Abstract
Pitx2 is a bicoid-related homeobox transcription factor implicated in regulating left-right patterning and organogenesis. However, only a limited number of Pitx2 downstream target genes have been identified and characterized. Here we demonstrate that Pitx2 is a transcriptional repressor of DEP domain containing 1B (DEPDC1B). The first intron of the human and mouse DEP domain containing 1B genes contains multiple consensus DNA-binding sites for Pitx2. Chromatin immunoprecipitation assays revealed that Pitx2, along with histone deacetylase 1, was recruited to the first intron of Depdc1b. In contrast, RNAi-mediated depletion of Pitx2 not only enhanced the acetylation of histone H4 in the first intron of Depdc1b, but also increased the protein level of Depdc1b. Luciferase reporter assays also showed that Pitx2 could repress the transcriptional activity mediated by the first intron of human DEPDC1B. The GAP domain of DEPDC1B interacted with nucleotide-bound forms of RAC1 in vitro. In addition, exogenous expression of DEPDC1B suppressed RAC1 activation and interfered with actin polymerization induced by the guanine nucleotide exchange factor TRIO. Moreover, DEPDC1B interacted with various signaling molecules such as U2af2, Erh, and Salm. We propose that Pitx2-mediated repression of Depdc1b expression contributes to the regulation of multiple molecular pathways, such as Rho GTPase signaling.
Keywords: GTPase signaling, transcriptional regulation, Pitx2, DEPDC1B
Introduction
Pitx2 is a bicoid-related homeobox transcription factor. Mutations of the Pitx2 gene in humans lead to Rieger’s syndrome, an autosomal dominant genetic human disease characterized by ocular, craniofacial, and umbilical abnormalities as well as defects in cardiac, limb, and pituitary development [1, 2]. As one of the downstream targets for Sonic Hedgehog and Nodal, Pitx2 also plays a critical role in controlling left-right patterning during embryonic development [3, 4]. Accumulating evidence further demonstrates that hypermethylation of the Pitx2 gene serves as a prognostic marker for human cancer [5, 6]. However, it is still poorly understood how Pitx2 mediates such a wide-range of cellular and developmental processes.
As a transcription factor, Pitx2 is likely to control left-right patterning, organogenesis, and tumor development through regulation of its downstream target genes at the transcriptional level. Pitx2 is a bicoid-related protein that is characterized by a lysine residue at position 50 in the third helix of the homeodomain. Like the bicoid protein in Drosophila, Pitx2 in various organisms also binds to the consensus TAATCC DNA-binding site in promoters and enhancers [7-10]. It has been demonstrated that Pitx2 can regulate the expression of a group of genes such as PLOD (procollagen lysyl hydroxylase), Dlx2, ANF (atrial natriuretic factor), cyclin D1/D2, p21, MYC, Amelogenin (Amel), and the T-box genes at the transcriptional level [9-16]. Through regulation of its downstream target genes, Pitx2 is implicated in controlling various biological processes such as cell migration and cell proliferation [12-14, 17]. In particular, fate-mapping studies in mice have shown that Pitx2 is required for the regulation of cell migration during craniofacial and hypothalamus development [18, 19].
Rho GTPase signaling has been implicated in the regulation of cell migration during embryonic development [20-25]. As the major components of Rho GTPase signaling, Rho GTPase proteins such as RHOA, RAC1, and CDC42 are primarily activated by guanine nucleotide exchange factors (GEFs) and inactivated by GTPase-activating proteins (GAPs) [26]. There are at least three Pitx2 isoforms (Pitx2a, 2b, and 2c) that are generated by alternative splicing [27]. We have reported previously that inducible expression of Pitx2a in HeLa cells leads to the activation of RAC1 and RHOA [28]. In this study, we have demonstrated that Pitx2 can inhibit the expression of a GTPase-activating protein termed DEP domain containing 1B (DEPDC1B) at the transcriptional level. In addition, exogenous expression of DEPDC1B leads to the inhibition of RAC1 activation. Furthermore, DEPDC1B interacts with various signaling molecules ranging from splicing regulators to transmembrane proteins, pointing to a direction that Pitx2-mediated repression of DEPDC1B expression is implicated in the regulation of multiple biological processes.
Materials and Methods
Reverse transcription polymerase chain reaction (RT-PCR) and Plasmids
Total RNAs were isolated from Pitx2a inducible HeLa cells using the TRI reagent according to the manufacturer’s instructions (Sigma). The superscript first-strand synthesis system was used to synthesize the human DEPDC1B cDNA according to the manufacturer’s instructions (Life technologies). The following primers were used to amplify the human DEPDC1B cDNA: 5′-ATGGAGCATCGCATCGTGGGG-3′ (forward primer) and 5′-TTACATTCGAAAACTTCTAGT-3′ (reverse primer). The full-length cDNA of human DEPDC1B was cloned into the pEGFP-C3 vector, generating pEGFP-C3-DEPDC1B. The cDNAs corresponding to amino acid residues 1-151, 151-529, and 171-400 of human DEPDC1B were cloned into pEGFP-C3, pEGFP-C1, and pET-28a vectors, respectively. The full-length cDNA of human DEPDC1B was also cloned into the pGEX-6 vector. The full-length cDNAs of mouse U2af2 and Erh were cloned into pCS3+MT and pGEX-6 vectors, respectively. The Salm1 plasmid was provided by Dr. Robert J. Wenthold (NIH). The TRIO plasmids were provided by Dr. Anne Blangy (Centre de Recherche de Biochimie Macromoléculaire, France).
Cell culture
Pitx2a inducible HeLa-Tet-On cell line was generated and maintained as described previously [28, 29]. U2OS, C2C12, and NIH3T3 cells were purchased from ATCC (American Type Culture Collection) and grown in DMEM supplemented with 10% fetal bovine serum. Lipofectamine was used for transfection with siRNA and plasmids according to the manufacturer’s instructions (Life Technologies). siRNA against mouse Pitx2 has been described previously [29].
Immunofluorescence analysis
Cells grown on coverslips were fixed with 4% paraformaldehyde for 12 min at 23°C. After blocking with 1% bovine serum albumin for 1 h at 23°C, the fixed cells were incubated with primary antibodies for 3 h at 23°C or overnight at 4°C. This was followed by an incubation with secondary antibodies for 40 min at 23°C. The primary antibody used for immunofluorescence analysis was mouse anti-MYC (1:1000; 9E10; catalogue number: sc-40; Santa Cruz). Alexa Fluor 594 goat anti-mouse IgG (1:500) and Alexa Fluor 350 goat anti-mouse IgG (1:250) were purchased from Life Technologies. Actin filaments were stained with Alexa Fluor 594 phalloidin (Life Technologies). The coverslips were mounted using a Prolong antifade kit (Life Technologies). Images were collected using the Nikon TiE Perfect Focus Digital Fluorescence Imaging System (Morrell Instrument Company, Inc.) with an Andor Zyla sCMOS 2560×2160 camera. The images were processed by deconvolution.
Quantification of actin filaments
To assess the effect of DEPDC1B on actin polymerization induced by TRIOGEFD1 (the RAC1-specific GEF domain of TRIO), the transfected HeLa cells were stained with Alexa Fluor 594 phalloidin and the fluorescence intensities in transfected cells were quantitated using the NIH ImageJ software [30, 31]. Briefly, the freehand selection tool was used to mark the cells of interest in the same field. Regions next to cells were also selected and served as background. The corrected total cell fluorescence (CTCF) was calculated based on the integrated density from transfected and surrounding untransfected cells as well as background readings: CTCF=integrated density – (area of selected cell × mean fluorescence of background readings). A greater than 2-fold increase in the CTCF of transfected cells as compared with that of surrounding untransfected cells was used as the threshold to indicate an increase in actin filament formation.
Luciferase reporter assays
C2C12 cells were transfected with the reporter plasmid I2.2-LUC, along with the β-galactosidase expression plasmid. 24 h after the transfection, the transfected cells were lysed in reporter lysis buffer (Promega), followed by centrifugation to remove cell debris. Luciferase activity was assessed using the Reporter™ Microplate Luminometer (Turner BioSystems). The light units were normalized to β-galactosidase activity.
Expression and purification of recombinant polypeptides
A bacterial expression system was used to express GST- and His-tagged recombinant polypeptides. BL21 bacterial cells expressing GST-tagged recombinant polypeptides were resuspended in PBS and homogenized by sonication. Triton X-100 was added to a final concentration of 1%. The bacterial lysates were then incubated with shaking at 23°C for 1 h. A glutathione-conjugated agarose (Sigma) column was used to purify GST-tagged recombinant polypeptides. The proteins were eluted with 100 mM Tris-HCl (pH 8.0), 10 mM glutathione and dialyzed against 50mM Tris-HCl (pH 7.5), 50 mM NaCl. BL21 bacterial cells expressing His-tagged recombinant polypeptides were resuspended in 50 mM sodium phosphate (pH 8.0), 300 mM NaCl and homogenized by sonication. A nickel-nitrilotriacetic acid (Ni-NTA) agarose (QIAGEN) column was used to purify His-tagged recombinant polypeptides. The proteins were eluted with 50 mM sodium phosphate (pH 8.0), 300 mM NaCl, 250 mM imidazole and dialyzed against 50mM Tris-HCl (pH 7.5), 50 mM NaCl.
