Abstract
Members of the multicopper oxidase (MCO) family of enzymes can be classified by their substrate specificity; for example, ferroxidases oxidize ferrous iron, ascorbate oxidases oxidize ascorbate, and laccases oxidize aromatic substrates such as diphenols. Our previous work on an insect multicopper oxidase, MCO1, suggested that it may function as a ferroxidase. This hypothesis was based on three lines of evidence: RNAi-mediated knock down of Drosophila melanogaster MCO1 (DmMCO1) affects iron homeostasis, DmMCO1 has ferroxidase activity, and DmMCO1 has predicted iron binding residues. In our current study, we expanded our focus to include MCO1 from Anopheles gambiae, Tribolium castaneum, and Manduca sexta. We verified that MCO1 orthologs have similar expression profiles, and that the MCO1 protein is located on the basal surface of cells where it is positioned to oxidize substrates in the hemolymph. In addition, we determined that RNAi-mediated knock down of MCO1 in A. gambiae affects iron homeostasis. To further characterize the enzymatic activity of MCO1 orthologs, we purified recombinant MCO1 from all four insect species and performed kinetic analyses using ferrous iron, ascorbate and two diphenols as substrates. We found that all of the MCO1 orthologs are much better at oxidizing ascorbate than they are at oxidizing ferrous iron or diphenols. This result is surpring because ascorbate oxidases are thought to be specific to plants and fungi. An analysis of three predicted iron binding residues in DmMCO1 revealed that they are not required for ferroxidase or laccase activity, but two of the residues (His374 and Asp380) influence oxidation of ascorbate. These two residues are conserved in MCO1 orthologs from insects and crustaceans; therefore, they are likely to be important for MCO1 function. The results of this study suggest that MCO1 orthologs function as ascorbate oxidases and influence iron homeostasis through an unknown mechanism.
Keywords: ferroxidase, ascorbate oxidase, iron homeostasis, insect, kinetic analysis, mutagenesis
1. Introduction
The multicopper oxidase (MCO) family of enzymes includes three main groups: laccases, ferroxidases and ascorbate oxidases (Sakurai and Kataoka, 2007). Most studied multicopper oxidases are categorized as laccases. They are present in invertebrates, plants, fungi and bacteria (Hoegger et al., 2006). Laccases oxidize a broad range of substrates, including diphenols and aromatic amines, and they have diverse functions (Nakamura and Go, 2005). A smaller number of multicopper oxidases are ferroxidases. The well-characterized ferroxidases are found in mammals, algae, and fungi (Kosman, 2010). They facilitate the transport of iron across cell membranes by oxidizing ferrous iron to ferric iron (Kosman, 2010). The third major group of multicopper oxidases are the ascorbate oxidases, which have been identified only in plants and fungi (Hoegger et al., 2006). They oxidize ascorbate to monodehydroascorbate (Farver and Pecht, 1992; Skotland and Ljones, 1980). Ascorbate oxidase activity changes the redox state of the plant apoplast and symplast, and thereby influences various cell signaling pathways (Foyer and Noctor, 2011; Pignocchi and Foyer, 2003; Pignocchi et al., 2003; Sanmartin et al., 2003).
Despite their diverse functions, multicopper oxidases are structurally similar (Zhukhlistova et al., 2008). A typical multicopper oxidase contains three contiguous cupredoxin-like domains and binds four copper atoms within two highly conserved copper centers (Sakurai and Kataoka, 2007). Conserved residues in domains I and III coordinate the four copper ions, and residues in domains II and III form the substrate binding pocket (Quintanar et al., 2007; Sakurai and Kataoka, 2007). During an oxidation event, an electron is transferred from the substrate to the T1 copper atom near the substrate binding site and then to the T2/T3 copper center (Kosman, 2010). Oxygen consumption occurs at the T2/T3 copper center, where molecular oxygen is reduced to water after the transfer of four electrons (Kosman, 2010).
The structural basis of substrate specificity in most multicopper oxidases is poorly understood (Kosman, 2010; Quintanar et al., 2007). Laccases have particularly low substrate specificity, and the substrate preferences of ferroxidases and ascorbate oxidases are not absolute; for example, ferroxidases can oxidize diphenols and ascorbate with low catalytic efficiency (Bielli and Calabrese, 2002; Osaki et al., 1964; Quintanar et al., 2007), and ascorbate oxidases can oxidize diphenols and ferrocyanide (Dayan and Dawson, 1976; O'Neill et al., 1984; Stark and Dawson, 1963).The structural basis of substrate preference is best understood for a yeast ferroxidase, Fet3p (Quintanar et al, 2007). The substrate binding pocket of Fet3p has three acidic residues that coordinate ferrous iron binding: Glu185, Asp283 and Asp409 (Stoj et al., 2006). A mutation in any of these three residues decreases the affinity of Fet3p for ferrous iron, while the E185A/D409A double mutant has greatly diminished affinity; in contrast, these mutations have no effect on the ability of Fet3p to oxidize hydroquinone (a diphenol) (Stoj et al., 2006). The effect of the double mutation on substrate specificity is dramatic: the wild-type enzyme has a 375,000 fold preference for ferrous iron, whereas the double mutant has a 170 fold preference for hydroquinone (Kosman, 2010). These kinetic analyses and other biochemical studies indicate that oxidation of ferrous iron by Fet3p entails binding of ferrous iron by the three acidic residues and transfer of an electron through either Glu185 or Asp409 to the T1 copper atom (Quintanar et al, 2007). Unfortunately, this type of detailed information about substrate-binding is not known for the laccases or ascorbate oxidases.
We have been interested in a conserved insect multicopper oxidase with unknown substrate specificity and unknown physiological functions. This enzyme, MCO1, is present in all of the insect genomes analyzed to date (Dittmer and Kanost, 2010). Our previous study of Drosophila melanogaster MCO1 suggested that MCO1 may function as a ferroxidase (Lang et al., 2012a). We found that DmMCO1 has ferroxidase activity, that weak knock down of DmMCO1 affects iron homeostasis, and that strong knock down is lethal (Lang et al., 2012a). An analysis of the DmMCO1 amino acid sequence suggested that Asp380 and Glu552 may bind to ferrous iron in the substrate binding pocket (Lang et al., 2012a). Immunohistochemistry experiments indicated that DmMCO1 is present on the basal surface of tissues, including midgut and Malphighian tubules (Lang et al., 2012a). Taken together, these results suggested that MCO1 may oxidize ferrous iron in the hemolymph and facilitate iron transport (Lang et al., 2012a).
The main goals of this study were to determine the substrate specificity of MCO1 orthologs and to test the hypothesis that Asp380 and Glu552 are required for ferroxidase activity. Although our previous studies of MCO1 have focused mainly on its function in D. melanogaster, we are interested in the role of MCO1 in a variety of insect species; therefore, we expanded our focus to include MCO1 from Anopheles gambiae (a hematophagous dipteran insect), Tribolium castaneum (a coleopteran species) and Manduca sexta (a lepidopteran species). We found that the MCO1 orthologs were similar in expression, protein localization, knock-down phenotype, and catalytic activity. These similarities indicate that MCO1 orthologs are likely to have similar physiological functions. Surprisingly, recombinant forms of MCO1 were much more efficient at oxidizing ascorbate than they were at oxidizing ferrous iron. In addition, we found that Asp380 and His374 have an influence on ascorbate oxidase activity but not ferroxidase activity. Taken together, our results suggest that MCO1 orthologs function as ascorbate oxidases and influence iron homeostasis through an unknown mechanism.
2. Materials and Methods
2.1. Insect culture
The w1118 strain of D. melanogaster was cultured at 25°C on K12 High Efficiency diet (USBiological). The G3 strain of A. gambiae was obtained from the Malaria Research and Reference Reagent Resource Center, and the mosquitoes were cultured as described previously (Lang et al., 2012b). The GA-1 strain of T. castaneum was reared at 30°C on wheat flour supplemented with 5% brewer's yeast. M. sexta larvae were cultured at 26°C on a wheat germ-based artificial diet.
2.2. Immune challenge experiments
Adult D. melanogaster (4 - 5 days old females) were inoculated with bacteria by pricking with a minuten pin dipped in a pellet of Escherichia coli (XL1 Blue) and Micrcococcus luteus (ATCC# 4698). Untreated flies were used as negative controls. At 24 hours post-inoculation, RNA was isolated from whole flies (eight flies per sample).
Larval M. sexta (one day old fifth instar) were injected with 100 μg freeze-dried M. luteus (ATCC# 4698) in sterile saline. Control larvae were injected with sterile saline. RNA was isolated from fat body collected 24 hours post-inoculation. Each sample was from one larva.
For each immune induction experiment, total RNA isolation, cDNA synthesis, and real-time PCR were done as described previously (Lang et al., 2012a). Real-time PCR reactions were monitored on an iCycler (BioRad) or CFX96 Real-Time PCR system (BioRad). The reference genes used were D. melanogaster ribosomal protein 49 (RP49) or M. sexta ribosomal protein S3 (RPS3). Three biological replicates were done, and differences in expression were assessed by performing an unpaired t test with GraphPad Prism 5.04 software. The primer sequences used for real-time PCR are listed in Table S1.
2.3. Developmental and tissue expression profiles of TcMCO1 and MsMCO1
TcMCO1 expression profiles were determined by qualitative RT-PCR as described by Peng et al. (2014). A developmental expression profile included RNA from eggs, early-, mid- and late-stage larvae, 4 day old pupae, and 5 day old adults. A tissue expression profile analyzed samples from adult females, including guts plus Malpighian tubules, ovaries, and abdominal carcass. Each sample included at least ten individuals or tissues from 16 insects. Ribosomal protein S3 (RPS3) was used as a reference gene. PCR primers are listed in Table S1.
MsMCO1 expression data was obtained from RNA sequence data (file 20140508RSEM_OGS2_Gene_FPKM.xlsx) that was downloaded from the Manduca Base website at the following link: ftp://ftp.bioinformatics.ksu.edu/pub/Manduca/OGS2/OSU_files/. The gene prediction number for MsMCO1 is Msex2.13042. The RNA sequencing was done by the Baylor College of Medicine - Human Genome Sequencing Center (www.hgsc.bcm.edu) and the Weill Cornel Medical College (weill.cornell.edu).
2.4. RNAi-mediated knock down of MCO1 in A. gambiae
Double stranded RNA that targets AgMCO1 or GFP (negative control) was synthesized with a MEGAscript RNAi kit (Life Technologies). To evaluate the dsRNA sequences for specificity of knock down, we used E-RNAi (Horne and Boutros, 2010). The dsRNAs used were predicted to have no off-targets. Newly eclosed adult females were injected with 2 μg dsRNA in 400 nl deionized water. To determine the efficiency of knock down at 4 days and 21 days after injection of dsRNA, total RNA was isolated, and cDNA synthesis and real-time PCR were done as described previously (Lang et al., 2012a). Ribosomal protein S7 (RPS7) was used as a reference gene. Three biological replicates were done, and differences in expression were assessed by performing an unpaired t test with GraphPad Prism 5.04 software. The primer sequences used for dsRNA template synthesis and real-time PCR are listed in Table S1.
