Abstract
The degree to which genetic factors influence brain connectivity is beginning to be understood. Large-scale efforts are underway to map the profile of genetic effects in various brain regions. The NIH-funded Human Connectome Project (HCP) is providing data valuable for analyzing the degree of genetic influence underlying brain connectivity revealed by state-of-the-art neuroimaging methods. We calculated the heritability of the fractional anisotropy (FA) measure derived from diffusion tensor imaging (DTI) reconstruction in 481 HCP subjects (194/287 M/F) consisting of 57/60 pairs of mono- and dizygotic twins, and 246 siblings. FA measurements were derived using (Enhancing NeuroImaging Genetics through Meta-Analysis) ENIGMA DTI protocols and heritability estimates were calculated using the SOLAR-Eclipse imaging genetic analysis package. We compared heritability estimates derived from HCP data to those publicly available through the ENIGMA-DTI consortium, which were pooled together from five-family based studies across the US, Europe, and Australia. FA measurements from the HCP cohort for eleven major white matter tracts were highly heritable (h2=0.53–0.90, p<10−5), and were significantly correlated with the joint-analytical estimates from the ENIGMA cohort on the tract and voxel-wise levels. The similarity in regional heritability suggests that the additive genetic contribution to white matter microstructure is consistent across populations and imaging acquisition parameters. It also suggests the overarching genetic influence provides an opportunity to define a common genetic search space for future gene-discovery studies. Uniquely, the measurements of additive genetic contribution performed in this study can be repeated using online genetic analysis tools provided by the HCP ConnectomeDB web application.
Introduction
Imaging genetics/genomics is an active research direction aimed at improving our understanding of the genetic underpinnings of brain structure, function, and connectivity in health and disease. The availability of data from a growing number of large-scale imaging projects enables meta-analyses that provide increased analytic power by combining data across projects. The ENIGMA (Enhancing Neuroimaging Genetics through Meta-Analysis) consortium was organized to facilitate this by bringing together genetic imaging researchers and developing methods for multi-site data harmonization and analyses (Thompson et al., 2014). The ENIGMA-DTI workgroup is focused on the analyses of Diffusion Tensor Imaging (DTI) data. Here, we compare the estimates of additive genetic contribution (heritability) to fractional anisotropy (FA) measurements previously reported for the ENIGMA-DTI (Kochunov et al., 2014) with comparably analyzed DTI data from the Human Connectome Project (HCP) (Van Essen et al., 2013). The HCP is a large-scale international collaboration aimed at elucidating the genetic and environmental sources of normal variability within the structural and functional connections of the human brain. The HCP is collecting and sharing data from a large cohort of healthy young adult twins and siblings using state of the art, high resolution, neuroimaging acquisition and analysis methods (Glasser et al., 2013; Van Essen et al., 2013). The HCP diffusion imaging data differs from these used in previous ENIGMA-DTI studies in several important ways, including higher spatial resolution (1.25 mm isotropic voxels vs. 2–3 mm for ENIGMA-DTI studies) and higher number of diffusion directions (270 vs. 30–100 for ENIGMA-DTI studies) (Sotiropoulos et al., 2013). Here, we tested whether the estimates of heritability obtained from the HCP data are comparable to published ENIGMA-DTI joint-analytic estimates and whether new insights and information emerge by analyzing the higher-resolution HCP data. Toward this aim, we compare regional and voxelwise heritability estimates for FA values in the current HCP public data sample with heritability estimates pooled from multiple sites across the world and published by the ENIGMA-DTI workgroup (http://enigma.ini.usc.edu) (Kochunov et al., 2014).
FA is a widely used quantitative measure of white matter microstructure (Basser et al., 1994; Basser and Pierpaoli, 1996) calculated from the diffusion tensor (DTI) model of water diffusion (Thomason and Thompson, 2011). This is an important biomarker in clinical studies, as it can sensitively track the white matter changes in Alzheimer’s disease (AD) (Clerx et al., 2012; Teipel et al., 2012), general cognitive function (Penke et al., 2010a; Penke et al., 2010b), and several neurological and psychiatric disorders (Barysheva et al., 2012; Carballedo et al., 2012; Kochunov et al., 2012; Mandl et al., 2012; Sprooten et al., 2011). The ENIGMA-DTI workgroup has developed a standardized protocol (http://enigma.ini.usc.edu/ongoing/dti-working-group/) for extraction and harmonization of phenotypes for genetic analyses of FA traits (Jahanshad et al., 2013; Kochunov et al., 2014). This protocol was previously evaluated in five family-based cohorts including 2248 children and adults (ages: 9–85). The findings were summarized in two ways. In the meta-analytic approach, heritability results across cohorts were normalized using a standard error (SE)-weighted model to yield meta-analytical estimates of heritability. In the mega-analytic approach, all the data was shared and synthesized pedigree was used to directly estimate heritability (Kochunov et al., 2014). Here, we applied the ENIGMA-DTI protocol to HCP DTI data to report the whole-brain and regional estimates heritability of FA values in the HCP sample in voxel-wise and region-of-interest based tests. Then, we compared the global and regional heritability estimates in HCP to the joint-analytic estimates previously reported by ENIGMA-DTI. Finally, we took advantage of the high spatial resolution of HCP acquisition to study the heritability pattern of the white matter periphery, where the common 2 mm or larger resolution of standard DTI scans leads to artificial lowering of FA magnitude in regions of diverging fibers due to partial voxel averaging effects (Basser et al., 1994; Basser and Pierpaoli, 1996).
This analysis is based on the previous studies of the ENIGMA-DTI workgroup that quantified heritability of the whole-brain and regional FA values in geographically and ethnically diverse cohorts (Jahanshad et al., 2013; Kochunov et al., 2014). It aimed to identify the “genetic search space” for FA measurements: a set of endophenotypes that are significantly heritable regardless of age, ethnicity and family structure to be used for follow-up genome-wide association (GWAS) analyses. To qualify as an endophenotype, a measurement must show a significant and reproducible heritability value across diverse cohorts. While significant heritability alone offers no guarantee that specific genetic variants associated to the trait will be discovered, measures that are not reliably heritable may be unstable and are unlikely to be influenced by genetic variants with effect sizes that are detectable in GWAS. In our prior work, the whole-brain average FA was found to be significantly heritable in all cohorts with tight confidence intervals. The regional FA measurements showed a variable additive genetic contribution (Kochunov et al., 2014) that suggested that there may be a consistent pattern of additive genetic contributions to variance in FA values across the brain regions assessed. Here, we extend this work by testing the reliability and generalizability of ENIGMA-DTI to the HCP cohort and attempt to take a deeper view on the spatial variability of heritability of FA values across brain regions. We demonstrate the consistency of heritability measurements across populations by showing that regional heritability estimates from an HCP cohort fall in line with the pooled estimates derived from independent populations.
METHODS
Subjects
ENIGMA-DTI processing of FA images and heritability analyses were performed in 481 (194/287 M/F) participants of the Human Connectome Project (HCP) for whom the scans and data were released in June 2014 (humanconnectome.org) after passing the HCP quality control and assurance standards (Marcus et al., 2013). The details of this release are available in the HCP reference manual1. The participants in the HCP study were recruited from the Missouri Family and Twin Registry that includes individuals born in Missouri (Van Essen et al., 2013). All HCP participants are in young adult sibships of average size 3–4 that include an MZ or DZ twin pair. Subjects ranged in age from 22 to 36 years (29.1±3.5 years). This age range is chosen because it corresponds to the period after neurodevelopment is completed and before the onset of neurodegenerative changes. This release included 117 twin pairs (57 monozygotic and 60 dizygotic pairs), and 246 of their siblings. The full set of inclusion and exclusion criteria is detailed elsewhere (Van Essen et al., 2013). In short, the HCP subjects are healthy individuals who are free from major psychiatric or neurological illnesses. They are drawn from ongoing longitudinal studies (Edens et al., 2010; Sartor et al., 2010), where they received extensive previous assessments including the history of drug use, emotional and behavioral problems. All subjects provided written informed consent on forms approved by the Institutional Review Board of Washington University in St Louis.
Diffusion data collection and preprocessing
Diffusion data was collected at Washington University St Louis using a customized Siemens Magnetom Connectome 3 Tesla scanner with a 100 mT/m maximum gradient strength and a 32 channel head coil. Details on the scanner, image acquisition and reconstruction are provided in (Ugurbil et al., 2013) and (https://www.humanconnectome.org/documentation/S500/HCP_S500_Release_Reference_Manual.pdf). Diffusion data were collected using a single-shot, single refocusing spin-echo, echo-planar imaging sequence with 1.25 mm isotropic spatial resolution (TE/TR=89.5/5520 ms, FOV=210×180 mm). Three gradient tables of 90 diffusion-weighted directions and six b=0 images each, were collected with right-to-left and left-to-right phase encoding polarities for each of the three diffusion weightings (b=1000b=2000, and 3000 s/mm2). The total imaging time for collection of diffusion data was approximately one hour.