Chromatin immunoprecipitation (ChIP) assays
The ChIP assays were conducted using the EZ ChIP Chromatin Immunoprecipitation Kit according to the manufacturer’s instructions (Millipore). Briefly, C2C12 cells were transfected with control or Pitx2 siRNA. 48 h following transfection, the transfected cells were washed twice with PBS and cross-linked with 1% formaldehyde for 10 min at room temperature. The cells were then resuspended in SDS lysis buffer containing protease inhibitors, followed by sonication 5 times (10 sec for each) using a QSONICA sonicator. The cell lysates were centrifuged for 10 min to remove cell debris and the supernatants were subjected to immunoprecipitation with antibodies specific for Pitx2 (Santa Cruz), acetylated histone H 4 (Millipore), or histone deacetylase 1 (Millipore). A freshly prepared elution buffer containing 1% SDS and 0.1 M NaHCO3 was used to elute immunoprecipitates. Eluates were heated at 65°C for 6 h to reverse the formaldehyde cross-linking. DNA fragments were recovered from the eluates and subjected to PCR analysis with following three pairs of primers to amplify three intronic regions that cover five of the consensus TAATCC DNA-binding sites: 5′-AAACCCTTTGAGCAGAGACC-3′ (forward primer) and 5′-TACTCCAACGAAGCCACAAC-3′ (reverse primers); 5′-TCAAGCTCAGCGTAGACT AATG-3′ (forward primer) and 5′-CTGGAAGTGGTGTTGAAAGAAAG-3′ (reverse primer); 5′-GTCTTGACATCTAGAGCCATCTG-3′ (forward primer) and 5′-AGTGCATATTGTCTCACTCA CTC-3′ (reverse primer).
Immunoblot analysis
Cell lysates or immunoprecipitates were separated on 10% or 4%-12% SDS-PAGE gels (Bio-Rad), transferred to an Immobilon-P transfer membrane (Millipore), blocked in 5% non-fat milk, and incubated with primary antibodies as indicated. The following primary antibodies were used: mouse anti-MYC (1:1000; 9E10; Santa Cruz), rabbit anti-β-tubulin (1:1,000; Santa Cruz), mouse anti-His (1:250; Bio-Rad), goat anti-Pitx2 (1:250; Santa Cruz); and rabbit anti-DEPDC1B (1:500). After three washes, the blots were incubated with horseradish peroxidase-conjugated secondary antibodies (1:5000; Santa Cruz) for 1 h at 23°C and visualized by SuperSignal West Pico Luminol/Enhancer solution (Pierce). Some of the immunoblots were processed with a ScanLater kit and then scanned by a SpectraMax i3 according to the manufacturer’s instructions (Molecular Devices). The DEPDC1B polyclonal antibody used in this study was generated using the following peptide as antigen: TDAKLSNKEKKKKLKQFQ. The peptide corresponds to amino acid residues 461-478 of human DEPDC1B.
GST pull-down assays
GST pull-down assays were done as described previously [32, 33]. Briefly, the immobilized GST-tagged polypeptides were incubated with in vitro translated polypeptides or transfected cell lysates overnight at 4°C. After washing four times with binding buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 0.05% Triton-X-100, 10% glycerol, 0.2 mM EDTA, and 1 mM DTT), the beads were resuspended in SDS loading buffer to elute the bound proteins. The in vitro translated MYC-U2af2 was synthesized using the TNT SP6 quick-coupled transcription/translation system (Promega) according to the manufacturer’s instructions.
Yeast two-hybrid screening
The cDNAs corresponding to amino acid residues 1-151 and 151-529 of human DEPDC1B were cloned into the pAS2-1 vector to generate bait plasmids. The bait plasmids were co-transformed into the yeast strain Y187 with a mouse 11-day embryo matchmaker cDNA library (Clontech). The positive yeast colonies that were able to grow in SD synthetic medium lacking leucine, tryptophan and histidine were further confirmed by a filter lift assay for β-galactosidase activity. The prey plasmids, which contain cDNAs corresponding to the potential DEPDC1B-interacting proteins, were then recovered from the positive yeast colonies and subjected to DNA sequencing.
Results
Regulation of DEPDC1B expression by Pitx2
We have reported previously that inducible expression of Pitx2a in HeLa cells promotes the activation of the Rho GTPase proteins RAC1 and RHOA, arrests cell cycle at G1 phase, and inhibits cell migration [28]. To understand how inducible expression of Pitx2a promotes RAC1 and RHOA activation, we conducted a DNA microarray analysis to identify the GEFs and GAPs that were regulated by Pitx2a in this HeLa cell line. Total mRNAs isolated from the inducible Pitx2a cell line were subjected to the DNA microarray analysis (Affymetrix). The microarray data showed that one of the GAPs termed DEPDC1B was down-regulated by the inducible expression of Pitx2a. To confirm the microarray data, RT-PCR was conducted to determine the mRNA levels of DEPDC1B in Pitx2a cells. RT-PCR showed that the mRNA levels of DEPDC1B decreased in the presence of doxycycline (Figure 1B; compare lane 1 with lane 2 in the top panel). It is of note that HeLa-Tet-On cells used to establish the Pitx2-inducible cell line do not express endogenous Pitx2 [28]. In contrast, C2C12 cells endogenously express both Pitx2 [12] and Depdc1b (see below). Therefore, in this study we used C2C12 cells to examine the regulation of Depdc1b expression by Pitx2.
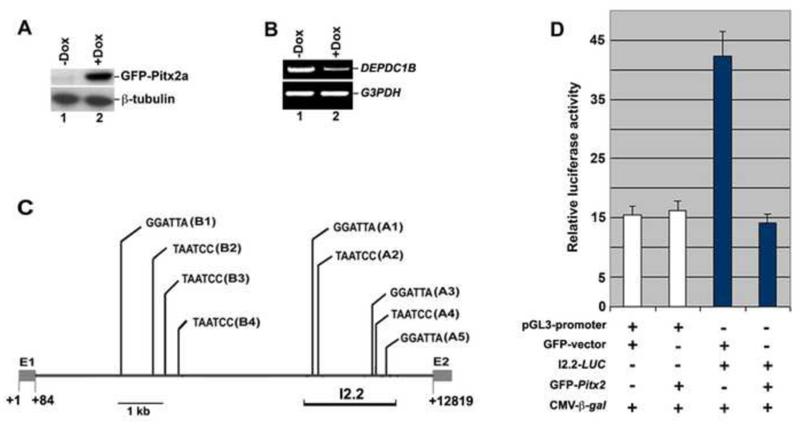
Figure 1. Pitx2 inhibits DEPDC1B expression at the transcriptional level.
(A) Immunoblot analysis showing that Pitx2a inducible HeLa cells express GFP-Pitx2a in the presence of doxycycline (Dox). (B) RT-PCR showing that inducible expression of GFP-Pitx2a decreases the mRNA levels of DEPDC1B. (C) Schematic showing that there are nine consensus TAATCC DNA-binding sites (A1-A5 and B1-B4) in the first intron of human DEPDC1B. The number below the diagram indicates the nucleotide residues. “GGATTA” and “TAATCC”, the consensus TAATCC DNA-binding site. E1, the first exon; E2, the second exon. (D) Luciferase reporter assays showing that exogenous expression of Pitx2a represses the transcriptional activity mediated by an intronic fragment of human DEPDC1B (I2.2). Three independent experiments were conducted and luciferase activity was normalized to β-galactosidase activity. β-gal, β-galactosidase.
Pitx2 is a transcription factor that regulates transcriptional activity by binding to the consensus TAATCC DNA-binding site in the promoters and enhancers of its target genes. To determine whether Pitx2 can regulate DEPDC1B expression at the transcriptional level, a 3-kb promoter region of human DEPDC1B was analyzed for the presence of consensus TAATCC DNA-binding sites. We did not identify any consensus TAATCC DNA-binding sites in the promoter region. In contrast, there are nine consensus TAATCC DNA-binding site in the first intron (11kb) of human DEPDC1B. As shown in Figure 1C, these consensus TAATCC DNA-binding sites (A1-A5 and B1-B4) are clustered in two regions of the first intron. A 2.2-kb intronic DNA fragment (I2.2) from the first intron of human DEPDC1B was cloned into a luciferase reporter vector to generate the reporter plasmid I2.2-LUC. To test whether Pitx2 inhibits the transcriptional activity mediated by the intronic DNA fragment, I2.2-LUC was co-transfected into C2C12 cells with a plasmid encoding Pitx2a. 24 h after transfection, the transfected C2C12 cell lysate was subjected to luciferase reporter assays. As shown in Figure 1D, expression of GFP-Pitx2a significantly reduced luciferase activity from I2.2-LUC, suggesting that Pitx2 inhibits intron-mediated transcriptional regulation of DEPDC1B.