2.5. Determination of iron content in A. gambiae
Many species of insects have midgut cells that contain a large amount of ferritin-bound iron, which can be detected by histological staining (Mehta et al., 2009). Briefly, this staining method involves fixing dissected midguts in 4% formaldehyde, permeabilizing with 1% Tween-20, and incubating with 2% K4Fe(CN)6 in 0.24 N HCl. We used this method to analyze midguts from adult female mosquitoes that were fed 10% sucrose supplemented with either 0.2 mM or 5 mM ferric ammonium citrate. Although we have used this method to detect ferritin-bound iron in the midguts of D. melanogaster (Lang et al., 2012a), we did not observe staining in A. gambiae midguts.
To determine the effect of MCO1 knock down in mosquitoes fed 10% sucrose, newly eclosed adult females were injected with dsRNA that targets AgMCO1 or GFP (2 (μg per insect), and insects were analyzed 9 days later. To determine the effect of MCO1 knock down in mosquitoes fed 10% sucrose supplemented with a toxic concentration of iron, injected mosquitoes were fed 10% sucrose for 3 days (to allow time for knock down to occur), then 10% sucrose supplemented with 5 mM ferric ammonium citrate for 5 days, then 10% sucrose for one day (to allow time for the high iron diet to be cleared from the crop and gut). To select against any mosquitoes retaining the high iron diet in the crop or midgut, the iron-supplemented sucrose contained green food coloring, which was visible in the crop and midgut. Whole body iron content was determined as described previously (Lang et al., 2012a). Briefly, mosquitoes were homogenized in lysis buffer, the protein concentration of each homogenate was determined using a bicinchoninic acid assay, and iron content was determined using a Ferrozine-based assay (Missirlis et al., 2006). The sample size was 5 mosquitoes for the experiment involving sucrose fed mosquitoes and 7 for the experiment involving mosquitoes fed iron supplemented sucrose. Three biological replicates were performed, and differences in iron content (in nmole iron per mg protein) were assessed by performing an unpaired t test with GraphPad Prism 5.04 software.
2.6. Generation of polyclonal antisera and immunoblot analysis
Production of polyclonal antiserum generated against DmMCO1 was described previously (Lang et al., 2012a). The same methods were used to generate antiserum against AgMCO1 (Asp414 to Gln782), TcMCO1 (Asp191 to Met470), and MsMCO1 (Ile1 to Pro126). Antisera were used at a 1:2,000 dilution for immunoblot analysis.
2.7. Immunohistochemistry
To verify specificity of the AgMCO1 antiserum, we confirmed that it did not cross-react with the other four A. gambiae MCOs (Gorman et al., 2008). This was accomplished by immunoblot analysis of 5 ng of the AgMCO1 antigen and 50 ng of AgMCO2, AgMCO3, AgMO4 and AgMCO5 antigens. Immunodetection using a 1:2,000 dilution of the polyclonal antiserum resulted in a prominent band in the AgMCO1 antigen lane, and no bands corresponding to the other MCOs (data not shown). To detect AgMCO1 in midguts and Malpighian tubules, immunohistochemistry was performed as described previously (Lang et al., 2012a). Because AgMCO1 expression is upregulated in response to blood feeding (Gorman et al., 2008), we analyzed tissues from mosquitoes that had taken a blood meal. The tissues analyzed were female adult midguts (two days after a blood meal) and whole Malpighian tubules (four days after a blood meal). The secondary antibody used was Alexa Fluor 488-conjugated goat anti-rabbit IgG. Nuclei were stained with DAPI. Sections and tissues were analyzed with a Zeiss LSM 510 META laser scanning confocal microscope using excitation wavelengths of 405 and 488 nm and a 40× Plan-Neofluar oil objective with a 1.3 numerical aperture.
2.8. Identification of signal peptides, transmembrane regions and GPI anchor sites
Signal sequences were predicted by Signal P (Bendtsen et al., 2004), and putative transmembrane regions were predicted with TMPred software (Hofmann and Stoffel, 1993). The presence or absence of GPI anchor sites was predicted with GPI-SOM (Frankhause and Maser, 2005).
2.9. Protein structure prediction
I-TASSER software (Roy et al., 2010; Zhang, 2008) was used to predict the structure of DmMCO1. The three predicted cupredoxin-like domains of DmMCO1 (residues Val229 through Arg898) were used for modeling. PyMOL (Schrödinger) was used to visualize the resulting homology model. The substrate binding pocket of a multicopper oxidase contains the three residues that coordinate the T1 copper atom, which accepts an electron from the substrate (Kosman, 2010). Based on this criterion, the pocket containing His379, Cys811 and His816 was predicted to be the substrate binding pocket of DmMCO1 (see section 3.9).
2.10. cDNAs used for recombinant protein expression
The amino acid sequence of DmMCO1 (NCBI Reference Sequence NP_609287.3) and the cDNA used for protein expression were described previously (Lang et al., 2012a). Some regions of DmMCO1 were excluded from the recombinant protein. Briefly, in an effort to circumvent problems associated with low protein expression and protein aggregation, we generated a truncated DmMCO1 cDNA that encoded only the cysteine-rich region and cupredoxin-like domains (and excluded the native signal peptide, amino-terminal von Willebrand domains, and carboxyl-terminal hydrophobic region). The truncated cDNA was cloned in-frame with the D. melanogaster Bip signal sequence so that the recombinant protein would be secreted. The recombinant enzyme that this cDNA encodes was designated DmMCO1T (Figure 1). We found that recombinant DmMCO1T in the cell culture medium had been cleaved by an unidentified protease; therefore, we used site-directed mutagenesis to create an Arg454Ala mutant form of DmMCO1T (DmMCO1T[R454A]). Site directed mutagenesis was accomplished with the use of the QuickChange Multi Site-Directed Mutagenesis Kit (Stratagene). Several additional mutant forms of DmMCO1T[R454A] were made: DmMCO1T[R454A, H374S], DmMCO1T[R454A, D380A], DmMCO1T[R454A, E552A], DmMCO1T[R454A, D380A, E552A], and DmMCO1T[R454A, H374S, D380A, E552A].
Figure 1.
Alignment of four MCO1 orthologs. Predicted signal peptides are indicated by bold italics. Von Willebrand factor domains are in purple text. Cysteine-rich regions are in green. Putative carboxyl-terminal transmembrane regions are in blue. Predicted copper binding residues are in orange. Three predicted cupredoxin-like domains are delineated with blue vertical lines. The sequences that were used to express recombinant enzymes are between red vertical lines. The extra residues present in the longer versions of TcMCO1 and MsMCO1 are in red. The arginine residue that was mutated in DmMCO1 and AgMCO1 to generate uncleaved recombinant protein is highlighted in black. Three putative iron binding residues are indicated by highlighting. Asp380 (magenta) and Glu552 (green) were identified by their alignment with iron binding residues in Fet3p. His374 (yellow) was identified by analyzing a homology model of DmMCO1 (see section 3.9).
The amino acid sequence of AgMCO1 (GenBank accession number AAN17505) is described in Gorman et al. (2008). The cDNAs used to express AgMCO1T and AgMCO1T[R542S] were analogous to the ones used to express DmMCO1T and DmMCO1T[R454A]; that is, the cysteine-rich region and cupredoxin-like domains were cloned in-frame with the D. melanogaster Bip signal peptide, and an Arg542Ser mutation was made to prevent proteolytic cleavage after residue 542 (Figure 1).
The amino acid sequence of TcMCO1 (synonym = TcLac1) was described previously (Arakane et al., 2005). We used RT-PCR to clone a cDNA that encodes full-length TcMCO1. The sequence was similar to NCBI Reference Sequence NP_001034514, but had several substitutions: Val198Ile, His429Tyr, Phe471Leu, Glu483Asp, Val554Ile, Lys613Arg, Val622Ile, Ser656Pro, Ser664Ala, His665Gln, and Thr668Ser. (The residue number refers to its position in the mature protein.) The Val198Ile substitution was identified in only one of the three cDNAs that we sequenced, but the other differences were present in all three cDNAs indicating that they were not errors. Two truncated forms of the TcMCO1 cDNA were made for protein expression: a shorter form, TcMCO1Ts, which encodes the start methionine through Val629, and a slightly longer form, TcMCO1Tl, which encodes the start methionine through Ser658 (Figure 1).
The amino acid sequence of MsMCO1 (synonym = MsLac1) was described by Dittmer et al. (2004). We used RT-PCR to clone a cDNA that encodes full-length MsMCO1. The sequence encodes a protein identical to GenBank accession number AAN17506 except for four amino acid substitutions: Ile1Phe, Ser7Phe, Ser181Asn, and Ile775Val. (The residue number refers to its position in the mature protein.) All of these substitutions are present in at least two independent cDNA clones, so they are not likely to be errors. Two truncated forms of the MsMCO1 cDNA were made for protein expression: a shorter form, MsMCO1Ts, which encodes the start methionine through Gln724, and a slightly longer form, MsMCO1Tl, which encodes the start methionine through Ser751 (Figure 1).
The cDNA used for TcLac2A expression is described in Gorman et al. (2012).
2.11. Recombinant baculovirus production
Production of recombinant baculoviruses was done as described previously (Lang et al., 2012a). Briefly, cDNAs were inserted into the pOET3 transfer plasmid, and the flashBAC GOLD system (Oxford Expression Technologies) was used to generate recombinant baculoviruses. The baculovirus used for AgMCO3 expression was described in Lang et al. (2012b).
2.12. Enzyme purification
Recombinant multicopper oxidases were expressed in cultured insect cells and purified as described previously (Lang et al, 2012a). Briefly, recombinant baculoviruses were used to infect cultured Sf9 cells, and the secreted enzymes were purified from the culture medium by concanavalin-A-Sepharose, Q-Sepharose, and Superdex 200 chromatography. The concentration of recombinant enzymes was determined as described previously (Lang et al., 2012b). Yields ranged from 0.4 - 2.2 mg per liter of cell culture.
Laccase-α from Trametes versicolor (TvLac-α) was purified from a commercial preparation of fungal laccases (Sigma, catalog number 51639). The sample was dialyzed against 20 mM Tris, pH 8.0 (4°C), and purified by Q-Sepharose and Superdex 200 chromatography. The Proteomics Center of the University of Nevada used peptide mass finger printing to identify the protein as TvLac-α (GenBank Accession Number = Q12718; Necochea et al., 2005; Figure S1). Briefly, the laccase was subjected to SDS-PAGE followed by trypsin digestion and LC/MS analysis. The LC/MS analysis was performed using a Thermo LTQ Orbitrap XL with ETD coupled to a Michrom Paradigm MDLC and Michrom CaptiveSpray. Protein identification analysis was performed on a SORCERER server using SEQUEST software. The yield of TvLac-α was 0.26 mg from 100 mg starting material.
2.13. Amino-terminal sequencing
Purified recombinant proteins were subjected to SDS-PAGE, transferred to a PVDF membrane, and stained with Coomassie blue R. Bands were excised from the membrane, and Edman protein sequencing was done by Dr. Kathleen Schegg at the Nevada Proteomics Center. An ABI 492 Procise sequencer was used to determine the first five residues of each polypeptide.
2.14. Laccase assay
Laccase activity assays, determination of pH optima, and calculation of kinetic constants were done as described previously (Gorman et al., 2012). Briefly, reactions were done by mixing 0.5 -5 μg enzyme with substrate in a total volume of 200 μl and detecting product formation with a microplate spectrophotometer. All of the recombinant forms of MCO1 had activity at pH 6.0 -7.5 (data not shown); therefore, pH 7.0 (the approximate pH of insect hemolymph) was used for determining kinetic constants. Two recombinant insect laccases, TcLac2A and AgMCO3, were used as positive controls (Gorman et al., 2012; Lang et al., 2012b).
