pathogens.
The fact that sRNA transport has been observed in plant-pathogen, plant-parasite, or plant-symbiotic interactions increases the possibility that beneficial fungi or unarmed pathogens (with essential virulence genes deleted) can be engineered to successfully manipulate plant physiology via trans-kingdom gene silencing (Figure 2E). Moreover, targeting pathogen mRNAs via harmless plant-interacting, organism-transmitted RNAi signals into associated plants has the potential to help defeat a broad range of pathogens and pests in a transgene-free plant framework. Thus, understanding the molecular mechanisms of RNA communications and transport between plants and interacting organisms will help improve RNA silencing-based technologies. While genetically modified crops remain a concern to some consumers, our advances in understanding cross-kingdom RNAi may help alleviate public concerns.
Other applications of mobile sRNAs in plants are currently being discussed in regards to metabolic engineering and systemic-induced resistance [75,76]. Last but not least, food RNAi might become an important component of plant food-based technologies in the future [77]. Feeding studies revealed that oral uptake of sRNA-containing nutrients led to accumulation of food-borne sRNAs in body fluids and organs, indicating that high-dosage sRNAs can partially survive the intestinal track [78]. It is currently under investigation and debate whether food-borne sRNAs have any negative or positive impacts on the physiology of individuals who consume foods containing abundant sRNAs [79–82].
Conclusions
RNAs are considered to have cell-autonomous functions in gene expression and protein synthesis. Despite the fact that RNAs are vulnerable targets for nucleases, they are able survive outside of a cell. Functional extracellular sRNAs move cell to cell and over long distances in plants, spread systemically in pests, and circulate via body fluids in mammals. Moreover, recent findings have demonstrated the uni- or bidirectional cellular exchange of sRNAs as silencing signals between hosts and pathogens/pests/parasites, or symbionts, in a phenomenon called cross-kingdom RNAi. Cross-kingdom RNAi influences host-pathogen interactions, e.g. sRNAs from the plant pathogen B. cinerea and the mammalian parasite helminth nematode H. polygyrus translocate into host cells and suppress host immunity genes. On the other hand, however, plant-produced RNAi signals silence pathogen and pest genes, providing host resistance in a transgenic approach called HIGS. We speculate that additional pathogens also produce sRNAs for host immune suppression, while plants produce natural mobile sRNAs for defense by silencing genes in the interacting organisms.
Cell-to-cell transport mechanisms must exist for cross-kingdom RNAi. Secretion pathways and cellular uptake of RNAs have been described in animals. The nematode C. elegans has evolved unique RNA transporters (SID-1, SID-2) that are required for dsRNA uptake and systemic silencing. In mammals, functional miRNAs circulate through body fluids, often encapsulated in vesicles called exosomes. Release and uptake of vesicular sRNAs is mediated via endocytosis and exocytosis. Circulating miRNAs are probably protected against degradation by RNA-associated proteins, such as silencing proteins, AGO2 and GW182. We speculate that similar sRNA transport mechanisms, perhaps vesicle-based, also exist in plants and fungi. Remarkably, the helminth H. polygyrus secretes miRNAs in exosomal-like vesicles that are taken up by mammalian cells. Released nematode miRNAs target immune-related mRNAs and potentially suppress host immunity. We hypothesize that additional parasites and pathogens also hijack conserved RNA transport mechanisms existing in their hosts to shuttle virulent sRNAs into host cells for immune suppression.
Several cases of cross-kingdom gene silencing have been observed, sometimes between interacting organisms that are phylogenetically unrelated, such as plants and fungi/insects/nematodes/symbionts, nematodes and bacteria, or mammals and parasites/nematodes. Some pathogen-secreted sRNAs mimic the endogenous sRNAs of their hosts, such as Botrytis sRNAs and Heligmosomoides miRNAs. Furthermore, RNA-mediated gene silencing is a ubiquitous phenomenon that exists in almost all eukaryotes, which always follows the principle of complementary nucleotide base pairing between regulatory sRNA and mRNA sequences. Despite the tremendous differences that are present in the structural features of regulatory RNAs (e.g. the differences between E. coli non-coding RNAs OxyS and DsrA and eukaryotic host siRNAs/miRNAs) and the completely unrelated or highly divergent RNA gene-silencing mechanisms and pathways that have evolved in diverse organisms, complementary sequence matches seem to be sufficient enough to trigger cross-kingdom gene-silencing. It seems that having intact RNAi machinery is not absolutely necessary. Cross-kingdom RNAi will be a valuable tool for future use in the development of novel therapeutic disease control and crop protection.