It is of note that the first intron (7.4 kb) of mouse Depdc1b also contains consensus TAATCC DNA-binding sites. Three and five consensus DNA-binding sites are clustered in a 1.6-kb and a 1.9-kb intronic region of mouse Depdc1b, respectively. Next, we used chromatin immunoprecipitation (ChIP) assays to examine whether Pitx2 binds to the first intron of mouse Depdc1b. As shown in Figure 2B, Pitx2 and histone deacetylase 1 (Hdac1), but not acetylated histone H4 (AcH4), were associated with the first intron of Depdc1b in C2C12 cells transfected with control siRNA (Figure 2B; compare lanes 3 and 4 with lane 5 in the top panel). In contrast, acetylated histone H4, but not Pitx2 and Hdac1, was present in the first intron of Depdc1b in C2C12 cells transfected with an siRNA against Pitx2 (Figure 2B; compare lane 5 with lanes 3 and 4 in the bottom panel). It has been well established that histone deacetylases and acetylated histones are associated with the inactive and active chromatin, respectively. Therefore, our findings suggest that Pitx2 can bind to the first intron of Depdc1b and inhibit the expression of Depdc1b at the transcriptional level.
Figure 2. Recruitment of Pitx2 to the first intron of mouse Depdc1b.
(A) Schematic showing that there are eight consensus TAATCC DNA-binding sites in the first intron of mouse Depdc1b. The number below the diagram indicates the nucleotide residues. Three intronic regions that can be amplified by primer pairs #1, #2, and #3 are shown. “GGATTA” and “TAATCC”, the consensus TAATCC DNA-binding site. E1, the first exon; E2, the second exon. (B) ChIP assays showing the interaction of Pitx2 with the first intron of mouse Depdc1b. The results of a typical ChIP assay using primer pair #1 are shown. Similar results were obtained with primer pairs #2 and #3.
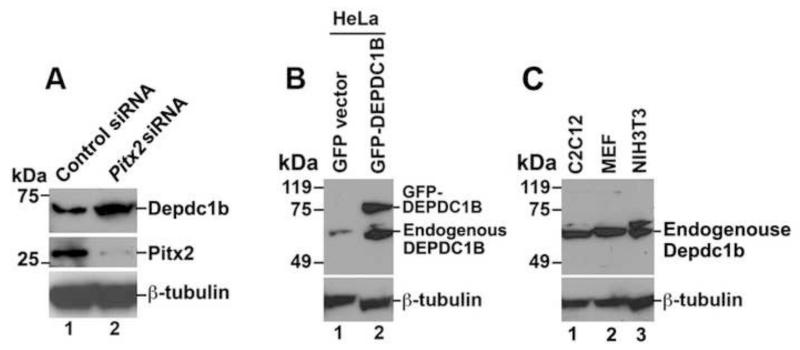
To determine whether Depdc1b expression is regulated by the endogenous Pitx2, C2C12 cells transfected with an siRNA specific for Pitx2 were subjected to immunoblot analysis with an antibody specific for DEPDC1B. As shown in Figure 3A, depletion of Pitx2 in C2C12 cells by RNAi increased the protein levels of Depdc1b (Figure 3A; compare lane 1 with lane 2 in the top panel). As shown in Figure 3B, the DEPDC1B peptide antibody could recognize both GFP-tagged and endogenous DEPDC1B in HeLa cells. In addition, the DEPDC1B peptide antibody also recognized endogenous Depdc1b that is expressed in C2C12, mouse embryonic fibroblast (MEF), and NIH3T3 cells (Figure 3C).
Figure 3. RNAi-mediated depletion of Pitx2 increases the protein levels of Depdc1b.
(A) C2C12 cells transfected with an siRNA against Pitx2 were subjected to immunoblot analysis with an antibody specific for Depdc1b. (B) Cell lysates from HeLa cells transfected with empty vector (lane 1) or a plasmid encoding GFP-DEPDC1B (lane 2) were subjected to immunoblot analysis with DEPDC1B antibody (top panels). (C) Cell lysates from untransfected C2C12 (lane 1), and MEF (lane 2), and NIH3T3 (lane 3) were subjected to immunoblot analysis with an antibody specific for DEPDC1B (top panels). The expected molecular mass for endogenous DEPDC1B is 61 kDa.
Binding of DEPDC1B to nucleotide-bound forms of RAC1
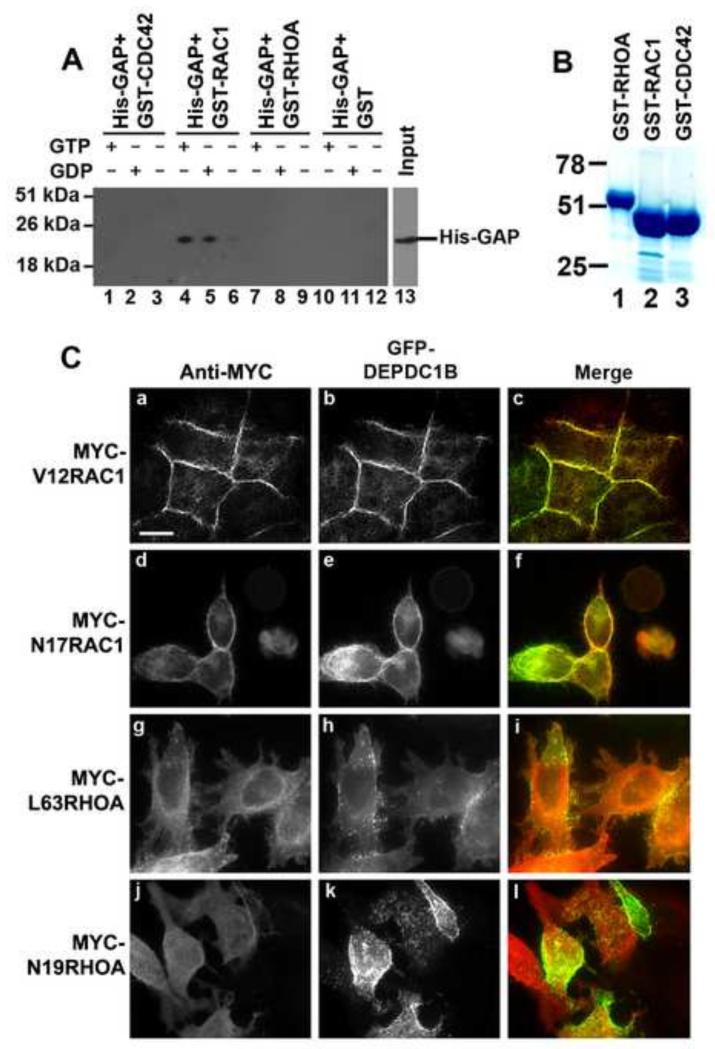
Data from the NCBI database show the GAP domain of DEPDC1B does not contain the critical arginine residue that is required for the GAP activity. In addition, recent study has demonstrated that the GAP domain of DEPDC1B does not have GAP activity toward RHOA [34]. Nonetheless, it has been shown that the conserved arginine residue in the GAP domain is not required for the binding of GAP domains to Rho GTPase proteins [35]. Furthermore, the GAP domain can act as a scaffold to bind to Rho GTPase proteins and serve as a downstream effector of Rho GTPase proteins, thus modulating Rho GTPase signaling [36-40]. Therefore, we asked whether the GAP domain of DEPDC1B could bind to Rho GTPase proteins. The GAP domain (residues 171-400) of DEPDC1B was expressed as a His-tagged polypeptide (His-GAP) using a bacterial expression system. The purified His-GAP was incubated with GST-RHOA, GST-RAC1 or GST-CDC42 in the absence of guanine nucleotides or in the presence of GDP or GTP. The mixtures were then subjected to GST pull-down assays. His-GAP co-precipitated with GST-RAC1 in the presence of either GDP or GTP, but did not co-precipitate with GST-RAC1 in the absence of guanine nucleotides (Figure 4A; compare lanes 4 and 5 with lane 6). In contrast, His-GAP did not co-precipitate with GST-RHOA or GST-CDC42 in the presence or absence of guanine nucleotides (Figure 4A; compare lanes 1-3 and 7-12 with lanes 4-6). These findings suggest that the GAP domain of DEPDC1B can bind to inactivated, GDP-bound and activated, GTP-bound forms of RAC1, but not to nucleotide-free RAC1. Next, we asked whether DEPDC1B and RAC1 colocalized in transfected HeLa cells. As shown in Figure 4C, GFP-DEPDC1B colocalized with a dominant active form of RAC1, V12RAC1, to the sites of cell–cell contacts (Figure 4C, a-c). GFP-DEPDC1B also colocalized with a dominant negative form of RAC1, N17RAC1, to the cell periphery (Figure 4C, d-f). In contrast, GFP-DEPDC1B did not colocalize with dominant active (L63RHOA) or dominant negative (N19RHOA) forms of RHOA in transfected HeLa cells (Figure 4C, g-i).
Figure 4. The GAP domain of DEPDC1B binds to nucleotide-bound forms of RAC1.