2.15. Ferroxidase assays
Two ferroxidase assays were used: a Ferrozine-based assay and an apo-transferrin-based assay. The Ferrozine-based assays were done as described previously (de Silva et al., 1997) with minor modifications. Briefly, reactions (100 μl) contained 5 μg enzyme and 0 - 240 μM ferrous ammonium sulfate in 100 mM sodium acetate, 100 μM sodium citrate, pH 6.0. Reactions were stopped at 0, 1, 2, 3, 4, or 5 min with the addition of 15 mM Ferrozine, which forms a stable complex with ferrous iron. This complex, representing the amount of unreacted substrate, was detected by measuring absorbance at 562 nm and using a molar extinction coefficient of 27,900 cm-1 M-1 (Stookey, 1970). The decrease in substrate concentration was assumed to be equal to the concentration of product (ferric iron) formed. An apo-transferrin-based assay was done as described previously (Osaki et al., 1966; Johnson et al., 1967) with minor modifications. Human apo-transferrin (Sigma) was dissolved in 50 mM Tris, 150 mM sodium chloride, 20 mM sodium bicarbonate, pH 7.3. Reactions (200 μl) contained 20 - 80 μg enzyme, 780 μg apo-transferrin, and ferrous ammonium sulfate (0 - 240 μM) in 100 mM sodium acetate, pH 6.0. The formation of holo-transferrin from apo-transferrin was determined by measuring absorbance at 460 nm and using a molar extinction coefficient of 2,500 cm-1 M-1. Human ceruloplasmin (1 μg per reaction) (Sigma, catalog number C4519) was used as a positive control. A fungal laccase (TvLac-α, 1 μg per reaction) and two insect laccases (TcLac2A and AgMCO3, 80 μg per reaction) were used as negative controls (Gorman et al., 2012; Lang et al., 2012b; Necochea et al., 2005). Kinetic curves were made by plotting activity versus substrate concentration. The data were fit to the Michaelis-Menten equation by non-linear regression, and kinetic constants were estimated from the fitted data. (An early kinetic analysis of ceruloplasmin suggested that it has both a high affinity and low affinity iron binding site (resulting in two Kms) (Osaki, 1966), but subsequent work suggested that this is not the case (Huber and Frieden, 1970), and more recent studies have used the Michaelis-Menten equation to characterize the kinetic properties of ceruloplasmin (Stoj and Kosman, 2003).)
2.16. Ascorbate oxidase assay
A spectrophotometric assay was used to detect ascorbate oxidase activity. Ascorbate concentration is commonly monitored by measuring absorbance at 265 nm (Oberbacher and Vines, 1963); however, to avoid excessively high absorbance readings when the concentration of ascorbate was greater than 500 μM, we monitored ascorbate concentration at 275 nm. We confirmed a linear relationship between absorbance at 275 nm and ascorbate concentration from 5 - 600 μM (R2 = 0.9997), and calculated a molar extinction coefficient of 12,000 M-1 cm-1 (data not shown). Reactions were done by mixing 0.1 or 0.2 μg MCO1 with ascorbic acid in 0.1 M sodium phosphate, pH 7.0, in a total volume of 100 μl and observing a decrease in substrate concentration over time with a microplate spectrophotometer. Blank reactions containing no enzyme measured the rate of non-enzymatic oxidation. Ascorbate oxidase (1.2 ng per reaction) from Curbita sp. (Sigma, catalog number A0157) was used as a positive control. Three laccases (0.2 μg per reaction) were used as negative controls: TvLac-α (Necochea et al., 2005), TcLac2A (Gorman et al., 2012), and AgMCO3 (Lang et al., 2012b). Assays were done in duplicate. Kinetic curves were made by plotting activity versus substrate concentration. The data were fit to the Michaelis-Menten and allosteric sigmoidal equations by non-linear regression using GraphPad Prism. Akaike's Information Criteria method was used to determine which equation fit the data better. Kinetic constants were estimated from the fitted data.
2.17. Phylogenetic analysis
To identify MCO genes in non-insect invertebrates, we used the MsMCO1 sequence as a query to perform a BLAST search of the National Center for Biotechnology Information (NCBI) non-redundant protein database excluding sequences from vertebrates, insects, fungi and plants. In addition, we searched the NCBI Transcriptome Shotgun Assembly to identify additional MCO genes from invertebrates. From the sequences we identified, we selected those that contained an amino-terminal cysteine-rich region and eleven predicted copper binding residues (Dittmer and Kanost, 2010). We used these sequences and MCO sequences from D. melanogaster, A. gambiae, T. castaneum and M. sexta to do a phylogenetic analysis. (The insect MCO sequences are described in Dittmer and Kanost, 2010.) Phylogenetic analysis was performed using MEGA 6.06 software. Sequences were aligned globally by using the Clustal W option with default settings. The phylogenetic tree was constructed by the Neighbor-Joining method with a Poisson correction model. Gaps were treated by the pairwise deletion method. Statistical analysis was carried out by the bootstrap method with 5000 replications; branch points with bootstrap values less than 50% were collapsed.
3. Results
3.1. MCO1 is a conserved insect gene
The predicted amino acid sequences of MCO1 orthologs from D. melanogaster, A. gambiae, T. castaneum and M. sexta are 47-55% identical. All of the MCO1 sequences contain three putative cupredoxin-like domains, which comprise the catalytic portion of multicopper oxidases, and eleven conserved copper coordinating residues (Figure 1). They each have a predicted signal peptide and carboxyl-terminal transmembrane region (Figure 1), and they are predicted to be GPI-anchored; therefore, MCO1 orthologs are expected to be attached to the exterior of the plasma membrane through a carboxyl-terminal transmembrane region or a GPI-anchor. Each of the four MCO1 sequences contains two acidic residues that were predicted to bind ferrous iron in the substrate binding pocket (Figure 1) (Lang et al., 2012a). The only notable difference among MCO1 sequences is that the dipteran sequences contain amino-terminal von Willebrand factor domains, whereas MCO1 sequences from non-dipteran insects do not (Figure 1). The function of the von Willebrand factor domains in the dipteran MCO1 orthologs is unknown (Dittmer and Kanost, 2010).
3.2. MCO1 orthologs have similar expression profiles
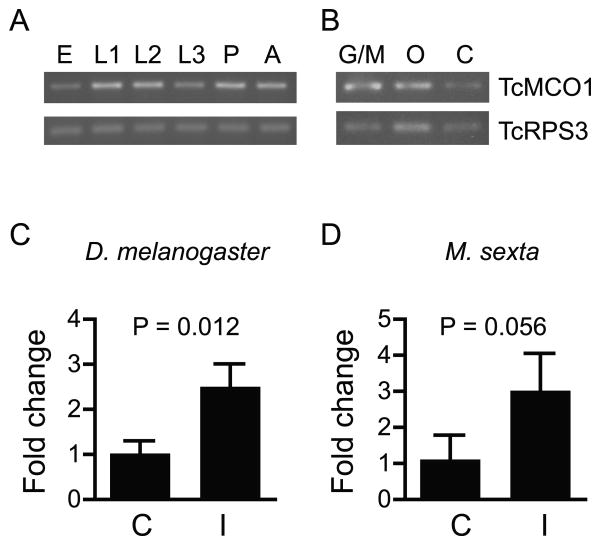
DmMCO1, AgMCO1 and MsMCO1 are expressed throughout development in many tissues, including the midgut and Malpighian tubules (Baker et al., 2011; Chintapalli et al., 2007; Dittmer et al, 2004; Gorman et al., 2008; Gravely et al., 2011; Lang et al., 2012a; www.agripestbase.org/manduca). Using qualitative RT-PCR, we found that, like the other MCO1 orthologs, TcMCO1 is expressed in all developmental stages and in each of the tissues analyzed (Figure 2). Upregulation in response to immune challenge has been observed for DmMCO1 and AgMCO1 (De Gregorio et al., 2001; Gorman et al., 2008), and RNA sequence data suggest that MsMCO1 may also be immune responsive (www.agripestbase.org/manduca). We used qPCR to determine the degree of upregulation of DmMCO1 and MsMCO1 after injection of bacteria. The results suggest that MCO1 transcript abundance increases 2-3 fold 24 hours after an immune challenge (Figure 2). Taken together, the gene expression data suggest that MCO1 orthologs have similar expression profiles and, thus, support the hypothesis that MCO1 orthologs have similar functions.
Figure 2.
Expression profiles of MCO1 orthologs. Qualitative RT-PCR was used to determine expression profiles of TcMCO1. TcMCO1 expression was detected in all of the developmental stages (A) and tissues (B) analyzed. Quantitative RT-PCR was used to determine the degree of immune-induced upregulation of DmMCO1 (C) and MsMCO1 (D). qPCR data were from three biological replicates. Means ± SD are shown. Differences in expression were assessed by performing an unpaired t test. (Abbreviations for A and B: E, eggs; L1, early-stage larvae; L2, mid-stage larvae; L3, late-stage larvae; P, pupae; A, adults; G/M, midguts plus Malpighian tubules; O, ovaries; C, abdominal carcass. Abbreviations for C and D: C, control; I = 24 hours post-immune induction.)
3.3. AgMCO1 is present on the basal surface of the midgut and Malpighian tubules
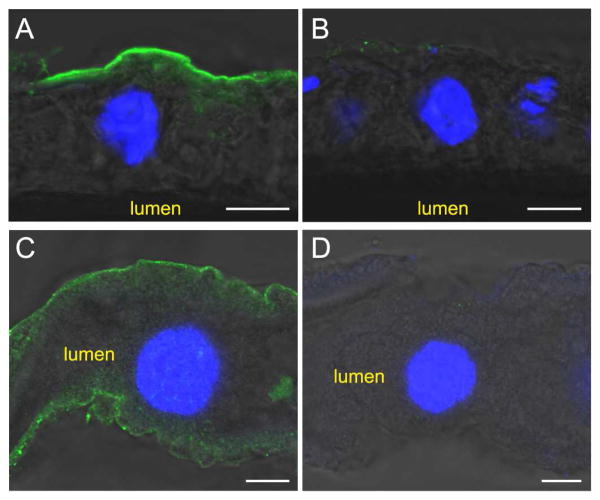
As described in section 3.1, MCO1 is predicted to be membrane bound, with the enzyme positioned to oxidize substrates in the hemolymph. We used immunohistochemistry to determine the location of AgMCO1 within adult mosquitoes. As we observed for DmMCO1 (Lang et al., 2012a), AgMCO1 was detected on the basal surface of the midgut and Malpighian tubules (Figure 3).
Figure 3.