Highlights.
sRNAs can trigger cell non-autonomous gene silencing.
Cross-kingdom RNAi is important for communication between interactive organisms.
HIGS is a powerful tool for engineering pathogen-resistant crops.
Pathogen-derived sRNAs are translocated into host cells to suppress host immunity.
Mobile sRNA-based tools have great potential in human health and crop protection.
Acknowledgments
We apologize that we would not be able to include and cite many related interesting studies due to the limited space. Work in Jin’s laboratory was supported by an NIH grant (R01 GM093008), an NSF Career Award (MCB-0642843), an NSF Award (IOS-1257576), California Department Food & Agriculture Award CDFA-SCB12057, Citrus Research Board Award (5100-131), and an AES-CE Award (PPA-7517H) to H.J. We thank Yifan E. Lii for critical reading of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Seo JK, Wu J, Lii Y, Li Y, Jin H. Contribution of small RNA pathway components in plant immunity. Mol Plant Microbe Interact. 2013;26:617–625. doi: 10.1094/MPMI-10-12-0255-IA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Staiger D, Korneli C, Lummer M, Navarro L. Emerging role for RNA-based regulation in plant immunity. New Phytol. 2013;197:394–404. doi: 10.1111/nph.12022. [DOI] [PubMed] [Google Scholar]
- 3.Yang L, Huang H. Roles of small RNAs in plant disease resistance. J Integr Plant Biol. 2014;56:962–970. doi: 10.1111/jipb.12200. [DOI] [PubMed] [Google Scholar]
- 4.Weiberg A, Wang M, Bellinger M, Jin H. Small RNAs: A New Paradigm in Plant-Microbe Interactions. Annu Rev Phytopathol. 2014;52:495–516. doi: 10.1146/annurev-phyto-102313-045933. [DOI] [PubMed] [Google Scholar]
- 5.Molnar A, Melnyk C, Baulcombe DC. Silencing signals in plants: a long journey for small RNAs. Genome Biol. 2011;12:215. doi: 10.1186/gb-2010-11-12-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melnyk CW, Molnar A, Baulcombe DC. Intercellular and systemic movement of RNA silencing signals. EMBO J. 2011;30:3553–3563. doi: 10.1038/emboj.2011.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brosnan CA, Voinnet O. Cell-to-cell and long-distance siRNA movement in plants: mechanisms and biological implications. Curr Opin Plant Biol. 2011;14:580–587. doi: 10.1016/j.pbi.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 8.Dunoyer P, Schott G, Himber C, Meyer D, Takeda A, Carrington JC, Voinnet O. Small RNA duplexes function as mobile silencing signals between plant cells. Science. 2010;328:912–916. doi: 10.1126/science.1185880. [DOI] [PubMed] [Google Scholar]
- 9.Molnar A, Melnyk CW, Bassett A, Hardcastle TJ, Dunn R, Baulcombe DC. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science. 2010;328:872–875. doi: 10.1126/science.1187959. [DOI] [PubMed] [Google Scholar]
- 10.Sarkies P, Miska EA. Small RNAs break out: the molecular cell biology of mobile small RNAs. Nat Rev Mol Cell Biol. 2014;15:525–535. doi: 10.1038/nrm3840. [DOI] [PubMed] [Google Scholar]
- 11.Whangbo JS, Hunter CP. Environmental RNA interference. Trends Genet. 2008;24:297–305. doi: 10.1016/j.tig.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Jose AM, Garcia GA, Hunter CP. Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Nat Struct Mol Biol. 2011;18:1184–1188. doi: 10.1038/nsmb.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *13.Mittelbrunn M, Sanchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol. 2012;13:328–335. doi: 10.1038/nrm3335. This opinion piece reviews the concept of cell-to-cell communication in mammals and exosomal miRNA transport and summarizes the modes of sRNA exchange. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knip M, Constantin ME, Thordal-Christensen H. Trans-kingdom Cross-Talk: Small RNAs on the Move. PLoS Genet. 2014;10:e1004602. doi: 10.1371/journal.pgen.1004602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang H, Zen K, Zhang J, Zhang CY, Chen X. New roles for microRNAs in cross-species communication. RNA Biol. 2013;10:367–370. doi: 10.4161/rna.23663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 17.Palauqui JC, Elmayan T, Pollien JM, Vaucheret H. Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 1997;16:4738–4745. doi: 10.1093/emboj/16.15.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voinnet O, Baulcombe DC. Systemic signalling in gene silencing. Nature. 1997;389:553. doi: 10.1038/39215. [DOI] [PubMed] [Google Scholar]