(A) Purified His-GAP (the GAP domain of DEPDC1B) were incubated with GST-CDC42 (lanes 1-3), GST-RAC1 (lanes 4-6), GST-RHOA (lanes 7-9), or GST alone (lanes 10-12) in the absence of guanine nucleotide (lanes 3, 6, 9, and 12) or in the presence of GTP (lanes 1, 4, 7, and 10) or GDP (lanes 2, 5, 8, and 11). The mixtures were then subjected to GST pull-down assays, followed by immunoblot analysis with an antibody specific for His-tag. (B) Coomassie Blue staining image of GST-RHOA (lane 1), GST-RAC1 (lane 2), and GST-CDC42 (lane 3) that were used in (A). (C) A plasmid encoding GFP-DEPDC1B (b, e, h, and k) were co-transfected into HeLa cells with plasmids encoding MYC-tagged V12RAC1 (a), N17RAC1 (d), L63RHOA (g), or N19RHOA (j). The transfected cells were subjected to immunofluorescence analysis with an antibody specific for MYC (a, d, g, and j). Scale bar = 20 μm.
Inhibition of RAC1 activation by DEPDC1B
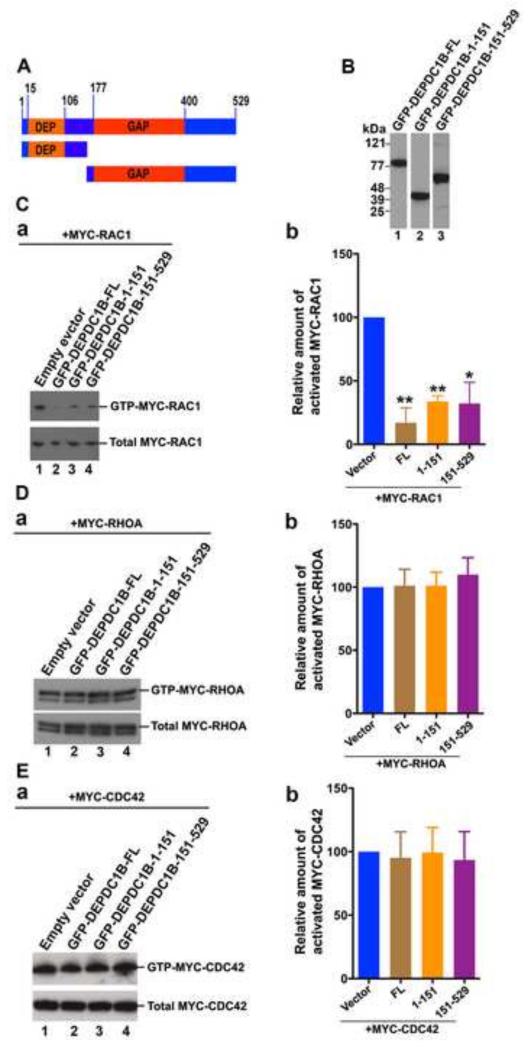
Although the critical arginine residue in the GAP domain is required for the catalytic activity of numerous GAPs, it has also been demonstrated that GAPs without the critical arginine residue utilize an alternative mechanism to promote GTP hydrolysis [41, 42]. Therefore, we asked whether exogenous expression of DEPDC1B in transfected cells had an impact on the activation of Rho GTPase proteins. As shown in Figure 5A, DEPDC1B contains a DEP domain (residues 15-106) and a GAP domain (residues 177-400). Plasmids encoding GFP-tagged full-length DEPDC1B or truncated DEPDC1B fragments were co-transfected into HeLa cells with plasmids encoding MYC-RAC1, MYC-RHOA, or MYC-CDC42. The transfected cells were then subjected to RBD (Rhotekin Rho-binding domain) and PBD (PAK1/p21 binding domain) pull-down assays to assess the levels of the activated Rho GTPase proteins RHOA and RAC1/CDC42, respectively. As shown in Figure 5C, a and b, exogenous expression of GFP-DEPDC1B-FL (full-length) in HeLa cells inhibited RAC1 activation (Figure 5Ca; compare lane 2 with lane 1 in the top panel). To determine which regions of DEPDC1B are required for DEPDC1B-mediated suppression of RAC1 activation, a plasmid encoding MYC-RAC1 was co-transfected into HeLa cells with a plasmid encoding either GFP-DEPDC1B-1-151 (containing the DEP domain) or GFP-DEPDC1B-151-529 (containing the GAP domain). The transfected cells were then subjected to PBD pull-down assays. As shown in Figure 5Ca, exogenous expression of either DEPDC1B-1-151 or DEPDC1B-151-529 also decreased RAC1 activation (Figure 5Ca; compare lanes 3 and 4 with lane 1 in the top panel). In contrast, exogenous expression of full-length or truncated DEPDC1B did not markedly affect the activation of RHOA or CDC42 (Figures 5D and 5E).
Figure 5. Exogenous expression of DEPDC1B suppresses RAC1 activation.
(A) Schematic of DEPDC1B polypeptides that were used in (B), (C), (D), and (E). The number above the diagram indicates the amino acid residues. (B) Immunoblots showing that GFP-tagged DEPDC1B full-length (lane 1) and fragments (lanes 2 and 3) are expressed in transfected HeLa cells. (C-E) Plasmids encoding MYC-RAC1 (C), MYC-RHOA (D), or MYC-CDC42 (E) were co-transfected into HeLa cells with empty vector or plasmids encoding GFP-DEPDC1B-FL, GFP-DEPDC1B-1-151, or GFP-DEPDC1B-151-529. The transfected cells were subjected to PBD pull-down assays to assess the levels of activated MYC-RAC1 (C) and MYC-CDC42 (E) or subjected to RBD pull-down assays to assess the levels of activated MYC-RHOA (D). Typical immunoblots for the activation assays of RAC1 (Ca), RHOA (Da), and CDC42 (Ea) were shown. Relative amount of activated MYC-RAC1 (Cb), MYC-RHOA (Db), and MYC-CDC42 (Eb) in each group of transfected cells were quantitated based on three independent experiments. The error bars indicate standard deviation around the mean. In (Cb), (Db), and (Eb), FL, 1-151, and 151-529 represent GFP-DEPDC1B-FL, GFP-DEPDC1B-1-151, and GFP-DEPDC1B-151-529, respectively. **, p<0.01; *, p<0.05.
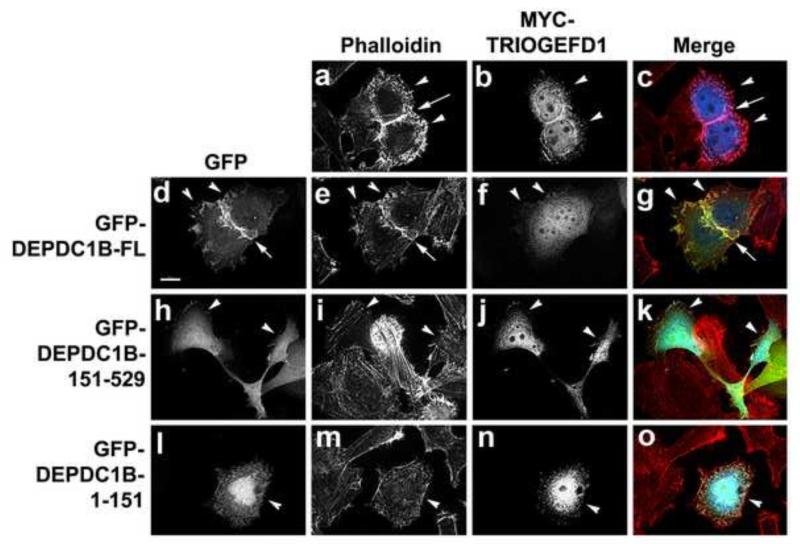
Disruption of TRIO-induced actin bundling by DEPDC1B
We have reported previously that inducible expression of Pitx2 not only activates RHOA and RAC1, but also stimulates the expression of the GEF TRIO, [28]. TRIO contains two functional GEF domains. One of the TRIO GEF domains (TRIOGEFD1) shows specific GEF activity toward RAC1, while another TRIO GEF domain (TRIOGEFD2) has specific GEF activity toward RHOA [43]. In the current study, our results showed that Pitx2 repressed the expression of DEPDC1B (Figures 1 and 2) and that exogenous expression of DEPDC1B in HeLa cells suppressed RAC1 activation (Figure 5C). Therefore, we asked whether DEPDC1B could inhibit actin polymerization induced by TRIOGEFD1. Exogenous expression of TRIOGEFD1 induced actin bundling at the cell periphery as well as at the sites of cell-cell contacts (Figure 6, a-c). Figure 6, d-g, show that exogenous expression of full-length DEPDC1B decreased TRIOGEFD1-induced actin bundling, especially at the cell periphery (n = 23/47 cells), although actin bundling was largely retained at the sites of cell-cell contacts (Figure 6; compare a with e). As shown in Figure 6, h-o, exogenous expression of truncated DEPDC1B fragments, either DEPDC1B-1-151 (n = 17/29 cells) or DEPDC1B-151-529 (n = 16/23 cells), also decreased TRIOGEFD1-induced actin polymerization. Therefore, our results indicate that DEPDC1B can antagonize TRIO-mediated RAC1 activation. However, exogenous expression of DEPDC1B did not affect TRIOGEFD2-induced actin polymerization (data not shown). These findings are consistent with our observations that exogenous expression of DEPDC1B did not have an obvious impact on RHOA activation and that the GAP domain of DEPDC1B did not interact with RHOA (Figures 4 and 5).
Figure 6. Exogenous expression of DEPDC1B suppresses TRIOGEFD1-induced actin polymerization.