Immunolocalization of AgMCO1 in adult female mosquitoes. Cryosections of midguts (A and B) and unsectioned Malpighian tubules (C and D) were immunostained with AgMCO1 antiserum (A and C) or pre-immune serum (B and D). Anti-MCO1 antibodies were detected with Alexa Fluor 488-conjugated anti-rabbit IgG antibodies (green). Nuclei were stained with DAPI (blue). AgMCO1 immunoreactivity was detected along the basal surface of the epithelial cells of the midgut and Malpighian tubules (A and C). No signal was observed in samples incubated with preimmune serum (B and D). (Scale bars: 10 μm)
3.4. AgMCO1 is required for iron homeostasis
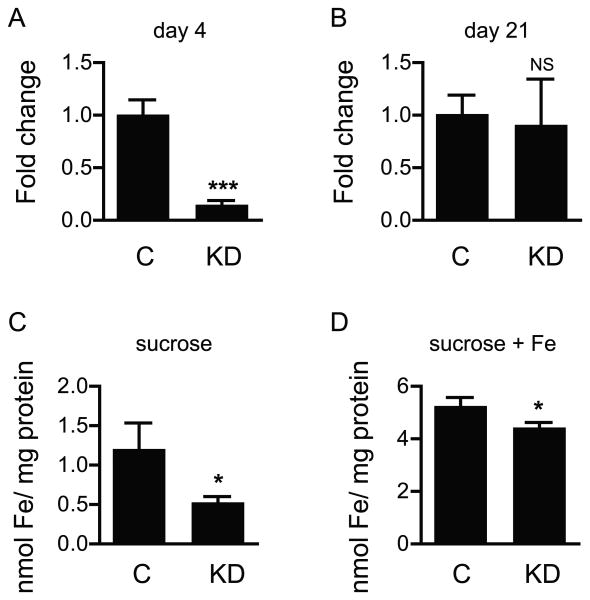
Our previous work demonstrated that RNAi-mediated knock down of DmMCO1 increases the longevity of flies fed a toxic concentration of iron, decreases the amount of iron in specialized midgut cells, and decreases whole body iron content. We wanted to know if knock down of MCO1 in A. gambiae affects iron homeostasis in a similar manner. First, qPCR was used to verify that injection of dsRNA would decrease the amount of AgMCO1 mRNA in adult female mosquitoes. RNAi was successful 4 days after injection of dsRNA, but by 21 days, the knock-down effect was gone (Figure 4). Because of the transient nature of the knock down, RNAi could not be used to evaluate the effect of MCO1 knock down on longevity. Next, we used a histological method to determine whether A. gambiae adult females have detectable ferritin-bound iron in the midgut. No staining of the midgut was observed (data not shown); therefore, this method could not be used to assess whether AgMCO1 activity affects the amount of ferritin-bound iron in midgut cells. To determine whether AgMCO1 knock down results in a decrease in whole body iron content, we used a Ferrozine-based method to estimate the amount of iron in homogenates of adult females. Newly eclosed mosquitoes were injected with dsRNA, and iron content was analyzed 9 days later. The experiment was done twice, once with insects fed 10% sucrose and once with insects fed 10% sucrose plus 5 mM ferric ammonium citrate. Under both conditions, knock down of AgMCO1 was associated with a decrease in iron content (Figure 4). This result suggests that AgMCO1, like DmMCO1, plays a role in iron homeostasis.
Figure 4.
Effect of MCO1 knock down on iron content in adult mosquitoes. Quantitative RT-PCR was used to determine the extent of knock down of AgMCO1 4 days (A) and 21 days (B) after injection of dsRNA. Knock down was apparent at 4 days but not 21 days post-injection. A Ferrozine-based assay was used to determine the iron content in control and AgMCO1 knock down mosquitoes. A decrease in iron content was observed for insects that were fed 10% sucrose (C) and for those that were fed 10% sucrose supplemented with 5 mM ferric ammonium citrate (D). Data were from three biological replicates. Means ± SD are shown. Differences in expression and in iron content were assessed by performing an unpaired t test. (*P < 0.05, ***P < 0.001, NS = no statistically significant difference.) (Abbreviations: C, control; KD, knock down.)
3.5. Expression and purification of recombinant forms of MCO1
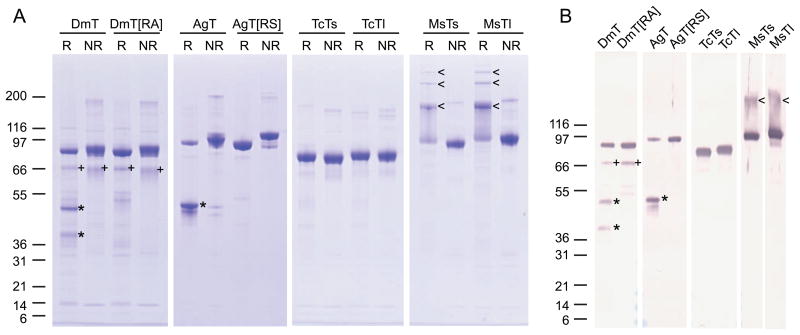
To evaluate the substrate specificity of MCO1 orthologs, we analyzed two recombinant forms of each of the four MCO1 orthologs (Figure 1). All of the recombinant enzymes lacked the predicted carboxyl-terminal transmembrane region so that they could be purified from the cell culture medium. DmMCO1T and AgMCO1T also lack the amino-terminal von Willebrand factor domains, which are unlikely to influence substrate specificity. We found that the two dipteran enzymes were cleaved by an unidentified protease (Figure 5). The cleavage site, which was identified by amino-terminal sequencing, was at an arginine residue that is conserved in dipteran MCO1 sequences (Figure 1). Uncleaved enzymes, DmMCO1T[R454A] and AgMCO1T[R542S], were made by mutating the arginine codon. Two recombinant variants of TcMCO1 and MsMCO1 were purified: a longer form that is missing only the predicted carboxyl-terminal transmembrane region (TcMCO1Tl and MsMCO1Tl) and a shorter form that is missing a slightly larger portion of the carboxyl-end (TcMCO1Ts and MsMCO1Ts) (Figure 1). The secreted proteins were purified from serum-free medium with the use of lectin affinity, anion exchange, and size-exclusion chromatography. SDS-PAGE analysis demonstrated that each of the recombinant enzymes was highly purified (Figure 5). The identity of the recombinant proteins was verified by immunoblot analysis (Figure 5). A comparison of the migration of DmMCO1T and AgMCO1T under reducing and non-reducing conditions indicated that the polypeptides generated by proteolytic cleavage were connected by a disulfide bond (Figure 5). Surprisingly, MsMCO1Tl and MsMCO1Ts formed stable multimers when they were heated in SDS sample buffer containing β-mercaptoethanol (Figure 5). We do not know what caused this unusual behavior.
Figure 5.
SDS-PAGE and immunoblot analysis of recombinant forms of MCO1. A) Purified recombinant forms of MCO1 were analyzed by SDS-PAGE using reducing (R) or non-reducing (NR) conditions followed by Coomassie staining. Each lane contains 1 μg protein. B) Purified recombinant forms of MCO1 were analyzed by SDS-PAGE using reducing conditions followed by immunodetection. Each lane contains 50 ng protein. DmMCO1T and AgMCO1T cleavage products that are linked by a disulfide bond under non-reducing conditions are indicated by an asterisk. A minor, uncharacterized cleavage product of DmMCO1 is indicated by a plus sign. Multimers of MsMCO1 that form after heating in reducing SDS-PAGE sample buffer are indicated by an arrow head. Note that the highest molecular mass multimers are not visible in panel B because proteins 200 kDa or greater did not transfer to the nitrocellulose membrane. The expected mass of each recombinant protein: DmMCO1T, 79.8 kDa, DmMCO1T amino-terminal cleavage product, 34.1 kDa; DmMCO1T carboxyl-terminal cleavage product, 45.7 kDa; DmMCO1T[R454A], 79.8 kDa; AgMCO1T, 81.3 kDa; AgMCO1T amino-terminal cleavage product, 35.3kDa (plus possible O-linked glycosylation, which may explain why this product co-migrates with the 46.1 kDa carboxyl-terminal product); AgMCO1T carboxyl-terminal cleavage product, 46.1 kDa; AgMCO1T[R542S], 81.3 kDa; TcMCO1Ts, 72.1 kDa; TcMCO1Tl, 75.3 kDa; MsMCO1Ts, 82.8 kDa; MsMCO1Tl, 86.0 kDa. (Abbreviations: DmT, DmMCO1T; DmT[RA], DmMCO1T[R454A]; AgT, AgMCO1T; AgT[RS], AgMCO1T[R542S]; TcTs, TcMCO1Ts; TcTl, TcMCO1Tl; MsTs, MsMCO1Ts; MsTl, MsMCO1Tl.)
3.6. MCO1 orthologs have low laccase activity
A kinetic analysis of DmMCO1T[R454A] demonstrated that it has very low laccase activity (Lang et al., 2012a). To analyze the laccase activity of the other MCO1 variants, we used two laccase substrates: hydroquinone and dopamine. Hydroquinone is a p-diphenol that is commonly used in studies of multicopper oxidases, and dopamine is an o-diphenol that is an endogenous substrate of insect laccase-2 (Gorman et al., 2012). All of the MCO1 variants had low affinity for the diphenols (with Kms ranging from 10 to 62 mM), and, thus, very low catalytic efficiency (Table 1). These results strongly suggest that MCO1 orthologs do not function as laccases.
Table 1.
Kinetic characterization of the laccase activity of MCO1 orthologs.
| Hydroquinone | Dopamine | |||||
|---|---|---|---|---|---|---|
|
Kcat (min-1) |
Km (mM) |
kcat/Km (min-1 mM-1) |
kcat (min-1) |
Km (mM) |
kcat/Km (min-1 mM-1) |
|
| DmMCO1 isoforms | ||||||
| DmMCO1T | 62 ± 2.1 | 14.4 ± 0.91 | 4 | 19.0 ± 1.1 | 7.0 ± 0.93 | 3 |
| DmMCO1T[R454A]a | 63 ± 2.1 | 10.0 ± 0.60 | 6 | 13.9 ± 0.8 | 7.1.± 0.91 | 2 |
| AgMCO1 isoforms | ||||||
| AgMCO1T | 95 ± 7.8 | 31.9 ± 3.75 | 3 | 15.5 ± 1.1 | 35.2 ± 3.59 | 0.4 |
| AgMCO1T[R542S] | 53 ± 2.4 | 34.2 ± 2.16 | 2 | 6.0 1.2 | 37.8 ± 10.48 | 0.2 |
| TcMCO1 isoforms | ||||||
| TcMCO1Ts | 311 ± 31 | 10.8 ± 2.19 | 29 | 19.2 ± 3.1 | 47.2 ± 9.97 | 0.4 |
| TcMCO1Tl | 301 ± 13 | 12.3 ± 1.00 | 24 | 5.1 ± 0.4 | 20.6 ± 2.38 | 0.2 |
| MsMCO1 isoforms | ||||||
| MsMCO1Ts | 44 ± 2.7 | 53.2 ± 4.19 | 0.8 | 31.9 ± 1.2 | 37.6 ± 2.01 | 0.8 |
| MsMCO1Tl | 48 ± 4.5 | 62.1 ± 7.10 | 0.8 | 30.8 ± 2.7 | 32.7 ± 4.18 | 0.9 |
| Insect laccases | ||||||
| AgMCO3b | 1,571 ± 123 | 5.2 ± 0.57 | 302 | 274 ± 4.0 | 1.6 ± 0.08 | 171 |
| TcLac2Ac | 213 ± 5.9 | 1.0 ± 0.11 | 213 | 41 ± 1.0 | 0.8 ± 0.06 | 51 |
| Fungal laccase | ||||||
| TvLac-α | 1,963 ± 73 | 0.038 ± 0.006 | 51,661 | 692 ± 10.8 | 0.055 ± 0.005 | 12,597 |
| Ferroxidase | ||||||
| Fet3pd | 131 ± 5.0 | 25.5 ± 2.5 | 5 | NDe | ND | ND |
Note: the kinetic constants reported are best-fit values with standard errors
Data are from Lang et al., 2012a
Data are from Lang et al., 2012b
Data are from Gorman et al., 2012
Data are from Stoj et al., 2006
ND = not determined
3.7. MCO1 orthologs have low ferroxidase activity
An in-gel ferroxidase assay indicated that DmMCO1T[R454A] has ferroxidase activity (Lang et al., 2012a), but to evaluate whether this activity is significant, we needed to use an assay that provides a quantitative measure of activity. The most accurate method for analyzing the activity of multicopper ferroxidases is to measure the consumption of oxygen during the reaction (Hassett et al., 1998); however, this method requires more enzyme than we can purify from cultured insect cells. A second commonly used method makes use of Ferrozine to detect the substrate (ferrous iron) remaining in the reaction (de Silva et al., 1997). This method works well when the substrate concentration is low, but we experienced poor reproducibility when ferrous iron concentrations above 100 μM were used (data not shown). A third method (Osaki et al., 1966) that makes use of apo-transferrin to detect product formation resulted in much better reproducibility. In addition, results obtained with the apo-transferrin method were similar to those obtained with the Ferrozine and oxygen consumption methods (see kinetic constants for DmMCO1T[R454A] and ceruloplasmin in Table 2). For these reasons, we used the apo-transferrin method for kinetic characterization of ferroxidase activity. The analysis of DmMCO1 activity confirmed that DmMCO1 does have ferroxidase activity, but its affinity for ferrous iron is relatively low (Table 2). The Km of DmMCO1T was estimated to be 260 μM, whereas the well-studied ferroxidases have Kms in the range of 2 - 20 μM (Griffiths et al., 2005; Stoj and Kosman, 2003; Stoj et al., 2006). The catalytic efficiency of DmMCO1T was estimated to be 160 min-1 mM-1 (Table 2). Kinetic characterization of the MCO1 orthologs from other species demonstrated that they had catalytic efficiencies ranging from 5-30 min-1 mM-1, values that were not much higher than those of two insect laccases (Table 2). These results suggest that MCO1 orthologs may not function as ferroxidases.