- 19.McEwan DL, Weisman AS, Hunter CP. Uptake of extracellular double-stranded RNA by SID-2. Mol Cell. 2012;47:746–754. doi: 10.1016/j.molcel.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shih JD, Hunter CP. SID-1 is a dsRNA-selective dsRNA-gated channel. RNA. 2011;17:1057–1065. doi: 10.1261/rna.2596511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heard E, Martienssen RA. Transgenerational epigenetic inheritance: myths and mechanisms. Cell. 2014;157:95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bond DM, Baulcombe DC. Small RNAs and heritable epigenetic variation in plants. Trends Cell Biol. 2014;24:100–107. doi: 10.1016/j.tcb.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 23.Luna E, Ton J. The epigenetic machinery controlling transgenerational systemic acquired resistance. Plant Signal Behav. 2012;7:615–618. doi: 10.4161/psb.20155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rasmann S, De Vos M, Jander G. Ecological role of transgenerational resistance against biotic threats. Plant Signal Behav. 2012;7:447–449. doi: 10.4161/psb.19525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D, Jr, Mello CC. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skopelitis DS, Husbands AY, Timmermans MC. Plant small RNAs as morphogens. Curr Opin Cell Biol. 2012;24:217–224. doi: 10.1016/j.ceb.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 28.Furuta K, Lichtenberger R, Helariutta Y. The role of mobile small RNA species during root growth and development. Curr Opin Cell Biol. 2012;24:211–216. doi: 10.1016/j.ceb.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 29.Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vaten A, Thitamadee S, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benkovics AH, Timmermans MC. Developmental patterning by gradients of mobile small RNAs. Curr Opin Genet Dev. 2014;27:83–91. doi: 10.1016/j.gde.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 31.Marin-Gonzalez E, Suarez-Lopez P. “And yet it moves”: cell-to-cell and long-distance signaling by plant microRNAs. Plant Sci. 2012;196:18–30. doi: 10.1016/j.plantsci.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 32.Saleh MC, Tassetto M, van Rij RP, Goic B, Gausson V, Berry B, Jacquier C, Antoniewski C, Andino R. Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature. 2009;458:346–350. doi: 10.1038/nature07712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhuang JJ, Hunter CP. RNA interference in Caenorhabditis elegans: uptake, mechanism, and regulation. Parasitology. 2012;139:560–573. doi: 10.1017/S0031182011001788. [DOI] [PubMed] [Google Scholar]
- 34.Salido-Guadarrama I, Romero-Cordoba S, Peralta-Zaragoza O, Hidalgo-Miranda A, Rodriguez-Dorantes M. MicroRNAs transported by exosomes in body fluids as mediators of intercellular communication in cancer. Onco Targets Ther. 2014;7:1327–1338. doi: 10.2147/OTT.S61562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan ML, Karlsson JM, Baty CJ, Gibson GA, Erdos G, Wang Z, et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119:756–766. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, Bernad A, Sanchez-Madrid F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol. 2014;14:195–208. doi: 10.1038/nri3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarkies P, Miska EA. Molecular biology. Is there social RNA? Science. 2013;341:467–468. doi: 10.1126/science.1243175. [DOI] [PubMed] [Google Scholar]