HeLa cells were transfected with the MYC-TRIOGEFD1 plasmid alone (a-c) or together with a plasmid encoding GFP-DEPDC1B-FL (d-g), GFP-DEPDC1B-151-529 (h-k), or GFP-DEPDC1B-1-151 (l-o). The transfected cells were subjected to immunofluorescence staining for actin filaments (a, e, i, and m) and MYC-TRIOGEFD1 (b, f, j, and n). Arrowheads indicate the transfected cells. Arrows in a, c, d, e, and g indicate the sites of cell-cell contacts. Scale bar = 20 μm.
Binding of DEPDC1B to both nuclear and transmembrane proteins
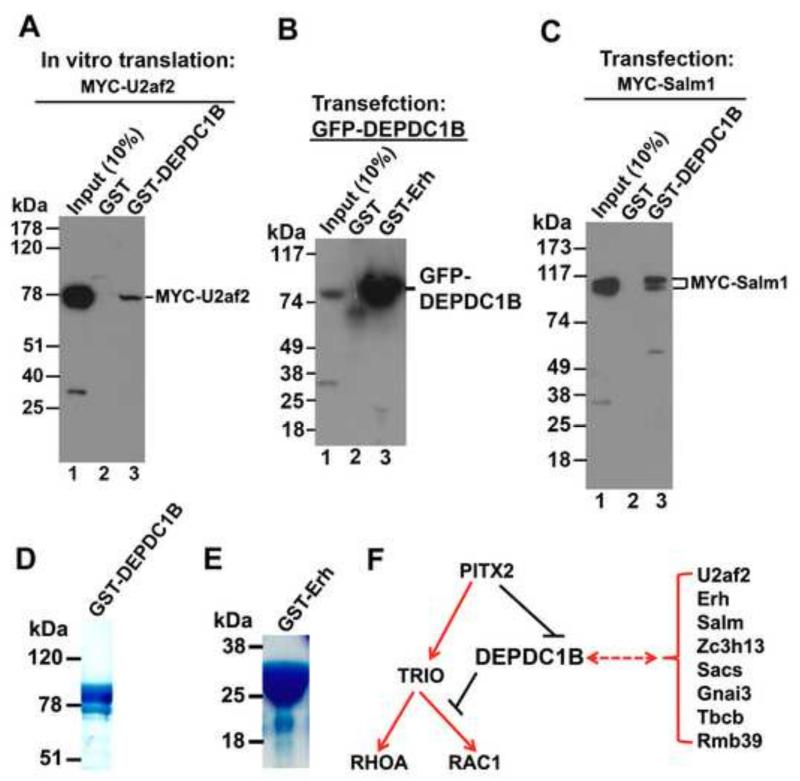
To better understand the biological functions of DEPDC1B, we used the yeast-hybrid screening to identify the proteins that interact with DEPDC1B. We used an amino-terminal fragment (residues 1-151; containing the DEP domain) and a carboxyl-terminal fragment (residues 151-529; containing the GAP domain) as baits to screen a 12-day-mouse embryo cDNA library. Although the screen with the 151-529 fragment as bait did not result in any positive clones, the screen with the 1-151 fragment as bait led to the identification of eight positive clones (Table 1): U2 small nuclear RNA auxiliary factor 2 (U2af2), enhancer of rudimentary homolog (Erh), synaptic adhesion-like molecule (Salm), Zc3h13 (zinc finger CCCH-type containing 13), sacsin molecular chaperone (Sacs; Spastic Ataxia Of Charlevoix-Saguenay), Gnai3 [guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3], tubulin folding cofactor B (Tbcb), and RNA binding motif protein 39 (Rbm39).
Table 1. DEPDC1B-interacting proteins identified by yeast-two hybrid screening.
| DEPDC1B- interacting proteins |
Functions | Subcellular localization |
References |
|---|---|---|---|
| U2af2 | Splicing | Nucleus | [60] |
| Erh | Splicing and transcriptional regulation |
Nucleus | [61, 63] |
| Salm | Neurite outgrowth and synapse formation |
Membrane | [64, 65] |
| Zc3h13 | Unknown | Unknown | [66] |
| Sacs | Mutations of the human SACS gene cause autosomal recessive spastic ataxia of Charlevoix- Saguenay (ARSACS) |
Unknown | [67] |
| Gnai3 | Cell migration, asymmetric cell division, and axial skeleton development |
Cytoplasm and cell periphery |
[68-70] |
| Tbcb | Microtubule assembly | Cytoplasm | [71, 72] |
| Rbm39 | Splicing and transcriptional regulation |
Nucleus | [73] |
Next, we used in vitro pull-down assays to confirm the interactions of DEPDC1B with U2af2, Erh, and Salm. We selected these three proteins for further analysis because U2af2 and Erh are nuclear proteins while Salm is a transmembrane protein, thus allowing us to confirm whether DEPDC1B can interact with both nuclear and transmembrane proteins. GST pull-down assays showed that GST-DEPDC1B co-precipitated with the in vitro translated MYC-U2af2 (Figure 7A). GST-Erh also co-precipitated with GFP-DEPDC1B from transfected HeLa cells (Figure 7B). In addition, Figure 7C shows that GST-DEPDC1B co-precipitated with MYC-Salm1 (a member of the Salm family proteins) from transfected HeLa cells. Therefore, our findings indicate that the DEP domain of DEPDC1B can bind to both nuclear (U2af2 and Erh) and transmembrane (Salm) proteins.
Figure 7. The DEP domain of DEPDC1B interacts with both nuclear and transmembrane proteins.
(A) GST pull-down assays showing that GST-DEPDC1B co-precipitates with the in vitro translated MYC-U2af2. (B) GST pull-down assays showing that GST-Erh co-precipitates with GFP-DEPDC1B from transfected HeLa cell lysates. (C) GST pull-down assays showing that GST-DEPDC1B co-precipitates with MYC-Salm1 from transfected HeLa cell lysates. (D) Coomassie Blue staining image of GST-DEPDC1B that was used in (A) and (C). (E) Coomassie Blue staining image of GST-Erh that was used in (B). (F) Schematic showing the coordinated regulation of RHOA and RAC1 activation by Pitx2, TRIO, and DEPDC1B. Pitx2 up-regulates the expression of TRIO that, in turn, activates both RHOA and RAC1. Pitx2 also down-regulates the expression of DEPDC1B, thus relieving DEPDC1B-mediated suppression of RAC1 activation. The DEP domain of DEPDC1B interacts with proteins, such as U2af2, Erh, Salm, Zc3h13, Sacs, Gnai3, Tbcb, and Rbm39.
Discussion
In this study, we have demonstrated that Pitx2 can inhibit DEPDC1B expression at the transcriptional level. In addition, DEPDC1B can bind to nucleotide-bound forms of RAC1, suppress RAC1 activation in transfected cells, and interfere with actin polymerization induced by the RAC1-specific GEF domain of TRIO. Furthermore, DEPDC1B interacts with various signaling molecules ranging from splicing regulators to transmembrane proteins. Our findings indicate that Pitx2-mediated repression of DEPDC1B expression contributes to the regulation of multiple signaling processes, such as Rho GTPase signaling.
Regulation of RAC1 activation by DEPDC1B
It is generally thought that a critical arginine residue in the GAP domain of GAPs plays a pivotal role in promoting the intrinsic GTPase activity of Rho GTPase proteins [44]. The conserved arginine residue is present in the GAP domain of most GAPs. In the absence of GAPs, Rho GTPase proteins only show low intrinsic activity of GTP hydrolysis. In contrast, in the presence of GAPs, the conserved arginine residue from the GAP domain of GAPs is inserted into the active site of Rho GTPase proteins to stabilize a conformation required for GTP hydrolysis, thus increasing the intrinsic GTPase activity of Rho GTPase proteins [45-47]. Although the critical arginine residue is absent in the GAP domain of DEPDC1B, our results showed that exogenous expression of DEPDC1B led to the suppression of RAC1 activation in transfected HeLa cells (Figure 5C). These findings raise the question of how DEPDC1B inhibits RAC1 activation. One possibility is that the GAP domain of DEPDC1B utilizes an alternative mechanism to promote the intrinsic GTPase activity without participation of an arginine residue. In support of this speculation, some GAPs lacking the critical arginine residue are also capable of enhancing GTP hydrolysis through alternative mechanisms [41, 42, 44, 48]. In particular, a GAP termed OCRL1 (the oculocerebrorenal syndrome of Lowe) does not contain the conserved arginine residue [38]. Yet, OCRL1 has GAP activity toward RAC1 in vitro [49]. In addition, when exogenously expressed in Swiss 3T3 cells, OCRL1 suppresses RAC1-specific cell phenotypes such as membrane ruffling [49]. In the current study, our results showed that exogenous expression of DEPDC1B disrupted actin polymerization induced by the RAC1-specific GEF domain of TRIO. Furthermore, our results showed that the GAP domain of DEPDC1B could bind to GTP-bound forms of RAC1 (Figure 4A). Therefore, our findings suggest that the GAP domain of DEPDC1B can bind to GTP-RAC1 and promote GTP hydrolysis. However, it should be noted that the GAP domain of DEPDC1B also interacted with GDP-bound forms of RAC1 (Figure 4A). Thus, binding of DEPDC1B to GDP-bound forms of RAC1 may also contribute to the inhibition of RAC1 activation in transfected HeLa cells. For instance, it is possible that the GAP domain of DEPDC1B binds to GDP-RAC1 and interferes with the conversion of GDP-RAC1 to GTP-RAC1, thus decreasing the overall levels of activated RAC1 in transfected cells. It is of note that some other GAPs such as OCRL1 and ArhGAP15 can also bind to both GDP- and GTP-bound forms of RAC1 [49]. In addition, mutation of the critical arginine to alanine does not interfere with the interaction between a CDC42-GAP and CDC42 [35], suggesting that the conserved arginine residue in the GAP domain is dispensable for the binding of GAP domains to Rho GTPase proteins.