Table 2.
Kinetic characterization of the ferroxidase activity of MCO1 orthologs.
| Ferrous iron | ||||
|---|---|---|---|---|
| method |
kcat (min-1) |
Km (mM) |
kcat/Km (min-1 mM-1) |
|
| DmMCO1 isoforms | ||||
| DmMCO1T | apoTf | 40 ± 2.4 | 0.26 ± 0.02 | 156 |
| DmMCO1T[R454A] | apoTf | 42 ± 2.7 | 0.12 ± 0.02 | 359 |
| Ferrozine | 20 ± 1.9 | 0.06 ± 0.01 | 351 | |
| AgMCO1 isoforms | ||||
| AgMCO1T | apoTf | 3 ± 0.2 | 0.12 ± 0.01 | 30 |
| AgMCO1T[R542S] | apoTf | 2 ± 0.2 | 0.11 ± 0.03 | 16 |
| TcMCO1 isoforms | ||||
| TcMCO1Ts | apoTf | 3 ± 0.7 | 0.45 ± 0.16 | 7 |
| TcMCO1Tl | apoTf | 9 ± 6.8 | 1.71 ± 1.49 | 5 |
| MsMCO1 isoforms | ||||
| MsMCO1Ts | apoTf | 3 ± 0.2 | 0.21 ± 0.02 | 16 |
| MsMCO1Tl | apoTf | 9 ± 1.9 | 1.41 ± 0.35 | 6 |
| Insect laccases | ||||
| AgMCO3 | apoTf | 0.4 ± 0.0 | 0.09 ± 0.02 | 5 |
| TcLac2A | apoTf | 1 ± 0.1 | 0.33 ± 0.05 | 3 |
| Fungal laccase | ||||
| TvLac-α | apoTf | 372 ± 6.6 | 0.15 ± 0.005 | 2,407 |
| Ferroxidases | ||||
| hCpa | O2 uptake | 30.3 ± 1.6 | 0.0083 ± 0.0015 | 3,650 |
| hCp | apoTf | 120 ± 8.0 | 0.023 ± 0.0052 | 5,106 |
| Fet3pb | O2 uptake | 40.1 ± 1.4 | 0.0049 ± 0.0008 | 8,000 |
Note: the kinetic constants reported are best-fit values with standard errors
Data are from Stoj and Kosman, 2003
Data are from Stoj et al., 2006
3.8. MCO1 orthologs are much better at oxidizing ascorbate than they are at oxidizing diphenols or ferrous iron
Almost all of the well-studied multicopper oxidases are laccases, ferroxidases or ascorbate oxidases (Nakamura and Go, 2005). Because the MCO1 orthologs had low laccase and ferroxidase activity, we decided to assess their ability to oxidize ascorbate. We used a slightly modified version of a spectrophotometric assay that detects the amount of ascorbate in the reaction (Oberbacher and Vines, 1963). To validate this method, we analyzed the activity of ascorbate oxidase from Cucurbita sp. and compared our results to those obtained with an oxygen consumption method (Wimalasena and Dharmasena, 1994). The kinetic constants estimated by both assays were similar (Table 3). Kinetic characterization of the MCO1 orthologs demonstrated that they all have significant ascorbate oxidase activity (Table 3). Most of the recombinant forms of MCO1 had kinetic curves that fit an allosteric sigmoidal equation slightly better than the Michaelis-Menten equation, so instead of calculating a Km for those variants, we calculated an equivalent kinetic constant, Khalf. (MCO1 orthologs may form functional dimers similar to plant ascorbate oxidases (Messerschmidt et al., 1989), and this property may explain the kinetic curves that we observed.) Recombinant forms of MCO1 had Km or Khalf values of 0.12 - 0.77 mM, kcat values of 570 - 12,900 min-1, and catalytic efficiencies of 2,780 - 39,770 min-1 mM-1. Collectively, the kinetic properties of MCO1 orthologs indicate that they are much better at oxidizing ascorbate than they are at oxidizing ferrous iron or diphenols (Table 4).
Table 3.
Kinetic characterization of the ascorbate oxidase activity of MCO1 orthologs.
| Ascorbate | ||||
|---|---|---|---|---|
| modela |
kcat (min-1) |
Khalf or Km (mM) |
kcat/Km (min-1 mM-1) |
|
| DmMCO1 isoforms | ||||
| DmMCO1T | AS | 2,570 ± 315 | 0.29 ± 0.05 | 8,806 |
| DmMCO1T[R454A] | AS | 1,480 ± 170 | 0.27 ± 0.05 | 5,516 |
| AgMCO1 isoforms | ||||
| AgMCO1T | AS | 916 ± 84 | 0.12 ± 0.02 | 7,559 |
| AgMCO1T[R542S] | MM | 568 ± 43 | 0.20 ± 0.04 | 2,785 |
| TcMCO1 isoforms | ||||
| TcMCO1Ts | AS | 12,912 ± 492 | 0.32 ± 0.01 | 39,766 |
| TcMCO1Tl | AS | 10,541 ± 504 | 0.42 ± 0.02 | 25,357 |
| MsMCO1 isoforms | ||||
| MsMCO1Ts | AS | 3,013 ± 497 | 0.62 ± 0.16 | 4,838 |
| MsMCO1Tl | AS | 4,059 ± 889 | 0.77 ± 0.27 | 5,244 |
| Insect laccases | ||||
| AgMCO3 | MM | 148 ± 36 | 0.64 ± 0.26 | 233 |
| TcLac2A | activity too low to characterize | |||
| Fungal laccase | ||||
| TvLac-α | MM | 29 ± 3.4 | 0.21 ± 0.06 | 135 |
| Ascorbate oxidase | ||||
| Curcubita AOb | 40,920 ± 2,040 | 0.20 ± 0.03 | 204,600 | |
| Curcubita AO | AS | 21,264 ± 996 | 0.17 ± 0.01 | 127,558 |
Note: the kinetic constants reported are best-fit values with standard errors
AS = allosteric sigmoidal; MM = Michaelis-Menten
Data are from Wimalasena and Dharmasena, 1994
Table 4.
Comparison of the catalytic efficiency of MCO1 orthologs with four substrates.
| Hydroquinone | Dopamine | Ferrous iron | Ascorbate | |
|---|---|---|---|---|
| DmMCO1 isoforms | ||||
| DmMCO1T | 4 | 3 | 156 | 8,806 |
| DmMCO1T[R454A] | 6 | 2 | 359 | 5,516 |
| AgMCO1 isoforms | ||||
| AgMCO1T | 3 | 0.4 | 30 | 7,559 |
| AgMCO1T[R542S] | 2 | 0.2 | 16 | 2,785 |
| TcMCO1 isoforms | ||||
| TcMCO1Ts | 29 | 0.4 | 7 | 39,766 |
| TcMCO1Tl | 24 | 0.3 | 5 | 25,357 |
| MsMCO1 isoforms | ||||
| MsMCO1Ts | 0.8 | 0.8 | 16 | 4,838 |
| MsMCO1Tl | 0.8 | 0.9 | 6 | 5,244 |
| Insect laccases | ||||
| AgMCO3 | 302 | 171 | 5 | 233 |
| TcLac2A | 213 | 51 | 3 | very low |
| Fungal laccase | ||||
| TvLac-α | 51,661 | 12,597 | 2,407 | 135 |
| Ferroxidase | ||||
| Fet3pa | 5 | NDb | 8,000 | ND |
| Ascorbate oxidase | ||||
| Curcubita AO | ND | ND | ND | 127,558 |
Note: catalytic efficiency is reported as min-1 mM-1
Data are from Stoj et al., 2006
ND = not determined
3.9. Predicted iron binding residues of DmMCO1 are involved in oxidation of ascorbate but not ferrous iron or diphenols
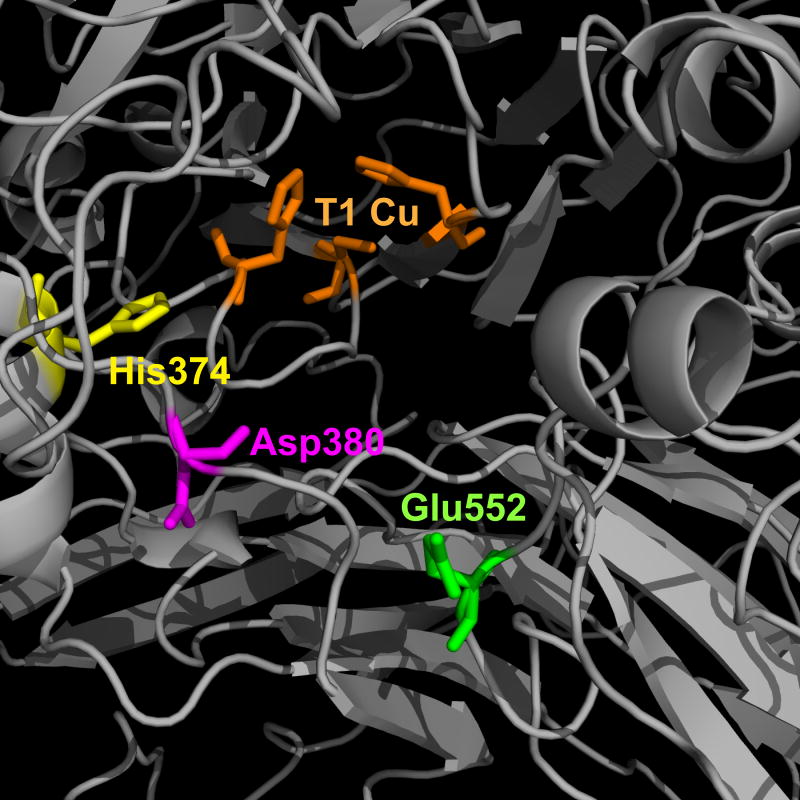
Our previous analysis of the DmMCO1 amino acid sequence suggested that Asp380 and Glu552 may bind to ferrous iron in the substrate binding pocket (Lang et al., 2012a). These residues align with the iron binding residues Glu185 and Asp283 in the yeast ferroxidase Fet3p, and they are conserved in MCO1 orthologs (Lang et al., 2012a). To identify additional putative iron binding residues, we generated a homology model of DmMCO1 (Figure 6) and searched the putative substrate binding pocket for residues that are known to bind iron (aspartic acid, glutamic acid, histidine and tyrosine). We assumed that substrate binding residues should be conserved, so we screened candidate residues for their presence in MCO1 sequences from 10 insect species. From this analysis, we identified His374 as a third putative binding residue (Figure 6).