- **39.Liu H, Wang X, Wang HD, Wu J, Ren J, Meng L, Wu Q, Dong H, Wu J, Kao TY, et al. Escherichia coli noncoding RNAs can affect gene expression and physiology of Caenorhabditis elegans. Nat Commun. 2012;3:1073. doi: 10.1038/ncomms2071. This papers describes the gene expression regulation of C. elegans mRNAs by two conserved endogenous E. coli non-coding RNAs through bacterial feeding resulting in physiological consequences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **40.LaMonte G, Philip N, Reardon J, Lacsina JR, Majoros W, Chapman L, Thornburg CD, Telen MJ, Ohler U, Nicchitta CV, et al. Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host Microbe. 2012;12:187–199. doi: 10.1016/j.chom.2012.06.007. This article illustrates how the human sickle cell erythrocyte-produced miRNA-451 is translocated into the malaria-causing parasite Plasmodium palcifarum and blocks translation of the Plasmodium PKA-R mRNA, resulting in reduced parasitemia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **41.Buck AH, Coakley G, Simbari F, McSorley HJ, Quintana JF, Le Bihan T, Kumar S, Abreu-Goodger C, Lear M, Harcus Y, et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat Commun. 2014;5:5488. doi: 10.1038/ncomms6488. This report provides genetic and microscopic evidence that the helminth nematode Heligmosomoides polygyrus secretes miRNA-loaded vesicles similar to mammalian exosomes that are internalized by mouse cells and suppress host immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 44.Garcia-Silva MR, das Neves RF, Cabrera-Cabrera F, Sanguinetti J, Medeiros LC, Robello C, Naya H, Fernandez-Calero T, Souto-Padron T, de Souza W, et al. Extracellular vesicles shed by Trypanosoma cruzi are linked to small RNA pathways, life cycle regulation, and susceptibility to infection of mammalian cells. Parasitol Res. 2014;113:285–304. doi: 10.1007/s00436-013-3655-1. [DOI] [PubMed] [Google Scholar]
- 45.Cheng G, Luo R, Hu C, Cao J, Jin Y. Deep sequencing-based identification of pathogen-specific microRNAs in the plasma of rabbits infected with Schistosoma japonicum. Parasitology. 2013;140:1751–1761. doi: 10.1017/S0031182013000917. [DOI] [PubMed] [Google Scholar]
- 46.Dang Y, Yang Q, Xue Z, Liu Y. RNA interference in fungi: pathways, functions, and applications. Eukaryot Cell. 2011;10:1148–1155. doi: 10.1128/EC.05109-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang SS, Zhang Z, Liu Y. RNA interference pathways in fungi: mechanisms and functions. Annu Rev Microbiol. 2012;66:305–323. doi: 10.1146/annurev-micro-092611-150138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee HC, Li L, Gu W, Xue Z, Crosthwaite SK, Pertsemlidis A, Lewis ZA, Freitag M, Selker EU, Mello CC, et al. Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol Cell. 2010;38:803–814. doi: 10.1016/j.molcel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin H, Zhu JK. How many ways are there to generate small RNAs? Mol Cell. 2010;38:775–777. doi: 10.1016/j.molcel.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nunes CC, Dean RA. Host-induced gene silencing: a tool for understanding fungal host interaction and for developing novel disease control strategies. Mol Plant Pathol. 2012;13:519–529. doi: 10.1111/j.1364-3703.2011.00766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koch A, Kogel KH. New wind in the sails: improving the agronomic value of crop plants through RNAi-mediated gene silencing. Plant Biotechnol J. 2014;12:821–831. doi: 10.1111/pbi.12226. [DOI] [PubMed] [Google Scholar]
- 52.Vega-Arreguin JC, Jalloh A, Bos JI, Moffett P. Recognition of an Avr3a homologue plays a major role in mediating nonhost resistance to Phytophthora capsici in Nicotiana species. Mol Plant Microbe Interact. 2014;27:770–780. doi: 10.1094/MPMI-01-14-0014-R. [DOI] [PubMed] [Google Scholar]
- *53.Yin C, Park JJ, Gang DR, Hulbert SH. Characterization of a tryptophan 2-monooxygenase gene from Puccinia graminis f. sp. tritici involved in auxin biosynthesis and rust pathogenicity. Mol Plant Microbe Interact. 2014;27:227–235. doi: 10.1094/MPMI-09-13-0289-FI. The authors used host-induced gene silencing as a reverse genetics tool to study the function of the pathogenicity-related gene Iaam, encoding for a putative tryptophan 2-monooxygenase involved in auxin production, in the non-transformable rust fungus Puccinia graminis. [DOI] [PubMed] [Google Scholar]