The DEP domain is a conserved sequence element containing approximately 90 amino acid residues that was first identified in Dishevelled, EGL-10, and Pleckstrin [50]. DEP domains can act as a scaffold to recruit signaling molecules to various subcellular locations such as the plasma membrane [50]. DEPDC1B-1-151 contains a DEP domain that corresponds to amino acid residues 15-106. Our results showed that exogenous expression of DEPDC1B-1-151 inhibited RAC1 activation and decreased actin polymerization induced by the RAC1-specific GEF domain of TRIO (Figures 5 and 6). However, in vitro pull-down assays showed that the 1-151 region of DEPDC1B did not interact with RAC1 (data not shown). Therefore, the DEP domain of DEPDC1B is likely to suppress RAC1 activation through regulation of the upstream regulators of RAC1. In the current study, our results showed that the DEP domain of DEPDC1B could bind to transcriptional regulators such as Erh and Rbm39 (Table 1). Therefore, one possibility is that the DEP domain of DEPDC1B is implicated in the transcriptional regulation of unknown signaling molecules that, in turn, regulate RAC1 activation. Our yeast-two hybrid screening assays also revealed that the DEP domain of DEPDC1B could bind to Tbcb, an important regulator of microtubule assembly (Table 1). Thus, the DEPDC1B-Tbcb interaction is likely to play a role in regulating microtubule dynamics. Importantly, a line of evidence has demonstrated that microtubule growth promotes cell migration through activation of RAC1 at the cell leading edge [51, 52]. Furthermore, microtubule assembly enhances the GEF activity of TRIO toward RAC1 [53]. In the current study, our results showed that exogenous expression of DEPDC1B-1-151 interfered with actin polymerization induced by the RAC1-specific GEF domain of TRIO (Figure 6). Therefore, it is possible that the DEP domain of DEPDC1B binds Tbcb and interferes with microtubule assembly, thus inhibiting RAC1 activation.
DEPDC1B as a downstream target of Pitx2
We have reported previously that inducible expression of Pitx2a in HeLa cells leads to the activation of RAC1 and RHOA [28]. In addition, we have also shown that Pitx2 stimulates the expression of TRIO [28]. TRIO is a GEF that can activate both RAC1 and RHOA [43]. In the current study, we found that Pitx2 could suppress DEPDC1B expression at the transcriptional level (Figures 1 and 2). Furthermore, exogenous expression of DEPDC1B could inhibit RAC1 activation and suppress actin polymerization induced by the RAC1-specific GEF domain of TRIO (Figures 5 and 6). Our findings suggest that DEPDC1B can antagonize RAC1 activation mediated by the Pitx2-TRIO axis. Therefore, we propose that Pitx2-mediated repression of DEPDC1B expression coordinates with Pitx2-mediated stimulation of TRIO expression to regulate the activation of RAC1 and RHOA (Figure 7F). A line of evidence has demonstrated that Rho GTPase proteins play a central role in the regulation of cell migration during embryonic development [54-56]. In addition, Pitx2 is required for the regulation of cell migration during organogenesis such as craniofacial and hypothalamus development [18, 19]. Therefore, our findings indicate that regulation of TRIO and DEPDC1B expression by Pitx2 leads to the activation of RAC1 and RHOA, thus contributing to the regulation of cell migration during embryonic development. Although a role for DEPDC1B in the regulation of cell migration during embryonic development remains to be elucidated, studies using cultured cells have demonstrated that DEPDC1B is implicated in the regulation of cell migration and cell proliferation [57-59].
DEPDC1B contains a GAP domain and a DEP domain. It is generally thought that the DEP domain acts as a scaffold to recruit signaling molecules to various subcellular locations such as the plasma membrane [50]. In the current study, a yeast two-hybrid screening with the 1-151 region (containing the DEP domain) of DEPDC1B as bait led to the identification of eight putative DEPDC1B-interacting proteins, which are implicated in various biological processes such as transcriptional regulation, RNA splicing, cell migration, microtubule assembly, and synapse formation (Table 1). In addition, the interactions of DEPDC1B with U2af2, Erh, and Salm1 were confirmed by GST pull-down assays (Figure 7). It is of note that both U2af2 and Erh have been implicated in the regulation of RNA splicing [60, 61]. Furthermore, U2af2 interacts with Rbm39 in yeast-two hybrid assays [62]. In the current study, Rbm39 was also identified as a DEPDC1B-interacting protein. Therefore, interactions of DEPDC1B with U2af2, Erh, and Rbm39 are likely to play a role in the regulation of RNA splicing. Future research will focus on understanding how DEPDC1B and its interacting partners are implicated in the regulation of Pitx2-mediated developmental processes.
Highlight.
Pitx2 represses DEPDC1B expression at the transcriptional level.
DEPDC1B binds to RAC1 and inhibits RAC1 activation.
DEPDC1B interacts with various signaling molecules such as U2af2, Erh, and Salm.
Pitx2-mediated repression of DEPDC1B expression may impact multiple signaling pathways.
Acknowledgments
This work was supported in part by National Institutes of Health Grant R15GM097702.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Semina EV, Reiter R, Leysens NJ, Alward WL, Small KW, Datson NA, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel BU, Carey JC, Murray JC. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet. 1996;14:392–399. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- [2].Alward WL. Axenfeld-Rieger syndrome in the age of molecular genetics. Am J Ophthalmol. 2000;130:107–115. doi: 10.1016/s0002-9394(00)00525-0. [DOI] [PubMed] [Google Scholar]
- [3].Nakamura T, Hamada H. Left-right patterning: conserved and divergent mechanisms. Development. 2012;139:3257–3262. doi: 10.1242/dev.061606. [DOI] [PubMed] [Google Scholar]
- [4].Vandenberg LN, Levin M. A unified model for left-right asymmetry? Comparison and synthesis of molecular models of embryonic laterality. Dev Biol. 2013;379:1–15. doi: 10.1016/j.ydbio.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Vinarskaja A, Schulz WA, Ingenwerth M, Hader C, Arsov C. Association of PITX2 mRNA down-regulation in prostate cancer with promoter hypermethylation and poor prognosis. Urologic oncology. 2013;31:622–627. doi: 10.1016/j.urolonc.2011.04.010. [DOI] [PubMed] [Google Scholar]
- [6].Nimmrich I, Sieuwerts AM, Meijer-van Gelder ME, Schwope I, Bolt-de Vries J, Harbeck N, Koenig T, Hartmann O, Kluth A, Dietrich D, Magdolen V, Portengen H, Look MP, Klijn JG, Lesche R, Schmitt M, Maier S, Foekens JA, Martens JW. DNA hypermethylation of PITX2 is a marker of poor prognosis in untreated lymph node-negative hormone receptor-positive breast cancer patients. Breast cancer research and treatment. 2008;111:429–437. doi: 10.1007/s10549-007-9800-8. [DOI] [PubMed] [Google Scholar]
- [7].Dave V, Zhao C, Yang F, Tung CS, Ma J. Reprogrammable recognition codes in bicoid homeodomain-DNA interaction. Mol Cell Biol. 2000;20:7673–7684. doi: 10.1128/mcb.20.20.7673-7684.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Espinoza HM, Cox CJ, Semina EV, Amendt BA. A molecular basis for differential developmental anomalies in Axenfeld-Rieger syndrome. Human molecular genetics. 2002;11:743–753. doi: 10.1093/hmg/11.7.743. [DOI] [PubMed] [Google Scholar]
- [9].Ganga M, Espinoza HM, Cox CJ, Morton L, Hjalt TA, Lee Y, Amendt BA. PITX2 isoform-specific regulation of atrial natriuretic factor expression: synergism and repression with Nkx2.5. J Biol Chem. 2003;278:22437–22445. doi: 10.1074/jbc.M210163200. [DOI] [PubMed] [Google Scholar]