Figure 6.
Homology model of the putative substrate binding pocket of DmMCO1. I-TASSER was used to create a homology model of DmMCO1. The three residues that are expected to coordinate the T1 copper are shown in orange. His374, Asp380 and Glu552 are conserved residues that were predicted to bind to iron in the putative substrate binding pocket.
To evaluate the requirement of the three putative iron binding residues for ferroxidase activity, we purified five mutant forms of DmMCO1T (DmMCO1T[R454A, H374S], DmMCO1T[R454A, D380A], DmMCO1T[R454A, E552A], DmMCO1T[R454A, D380A, E552A], and DmMCO1T[R454A, H374S, D380A, E552A]). We then compared their activity with the activity of the wild-type form (DmMCO1T[R454A]). As expected, the mutations had no detectable effect on laccase activity (Table S2). Surprisingly, the mutations also had no detectable effect on ferroxidase activity (Table 5). Even the triple mutant had kinetic constants that were similar to those of the wild-type enzyme. These results strongly suggest that the three putative iron binding residues do not bind iron and are not required for ferroxidase activity. The substrate binding residues in ascorbate oxidases are unknown, but because Asp, Glu and His can form bonds with ascorbate, we predicted that the three conserved residues may influence the affinity of DmMCO1 for ascorbate. The mutants showed no difference in affinity (i.e., the Km values for the mutants were not higher than that of the wild-type enzyme); however, some unexpected differences in activity were observed for two of the mutants (Table 5). The H374S mutation resulted in a five fold decrease in the turnover number (kcat) and a four fold decrease in catalytic efficiency (kcat/Km). The D380A mutant had a six fold lower kcat and five fold lower Km, resulting in a catalytic efficiency that was similar to that of the wild-type enzyme. These results suggest that His374 and Asp380 are involved in oxidation of ascorbate. The E552A mutation had little effect on activity, as did the D380A/E552A double mutation; however, the triple mutant had no detectable ascorbate oxidase activity. Taken together, our kinetic analyses suggest that His374 and Asp380 influence oxidation of ascorbate but not ferrous iron or diphenols.
Table 5.
Kinetic characterization of the ferroxidase and ascorbate oxidase activity of DmMCO1 mutant isoforms.
| Ferrous iron | Ascorbate | |||||
|---|---|---|---|---|---|---|
| DmMCO1T mutant isoform |
Kcat (min-1) |
Km (mM) |
kcat/Km (min-1 mM-1) |
kcat (min-1) |
Km (mM) |
kcat/Km (min-1 mM-1) |
| [R454A]a | 42 ± 2.7 | 0.12 ± 0.02 | 359 | 1,480 ± 170 | 0.27 ± 0.05 | 5,516 |
| [R454A, H374S] | 35 ± 4.8 | 0.10 ± 0.03 | 350 | 294 ± 34 | 0.20 ± 0.06 | 1,487 |
| [R454A, D380A] | 54 ± 14 | 0.38 ± 0.15 | 139 | 243 ± 12 | 0.05 ± 0.01 | 4,470 |
| [R454A, E552A] | 48 ± 7.8 | 0.17 ± 0.05 | 276 | 747 ± 129 | 0.24 ± 0.06 | 3,135 |
| [R454A, D380A, E552A] | 39 ± 1.8 | 0.13 ± 0.01 | 295 | 765 ± 145 | 0.36 ± 0.14 | 2,104 |
| [R454A, H374S, D380A, E552A] | 40 ± 3.3 | 0.21 ± 0.03 | 191 | no detectable activity | ||
Note: the kinetic constants reported are best-fit values with standard errors
Data are from Lang et al., 2012a
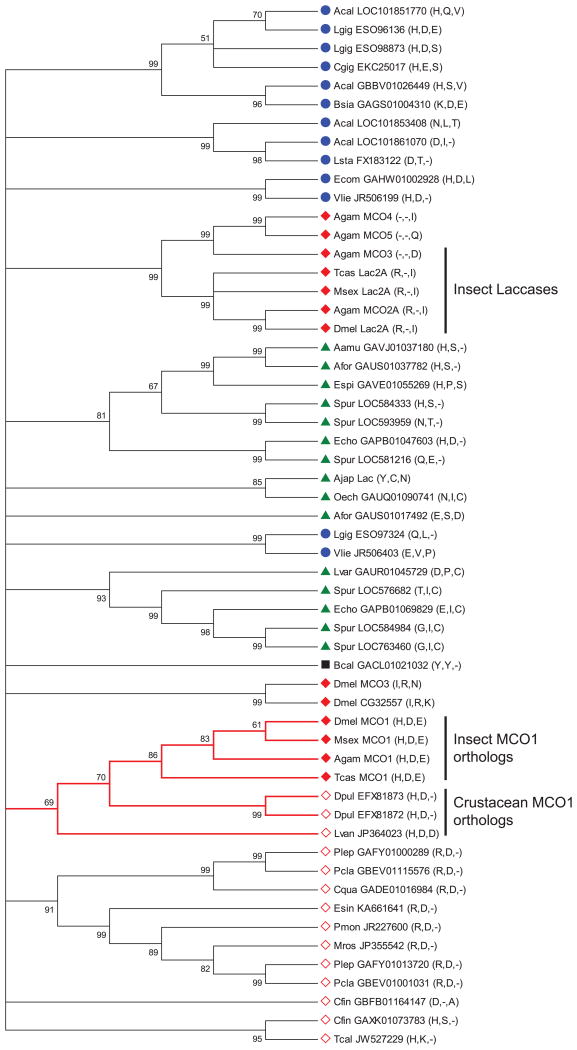
3.10. Putative MCO1 orthologs are present in crustacean genomes
Recently, MCO1-like genes were identified in invertebrates other than insects (Asano, 2014). We were interested in what types of non-insect invertebrates have MCO1 orthologs and whether the three residues described above (His374, Asp380, and Glu552) are conserved. Our phylogenetic analysis suggests that there are orthologous genes in some crustaceans (two in Daphnia pulex and one in Litopenaeus vannamei), but no definitive orthologs in the other invertebrates analyzed (Figure 7). Of the three residues discussed in Section 3.9, the histidine and aspartate residues are conserved in the putative crustacean MCO1 orthologs (Figure 7). This result further supports the hypothesis that His374 and Asp380 are important to the function of MCO1. Interestingly, there is a cluster of multicopper oxidases in crustaceans that have the conserved aspartate but an arginine in place of the histidine (Figure 7). Because arginine can sometimes act as a conservative substitution for histidine, this group of multicopper oxidases may have a catalytic function similar to that of the MCO1 orthologs. Some additional multicopper oxidases of interest include mollusc sequences with the conserved histidine and aspartate, and crustacean, mollusc, and echinoderm sequences with only the conserved histidine. The evolutionary relationship between these proteins and MCO1 orthologs is not clear.
Figure 7.
Phylogenetic analysis of MCO genes from insects and other invertebrates. A bootstrap consensus tree constructed by the neighbor-joining method is shown. The numbers above the branches are bootstrap values expressed as a percentage. All branches with bootstrap values less than 50 were collapsed. A cluster of putative MCO1 orthologs is indicated by red branches. The residues corresponding to His374, Asp380 and Glu552 in DmMCO1 are listed after each gene name. Symbols indicate sequences from crustaceans (open red diamond), echinoderms (green triangle), insects (closed red diamond), molluscs (blue circle), or rotifers (black square). Abbreviations used are: Acal, Aplysia californica; Ajap, Apostichopus japonicas; Aamu, Asterias amurensis; Afor, Asterias forbesi; Bsia, Bithynia siamensis goniomphalos; Bcal, Brachionus calyciflorus; Cfin, Calanus finmarchicus; Cqua, Cherax quadricarinatus; Cgig, Crassostrea gigas; Dpul, Daphnia pulex; Espi, Echinaster spinulosus; Ecom, Elliptio complanata; Esin, Eriocheir sinensis; Echo, Evechinus chloroticus; Lvan, Litopenaeus vannamei; Lgig, Lottia gigantean; Lsta, Lymnaea stagnalis; Lvar, Lytechinus variegatus; Mros, Macrobrachium rosenbergii; Oech, Ophiocoma echinata; Pmon, Penaeus monodon; Plep, Pontastacus leptodactylus; Pcla, Procambarus clarkii; Spur, Strongylocentrotus purpuratus; Tcal, Tigriopus californicus; Vlie, Villosa lienosa.
4. Discussion
Before undertaking this study, our working model of MCO1 was that it functions as a ferroxidase. This model was based on the knowledge that DmMCO1 has predicted iron binding residues and detectable ferroxidase activity, and that RNAi-mediated knock down of DmMCO1 affects iron homeostasis (Lang et al., 2012a). Consistent with this model is our finding that RNAi-mediated knock down of MCO1 affects iron homeostasis in A. gambiae. However, the kinetic characterization of MCO1 orthologs from four species of insects strongly suggests that MCO1 is not a ferroxidase. Compared with the activity of known ferroxidases, all of the MCO1 variants had low ferroxidase activity due to high Km values, ranging from 0.1 - 1.7 mM. The concentration of ferrous iron in hemolymph (where MCO1 is situated) is unknown, but ferrous iron concentrations in biological fluids are extremely low. The high Kms seem to be inconsistent with the hypothesis that MCO1 oxidizes iron in the hemolymph. In addition, we found that the putative iron binding residues in DmMCO1 are not required for ferroxidase activity and are, thus, not likely to bind ferrous iron.
In contrast, the results of our kinetic analyses are consistent with the hypothesis that MCO1 functions as an ascorbate oxidase. MCO1 from all four species oxidized ascorbate with a much higher catalytic efficiency than they oxidized ferrous iron or diphenols, and they had Km or Khalf values ranging from 0.1 - 0.8 mM ascorbate. The concentration of ascorbate in the hemolymph of most insects is unknown, but the concentration is approximately 2.5 mM in the hemolymph of M. sexta larvae and 0.06 - 0.3 mM in the hemolymph of Schistocerca gregaria (Kramer and Seib, 1982); therefore, the Km and Khalf values are compatible with the concentration of ascorbate in hemolymph. In addition, two of the conserved residues in the predicted substrate binding pocket of DmMCO1 have considerable influence on ascorbate oxidase activity. These results support the hypothesis that MCO1 orthologs may function as ascorbate oxidases. This result is somewhat surprising because ascorbate oxidases from animals have not been reported.