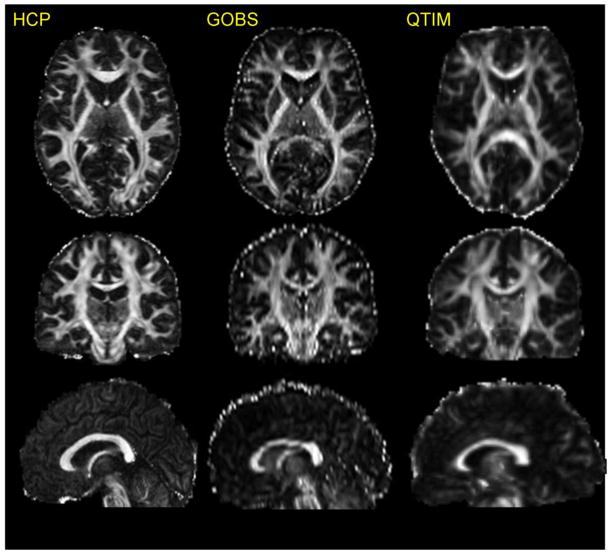
Diffusion data were preprocessed using the HCP Diffusion pipeline (Glasser et al., 2013; Sotiropoulos et al., 2013) that included: normalization of b0 image intensity across runs; correction for EPI susceptibility and eddy-current-induced distortions, gradient-nonlinearities, subject motion and application of a brain mask. FA maps were obtained by fitting diffusion tensor model using FSL-FDT toolkit (Behrens et al., 2003). For visual comparison purposes, an FA image for a random HCP subject is shown next to corresponding FA maps from age-matched individuals scanned with the DTI protocols used by the Genetics Of Brain Structure (GOBS) study (Kochunov et al., 2010b) and the Queensland Twin IMaging (QTIM) studies (Figure 1). The FA image collected using the HCP protocol shows a finer spatial resolution and an improved signal-to-noise ratio (SNR) throughout, including thin white matter blades underlying convoluted cortex (Figure 1).
Figure 1.
An FA image collected using HCP protocl is shown next to the images of age-and-gender matched subjects from the two conventional DTI protocols GOBS (1.71×1.71×3mm, 55 direction) and QTIM (1.8×1.8×2mm isotropic resolution, 94 direction).
ENIGMA-DTI processing
ENIGMA-DTI protocols for extraction of tract-wise average FA values were used. These protocols are detailed elsewhere (Jahanshad et al., 2013) and are available online http://enigma.ini.usc.edu/protocols/dti-protocols/. Briefly, FA images from HCP subjects were nonlinearly registered to the ENIGMA-DTI target brain using FSL’s FNIRT (Jahanshad et al., 2013). This target was created as a “minimal deformation target” based on images from the participating studies as previously described (Jahanshad et al., 2013). The data were then processed using FSL’s tract-based spatial statistics (TBSS; http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/TBSS) analytic method (Smith et al., 2006) modified to project individual FA values on the hand-segmented ENIGMA-DTI skeleton mask. After extracting the skeletonized white matter and the projection of individual FA values, ENIGMA tract-wise regions of interest, derived from the Johns Hopkins University (JHU) white matter parcellation atlas (Mori et al., 2008), were transferred to extract the mean FA across the full skeleton and average FA values for eleven major white matter tracts, with subdivision of the corpus callosum into 3 regions, for a total of 15 regions of interest (ROIs) (Table 1). The whole brain average FA values were calculated to include all voxels in the ENIGMA-DTI skeleton. The protocol, target brain, ENIGMA-DTI skeleton mask, source code and executables, are all publicly available (http://enigma.ini.usc.edu/ongoing/dti-working-group/). Finally, we analyzed the voxelwise FA values along the ENIGMA skeleton mask.
Table 1.
Results of the additive analysis for the whole average and regional FA values in HCP subjects, including the heritability values (first column) and significance values for each of the five covariates. Regions of interest (ROIs) examined along the ENIGMA-DTI skeleton as defined by the JHU white matter parcellation atlas (Mori et al., 2008).
| Trait | h2±SE (p) | Covariates | |||||
|---|---|---|---|---|---|---|---|
| Age (p) | Age2(p) | Sex(p) | Age*Sex(p) | Age2*Sex (p) | Variance explained (%) | ||
| Average FA | 0.88±0.03 (10−25) | 0.60 | 0.75 | 6.6·10−8 | 0.48 | 0.56 | 10.9 |
| Genu of the corpus callosum (GCC) | 0.89±0.02 (10−30) | 0.19 | 0.23 | 8.6·10−5 | 0.48 | 0.08 | 4.6 |
| Body of the corpus callosum (BCC) | 0.90±0.02 (10−25) | 0.32 | 0.44 | 2.9·10−8 | 0.92 | 0.36 | 11.1 |
| Splenium of corpus callosum (SCC) | 0.90±0.02 (10−26) | 0.40 | 0.57 | 1.8·10−9 | 0.92 | 0.47 | 12.9 |
| Fornix (FX) | 0.53±0.08 (10−9) | 0.50 | 0.73 | 3.8·10−8 | 0.26 | 0.62 | 14.7 |
| Cingulum (cingulate gyrus) - L and R combined (CGC) | 0.81±0.04 (10−22) | 0.45 | 0.46 | 8.0·10−8 | 0.30 | 0.18 | 9.2 |
| Corona radiata - L and R anterior, superior and posterior sections combined (CR) | 0.87±0.03 (10−23) | 0.64 | 0.62 | 2.0·10−3 | 0.98 | 0.86 | 3.8 |
| External capsule - L and R combined (EC) | 0.82±0.05 (10−15) | 0.35 | 0.87 | 7.9·10−5 | 0.44 | 0.78 | 6.5 |
| Internal capsule - L and R anterior limb, posterior limb, and retrolenticular parts combined (IC) | 0.86±0.03 (10−28) | 0.53 | 0.67 | 3.3·10−13 | 0.94 | 0.58 | 19.1 |
| Inferior fronto-occipital fasciculus - L and R combined (IFO) | 0.78±0.06 (10−13) | 0.15 | 0.41 | 0.3 | 0.89 | 0.77 | 1.6 |
| Posterior thalamic radiation - L and R combined (PTR) | 0.85±0.03 (10−23) | 0.89 | 0.22 | 2.1·10−7 | 0.87 | 0.23 | 8.6 |
| Superior fronto-occipital fasciculus - L and R combined (SFO) | 0.76±0.05 (10−18) | 0.89 | 0.41 | 0.2 | 0.89 | 0.65 | 1.3 |
| Superior longitudinal fasciculus - L and R combined (SLF) | 0.87±0.03 (10−28) | 0.32 | 0.34 | 1.5·10−3 | 0.75 | 0.40 | 3.1 |
| Sagittal stratum (include inferior longitudinal fasciculus and inferior fronto-occipital fasciculus) - L and R combined (SS) | 0.81±0.05 (10−19) | 0.33 | 0.24 | 6.2·10−10 | 0.73 | 0.18 | 12.8 |
| Corticospinal tract - L and R combined (CST) | 0.66±0.05 (10−18) | 0.20 | 0.74 | 7.7·10−7 | 0.29 | 0.67 | 9.3 |
Heritability measurements: Analysis of additive genetic variance
The variance components method, as implemented in the Sequential Oligogenic Linkage Analysis Routines (SOLAR)-Eclipse software package (http://www.nitrc.org/projects/se_linux) was used for all individual cohort heritability estimations. In short, the algorithms in SOLAR employ maximum likelihood variance decomposition methods and are an extension of the strategy developed by (Amos, 1994). The covariance matrix Ω for a pedigree of individuals is given by:
| (1) |
where σg2 is the genetic variance due to the additive genetic factors, Φ is the kinship matrix representing the pair-wise kinship coefficients among all individuals, σe2 is the variance due to individual-specific environmental effects, and I is an identity matrix (under the assumption that all environmental effects are uncorrelated among family members). Narrow sense heritability is defined as the fraction of phenotypic variance σP2 attributable to additive genetic factors,
| (2) |
The variance parameters are estimated by comparing the observed phenotypic covariance matrix with the covariance matrix predicted by kinship (Almasy and Blangero, 1998). Significance of heritability is tested by comparing the likelihood of the model in which σg2 is constrained to zero with that of a model in which σg2 is estimated. Twice the difference between the loge likelihoods of these models yields a test statistic, which is asymptotically distributed as a 1/2:1/2 mixture of a χ2 variable with 1 degree-of-freedom and a point mass at zero. Prior to testing for the significance of heritability, phenotype values for each individual within the cohort were adjusted for covariates including sex, age, age2, age x sex interaction, and age2 x sex interaction. Inverse Gaussian transformation was also applied to ensure normality of the measurements. Outputs from SOLAR include the heritability value, the significance value (p), and the standard error for each trait (ROI or voxel). All heritability analyses were conducted with age, sex, age*sex, age2, and age2*sex included as covariates. Registered HCP users can replicate our analyses using the web version of SOLAR-Eclipse software (www.humanconnectome.org)
Variation in heritability estimates in HCP vs. ENIGMA
Meta-SE and mega-genetic joint-analytic heritability estimates from ENIGMA-DTI were compared to and evaluated as predictors of heritability estimates in HCP subjects. Specifically, we used a z-test to evaluate whether the heritability estimates for the HCP subjects fell within the confidence intervals for the estimates of heritability derived by meta and mega analysis pooling methods. Next, we tested whether variability in regional heritability estimates for HCP subjects could be predicted from the regional heritability estimates derived for the pooling methods. This analysis was performed for both the tract-wise average FA and the voxel-wise FA values. Finally, we tested whether the variability in the voxel-wise heritability estimates in HCP subjects could be predicted based on the voxel-wise FA values and the distance from the center of the MNI brain array space (voxel position x=91, y=109, z=91 mm). This was tested using two regression analyses, including testing two predictors (eq. 3) and their interaction (eq. 4)
| (3) |
| (4) |
where h2 is a heritability, the FA is the fractional anisotropy value and d is the Euclidean distance from the center of the MNI brain space and to a voxel (i,j,k). In the HCP cohort, the voxel-wise average FA map was obtained by averaging FA maps for individual subjects. This modeling was performed with the [R] package(R-Development-Core-Team, 2009) using the linear effects model library and the maximum likelihood estimation algorithm (Pinheiro et al., 2008).