- [10].Hjalt TA, Amendt BA, Murray JC. PITX2 regulates procollagen lysyl hydroxylase (PLOD) gene expression: implications for the pathology of Rieger syndrome. J Cell Biol. 2001;152:545–552. doi: 10.1083/jcb.152.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Green PD, Hjalt TA, Kirk DE, Sutherland LB, Thomas BL, Sharpe PT, Snead ML, Murray JC, Russo AF, Amendt BA. Antagonistic regulation of Dlx2 expression by PITX2 and Msx2: implications for tooth development. Gene Expr. 2001;9:265–281. doi: 10.3727/000000001783992515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Baek SH, Kioussi C, Briata P, Wang D, Nguyen HD, Ohgi KA, Glass CK, Wynshaw-Boris A, Rose DW, Rosenfeld MG. Regulated subset of G1 growth-control genes in response to derepression by the Wnt pathway. Proc Natl Acad Sci U S A. 2003;100:3245–3250. doi: 10.1073/pnas.0330217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, Herman T, Ohgi KA, Lin C, Gleiberman A, Wang J, Brault V, Ruiz-Lozano P, Nguyen HD, Kemler R, Glass CK, Wynshaw-Boris A, Rosenfeld MG. Identification of a Wnt/Dvl/beta-Catenin --> Pitx2 pathway mediating cell-type-specific proliferation during development. Cell. 2002;111:673–685. doi: 10.1016/s0092-8674(02)01084-x. [DOI] [PubMed] [Google Scholar]
- [14].Cao H, Florez S, Amen M, Huynh T, Skobe Z, Baldini A, Amendt BA. Tbx1 regulates progenitor cell proliferation in the dental epithelium by modulating Pitx2 activation of p21. Dev Biol. 2010;347:289–300. doi: 10.1016/j.ydbio.2010.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hilton T, Gross MK, Kioussi C. Pitx2-dependent occupancy by histone deacetylases is associated with T-box gene regulation in mammalian abdominal tissue. J Biol Chem. 2010;285:11129–11142. doi: 10.1074/jbc.M109.087429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li X, Venugopalan SR, Cao H, Pinho FO, Paine ML, Snead ML, Semina EV, Amendt BA. A model for the molecular underpinnings of tooth defects in Axenfeld-Rieger syndrome. Human molecular genetics. 2014;23:194–208. doi: 10.1093/hmg/ddt411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Campbell AL, Shih HP, Xu J, Gross MK, Kioussi C. Regulation of motility of myogenic cells in filling limb muscle anlagen by Pitx2. PloS one. 2012;7:e35822. doi: 10.1371/journal.pone.0035822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Liu W, Selever J, Lu MF, Martin JF. Genetic dissection of Pitx2 in craniofacial development uncovers new functions in branchial arch morphogenesis, late aspects of tooth morphogenesis and cell migration. Development. 2003;130:6375–6385. doi: 10.1242/dev.00849. [DOI] [PubMed] [Google Scholar]
- [19].Skidmore JM, Cramer JD, Martin JF, Martin DM. Cre fate mapping reveals lineage specific defects in neuronal migration with loss of Pitx2 function in the developing mouse hypothalamus and subthalamic nucleus. Mol Cell Neurosci. 2008;37:696–707. doi: 10.1016/j.mcn.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lu Y, Settleman J. The Drosophila Pkn protein kinase is a Rho/Rac effector target required for dorsal closure during embryogenesis. Genes Dev. 1999;13:1168–1180. doi: 10.1101/gad.13.9.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Endo Y, Wolf V, Muraiso K, Kamijo K, Soon L, Uren A, Barshishat-Kupper M, Rubin JS. Wnt-3a-dependent cell motility involves RhoA activation and is specifically regulated by dishevelled-2. J Biol Chem. 2005;280:777–786. doi: 10.1074/jbc.M406391200. [DOI] [PubMed] [Google Scholar]
- [22].Miyakoshi A, Ueno N, Kinoshita N. Rho guanine nucleotide exchange factor xNET1 implicated in gastrulation movements during Xenopus development. Differentiation. 2004;72:48–55. doi: 10.1111/j.1432-0436.2004.07201004.x. [DOI] [PubMed] [Google Scholar]
- [23].Matsui T, Raya A, Kawakami Y, Callol-Massot C, Capdevila J, Rodriguez-Esteban C, Izpisua Belmonte JC. Noncanonical Wnt signaling regulates midline convergence of organ primordia during zebrafish development. Genes Dev. 2005;19:164–175. doi: 10.1101/gad.1253605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Habas R, Kato Y, He X. Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell. 2001;107:843–854. doi: 10.1016/s0092-8674(01)00614-6. [DOI] [PubMed] [Google Scholar]
- [25].Habas R, Dawid IB, He X. Coactivation of Rac and Rho by Wnt/Frizzled signaling is required for vertebrate gastrulation. Genes Dev. 2003;17:295–309. doi: 10.1101/gad.1022203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Schmidt A, Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes & development. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- [27].Cox CJ, Espinoza HM, McWilliams B, Chappell K, Morton L, Hjalt TA, Semina EV, Amendt BA. Differential regulation of gene expression by PITX2 isoforms. J Biol Chem. 2002;277:25001–25010. doi: 10.1074/jbc.M201737200. [DOI] [PubMed] [Google Scholar]
- [28].Wei Q, Adelstein RS. Pitx2a expression alters actin-myosin cytoskeleton and migration of HeLa cells through Rho GTPase signaling. Mol Biol Cell. 2002;13:683–697. doi: 10.1091/mbc.01-07-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wei Q. Pitx2a binds to human papillomavirus type 18 E6 protein and inhibits E6-mediated P53 degradation in HeLa cells. J Biol Chem. 2005;280:37790–37797. doi: 10.1074/jbc.M502974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wu D, Jiao M, Zu S, Sollecito CC, Jimenez-Cowell K, Mold AJ, Kennedy RM, Wei Q. Intramolecular Interactions between the Dbl Homology (DH) Domain and the Carboxyl-terminal region of Myosin II-interacting Guanine Nucleotide Exchange Factor (MyoGEF) Act as an Autoinhibitory Mechanism for the Regulation of MyoGEF Functions. J Biol Chem. 2014;289:34033–34048. doi: 10.1074/jbc.M114.607267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Asiedu M, Wu D, Matsumura F, Wei Q. Centrosome/Spindle Pole-associated Protein Regulates Cytokinesis via Promoting the Recruitment of MyoGEF to the Central Spindle. Mol Biol Cell. 2009;20:1428–1440. doi: 10.1091/mbc.E08-01-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pal D, Wu D, Haruta A, Matsumura F, Wei Q. Role of a novel coiled-coil domain-containing protein CCDC69 in regulating central spindle assembly. Cell cycle. 2010;9:4117–4129. doi: 10.4161/cc.9.20.13387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Marchesi S, Montani F, Deflorian G, D’Antuono R, Cuomo A, Bologna S, Mazzoccoli C, Bonaldi T, Di Fiore PP, Nicassio F. DEPDC1B Coordinates De-adhesion Events and Cell-Cycle Progression at Mitosis. Dev Cell. 2014;31:420–433. doi: 10.1016/j.devcel.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Leonard DA, Lin R, Cerione RA, Manor D. Biochemical studies of the mechanism of action of the Cdc42-GTPase-activating protein. J Biol Chem. 1998;273:16210–16215. doi: 10.1074/jbc.273.26.16210. [DOI] [PubMed] [Google Scholar]
- [36].Yoon HY, Miura K, Cuthbert EJ, Davis KK, Ahvazi B, Casanova JE, Randazzo PA. ARAP2 effects on the actin cytoskeleton are dependent on Arf6-specific GTPase-activating-protein activity and binding to RhoA-GTP. J Cell Sci. 2006;119:4650–4666. doi: 10.1242/jcs.03237. [DOI] [PubMed] [Google Scholar]
- [37].Musacchio A, Cantley LC, Harrison SC. Crystal structure of the breakpoint cluster region-homology domain from phosphoinositide 3-kinase p85 alpha subunit. Proc Natl Acad Sci U S A. 1996;93:14373–14378. doi: 10.1073/pnas.93.25.14373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Peck J, Douglas G.t., Wu CH, Burbelo PD. Human RhoGAP domain-containing proteins: structure, function and evolutionary relationships. FEBS Lett. 2002;528:27–34. doi: 10.1016/s0014-5793(02)03331-8. [DOI] [PubMed] [Google Scholar]
- [39].Chiang SH, Hwang J, Legendre M, Zhang M, Kimura A, Saltiel AR. TCGAP, a multidomain Rho GTPase-activating protein involved in insulin-stimulated glucose transport. EMBO J. 2003;22:2679–2691. doi: 10.1093/emboj/cdg262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kozma R, Ahmed S, Best A, Lim L. The GTPase-activating protein n-chimaerin cooperates with Rac1 and Cdc42Hs to induce the formation of lamellipodia and filopodia. Mol Cell Biol. 1996;16:5069–5080. doi: 10.1128/mcb.16.9.5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Seewald MJ, Korner C, Wittinghofer A, Vetter IR. RanGAP mediates GTP hydrolysis without an arginine finger. Nature. 2002;415:662–666. doi: 10.1038/415662a. [DOI] [PubMed] [Google Scholar]
- [42].Brinkmann T, Daumke O, Herbrand U, Kuhlmann D, Stege P, Ahmadian MR, Wittinghofer A. Rap-specific GTPase activating protein follows an alternative mechanism. J Biol Chem. 2002;277:12525–12531. doi: 10.1074/jbc.M109176200. [DOI] [PubMed] [Google Scholar]