- *54.Helber N, Wippel K, Sauer N, Schaarschmidt S, Hause B, Requena N. A versatile monosaccharide transporter that operates in the arbuscular mycorrhizal fungus Glomus sp is crucial for the symbiotic relationship with plants. Plant Cell. 2011;23:3812–3823. doi: 10.1105/tpc.111.089813. This is the first report using host-induced gene silencing to study gene function in an arbuscluar mycorrhiza fungus, Glomus sp. The silencing a monosaccharide transporter resulted in impaired mycorrhiza formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **55.Koch A, Kumar N, Weber L, Keller H, Imani J, Kogel KH. Host-induced gene silencing of cytochrome P450 lanosterol C14alpha-demethylase-encoding genes confers strong resistance to Fusarium species. Proc Natl Acad Sci U S A. 2013;110:19324–19329. doi: 10.1073/pnas.1306373110. This article describes that classical fungicide targets are well-chosen targets for host-induced gene silencing to engineer robust disease resistance. Futhermore, an in vitro feedling assay of antisense RNA molecules demonstrates the capability to induce enviromental RNAi in fungi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **56.Weiberg A, Wang M, Lin FM, Zhao H, Zhang Z, Kaloshian I, Huang HD, Jin H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science. 2013;342:118–123. doi: 10.1126/science.1239705. This work demonstrates that the fungal plant pathogen Botrytis cinerea uses mobile siRNAs that mimic plant siRNAs to suppress host mRNAs involved in plant immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raman V, Simon SA, Romag A, Demirci F, Mathioni SM, Zhai J, Meyers BC, Donofrio NM. Physiological stressors and invasive plant infections alter the small RNA transcriptome of the rice blast fungus, Magnaporthe oryzae. BMC Genomics. 2013;14:326. doi: 10.1186/1471-2164-14-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *58.Martinez F, Dubos B, Fermaud M. The Role of Saprotrophy and Virulence in the Population Dynamics of Botrytis cinerea in Vineyards. Phytopathology. 2005;95:692–700. doi: 10.1094/PHYTO-95-0692. In this study transposon-containing isolates of Botrytis cinerea (transposa) caused significantly more disease incidence and disease severity on tested grape berries compared to transposon-free Botrytis isolates (vacuma). This observation has been confirmed in terms of host preferences and elevated aggressiveness of transposa isolates among independent genetical studies of Botrytis field populations, worldwide. [DOI] [PubMed] [Google Scholar]
- 59.Raffaele S, Kamoun S. Genome evolution in filamentous plant pathogens: why bigger can be better. Nat Rev Microbiol. 2012;10:417–430. doi: 10.1038/nrmicro2790. [DOI] [PubMed] [Google Scholar]
- 60.Katoch R, Sethi A, Thakur N, Murdock LL. RNAi for insect control: current perspective and future challenges. Appl Biochem Biotechnol. 2013;171:847–873. doi: 10.1007/s12010-013-0399-4. [DOI] [PubMed] [Google Scholar]
- 61.Zhang H, Li HC, Miao XX. Feasibility, limitation and possible solutions of RNAi-based technology for insect pest control. Insect Sci. 2013;20:15–30. doi: 10.1111/j.1744-7917.2012.01513.x. [DOI] [PubMed] [Google Scholar]
- 62.Lilley CJ, Davies LJ, Urwin PE. RNA interference in plant parasitic nematodes: a summary of the current status. Parasitology. 2012;139:630–640. doi: 10.1017/S0031182011002071. [DOI] [PubMed] [Google Scholar]
- 63.Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ, Huang YP, Chen XY. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol. 2007;25:1307–1313. doi: 10.1038/nbt1352. [DOI] [PubMed] [Google Scholar]
- 64.Nolte-’t Hoen EN, Buermans HP, Waasdorp M, Stoorvogel W, Wauben MH, t Hoen PA. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40:9272–9285. doi: 10.1093/nar/gks658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guduric-Fuchs J, O’Connor A, Camp B, O’Neill CL, Medina RJ, Simpson DA. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics. 2012;13:357. doi: 10.1186/1471-2164-13-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *66.Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, van Eijndhoven MA, Sadek P, Sie D, Zini N, Middeldorp JM, Ylstra B, de Menezes RX, et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014;8:1649–1658. doi: 10.1016/j.celrep.2014.08.027. This article revealed a criteria for sorting small RNAs with 3′uridylation into exosomes by bioinformatical and statiscitcal analysis of whole cell versus exosomal sRNA sequencing of human B-cells. [DOI] [PubMed] [Google Scholar]