Results
Heritability estimates for whole-brain averaged and by-tract FA values are shown in Table 1. The whole-brain average and regional FA values in the HCP subjects were significantly heritable (p<0.001) (Figure 2). The covariates (age, sex, age2, age-x-sex, and age2-x-sex) explained 10.9% of the phenotypic variance in the whole-brain averaged FA values (Table 1). Sex was the only significant covariate (p=6.6·10−8) and female subjects showed ~2% higher average FA values (FA=0.40±0.12 vs 0.39±0.14 for females and males, respectively). Additive genetic factors explained 88% of the residual (or 78% of the total) phenotypic variance in the whole brain FA values (h2=0.88, p<10−10). Together, covariates and additive genetic factors explained 89% of the total variance, leaving 11% of unexplained variance that was attributed to environmental factors.
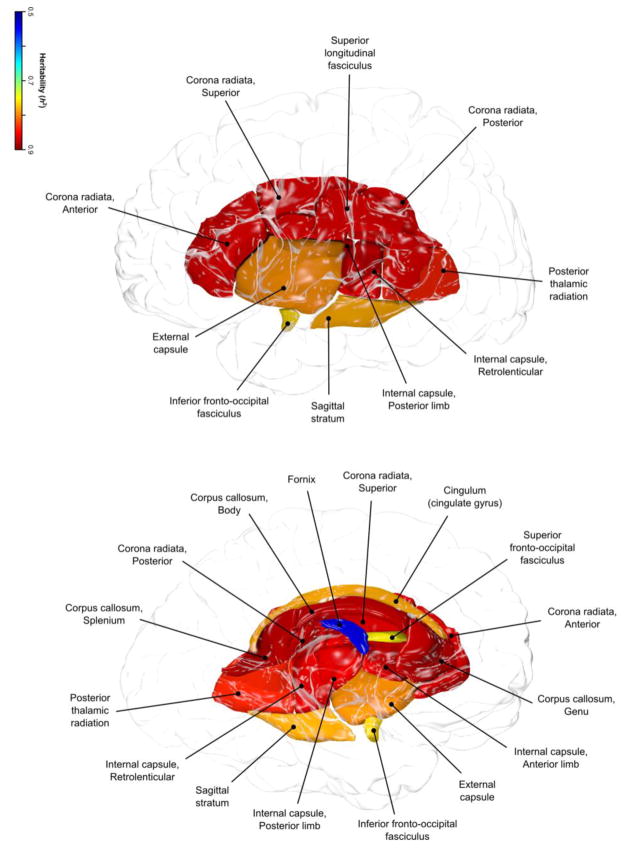
Figure 2.
Regional heritability pattern in HCP sample is shown for eleven tract-wise measurements of FA values.
For regional FA measurements, the highest heritability was observed for the body (BCC) and splenium (SCC) of corpus callosum (h2=0.90, p<10−10). The lowest heritability was observed for the fornix (FX) and corticospinal tract (CST) (h2=0.53 and 0.66, respectively) and both were significantly lower (p<0.001) than the next lowest heritability estimate h2=0.76 for superior longitudinal fasciculus (SLF). Covariates explained, on average, 8.5±5.3% of the variance. Sex was the only significant covariate for all regional FA measurements but for the inferior (IFO) and superior frontal occipital (SFO) tracts (Table 1). Sex explained the largest proportion of variance in the internal capsule (IC) and fornix (FX) tracts (19.1% and 14%, respectively). The lowest proportion of variance explained by covariates was observed for the SFO and IFO tracts (1.3% and 1.6%, respectively).
We further explored the effect of sex on heritability in HCP by calculating heritability of the female and male participants separately. Average FA was highly heritable in both sexes, with females (N=287) having 85.7% (p=1.9×10−15) of the overall average FA variance explained by additive genetic factors, and males (N=194) 91.9% (p=4.7×10−11). In females, 0.15% of the variance was attributable to the linear and nonlinear effects of age, whereas this proportion was modestly higher (1.5%) in males.
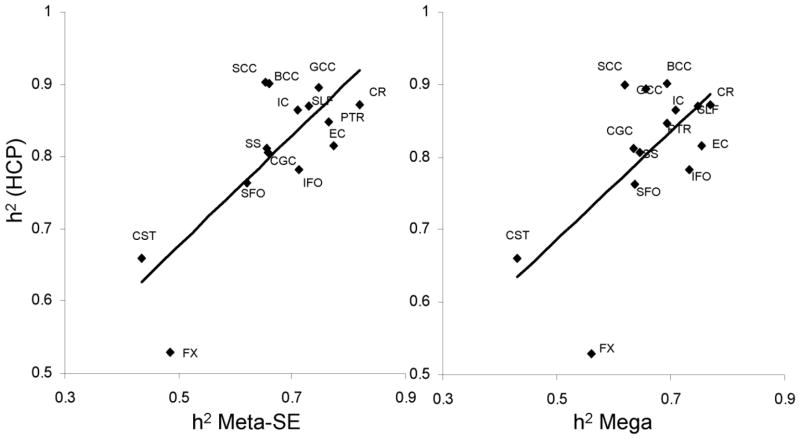
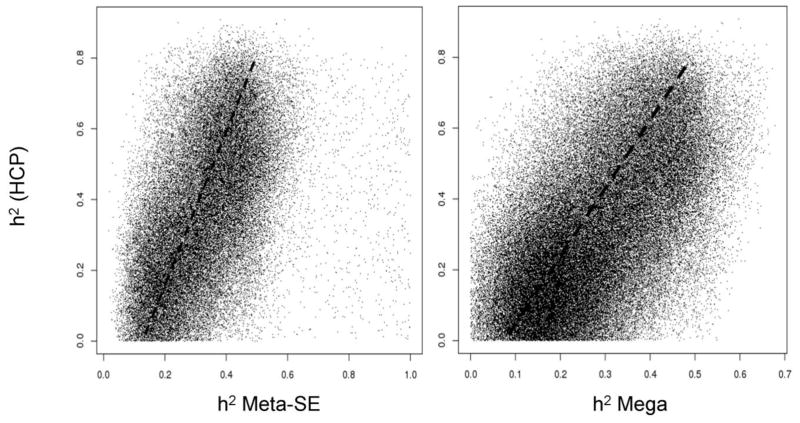
The heritability estimate for the average FA values in HCP cohort was significantly (p<10−4) higher than the joint-analytic estimates reported by ENIGMA-DTI (Figure 3A). Likewise, the tract-average FA heritability estimates for HCP cohort were higher than those reported by ENIGMA-DTI (Figure 3B). These differences were significant (p<0.0035, after correcting for fourteen comparisons) for the following tracts from the ENIGMA-DTI meta-analysis SE estimate: GCC, BCC, SCC, CR, IC and SS. HCP heritability estimates were significantly higher for all but three tract-wise estimates of heritability for the mega-analysis (FX, EC, IFO) analytical estimates of heritability (Figure 3B). The overall regional patterns of heritability for the ENIGMA-DTI estimation methods significantly predicted the regional pattern of heritability in the HCP cohort (r=0.79 and 0.64; p<0.01, for meta- and mega-analysis estimates, respectively; Figure 4). Likewise, aggregated voxelwise heritability estimates from ENIGMA-DTI were significantly predictive of the voxel-wise heritability measurements in HCP (r=0.51 and 0.62; p<10−10, for meta- and mega-analysis estimates, respectively; Figure 5).
Figure 3.
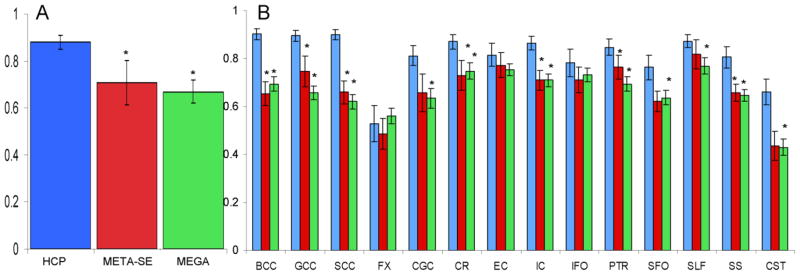
Heritability estimates for the whole-brain (A) and tract-wise average FA values (B) for the HCP and Meta-SE and Mega-genetic estimate derived by ENIGMA-DTI study. Standard error of measurement is represented by the error bars. *Pooled estimate was significantly (p<0.0035) higher for the HCP sample.
Figure 4.
Scatterplot of heritability estimate for tract-wise average FA measurements plotted for the HCP sample versus the Meta-SE and Mega-genetic heritability estimates derived by ENIGMA-DTI study
Figure 5.