- [43].Bellanger JM, Lazaro JB, Diriong S, Fernandez A, Lamb N, Debant A. The two guanine nucleotide exchange factor domains of Trio link the Rac1 and the RhoA pathways in vivo. Oncogene. 1998;16:147–152. doi: 10.1038/sj.onc.1201532. [DOI] [PubMed] [Google Scholar]
- [44].Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- [45].Nassar N, Hoffman GR, Manor D, Clardy JC, Cerione RA. Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP. Nat Struct Biol. 1998;5:1047–1052. doi: 10.1038/4156. [DOI] [PubMed] [Google Scholar]
- [46].Rittinger K, Walker PA, Eccleston JF, Nurmahomed K, Owen D, Laue E, Gamblin SJ, Smerdon SJ. Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP. Nature. 1997;388:693–697. doi: 10.1038/41805. [DOI] [PubMed] [Google Scholar]
- [47].Rittinger K, Walker PA, Eccleston JF, Smerdon SJ, Gamblin SJ. Structure at 1.65 A of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature. 1997;389:758–762. doi: 10.1038/39651. [DOI] [PubMed] [Google Scholar]
- [48].Scrima A, Thomas C, Deaconescu D, Wittinghofer A. The Rap-RapGAP complex: GTP hydrolysis without catalytic glutamine and arginine residues. EMBO J. 2008;27:1145–1153. doi: 10.1038/emboj.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Faucherre A, Desbois P, Satre V, Lunardi J, Dorseuil O, Gacon G. Lowe syndrome protein OCRL1 interacts with Rac GTPase in the trans-Golgi network. Hum Mol Genet. 2003;12:2449–2456. doi: 10.1093/hmg/ddg250. [DOI] [PubMed] [Google Scholar]
- [50].Consonni SV, Maurice MM, Bos JL. DEP domains: structurally similar but functionally different. Nat Rev Mol Cell Biol. 2014;15:357–362. doi: 10.1038/nrm3791. [DOI] [PubMed] [Google Scholar]
- [51].Waterman-Storer CM, Worthylake RA, Liu BP, Burridge K, Salmon ED. Microtubule growth activates Rac1 to promote lamellipodial protrusion in fibroblasts. Nat Cell Biol. 1999;1:45–50. doi: 10.1038/9018. [DOI] [PubMed] [Google Scholar]
- [52].Siegrist SE, Doe CQ. Microtubule-induced cortical cell polarity. Genes Dev. 2007;21:483–496. doi: 10.1101/gad.1511207. [DOI] [PubMed] [Google Scholar]
- [53].Blangy A, Vignal E, Schmidt S, Debant A, Gauthier-Rouviere C, Fort P. TrioGEF1 controls Rac- and Cdc42-dependent cell structures through the direct activation of rhoG. J Cell Sci. 2000;113(Pt 4):729–739. doi: 10.1242/jcs.113.4.729. [DOI] [PubMed] [Google Scholar]
- [54].Sugihara K, Nakatsuji N, Nakamura K, Nakao K, Hashimoto R, Otani H, Sakagami H, Kondo H, Nozawa S, Aiba A, Katsuki M. Rac1 is required for the formation of three germ layers during gastrulation. Oncogene. 1998;17:3427–3433. doi: 10.1038/sj.onc.1202595. [DOI] [PubMed] [Google Scholar]
- [55].Migeotte I, Grego-Bessa J, Anderson KV. Rac1 mediates morphogenetic responses to intercellular signals in the gastrulating mouse embryo. Development. 2011;138:3011–3020. doi: 10.1242/dev.059766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Yue Q, Wagstaff L, Yang X, Weijer C, Munsterberg A. Wnt3a-mediated chemorepulsion controls movement patterns of cardiac progenitors and requires RhoA function. Development. 2008;135:1029–1037. doi: 10.1242/dev.015321. [DOI] [PubMed] [Google Scholar]
- [57].Boudreau HE, Broustas CG, Gokhale PC, Kumar D, Mewani RR, Rone JD, Haddad BR, Kasid U. Expression of BRCC3, a novel cell cycle regulated molecule, is associated with increased phospho-ERK and cell proliferation. Int J Mol Med. 2007;19:29–39. [PubMed] [Google Scholar]
- [58].Yang Y, Liu L, Cai J, Wu J, Guan H, Zhu X, Yuan J, Li M. DEPDC1B enhances migration and invasion of non-small cell lung cancer cells via activating Wnt/beta-catenin signaling. Biochem Biophys Res Commun. 2014;450:899–905. doi: 10.1016/j.bbrc.2014.06.076. [DOI] [PubMed] [Google Scholar]
- [59].Su YF, Liang CY, Huang CY, Peng CY, Chen C, Lin MC, Lin RK, Lin WW, Chou MY, Liao PH, Yang JJ. A putative novel protein, DEPDC1B, is overexpressed in oral cancer patients, and enhanced anchorage-independent growth in oral cancer cells that is mediated by Rac1 and ERK. Journal of biomedical science. 2014;21:67. doi: 10.1186/s12929-014-0067-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kielkopf CL, Lucke S, Green MR. U2AF homology motifs: protein recognition in the RRM world. Genes Dev. 2004;18:1513–1526. doi: 10.1101/gad.1206204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Weng MT, Luo J. The enigmatic ERH protein: its role in cell cycle. RNA splicing and cancer, Protein & cell. 2013;4:807–812. doi: 10.1007/s13238-013-3056-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Prigge JR, Iverson SV, Siders AM, Schmidt EE. Interactome for auxiliary splicing factor U2AF(65) suggests diverse roles. Biochim Biophys Acta. 2009;1789:487–492. doi: 10.1016/j.bbagrm.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Weng MT, Lee JH, Wei SC, Li Q, Shahamatdar S, Hsu D, Schetter AJ, Swatkoski S, Mannan P, Garfield S, Gucek M, Kim MK, Annunziata CM, Creighton CJ, Emanuele MJ, Harris CC, Sheu JC, Giaccone G, Luo J. Evolutionarily conserved protein ERH controls CENP-E mRNA splicing and is required for the survival of KRAS mutant cancer cells. Proc Natl Acad Sci U S A. 2012;109:E3659–3667. doi: 10.1073/pnas.1207673110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Wang PY, Seabold GK, Wenthold RJ. Synaptic adhesion-like molecules (SALMs) promote neurite outgrowth. Mol Cell Neurosci. 2008;39:83–94. doi: 10.1016/j.mcn.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Nam J, Mah W, Kim E. The SALM/Lrfn family of leucine-rich repeat-containing cell adhesion molecules. Semin Cell Dev Biol. 2011;22:492–498. doi: 10.1016/j.semcdb.2011.06.005. [DOI] [PubMed] [Google Scholar]
- [66].Duhoux FP, Ameye G, Lambert C, Herman M, Iossifidis S, Constantinescu SN, Libouton JM, Demoulin JB, Poirel HA. Novel head-to-head gene fusion of MLL with ZC3H13 in a JAK2 V617F-positive patient with essential thrombocythemia without blast cells. Leukemia research. 2012;36:e27–30. doi: 10.1016/j.leukres.2011.09.003. [DOI] [PubMed] [Google Scholar]
- [67].Grieco GS, Malandrini A, Comanducci G, Leuzzi V, Valoppi M, Tessa A, Palmeri S, Benedetti L, Pierallini A, Gambelli S, Federico A, Pierelli F, Bertini E, Casali C, Santorelli FM. Novel SACS mutations in autosomal recessive spastic ataxia of Charlevoix-Saguenay type. Neurology. 2004;62:103–106. doi: 10.1212/01.wnl.0000104491.66816.77. [DOI] [PubMed] [Google Scholar]
- [68].Ghosh P, Garcia-Marcos M, Bornheimer SJ, Farquhar MG. Activation of Galphai3 triggers cell migration via regulation of GIV. J Cell Biol. 2008;182:381–393. doi: 10.1083/jcb.200712066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Plummer NW, Spicher K, Malphurs J, Akiyama H, Abramowitz J, Nurnberg B, Birnbaumer L. Development of the mammalian axial skeleton requires signaling through the Galpha(i) subfamily of heterotrimeric G proteins. Proc Natl Acad Sci U S A. 2012;109:21366–21371. doi: 10.1073/pnas.1219810110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Williams SE, Ratliff LA, Postiglione MP, Knoblich JA, Fuchs E. Par3-mInsc and Galphai3 cooperate to promote oriented epidermal cell divisions through LGN. Nat Cell Biol. 2014;16:758–769. doi: 10.1038/ncb3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Kortazar D, Fanarraga ML, Carranza G, Bellido J, Villegas JC, Avila J, Zabala JC. Role of cofactors B (TBCB) and E (TBCE) in tubulin heterodimer dissociation. Experimental cell research. 2007;313:425–436. doi: 10.1016/j.yexcr.2006.09.002. [DOI] [PubMed] [Google Scholar]
- [72].Tian G, Jaglin XH, Keays DA, Francis F, Chelly J, Cowan NJ. Disease-associated mutations in TUBA1A result in a spectrum of defects in the tubulin folding and heterodimer assembly pathway. Human molecular genetics. 2010;19:3599–3613. doi: 10.1093/hmg/ddq276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Dowhan DH, Hong EP, Auboeuf D, Dennis AP, Wilson MM, Berget SM, O’Malley BW. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERalpha and CAPERbeta. Mol Cell. 2005;17:429–439. doi: 10.1016/j.molcel.2004.12.025. [DOI] [PubMed] [Google Scholar]