We identified His374, Asp380, and Glu552 as possible iron binding residues in DmMCO1. All three residues are conserved in insect MCO1 sequences; His374 and Asp380 are conserved in crustaceans. Because we expected the three residues to be involved in iron binding, we were surprised that a triple mutant had essentially wild-type ferroxidase activity. These results strongly suggest that the three putative iron binding residues do not bind iron; therefore, an iron binding site remains unidentified. Given that the oxidation of ferrous iron by DmMCO1 is very inefficient, it is possible that MCO1 does not have a specific iron binding site. The substrate binding residues in ascorbate oxidases are unknown, but because His, Asp, and Glu can form bonds with ascorbate, we decided to evaluate the mutants for ascorbate oxidase activity. The E552A mutation had little effect on activity, but the H374S and D380A mutants had a lower turnover number (kcat). In addition, the triple mutant had no detectable ascorbate oxidase activity. These results demonstrate that His374 and Asp380 influence ascorbate oxidase activity; however, because the single mutants retained considerable activity, neither residue is essential for full catalytic activity. Mutating charged residues may disrupt the microenvironment of the substrate binding pocket, leading to a conformational change that interferes with catalytic activity when ascorbate is the substrate.
Ascorbate has multiple functions in animals and plants, including acting as an antioxidant, serving as a cofactor for various enzymes, and participating in cell signalling (Corti et al., 2010; Foyer and Noctor, 2011; Goggin et al., 2010; Pignocchi and Foyer, 2003). Ascorbate is present in animals and plants as ascorbate (reduced form), monodehydroascorbate (a radical that is formed after the loss of one electron), and dehydroascorbate (a somewhat more stable oxidized form that is created by the loss of two electrons). All three forms of ascorbate have biological activity (Corti et al, 2010; Gest et al., 2013; Smirnoff, 2000). Ascorbate oxidase removes one electron from ascorbate to produce monodehydroascorbate (Farver and Pecht, 1992; Skotland and Ljones, 1980). Monodehydroascorbate is an unstable, short-lived radical that can undergo disproportionation to produce dehydroascorbate and ascorbate; therefore, dehydroascorbate is often considered the final product of ascorbate oxidase activity. In mammals, which have no known ascorbate oxidase, ascorbate is oxidized when it participates in antioxidant or enzyme-mediated reactions (Corti et al., 2010). In mammals and plants, reductases reduce monodehydroascorbate and dehydroascorbate to ascorbate (Corti, et al., 2010; Smirnoff, 2000).
It is not obvious what the physiological functions of an ascorbate oxidase might be in insect hemolymph. Like MCO1, plant ascorbate oxidase is an extracellular enzyme. It is present in the apoplast, where ascorbate is present at millimolar concentrations (Pignocchi and Foyer, 2003). Plants that over express ascorbate oxidase have more dehydroascorbate than ascorbate in the apoplast, whereas plants with decreased ascorbate oxidase activity have the reverse phenotype (Pignocchi et al., 2003). The resulting changes in the apoplastic redox state have been implicated in cell signalling events that influence plant growth, defense and response to stress (Foyer and Noctor, 2011; Pignocchi and Foyer, 2003; Smirnoff, 2000). Whether the redox state of ascorbate in hemolymph influences cell signalling in insects is unknown. A second hypothesis about MCO1 function in insects is based on differences in ascorbate and dehydroascorbate transport in mammals. In mammals, the reduced and oxidized forms of ascorbate are transported into cells by distinct transporters that have different expression patterns (Corti et al., 2010). It is unknown how ascorbate influx occurs in insects, but if some insect cells take up dehydroascorbate rather than ascorbate, then MCO1 may participate in ascorbate uptake by producing dehydroascorbate.
Several experiments have demonstrated that knock down of MCO1 affects iron homeostasis in D. melanogaster and A. gambiae. Our original interpretation of these results was that the disruption in iron homeostasis was related to the ferroxidase activity of MCO1 (Lang et al., 2012a). Given the newly determined kinetic properties of MCO1, we now think this interpretation is unlikely. It seems much more likely that MCO1 knock down affects iron homeostasis indirectly, possibly through a disturbance in ascorbate metabolism. Iron homeostasis and ascorbate homeostasis are linked via redox homeostasis (Foyer and Noctor, 2011; Valko et al., 2005); therefore, a disturbance in ascorbate metabolism due to MCO1 knock down could lead indirectly to a change in iron metabolism. On the other hand, recent studies in plants and animals have identified putative iron transport mechanisms that involve ascorbate (Grillet et al., 2014; Lane and Richardson, 2014); therefore, another possibility is that the changes in iron homeostasis are a direct result of changes in ascorbate redox state. Additional experiments are needed to determine whether MCO1 does function as an ascorbate oxidase, and, if so, how this function is related to iron homeostasis.
Supplementary Material
Figure S1. Peptide mass finger printing of TvLac-α. Peptide mass finger printing was used to identify a laccase purified from a commercial preparation of T. versicolor laccases. The amino acid sequence of TvLac-α is shown. Eleven matching peptides are highlighted in yellow. The green highlighted methionine was oxidized. The peptides were matched using MS/MS data, with a probability of ≥ 95%. Peptide coverage was 32% (168/519 amino acids).
Table S1. Primers used for expression analysis and amplification of templates for dsRNA synthesis.
Table S2. Kinetic characterization of the laccase activity of DmMCO1 mutants.
Highlights.
Multicopper oxidase-1 (MCO1) is a conserved insect multicopper oxidase
MCO1 is located on the basal surface of cells where it is positioned to oxidize substrates in the hemolymph
MCO1 oxidizes ascorbate with a much higher efficiency than it oxidizes ferrous iron or diphenols
Two conserved residues, a histidine and an aspartate, influence oxidation of ascorbate
MCO1 activity affects iron homeostasis, presumably through its ascorbate oxidase activity
Acknowledgments
We thank Yoonseong Park and Ramaswamy Krishnamoorthi for helpful suggestions regarding this work, Zhen Li and Jayne Christen for providing cDNAs, Stewart Gardner for help with mutagenesis and protein purification, Sandi Yungeberg for generation of MsMCO1 antiserum, Yasuaki Hiromasa for peptide mass fingerprinting, and Renzhi Cao and Jianlin Cheng for help with homology modeling. We thank Haobo Jiang, Gary Bissard, and Xiaolong Cao for making Manduca sexta transcriptomics data available prior to publication. We thank Joel Sanneman for help with cryosectioning and confocal microscopy, and Philine Wangemann for advice and the use of the Center of Biomedical Research Excellence (COBRE) Confocal Microfluorometry and Microscopy Core Facility. The COBRE Confocal Microfluorometry and Microscopy Core Facility was supported by Kansas State University, the College of Veterinary Medicine, and the National Institutes of Health Grant P20-RR017686. Edman protein sequencing was done by Kathleen Schegg and LC/MS analysis was done by Rebekah Woolsey, both at the Nevada Proteomics Center, which is supported by a grant from the National Institute of General Medical Sciences (P20GM103440). This work was supported by National Institute of Allergy and Infectious Diseases grant R01 AI070864 and National Institute of General Medical Sciences grant R37 GM41247. This is contribution 15-201-J from the Kansas Agricultural Experiment Station.
Abbreviations
- dsRNA
double-stranded RNA
- GFP
green fluorescent protein
- Lac
laccase
- LC/MS
liquid chromatography/ mass spectrometry
- MCO1
multicopper oxidase-1
- NCBI
National Center for Biotechnology Information
- RPS
ribosomal protein subunit
- RNAi
RNA interference
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Zeyu Peng, Email: zeyupeng@ksu.edu.
Neal T. Dittmer, Email: ndittmer@ksu.edu.
Minglin Lang, Email: langml@ucas.ac.cn.
Lisa M. Brummett, Email: lmbrumme@ksu.edu.
Caroline L. Braun, Email: cbraun@ksu.edu.
Lawrence C. Davis, Email: ldavis@ksu.edu.
Michael R. Kanost, Email: kanost@ksu.edu.
Maureen J. Gorman, Email: mgorman@ksu.edu.