Scatterplot of voxel-wise heritability values for HCP subject plotted versus the Meta-SE and Mega-genetic voxel-wise heritability derived by the ENIGMA-DTI study. Dash lines show significant linear correlation between voxel HCP heritability values and two joint ENIGMA-DTI estimates (r=0.51 and 0.62; p<10−10, for Meta-SE and Mega-genetic analytic estimates, respectively)
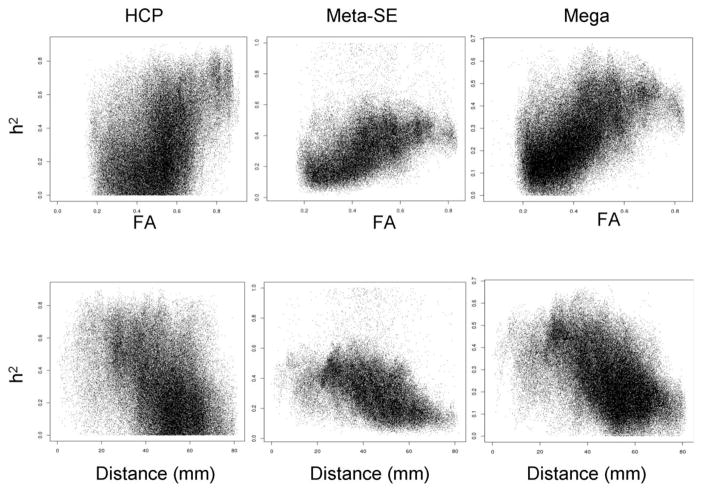
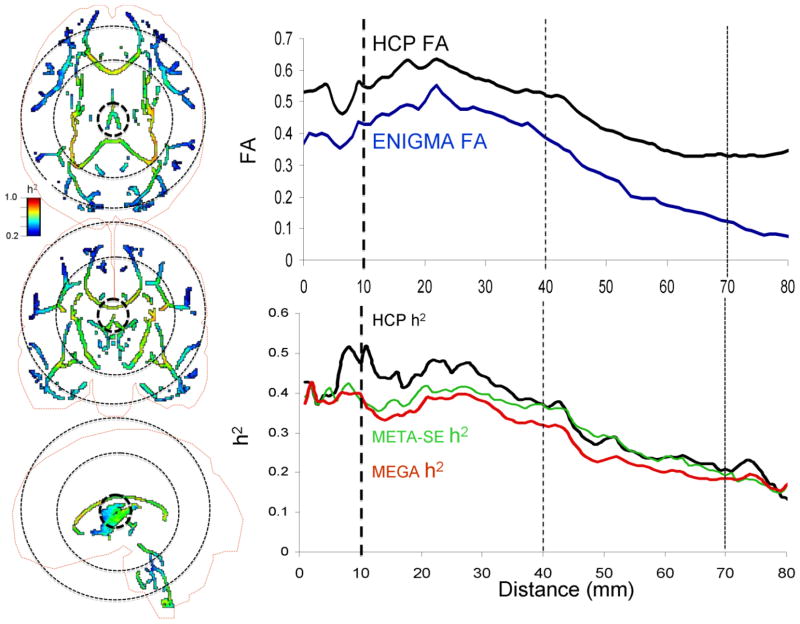
Voxel-wise variations in heritability were tested as a function of FA magnitude, distance and their interaction (eq 3 and 4). FA and distance were significant predictors of voxelwise heritability values in both HCP and ENIGMA cohorts (Table 2). Higher heritability values corresponded to higher FA values (Figure 6, top row) and voxels located near the center of the MNI array space (Figure 6, bottom row). Together, these factors explained 25% of variability in heritability values in HCP subjects and 32% and 41% of variability in the meta- and mega-genetic estimates of heritability in the ENIGMA-DTI dataset, respectively. Testing of the interaction model (eq 4) demonstrated that FA x distance term was not significant (p=0.75) in the HCP cohort. In contrast, this interaction was highly significant in the ENIGMA-DTI sample (Table 2). When comparing the plot of FA values versus distance from the center of MNI array space, the HCP data showed higher FA values throughout the brain and especially at the periphery (distance: 40–70 mm) than average ENIGMA-DTI FA values (Figures 6 and 7). In contrast, voxel-wise heritability values in HCP subjects were only higher for the central regions (distance 10–40 mm; p>0.001) and showed no difference from the meta-and-mega genetic ENIGMA-DTI estimates in peripheral regions (distance 40–70 mm, p=0.3; Figure 7).
Table 2.
Results of the testing of two predictive models for regional heritability values.
| Model 1 (h2=FA + d) | HCP | Meta-SE | Mega |
|---|---|---|---|
| βFA ± sd (t-value, P-value) | 0.40±0.004 (90.3, p<10−16) | 0.35±0.004 (91.2, p<10−16) | 0.42±0.002 (169.7, p<10−16) |
| βd ± sd (t-value, P-value) | −.004±4.3·10−5 (101.4, p<10−16) | −0.031±3.8·10−5 (80.9, p<10−16) | −0.026±2.4·10−5 (106.6, p<10−16) |
| R2 (t-value, P-value) | 0.25 (p<10−16) | 0.32 (p<10−16) | 0.41 (p<10−16) |
| Model 2 (h2=FA + d+FA·d) | HCP | Meta-SE | Mega |
| βFA ± sd (t-value, P-value) | 0.39±0.001 (32.9, p<10−16) | 0.03±0.01 (2.8, p=0.004) | 0.26±0.01 (35.5, p<10−16) |
| βd ± sd (t-value, P-value) | −0.004±0.0001 (65.3, p<10−16) | −0.007±0.0001 (67.3, p<10−16) | −0.004±0.0001 (65.3, p<10−16) |
| βFA*d ± sd (t-value, P-value) | −0.0008±0.002 ( 0.3, 0.75) | 0.009±0.0002 (40.9, p<10−16) | 0.004±0.0001 (28.4, p<10−16) |
| R2 (t-value, P-value) | 0.25 (p<10−16) | 0.33 (p<10−16) | 0.42 (p<10−16) |
Figure 6.
Scatterplot of voxel-wise heritability plotted versus FA value (top row) or distance from the center of the brain (bottom row) constituted testing of model 1 (Table 2).
Figure 7.
Left panels: Voxel-wise heritability (h2) values for HCP sample shown on the ENIGMA-DTI skeleton with the cortical outline (axial, coronal, and sagittal views). Right panels: voxel-wise FA and h2 plotted versus the distance from the center of the MNI space. The dotted circles (left panel) and lines (right panel) represent distance of 10, 40 and 70 mm from the center of the MNI space. The FA values were significantly higher (p<0.001) for both proximal (10–40 mm) and distal (40–70 mm) voxels in HCP vs. ENIGMA sample. HCP heritability values were significantly higher for proximal (p=0.001) but not distal voxels (p=0.32) (bottom row, right column).
Discussion
In this study, we performed three analyses: (1) A comprehensive heritability analysis of whole-brain and regional FA values in the HCP cohort indicated that FA measurements extracted using the ENIGMA-DTI protocol were highly heritable, with ~70–80% of the total phenotypic variance explained by additive genetic factors. (2) When compared to meta-and-mega-genetic estimates of heritability, the heritability measurements in HCP cohort were generally higher. Nonetheless, the agreement between the joint-analytical estimates and HCP heritability measurements indicates that the overall regional genetic contributions for tract-wise and voxel-wise levels is similar among independent cohorts. (3) The additive genetic contribution to voxel-wise FA values is modulated by the magnitude of FA and the distance away from the core of the brain. The trends in the HCP and ENIGMA-DTI cohorts were similar but for one substantive difference: the interaction between FA and distance was significant in ENIGMA, but not so in HCP sample. Overall, our study demonstrated highly robust estimates of additive genetic variability in HCP data. The joint-analytic estimates of heritability derived by ENIGMA-DTI group were highly predictive of the variance in regional heritability estimates in the data collected by HCP. Together this suggests the consistency of additive genetic contribution to FA values. This posits FA as a promising phenotype for future gene discovery studies. Replication of our analyses and further genetic analyses in the HCP subjects can be performed using a web-version of SOLAR-Eclipse available at HCP web-based analysis portal.
The heritability estimates for the whole-brain (h2=0.88) and all tract-wise average FA values (h2=0.53–0.90) calculated for HCP subjects were highly significant (p=10−9–10−26). Findings of significant heritability for average and regional FA measurements have withstood several independent replications (Chiang et al., 2008; Chiang et al., 2011; Duarte-Carvajalino et al., 2011; Jahanshad et al., 2013; Jahanshad et al., 2010; Kochunov et al., 2010b). Heritability is the proportion of the variance that is attributed to the additive genetic variance after correction for covariates. In HCP sample, we explored this relation further and found that sex was the only significant covariate. The HCP recruitment was designed to reduce the effects of age on the brain measurements by limiting recruitment to age-range that corresponds to a plateau in FA-aging trend (22–35 years) (Kochunov et al., 2011; Van Essen et al., 2013). Significant sex differences in the average and regional FA values are commonly reported (Bava et al., 2010; Menzler et al., 2010; O’Dwyer et al., 2013; Wang et al., 2012), however, the direction and size of this effect vary from study to study (den Braber et al., 2013). High heritability values and modest effects of covariates posit HCP cohort for further studies of genetic effects on normal variability in cerebral white matter.
The heritability estimate for the average FA in HCP cohort was higher than the joint analytic estimate calculated for the ENIGMA-DTI sample. The degree of additive genetic variation (i.e., heritability) depends on study design, sample characteristics, and the fidelity and ‘closeness’ of the trait to underlying genetic processes. The higher heritability of the whole-brain average FA values in the HCP cohort is likely to be the product of three factors: study design, recruitment strategy and the quality of the imaging data. The HCP study uses a twin-sibling study design and heritability estimates obtained using this design can be higher than those calculated in extended families (Kochunov et al., 2014). The shared environmental factors were not evaluated here as to maintain the same design as our previous ENIGMA-DTI studies (Jahanshad et al., 2013). The lack of aging effects on FA in HCP subjects is another likely contributor to the higher heritability estimates. The age-by-genotype interaction during maturation and aging, observed in studies that recruit subjects across the lifespan, can reduce heritability estimates (Batouli et al., 2013; Brouwer et al., 2012; Glahn et al., 2013). The ethnic diversity in the HCP sample may also add shared environmental aspects of FA variance to the apparent genetic influence. Lastly, the higher quality of the HCP DTI data likely reduces the measurement error and thus contributes to higher heritability estimates. Further research will be needed to assess the specific contributions to high heritability from these three factors. That being said, the heritability estimates in HCP are compatible with those collected in cohorts with similar study design (Kochunov et al., 2014).