- 67.Villarroya-Beltri C, Baixauli F, Gutierrez-Vazquez C, Sanchez-Madrid F, Mittelbrunn M. Sorting it out: regulation of exosome loading. Semin Cancer Biol. 2014;28:3–13. doi: 10.1016/j.semcancer.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *68.Kim G, LeBlanc ML, Wafula EK, dePamphilis CW, Westwood JH. Plant science. Genomic-scale exchange of mRNA between a parasitic plant and its hosts. Science. 2014;345:808–811. doi: 10.1126/science.1253122. The authors demonstrate massive biodirectional exchange of mRNAs between the parasitic plant Cuscuta and its hosts during parasitism. [DOI] [PubMed] [Google Scholar]
- 69.Leblanc M, Kim G, Westwood JH. RNA trafficking in parasitic plant systems. Front Plant Sci. 2012;3:203. doi: 10.3389/fpls.2012.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.David-Schwartz R, Runo S, Townsley B, Machuka J, Sinha N. Long-distance transport of mRNA via parenchyma cells and phloem across the host-parasite junction in Cuscuta. New Phytol. 2008;179:1133–1141. doi: 10.1111/j.1469-8137.2008.02540.x. [DOI] [PubMed] [Google Scholar]
- 71.Alakonya A, Kumar R, Koenig D, Kimura S, Townsley B, Runo S, Garces HM, Kang J, Yanez A, David-Schwartz R, et al. Interspecific RNA interference of SHOOT MERISTEMLESS-like disrupts Cuscuta pentagona plant parasitism. Plant Cell. 2012;24:3153–3166. doi: 10.1105/tpc.112.099994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *72.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110–2116. doi: 10.1073/pnas.1209414109. This is the first report showing the binding of exosomal miRNAs to TLR receptor protein family members using confocal co-localization and TLR-specific immunoprecipitation of miRNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **73.Oldenburg M, Kruger A, Ferstl R, Kaufmann A, Nees G, Sigmund A, Bathke B, Lauterbach H, Suter M, Dreher S, et al. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science. 2012;337:1111–1115. doi: 10.1126/science.1220363. This study reveals for the first time that conserved nucleic acids such as rRNA molecules of microbial pathogens elicit pathogen-associated molecular pattern (PAMP)-triggered immunity via recognition by a TLR receptor. [DOI] [PubMed] [Google Scholar]
- 74.Yakushiji S, Ishiga Y, Inagaki Y, Toyoda K, Shiraishi T, YI Bacterial DNA activates immunity in Arabidopsis thaliana. Journal of General Plant Pathology. 2009;75:227–234. [Google Scholar]
- 75.McGarry RC, Kragler F. Phloem-mobile signals affecting flowers: applications for crop breeding. Trends Plant Sci. 2013;18:198–206. doi: 10.1016/j.tplants.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 76.Saurabh S, Vidyarthi AS, Prasad D. RNA interference: concept to reality in crop improvement. Planta. 2014;239:543–564. doi: 10.1007/s00425-013-2019-5. [DOI] [PubMed] [Google Scholar]
- 77.Hirschi KD. New foods for thought. Trends Plant Sci. 2012;17:123–125. doi: 10.1016/j.tplants.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 78.Liang G, Zhu Y, Sun B, Shao Y, Jing A, Wang J, Xiao Z. Assessing the survival of exogenous plant microRNA in mice. Food Sci Nutr. 2014;2:380–388. doi: 10.1002/fsn3.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang L, Hou D, Chen X, Li D, Zhu L, Zhang Y, Li J, Bian Z, Liang X, Cai X, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22:107–126. doi: 10.1038/cr.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Melnik BC, John SM, Schmitz G. Milk is not just food but most likely a genetic transfection system activating mTORC1 signaling for postnatal growth. Nutr J. 2013;12:103. doi: 10.1186/1475-2891-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen X, Zen K, Zhang CY. Reply to Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat Biotechnol. 2013;31:967–969. doi: 10.1038/nbt.2741. [DOI] [PubMed] [Google Scholar]
- 82.Dickinson B, Zhang Y, Petrick JS, Heck G, Ivashuta S, Marshall WS. Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat Biotechnol. 2013;31:965–967. doi: 10.1038/nbt.2737. [DOI] [PubMed] [Google Scholar]