References
- Arakane Y, Muthukrishnan S, Beeman RW, Kanost MR, Kramer KJ. Laccase 2 is the phenoloxidase required for beetle cuticle tanning. Proc Natl Acad Sci U S A. 2005;102:11337–11342. doi: 10.1073/pnas.0504982102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T. Re-examination of a α-chymotrypsin-solubilized laccase in pupal cuticle of the silkworm, Bombyx mori: insights into the regulation system for laccase activation during the ecdysis process. Insect Biochem Mol Biol. 2014;5:61–69. doi: 10.1016/j.ibmb.2014.10.004. [DOI] [PubMed] [Google Scholar]
- Baker DA, Nolan T, Fischer B, Pinder A, Crisanti A, Russell S. A comprehensive gene expression atlas of sex- and tissue-specificity in the malaria mosquito, Anopheles gambiae. BMC Genomics. 2011;12:296. doi: 10.1186/1471-2164-12-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: signalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bielli P, Calabrese L. Structure to function relationships in ceruloplasmin: a moonlighting protein. Cell Mol Life Sci. 2002;59:1413–1427. doi: 10.1007/s00018-002-8519-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JAT. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Corti A, Casini AF, Pompella A. Cellular pathways for transport and efflux of ascorbate and dehydroascorbate. Arch Biohem Biophys. 2010;500:107–115. doi: 10.1016/j.abb.2010.05.014. [DOI] [PubMed] [Google Scholar]
- Dayan J, Dawson CR. Substrate specificity of ascorbate oxidase. Biochem Biophys Res Comm. 1976;73:451–458. doi: 10.1016/0006-291x(76)90728-2. [DOI] [PubMed] [Google Scholar]
- De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci U S A. 2001;98:12590–12595. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva D, Davis-Kaplan S, Fergestad J, Kaplan J. Purification and characterization of Fet3 protein, a yeast homologue of ceruloplasmin. J Biol Chem. 1997;272:14208–14213. doi: 10.1074/jbc.272.22.14208. [DOI] [PubMed] [Google Scholar]
- Dittmer NT, Kanost MR. Insect multicopper oxidases: diversity, properties and physiological roles. Insect Biochem Mol Biol. 2010;40:179–188. doi: 10.1016/j.ibmb.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Dittmer NT, Suderman RJ, Jiang H, Zhu YC, Gorman MJ, Kramer KJ, Kanost MR. Characterization of cDNAs encoding putative laccase-like multicopper oxidases and developmental expression in the tobacco hornworm, Manduca sexta, and the malaria mosquito, Anopheles gambiae. Insect Biochem Mol Biol. 2004;34:29–41. doi: 10.1016/j.ibmb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Farver O, Pecht I. Low activation barriers characterize intramolecular electron transfer in ascorbate oxidase. Proc Natl Acad Sci USA. 1992;89:8283–8287. doi: 10.1073/pnas.89.17.8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. Ascorbate and glutathione: the heart of the redox hub. Plant Physiol. 2011;155:2–18. doi: 10.1104/pp.110.167569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankhauser N, Maser P. Identification of GPI anchor attachment signals by a Kohonen self-organizing map. Bioinformatics. 2005;21:1846–1852. doi: 10.1093/bioinformatics/bti299. [DOI] [PubMed] [Google Scholar]
- Gest N, Gautier H, Stevens R. Ascorbate as seen through plant evolution: the rise of a successful molecule? J Exp Bot. 2013;64:33–53. doi: 10.1093/jxb/ers297. [DOI] [PubMed] [Google Scholar]
- Goggin FL, Avila CA, Lorence A. Vitamin C content in plants is modified by insects and influences susceptibility to herbivory. Bioessays. 2010;32:777–790. doi: 10.1002/bies.200900187. [DOI] [PubMed] [Google Scholar]
- Gorman MJ, Dittmer NT, Marshall JL, Kanost MR. Characterization of the multicopper oxidase gene family in Anopheles gambiae. Insect Biochem Mol Biol. 2008;38:817–824. doi: 10.1016/j.ibmb.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman MJ, Sullivan LI, Nguyen TDT, Dai H, Arakane Y, Dittmer NT, Syed LU, Li J, Hua DH, Kanost MR. Kinetic properties of alternatively spliced isoforms of laccase-2 from Tribolium castaneum and Anopheles gambiae. Insect Biochem Mol Biol. 2012;42:193–202. doi: 10.1016/j.ibmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravely BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths TAM, Mauk AG, MacGillivray RTA. Recombinant expression and functional characterization of human hephaestin: a multicopper oxidase with ferroxidase activity. Biochemistry. 2005;44:14725–14731. doi: 10.1021/bi051559k. [DOI] [PubMed] [Google Scholar]
- Grillet L, Ouerdane L, Flis P, Hoang MTT, Isaure MP, Lobinski R, Curie C, Mari S. Ascorbate efflux as a new strategy for iron reduction and transport in plants. J Biol Chem. 2014;289:2515–2525. doi: 10.1074/jbc.M113.514828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassett RF, Yuan DS, Kosman DJ. Spectral and kinetic properties of the Fet3 protein from Saccharomyces cerevisiae, a multinuclear copper ferroxidase enzyme. J Biol Chem. 1998;273:23274–23282. doi: 10.1074/jbc.273.36.23274. [DOI] [PubMed] [Google Scholar]
- Hoegger PJ, Kilaru S, James TY, Thacker JR, Kues U. Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J. 2006;273:2308–2326. doi: 10.1111/j.1742-4658.2006.05247.x. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMbase - A database of membrane spanning proteins segments Biol. Chem Hoppe-Seyler. 1993;347:166. [Google Scholar]
- Horne T, Boutros M. E-RNAi: a web application for the multi-species design of RNAi reagents - 2010 update. Nucleic Acids Res. 2010;38:W332–W339. doi: 10.1093/nar/gkq317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber CT, Frieden E. Substrate activation and the kinetics of ferroxidase. J Biol Chem. 1970;245:3973–3978. [PubMed] [Google Scholar]
- Johnson DA, Osaki S, Frieden E. A micromethod for the determination of ferroxidase (ceruloplasmin) in human serums. Clin Chem. 1967;13:142–150. [PubMed] [Google Scholar]
- Kosman DJ. Multicopper oxidases: a workshop on copper coordination chemistry, electron transfer, and metallophysiology. J Biol Inorg Chem. 2010;15:15–28. doi: 10.1007/s00775-009-0590-9. [DOI] [PubMed] [Google Scholar]
- Kramer KJ, Seib PA. Ascorbic acid and the growth and development of insects. Adv Chem Ser. 1982;200:275–291. [Google Scholar]
- Lane DJR, Richardson DR. The active role of vitamin C in mammalian iron metabolism: much more than just enhanced iron absorption. Free Radic Biol Med. 2014;75:69–83. doi: 10.1016/j.freeradbiomed.2014.07.007. [DOI] [PubMed] [Google Scholar]
- Lang M, Braun CL, Kanost MR, Gorman MJ. Multicopper oxidase-1 is a ferroxidase essential for iron homeostasis in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2012a;109:13337–13342. doi: 10.1073/pnas.1208703109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang M, Kanost MR, Gorman MJ. Multicopper oxidase-3 is a laccase associated with the peritrophic matrix of Anopheles gambiae. PLoS ONE. 2012b;7:e33985. doi: 10.1371/journal.pone.0033985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, Deshpande A, Bettedi L, Missirlis F. Ferritin accumulation under iron scarcity in Drosophila iron cells. Biochimie. 2009;91:1331–1334. doi: 10.1016/j.biochi.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Messerschmidt A, Rossi A, Ladenstein R, Huber R, Bolognesi M, Gatti G, Marchesini A, Petruzzelli R, Finazzi-Agro A. X-ray crystal structure of the blue oxidase ascorbate oxidase from zucchini. J Mol Biol. 1989;206:513–529. doi: 10.1016/0022-2836(89)90498-1. [DOI] [PubMed] [Google Scholar]
- Missirlis F, Holmberg S, Georgieva T, Dunkov BC, Rouault TA, Law JH. Characterization of mitochondrial ferritin in Drosophila. Proc Natl Acad Sci U S A. 2006;103:5893–5989. doi: 10.1073/pnas.0601471103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Go N. Function and molecular evolution of multicopper blue proteins. Cell Mol Life Sci. 2005;62:2050–2066. doi: 10.1007/s00018-004-5076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necochea R, Valderrama B, Diaz-Sandoval S, Folch-Mallol JL, Vazquez-Duhalt R, Iturriaga G. Phylogenetic and biochemical characterization of a recombinant laccase from Trametes versicolor. FEMS Microbiol Lett. 2005;244:235–241. doi: 10.1016/j.femsle.2005.01.054. [DOI] [PubMed] [Google Scholar]
- Oberbacher MF, Vines HM. Spectrophotometric assay of ascorbic acid oxidase. Nature. 1963;197:1203–1204. doi: 10.1038/1971203a0. [DOI] [PubMed] [Google Scholar]
- O'Neill P, Fielden EM, Morpurgo L, Agostinelli E. Pulse-radiolysis studies on the interaction of one-electron reduced species with blue oxidases. Biochem J. 1984;222:71–76. doi: 10.1042/bj2220071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaki S. Kinetic studies of ferrous iron oxidation with crystalline human ferroxidase (ceruloplasmin) J Biol Chem. 1966;241:5053–5059. [PubMed] [Google Scholar]
- Osaki S, McDermott JA, Frieden E. Proof for the ascorbate oxidase activity of ceruloplasmin. J Biol Chem. 1964;239:3570–3575. [PubMed] [Google Scholar]
- Osaki S, Johnson DA, Frieden E. The possible significance of the ferrous oxidase activity of ceruloplasmin in normal human serum. J Biol Chem. 1966;241:2746–2751. [PubMed] [Google Scholar]
- Peng Z, Green PG, Arakane Y, Kanost MR, Gorman MJ. A multicopper oxidase-related protein is essential for insect viability, longevity and ovary development. PLoS ONE. 2014;9:e111344. doi: 10.1371/journal.pone.0111344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignocchi C, Foyer CH. Apoplastic ascorbate metabolism and its role in the regulation of cell signalling. Curr Opin Plant Biol. 2003;6:379–389. doi: 10.1016/s1369-5266(03)00069-4. [DOI] [PubMed] [Google Scholar]
- Pignocchi C, Fletcher JM, Wilkinson JE, Barnes JD, Foyer CH. The function of ascorbate oxidase in tobacco. Plant Physiol. 2003;132:1631–1641. doi: 10.1104/pp.103.022798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintanar L, Stoj C, Taylor AB, Hart PJ, Kosman DJ, Solomon EI. Shall we dance? How a multicopper oxidase chooses its electron transfer partner Acc Chem Res. 2007;40:445–452. doi: 10.1021/ar600051a. [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Kataoka K. Basic and applied features of multicopper oxidases, CueO, bilirubin oxidase, and laccase. Chem Rec. 2007;7:220–229. doi: 10.1002/tcr.20125. [DOI] [PubMed] [Google Scholar]
- Sanmartin M, Drogoudi PD, Lyons T, Pateraki I, Barnes J, Kanellis AK. Over-expression of ascorbate oxidase in the apoplast of transgenic tobacco results in altered ascorbate and glutathione redox states and increased sensitivity to ozone. Planta. 2003;216:918–928. doi: 10.1007/s00425-002-0944-9. [DOI] [PubMed] [Google Scholar]
- Skotland T, Ljones T. Direct spectrophotometric detection of ascorbate free radical formed by dopamine-β-monooxygenase and by ascorbate oxidase. Biochim Biophys Acta. 1980;630:30–35. doi: 10.1016/0304-4165(80)90134-8. [DOI] [PubMed] [Google Scholar]
- Smirnoff N. Ascorbic acid: metabolism and functions of a multi-faceted molecule. Curr Opin Plant Biol. 2000;3:229–235. [PubMed] [Google Scholar]
- Stark GR, Dawson CR. Ascorbic acid oxidase. In: Boyer PD, Lardy H, Myrback K, editors. The Enzymes. Vol. 8. Academic Press; New York: 1963. pp. 297–311. [Google Scholar]
- Stoj C, Kosman DJ. Cuprous oxidase activity of yeast Fet3p and human ceruloplasmin: implication for function. FEBS Lett. 2003;554:422–426. doi: 10.1016/s0014-5793(03)01218-3. [DOI] [PubMed] [Google Scholar]
- Stoj CS, Augustine AJ, Zeigler L, Solomon EI, Kosman DJ. Structural basis of the ferrous iron specificity of the yeast ferroxidase, Fet3p. Biochemistry. 2006;45:12741–12749. doi: 10.1021/bi061543+. [DOI] [PubMed] [Google Scholar]
- Stookey LL. Ferrozine - a new spectrophotometric reagent for iron. Anal Chem. 1970;42:779–781. [Google Scholar]
- Valko M, Morris H, Cronin MTD. Metals, toxicity and oxidative stress. Curr Med Chem. 2005;12:1161–1208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- Wimalasena K, Dharmasena S. Substrate specificity of ascorbate oxidase: unexpected similarity to the reduction of dopamine β-monooxygenase. Biochem Biophys Res Commun. 1994;203:1471–1476. doi: 10.1006/bbrc.1994.2350. [DOI] [PubMed] [Google Scholar]
- [last accessed October, 2014]; www.agripestbase.org/manduca.
- Zheng Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhukhlistova NE, Zhukova YN, Lyashenko AV, Zaitsev VN, Mikhailov AM. Three-dimensional organization of three-domain copper oxidases: a review. Crystallogr Rep. 2008;53:92–109. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Peptide mass finger printing of TvLac-α. Peptide mass finger printing was used to identify a laccase purified from a commercial preparation of T. versicolor laccases. The amino acid sequence of TvLac-α is shown. Eleven matching peptides are highlighted in yellow. The green highlighted methionine was oxidized. The peptides were matched using MS/MS data, with a probability of ≥ 95%. Peptide coverage was 32% (168/519 amino acids).
Table S1. Primers used for expression analysis and amplification of templates for dsRNA synthesis.
Table S2. Kinetic characterization of the laccase activity of DmMCO1 mutants.