Importantly, the joint estimates of heritability from the ENIGMA-DTI studies were predictive of the regional pattern of additive genetic contribution to FA values in the HCP subjects. This suggests that the regional pattern of additive genetic variance in FA values is similar across populations. Our examination of the heritability of the FA along parcellated white matter tracts and voxel-wise FA showed a pattern of inheritance that was similar across cohorts. Specifically, heritability values for the fornix (FX) and corticospinal tract (CST) and were consistently low across cohorts. Confirmation of the previous ENIGMA-DTI findings of low heritability in the FX and CST regions in the HCP sample suggests that GWAS findings in these regions should be interpreted with caution. However, the correlation between tract-wise heritability estimates remained significant even when excluding CST and FX. Likewise, the voxel-wise correlation analyses showed that HCP shared 32% and 41% of the regional inheritance pattern with the joint-analytic estimates of heritability. The sources of regional variability in the FA heritability across the white matter skeleton are not clear. The regions with the greatest heritability overall are the regions that were the core brain tracts had simplified fiber architecture, including the three regions of the corpus callosum, corona radiata, superior longitudinal fasciculus and internal capsule.
Furthermore, we observed that both the magnitude of FA values and the distance from the center were significant predictors of heritability estimates for voxel-wise FA values in both HCP and ENIGMA estimates. An important observation was the difference in the FA-by-distance interaction in explaining the spatial pattern of voxel-wise heritability. This interaction was significant in ENIGMA but not in HCP data. The significance of FA by distance interaction in ENIGMA sample suggests that reduced additive genetic contribution was observed for voxels where FA magnitude was reduced due to spatial divergence of the tracts as they approach cerebral cortex. Reduced partial voxel produced higher FA values at the periphery of the brain in the HCP subjects. Yet in contrast, the higher peripheral FA values in the HCP data did not contribute to higher peripheral estimates of heritability, suggesting a rise in unexplained variance as the fiber tracts diverge toward cerebral cortex. Nonetheless, we observed an excellent agreement between the pattern of additive genetic variance in the HCP cohort and joint analytical estimate from five other cohorts. This does not imply that the same genes were responsible for the similar patterns of heritability in the different populations. Instead, regions where consistently high heritability is observed among populations provide reliable phenotypes for discovery of genetic factors that exert control over cerebral white matter structure and integrity.
Overall, this work posits FA as a useful phenotype for further genetic analyses, with some caveats. Both biological and methodological factors are likely to contribute to the findings of lower heritability in voxels more distant from the center of the image and those in the fornix and corticospinal tracts. The most significant biological factor that contributes to lower heritability is the residual intersubject variability, especially in cortical structures (Kochunov et al., 2002; Kochunov et al., 2001; Kochunov et al., 1999). Substantial variability in the cortical landscape is present even in monozygotic twins (Van Essen et al., 2013) and this leads to modest (30–50%) heritability estimates for cortical measurements (Van Essen et al., 2014 OHBM). Cerebral morphology in adults is the product of the primary, secondary and tertiary gyrogenesis processes that are driven by genetic and environmental factors (Kochunov et al., 2010a; Kochunov et al., 2009). These processes fold the cerebral cortex into an intricate pattern of sulci and gyri with variable function-structure relationships among cortical structures, underlying white matter tracts and functional areas (Fischl et al., 2007; Van Essen, 1997; Van Essen, 2004). Current intersubject alignment methods (even high-dimensional nonlinear registration) may not consistently align cortical and functional areas, especially in frontal and parietal areas where tertiary and anastomotic sulci add uniqueness to cortical gyrification patterns (Fischl et al., 2007; Kochunov et al., 2009; Kochunov et al., 2005). High spatial resolution in HCP protocol reduced partial voxel voluming effect, as signified by the higher FA values in the peripheral white matter on a voxel-by-voxel level. However, the complex organization and spatial distribution (including crossings) of white matter tracts in the periphery likely introduced errors in the FA projection step. Thus, there was no corresponding rise in heritability estimates, suggesting an increase in individualized variance that could not be explained by genetic factors.
Our findings of reduced additive genetic contribution towards the periphery may also reflect methodological limitations. Recent work by Bach and colleagues (Bach et al., 2014), built upon previous work by others (Edden and Jones, 2011; Keihaninejad et al., 2012; Zalesky, 2011), discusses two limitations of multi-subject analyses of FA values: spatial registration and FA projection. The multi-subjects spatial alignment was performed based on the voxel-wise maps of FA values (Jahanshad et al., 2013). Advanced, diffusion tensor-based spatial registration techniques have been advocated as an alternative approach to register DTI data (Wang et al., 2011)(Zhang and Arfanakis, 2013). Tensor-based alignment techniques such as those implemented in (http://dti-tk.sourceforge.net) use the similarity in voxel-wise diffusion tensors to drive multi-subject alignment. We studied if the use of this advanced warping approach might lead to high heritability estimates by repeating voxel-wise heritability analyses using the DTI-TK tensor-driven TBSS approach. This was executed using standard parameters and 1-mm isotropic resolution. DTI-TK uses the full tensor model for spatial alignment, and FA values are calculated at the final step. We observed that the tensor-based warping approach produced significantly (p<10−6) higher FA values at the core (distance=10–40mm from center) and at the periphery of the brain (distance=40–80mm) (Figure 1S). However, the tensor-based warping produced significantly lower voxel-wise heritability estimates compared to the FA-warping approach (average h2=0.20 vs. 0.34, p<10−10). Moreover, its voxel-wise heritability values showed a strong decline with distance (Figure 1S) and FA magnitude explained 64% of the variance. Clearly, tensor-based registration led to higher average FA estimate, but it did not result in a better intersubject overlap especially in areas of more complex fiber geometry near the periphery of the brain. This tensor-based analysis was only performed in the HCP sample because additional research is needed to demonstrate the stability of the this registration techniques when aligning multi-site data collected using DTI protocols with variable spatial resolutions and number of directions.
Additional methodological limitations such as partial voluming effects or intersubject misregistration can cause errors during projection of FA values of the small, tubular white matter structures such as the fornix and corticospinal tract on the group-wise skeleton (Bach et al., 2014). Contributing to this, the spatial course of the fornix parallels that of the stria terminalis and the projection-based techniques may not be sufficiently sensitive to separate these tracts in individual subjects (Vasung et al., 2010). In agreement with these limitations, we consistently observed low heritability estimate for FA values from these two regions (Kochunov et al., 2014). Our study reveals that despite the greatly improved spatial resolution of HCP data, this limitation remains. Therefore the results from fornix should be interpreted with caution, particularly in lower powered studies with reduced spatial resolution. Similar errors are also likely to be observed in the areas of high intersubject variability such as near periphery of the brain. Bach et al. offered several recommendations, including the use of a study-specific template and manual review and editing of the skeleton image. Both of these recommendations were implemented in ENIGMA-DTI protocol(Jahanshad et al., 2013). Overall, the manuscript affirms the validity of ENIGMA-DTI approach in HCP data collected with a highly advanced diffusion weighted imaging protocol. However, it is important to understand the methodological caveats. This study illuminated these limitations from the genetic imaging perspective and our findings can help to define the set of regional phenotypes that can be reliably extracted from multi-site data collected with diverse imaging protocols. Our approach ranks FA-based phenotypes based on the degree of observed variance attributable to additive genetic factors and can serve to limit future genetic analyses to brain regions where this is high and stable across populations and cohorts regardless of acquisition method.
CONCLUSION
The ENIGMA-DTI FA homogenization protocol was tested in the state-of-the-art data collected by the HCP. This research helps to define the genetic search space for future localization of risk factors that affect white matter integrity in behavioral, neurological, and neuropsychiatric disorders. Limiting genetic searches to the traits that show significant and replicable heritability will improve confidence in outcomes of these analyses and reduce the number of degrees of freedom. In agreement, we showed that both global and regional heritability estimates from pooled approaches were highly predictive of the heritability pattern in a new cohort derived using state of the art neuroimaging methods. We also demonstrated that genetic contribution is replicable and high for the core white matter areas and that environmental contributions are greater in the vicinity of the variable convolutions of cerebral cortex. We provide our heritability results online at http://enigma.ini.usc.edu/ongoing/dti-working-group/ to define voxelwise additive genetic contribution for future genetic studies. Unique to this study is the ability to repeat our genetic analyses using the registered users of the HCP online version of the genetic analysis tools.
Supplementary Material
Highlights.
Data from 488 HCP subjects were processed using ENIGMA-DTI protocols
Heritability in HCP sample were compared to ENIGMA-DTI joint-analytical estimates
Estimates from HCP and ENIGMA-DTI were highly correlated
Genetic contribution to white matter integrity is consistent across population
Defines common genetic search space for future gene-discovery studies.
Acknowledgments
This study was supported by R01 EB015611 to PK, R01 HD050735 to PT, MH0708143 and MH083824 grants to DCG and by MH078111 and MH59490 to JB. Additional support for algorithm development was provided by NIH R01 grants EB008432, EB008281, and EB007813 (to PT). JES is supported by a Clinical Research Training Fellowship from the Wellcome Trust (087727/Z/08/Z). AMM is supported by a NARSAD Independent Investigator Award and by a Scottish Funding Council Senior Clinical Fellowship.
This work was supported in part by a Consortium grant (U54 EB020403) from the NIH Institutes contributing to the Big Data to Knowledge (BD2K) Initiative, including the NIBIB and NCI.
Data were provided by the Human Connectome Project, WU-Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University.”
The GOBS study (PI DG and JB) was supported by the National Institute of Mental Health Grants MH0708143 (Principal Investigator [PI]: DCG), MH078111 (PI: JB), and MH083824 (PI: DCG & JB).
The QTIM study was supported by National Health and Medical Research Council (NHMRC 486682), Australia. GdZ is supported by an ARC Future Fellowship (FT0991634).
The TAOS study (PI DEW) was supported by the National Institute on Alcohol Abuse and Alcoholism (R01AA016274) - “Affective and Neurobiological Predictors of Adolescent-Onset AUD” and the Dielmann Family.
The NTR study (PI DvtE) was supported by the The Netherlands Organisation for Scientific Research (NWO) [Medical Sciences (MW): grant no. 904-61-193; Social Sciences (MaGW): grant no. 400-07-080; Social Sciences (MaGW): grant no. 480-04-004].
The BrainSCALE study (PI HH and DB) was supported by grants from the Dutch Organization for Scientific Research (NWO) to HEH (051.02.061) and HEH, DIB and RSK (051.02.060).
Data collection for the Bipolar Family Study was supported by an Academy of Medical Sciences/Health Foundation Clinician Scientist Fellowship to AMM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Peter Kochunov, Maryland Psychiatric Research Center, University of Maryland School of Medicine, Baltimore, USA.
Neda Jahanshad, Imaging Genetics Center, Institute for Neuroimaging and Informatics, Department of Neurology Keck School of Medicine University of Southern California, Marina del Rey, USA.
Daniel Marcus, Department of Radiology, Washington University School of Medicine, St. Louis, USA.
Anderson Winkler, FMRIB Centre, Oxford University, Oxford, UK.
Emma Sprooten, Olin Neuropsychiatry Research Center, Institute of Living, Hartford Hospital, Hartford, USA.
Thomas E. Nichols, Department of Statistics, University of Warwick, Warwick, UK
Susan N Wright, Maryland Psychiatric Research Center, University of Maryland School of Medicine, Baltimore, USA.
L Elliot Hong, Maryland Psychiatric Research Center, University of Maryland School of Medicine, Baltimore, USA.
Binish Patel, Maryland Psychiatric Research Center, University of Maryland School of Medicine, Baltimore, USA.
Timothy Behrens, FMRIB Centre, Oxford University, Oxford, UK.
Saad Jbabdi, FMRIB Centre, Oxford University, Oxford, UK.
Jesper Andersson, FMRIB Centre, Oxford University, Oxford, UK.
Christophe Lenglet, Center for Magnetic Resonance Research, Department of Radiology, University of Minnesota Medical School, Minneapolis, USA.
Essa Yacoub, Center for Magnetic Resonance Research, Department of Radiology, University of Minnesota Medical School, Minneapolis, USA.
Steen Moeller, Center for Magnetic Resonance Research, Department of Radiology, University of Minnesota Medical School, Minneapolis, USA.
Eddie Auerbach, Center for Magnetic Resonance Research, Department of Radiology, University of Minnesota Medical School, Minneapolis, USA.
Kamil Ugurbil, Center for Magnetic Resonance Research, Department of Radiology, University of Minnesota Medical School, Minneapolis, USA.
Stamatios N Sotiropoulos, FMRIB Centre, Oxford University, Oxford, UK.
Rachel M. Brouwer, University Medical Center Utrecht, Utrecht, The Netherlands
Bennett Landman, Vanderbilt University, Nashville, TN.
Hervé Lemaitre, INSERM-CEA-Faculté de Médecine Paris-Sud, Orsay, France.
Anouk den Braber, VU University, Amsterdam, The Netherlands.
Marcel P. Zwiers, Radboud University, Nijmegen, The Netherlands
Stuart Ritchie, University of Edinburgh, Edinburgh, UK.
Kimm vanHulzen, Radboud University, Nijmegen, The Netherlands.
Laura Almasy, Texas Biomedical Research Institute, San Antonio, TX.
Joanne Curran, Texas Biomedical Research Institute, San Antonio, TX.
Greig I deZubicaray, University of Queensland, Brisbane, Australia.
Ravi Duggirala, Texas Biomedical Research Institute, San Antonio, TX.
Peter Fox, University of Texas Health Science Center San Antonio, San Antonio, TX.
Nicholas G. Martin, QIMR Berghofer, Brisbane, Australia
Katie L. McMahon, University of Queensland, Brisbane, Australia
Braxton Mitchell, University of Maryland, Baltimore, MD USA.
Rene L Olvera, University of Texas Health Science Center San Antonio, San Antonio, TX.
Charles Peterson, Texas Biomedical Research Institute, San Antonio, TX.
John Starr, University of Edinburgh, Edinburgh, UK.
Jessika Sussmann, University of Edinburgh, Edinburgh, UK.
Joanna Wardlaw, University of Edinburgh, Edinburgh, UK.
Margie Wright, QIMR Berghofer, Brisbane, Australia.
Dorret I. Boomsma, VU University, Amsterdam, The Netherlands
Rene Kahn, University Medical Center Utrecht, Utrecht, The Netherlands.
Eco JC de Geus, VU University, Amsterdam, The Netherlands.
Douglas E Williamson, University of Texas Health Science Center San Antonio, San Antonio, TX.
Ahmad Hariri, Duke University, Durahm, NC.
Dennis van t Ent, VU University, Amsterdam, The Netherlands.
Mark E. Bastin, University of Edinburgh, Edinburgh, UK
Andrew McIntosh, University of Edinburgh, Edinburgh, UK.
Ian J. Deary, University of Edinburgh, Edinburgh, UK
Hilleke E. Hulshoff pol, University Medical Center Utrecht, Utrecht, The Netherlands
John Blangero, Texas Biomedical Research Institute, San Antonio, TX.
Paul M. Thompson, Imaging Genetics Center, Institute for Neuroimaging and Informatics, Department of Neurology Keck School of Medicine University of Southern California, Marina del Rey, USA
David C. Glahn, Olin Neuropsychiatry Research Center, Institute of Living, Hartford Hospital, Hartford, USA.
David C. Van Essen, Anatomy & Neurobiology Department at Washington University in St. Louis, St. Louis, USA.
References
- Amos CI. Robust variance-components approach for assessing genetic linkage in pedigrees. Am J Hum Genet. 1994;54:535–543. [PMC free article] [PubMed] [Google Scholar]
- Bach M, Laun FB, Leemans A, Tax CM, Biessels GJ, Stieltjes B, Maier-Hein KH. Methodological considerations on tract-based spatial statistics (TBSS) Neuroimage. 2014;100C:358–369. doi: 10.1016/j.neuroimage.2014.06.021. [DOI] [PubMed] [Google Scholar]
- Barysheva M, Jahanshad N, Foland-Ross L, Altshuler LL, Thompson PM. White matter microstructural abnormalities in bipolar disorder: A whole brain diffusion tensor imaging study. 2012. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser PJ, Mattiello J, LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophysical Journal. 1994;66:259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser PJ, Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. J Magn Reson B. 1996;111:209–219. doi: 10.1006/jmrb.1996.0086. [DOI] [PubMed] [Google Scholar]
- Batouli SA, Sachdev PS, Wen W, Wright MJ, Ames D, Trollor JN. Heritability of brain volumes in older adults: the Older Australian Twins Study. Neurobiol Aging. 2013;35:937.e935–918. doi: 10.1016/j.neurobiolaging.2013.10.079. [DOI] [PubMed] [Google Scholar]
- Bava S, Boucquey V, Goldenberg D, Thayer RE, Ward M, Jacobus J, Tapert SF. Sex differences in adolescent white matter architecture. Brain Res. 2010;1375:41–48. doi: 10.1016/j.brainres.2010.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens TE, Woolrich MW, Jenkinson M, Johansen-Berg H, Nunes RG, Clare S, Matthews PM, Brady JM, Smith SM. Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magn Reson Med. 2003;50:1077–1088. doi: 10.1002/mrm.10609. [DOI] [PubMed] [Google Scholar]
- Brouwer RM, Mandl RC, Schnack HG, van Soelen IL, van Baal GC, Peper JS, Kahn RS, Boomsma DI, Hulshoff Pol HE. White matter development in early puberty: a longitudinal volumetric and diffusion tensor imaging twin study. PLoS One. 2012;7:e32316. doi: 10.1371/journal.pone.0032316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballedo A, Amico F, Ugwu I, Fagan AJ, Fahey C, Morris D, Meaney JF, Leemans A, Frodl T. Reduced fractional anisotropy in the uncinate fasciculus in patients with major depression carrying the met-allele of the Val66Met brain-derived neurotrophic factor genotype. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:537–548. doi: 10.1002/ajmg.b.32060. [DOI] [PubMed] [Google Scholar]
- Chiang MC, Barysheva M, Lee AD, Madsen S, Klunder AD, Toga AW, McMahon KL, de Zubicaray GI, Meredith M, Wright MJ, Srivastava A, Balov N, Thompson PM. Brain fiber architecture, genetics, and intelligence: a high angular resolution diffusion imaging (HARDI) study. Med Image Comput Comput Assist Interv. 2008;11:1060–1067. doi: 10.1007/978-3-540-85988-8_126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang MC, McMahon KL, de Zubicaray GI, Martin NG, Hickie I, Toga AW, Wright MJ, Thompson PM. Genetics of white matter development: a DTI study of 705 twins and their siblings aged 12 to 29. Neuroimage. 2011;54:2308–2317. doi: 10.1016/j.neuroimage.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerx L, Visser PJ, Verhey F, Aalten P. New MRI markers for Alzheimer’s disease: a meta-analysis of diffusion tensor imaging and a comparison with medial temporal lobe measurements. J Alzheimers Dis. 2012;29:405–429. doi: 10.3233/JAD-2011-110797. [DOI] [PubMed] [Google Scholar]
- den Braber A, van’t Ent D, Stoffers D, Linkenkaer-Hansen K, Boomsma DI, de Geus EJC. Sex differences in gray and white matter structure in age-matched unrelated males and females and opposite-sex siblings. 2013. [Google Scholar]
- Duarte-Carvajalino JM, Jahanshad N, Lenglet C, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, Thompson PM, Sapiro G. Hierarchical topological network analysis of anatomical human brain connectivity and differences related to sex and kinship. Neuroimage. 2011;59:3784–3804. doi: 10.1016/j.neuroimage.2011.10.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edden RA, Jones DK. Spatial and orientational heterogeneity in the statistical sensitivity of skeleton-based analyses of diffusion tensor MR imaging data. J Neurosci Methods. 2011;201:213–219. doi: 10.1016/j.jneumeth.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edens EL, Glowinski AL, Pergadia ML, Lessov-Schlaggar CN, Bucholz KK. Nicotine addiction in light smoking African American mothers. J Addict Med. 2010;4:55–60. doi: 10.1097/ADM.0b013e3181af111e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Rajendran N, Busa E, Augustinack J, Hinds O, Yeo BT, Mohlberg H, Amunts K, Zilles K. Cortical Folding Patterns and Predicting Cytoarchitecture. Cerebral cortex. 2007 doi: 10.1093/cercor/bhm225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC, Kent JW, Jr, Sprooten E, Diego VP, Winkler AM, Curran JE, McKay DR, Knowles EE, Carless MA, Goring HH, Dyer TD, Olvera RL, Fox PT, Almasy L, Charlesworth J, Kochunov P, Duggirala R, Blangero J. Genetic basis of neurocognitive decline and reduced white-matter integrity in normal human brain aging. Proc Natl Acad Sci U S A. 2013;110:19006–19011. doi: 10.1073/pnas.1313735110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR, Van Essen DC, Jenkinson M. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage. 2013;80:105–124. doi: 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshad N, Kochunov P, Sprooten E, Mandl RC, Nichols TE, Almassy L, Blangero J, Brouwer RM, Curran JE, de Zubicaray GI, Duggirala R, Fox PT, Hong LE, Landman BA, Martin NG, McMahon KL, Medland SE, Mitchell BD, Olvera RL, Peterson CP, Starr JM, Sussmann JE, Toga AW, Wardlaw JM, Wright MJ, Hulshoff Pol HE, Bastin ME, McIntosh AM, Deary IJ, Thompson PM, Glahn DC. Multi-site genetic analysis of diffusion images and voxelwise heritability analysis: A pilot project of the ENIGMA-DTI working group. Neuroimage. 2013 doi: 10.1016/j.neuroimage.2013.04.061. pii: S1053-8119(13)00408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshad N, Lee AD, Barysheva M, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, Toga AW, Thompson PM. Genetic influences on brain asymmetry: a DTI study of 374 twins and siblings. Neuroimage. 2010;52:455–469. doi: 10.1016/j.neuroimage.2010.04.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keihaninejad S, Ryan NS, Malone IB, Modat M, Cash D, Ridgway GR, Zhang H, Fox NC, Ourselin S. The importance of group-wise registration in tract based spatial statistics study of neurodegeneration: a simulation study in Alzheimer’s disease. PLoS One. 2012;7:e45996. doi: 10.1371/journal.pone.0045996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Castro C, Davis D, Dudley D, Brewer J, Zhang Y, Kroenke CD, Purdy D, Fox PT, Simerly C, Schatten G. Mapping primary gyrogenesis during fetal development in primate brains: high-resolution in utero structural MRI of fetal brain development in pregnant baboons. Front Neurosci. 2010a;4:20. doi: 10.3389/fnins.2010.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Glahn D, Fox PT, Lancaster J, Saleem K, Shelledy W, Zilles K, Thompson P, Coulon O, Blangero J, Fox P., JR Genetics of primary cerebral gyrification: Heritability of length, depth and area of primary sulci in an extended pedigree of Papio baboons. Neuroimage. 2009;15:1126–1132. doi: 10.1016/j.neuroimage.2009.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Glahn D, Lancaster J, Wincker P, Smith S, Thompson P, Almasy L, Duggirala R, Fox P, Blangero J. Genetics of microstructure of cerebral white matter using diffusion tensor imaging. Neuroimage. 2010b;15:1109–1116. doi: 10.1016/j.neuroimage.2010.01.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Glahn DC, LMR, Olvera R, Wincker P, Yang D, Sampath H, Carpenter W, Duggirala R, Curran J, Blangero J, Hong LE. Testing the hypothesis of accelerated cerebral white matter aging in schizophrenia and major depression. Biol Psychiatry. 2012 doi: 10.1016/j.biopsych.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Glahn DC, Lancaster J, Thompson PM, Kochunov V, Rogers B, Fox P, Blangero J, Williamson DE. Fractional anisotropy of cerebral white matter and thickness of cortical gray matter across the lifespan. Neuroimage. 2011;58:41–49. doi: 10.1016/j.neuroimage.2011.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Jahanshad N, Sprooten E, Nichols TE, Mandl RC, Almasy L, Booth T, Brouwer RM, Curran JE, de Zubicaray GI, Dimitrova R, Duggirala R, Fox PT, Elliot Hong L, Landman BA, Lemaitre H, Lopez LM, Martin NG, McMahon KL, Mitchell BD, Olvera RL, Peterson CP, Starr JM, Sussmann JE, Toga AW, Wardlaw JM, Wright MJ, Wright SN, Bastin ME, McIntosh AM, Boomsma DI, Kahn RS, den Braber A, de Geus EJ, Deary IJ, Hulshoff Pol HE, Williamson DE, Blangero J, van ‘t Ent D, Thompson PM, Glahn DC. Multi-site study of additive genetic effects on fractional anisotropy of cerebral white matter: Comparing meta and megaanalytical approaches for data pooling. Neuroimage. 2014;95C:136–150. doi: 10.1016/j.neuroimage.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Lancaster J, Thompson P, Toga AW, Brewer P, Hardies J, Fox P. An optimized individual target brain in the Talairach coordinate system. Neuroimage. 2002;17:922–927. [PubMed] [Google Scholar]
- Kochunov P, Lancaster JL, Thompson P, Woods R, Mazziotta J, Hardies J, Fox P. Regional spatial normalization: toward an optimal target. J Comput Assist Tomogr. 2001;25:805–816. doi: 10.1097/00004728-200109000-00023. [DOI] [PubMed] [Google Scholar]
- Kochunov P, Mangin JF, Coyle T, Lancaster J, Thompson P, Riviere D, Cointepas Y, Regis J, Schlosser A, Royall DR, Zilles K, Mazziotta J, Toga A, Fox PT. Age-related morphology trends of cortical sulci. Hum Brain Mapp. 2005 doi: 10.1002/hbm.20198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov PV, Lancaster JL, Fox PT. Accurate high-speed spatial normalization using an octree method. Neuroimage. 1999;10:724–737. doi: 10.1006/nimg.1999.0509. [DOI] [PubMed] [Google Scholar]
- Mandl RC, Rais M, van Baal GC, van Haren NE, Cahn W, Kahn RS, Hulshoff Pol HE. Altered white matter connectivity in never-medicated patients with schizophrenia. Hum Brain Mapp. 2012 doi: 10.1002/hbm.22075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus DS, Harms MP, Snyder AZ, Jenkinson M, Wilson JA, Glasser MF, Barch DM, Archie KA, Burgess GC, Ramaratnam M, Hodge M, Horton W, Herrick R, Olsen T, McKay M, House M, Hileman M, Reid E, Harwell J, Coalson T, Schindler J, Elam JS, Curtiss SW, Van Essen DC. Human Connectome Project informatics: quality control, database services, and data visualization. Neuroimage. 2013;80:202–219. doi: 10.1016/j.neuroimage.2013.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzler K, Belke M, Wehrmann E, Krakow K, Lengler U, Jansen A, Hamer HM, Oertel WH, Rosenow F, Knake S. Men and women are different: diffusion tensor imaging reveals sexual dimorphism in the microstructure of the thalamus, corpus callosum and cingulum. Neuroimage. 2010;54:2557–2562. doi: 10.1016/j.neuroimage.2010.11.029. [DOI] [PubMed] [Google Scholar]
- O’Dwyer L, Lamberton F, Bokde AL, Ewers M, Faluyi YO, Tanner C, Mazoyer B, O’Neill D, Bartley M, Collins R, Coughlan T, Prvulovic D, Hampel H. Sexual dimorphism in healthy aging and mild cognitive impairment: a DTI study. PLoS One. 2013;7:e37021. doi: 10.1371/journal.pone.0037021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penke L, Munoz Maniega S, Houlihan LM, Murray C, Gow AJ, Clayden JD, Bastin ME, Wardlaw JM, Deary IJ. White matter integrity in the splenium of the corpus callosum is related to successful cognitive aging and partly mediates the protective effect of an ancestral polymorphism in ADRB2. Behav Genet. 2010a;40:146–156. doi: 10.1007/s10519-009-9318-4. [DOI] [PubMed] [Google Scholar]
- Penke L, Munoz Maniega S, Murray C, Gow AJ, Hernandez MC, Clayden JD, Starr JM, Wardlaw JM, Bastin ME, Deary IJ. A general factor of brain white matter integrity predicts information processing speed in healthy older people. J Neurosci. 2010b;30:7569–7574. doi: 10.1523/JNEUROSCI.1553-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D. nlme: Linear and Nonlinear Mixed Effects Models. 2008. [Google Scholar]
- R-Development-Core-Team. R: A Language and Environment for Statistical Computing. 2009. [Google Scholar]
- Sartor CE, McCutcheon VV, Pommer NE, Nelson EC, Grant JD, Duncan AE, Waldron M, Bucholz KK, Madden PA, Heath AC. Common genetic and environmental contributions to post-traumatic stress disorder and alcohol dependence in young women. Psychol Med. 2010;41:1497–1505. doi: 10.1017/S0033291710002072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Johansen-Berg H, Rueckert D, Nichols TE, Mackay CE, Watkins KE, Ciccarelli O, Cader MZ, Matthews PM, Behrens TE. Tract-based spatial statistics: Voxelwise analysis of multi-subject diffusion data. Neuroimage. 2006;31:1487–1505. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Sotiropoulos SN, Jbabdi S, Xu J, Andersson JL, Moeller S, Auerbach EJ, Glasser MF, Hernandez M, Sapiro G, Jenkinson M, Feinberg DA, Yacoub E, Lenglet C, Van Essen DC, Ugurbil K, Behrens TE. Advances in diffusion MRI acquisition and processing in the Human Connectome Project. Neuroimage. 2013;80:125–143. doi: 10.1016/j.neuroimage.2013.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprooten E, Sussmann JE, Clugston A, Peel A, McKirdy J, Moorhead TW, Anderson S, Shand AJ, Giles S, Bastin ME, Hall J, Johnstone EC, Lawrie SM, McIntosh AM. White matter integrity in individuals at high genetic risk of bipolar disorder. Biol Psychiatry. 2011;70:350–356. doi: 10.1016/j.biopsych.2011.01.021. [DOI] [PubMed] [Google Scholar]
- Teipel SJ, Wegrzyn M, Meindl T, Frisoni G, Bokde AL, Fellgiebel A, Filippi M, Hampel H, Kloppel S, Hauenstein K, Ewers M. Anatomical MRI and DTI in the Diagnosis of Alzheimer’s Disease: A European Multicenter Study. J Alzheimers Dis. 2012 doi: 10.3233/JAD-2012-112118. [DOI] [PubMed] [Google Scholar]
- Thomason ME, Thompson PM. Diffusion imaging, white matter, and psychopathology. Annu Rev Clin Psychol. 2011;7:63–85. doi: 10.1146/annurev-clinpsy-032210-104507. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Stein JL, Medland SE, Hibar DP, Vasquez AA, Renteria ME, Toro R, Jahanshad N, Schumann G, Franke B, Wright MJ, Martin NG, Agartz I, Alda M, Alhusaini S, Almasy L, Almeida J, Alpert K, Andreasen NC, Andreassen OA, Apostolova LG, Appel K, Armstrong NJ, Aribisala B, Bastin ME, Bauer M, Bearden CE, Bergmann O, Binder EB, Blangero J, Bockholt HJ, Boen E, Bois C, Boomsma DI, Booth T, Bowman IJ, Bralten J, Brouwer RM, Brunner HG, Brohawn DG, Buckner RL, Buitelaar J, Bulayeva K, Bustillo JR, Calhoun VD, Cannon DM, Cantor RM, Carless MA, Caseras X, Cavalleri GL, Chakravarty MM, Chang KD, Ching CR, Christoforou A, Cichon S, Clark VP, Conrod P, Coppola G, Crespo-Facorro B, Curran JE, Czisch M, Deary IJ, de Geus EJ, den Braber A, Delvecchio G, Depondt C, de Haan L, de Zubicaray GI, Dima D, Dimitrova R, Djurovic S, Dong H, Donohoe G, Duggirala R, Dyer TD, Ehrlich S, Ekman CJ, Elvsashagen T, Emsell L, Erk S, Espeseth T, Fagerness J, Fears S, Fedko I, Fernandez G, Fisher SE, Foroud T, Fox PT, Francks C, Frangou S, Frey EM, Frodl T, Frouin V, Garavan H, Giddaluru S, Glahn DC, Godlewska B, Goldstein RZ, Gollub RL, Grabe HJ, Grimm O, Gruber O, Guadalupe T, Gur RE, Gur RC, Goring HH, Hagenaars S, Hajek T, Hall GB, Hall J, Hardy J, Hartman CA, Hass J, Hatton SN, Haukvik UK, Hegenscheid K, Heinz A, Hickie IB, Ho BC, Hoehn D, Hoekstra PJ, Hollinshead M, Holmes AJ, Homuth G, Hoogman M, Hong LE, Hosten N, Hottenga JJ, Hulshoff Pol HE, Hwang KS, Jack CR, Jr, Jenkinson M, Johnston C, Jonsson EG, Kahn RS, Kasperaviciute D, Kelly S, Kim S, Kochunov P, Koenders L, Kramer B, Kwok JB, Lagopoulos J, Laje G, Landen M, Landman BA, Lauriello J, Lawrie SM, Lee PH, Le Hellard S, Lemaitre H, Leonardo CD, Li CS, Liberg B, Liewald DC, Liu X, Lopez LM, Loth E, Lourdusamy A, Luciano M, Macciardi F, Machielsen MW, Macqueen GM, Malt UF, Mandl R, Manoach DS, Martinot JL, Matarin M, Mather KA, Mattheisen M, Mattingsdal M, Meyer-Lindenberg A, McDonald C, McIntosh AM, McMahon FJ, McMahon KL, Meisenzahl E, Melle I, Milaneschi Y, Mohnke S, Montgomery GW, Morris DW, Moses EK, Mueller BA, Munoz Maniega S, Muhleisen TW, Muller-Myhsok B, Mwangi B, Nauck M, Nho K, Nichols TE, Nilsson LG, Nugent AC, Nyberg L, Olvera RL, Oosterlaan J, Ophoff RA, Pandolfo M, Papalampropoulou-Tsiridou M, Papmeyer M, Paus T, Pausova Z, Pearlson GD, Penninx BW, Peterson CP, Pfennig A, Phillips M, Pike GB, Poline JB, Potkin SG, Putz B, Ramasamy A, Rasmussen J, Rietschel M, Rijpkema M, Risacher SL, Roffman JL, Roiz-Santianez R, Romanczuk-Seiferth N, Rose EJ, Royle NA, Rujescu D, Ryten M, Sachdev PS, Salami A, Satterthwaite TD, Savitz J, Saykin AJ, Scanlon C, Schmaal L, Schnack HG, Schork AJ, Schulz SC, Schur R, Seidman L, Shen L, Shoemaker JM, Simmons A, Sisodiya SM, Smith C, Smoller JW, Soares JC, Sponheim SR, Sprooten E, Starr JM, Steen VM, Strakowski S, Strike L, Sussmann J, Samann PG, Teumer A, Toga AW, Tordesillas-Gutierrez D, Trabzuni D, Trost S, Turner J, Van den Heuvel M, van der Wee NJ, van Eijk K, van Erp TG, van Haren NE, van ‘t Ent D, van Tol MJ, Valdes Hernandez MC, Veltman DJ, Versace A, Volzke H, Walker R, Walter H, Wang L, Wardlaw JM, Weale ME, Weiner MW, Wen W, Westlye LT, Whalley HC, Whelan CD, White T, Winkler AM, Wittfeld K, Woldehawariat G, Wolf C, Zilles D, Zwiers MP, Thalamuthu A, Schofield PR, Freimer NB, Lawrence NS, Drevets W Alzheimer’s Disease Neuroimaging Initiative ECICSYSG. The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav. 2014;8:153–182. doi: 10.1007/s11682-013-9269-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugurbil K, Xu J, Auerbach EJ, Moeller S, Vu AT, Duarte-Carvajalino JM, Lenglet C, Wu X, Schmitter S, Van de Moortele PF, Strupp J, Sapiro G, De Martino F, Wang D, Harel N, Garwood M, Chen L, Feinberg DA, Smith SM, Miller KL, Sotiropoulos SN, Jbabdi S, Andersson JL, Behrens TE, Glasser MF, Van Essen DC, Yacoub E, Consortium WUMH. Pushing spatial and temporal resolution for functional and diffusion MRI in the Human Connectome Project. Neuroimage. 2013;80:80–104. doi: 10.1016/j.neuroimage.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC. A tension-based theory of morphogenesis and compact wiring in the central nervous system. Nature. 1997;385:313–318. doi: 10.1038/385313a0. [DOI] [PubMed] [Google Scholar]
- Van Essen DC. Surface-based approaches to spatial localization and registration in primate cerebral cortex. Neuroimage. 2004;23(Suppl 1):S97–107. doi: 10.1016/j.neuroimage.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Ugurbil K, Auerbach E, Barch D, Behrens TE, Bucholz R, Chang A, Chen L, Corbetta M, Curtiss SW, Della Penna S, Feinberg D, Glasser MF, Harel N, Heath AC, Larson-Prior L, Marcus D, Michalareas G, Moeller S, Oostenveld R, Petersen SE, Prior F, Schlaggar BL, Smith SM, Snyder AZ, Xu J, Yacoub E. The Human Connectome Project: a data acquisition perspective. Neuroimage. 2013;62:2222–2231. doi: 10.1016/j.neuroimage.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasung L, Huang H, Jovanov-Milosevic N, Pletikos M, Mori S, Kostovic I. Development of axonal pathways in the human fetal fronto-limbic brain: histochemical characterization and diffusion tensor imaging. J Anat. 2010;217:400–417. doi: 10.1111/j.1469-7580.2010.01260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Adamson C, Yuan W, Altaye M, Rajagopal A, Byars AW, Holland SK. Sex differences in white matter development during adolescence: a DTI study. Brain Res. 2012;1478:1–15. doi: 10.1016/j.brainres.2012.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gupta A, Liu Z, Zhang H, Escolar ML, Gilmore JH, Gouttard S, Fillard P, Maltbie E, Gerig G, Styner M. DTI registration in atlas based fiber analysis of infantile Krabbe disease. Neuroimage. 2011;55:1577–1586. doi: 10.1016/j.neuroimage.2011.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalesky A. Moderating registration misalignment in voxelwise comparisons of DTI data: a performance evaluation of skeleton projection. Magn Reson Imaging. 2011;29:111–125. doi: 10.1016/j.mri.2010.06.027. [DOI] [PubMed] [Google Scholar]
- Zhang S, Arfanakis K. Role of standardized and study-specific human brain diffusion tensor templates in inter-subject spatial normalization. J Magn Reson Imaging. 2013;37:372–381. doi: 10.1002/jmri.23842. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.