Abstract
Cellular transformation is associated with altered glutamine (Gln) metabolism. Tumor cells utilize Gln in the tricarboxylic acid (TCA) cycle to maintain sufficient pools of biosynthetic precursors to support rapid growth and proliferation. However, Gln metabolism also generates NADPH, and Gln-derived glutamate is used for synthesis of glutathione (GSH). Since both NADPH and GSH are antioxidants, Gln may also contribute to redox balance in transformed cells. The Hace1 E3 ligase is a tumor suppressor inactivated in diverse human cancers. Hace1 targets the Rac1 GTPase for degradation at Rac1-dependent NADPH oxidase complexes, blocking superoxide generation by the latter. Consequently, loss of Hace1 increases reactive oxygen species (ROS) levels in vitro and in vivo. Given the link between Hace1 loss and increased ROS, we investigated whether genetic inactivation of Hace1 alters Gln metabolism. We demonstrate that mouse embryonic fibroblasts (MEFs) derived from Hace1-/- mice are highly sensitive to Gln withdrawal, leading to enhanced cell death compared to wild type (wt) MEFs, and Gln depletion or chemical inhibition of Gln uptake block soft agar colony formation by Hace1-/- MEFs. Hace1-/- MEFs exhibit increased Gln uptake and ammonia secretion, and metabolic labeling using 13C-Gln revealed that Hace1 loss increases incorporation of Gln carbons into TCA cycle intermediates. Gln starvation markedly increases ROS levels in Hace1-/- but not in wt MEFs, and treatment with the antioxidant N-acetyl cysteine (NAC) or the TCA cycle intermediate oxaloacetate efficiently rescues Gln starvation-induced ROS elevation and cell death in Hace1-/- MEFs. Finally, Gln starvation increases superoxide levels in Hace1-/- MEFs, and NADPH oxidase inhibitors block the induction of superoxide and cell death by Gln starvation. Together, these results suggest that increased ROS production due to Hace1 loss leads to Gln addiction as a mechanism to cope with increased ROS-induced oxidative stress.
Keywords: Hace1, glutamine, reactive oxygen species, oxidative stress, metabolism
INTRODUCTION
Hace1 (HECT domain and ankyrin repeat containing E3 ubiquitin-protein ligase 1) is a HECT family E3 ligase that was first identified as a tumor suppressor in human Wilms’ tumors, the most common kidney cancer in children (1, 2). Recent studies demonstrated epigenetic inactivation of the Hace1 gene in multiple other human tumors (3-8). Hace1 knockout mice develop spontaneous late onset tumors of diverse phenotypes, highlighting Hace1 as a bona fide tumor suppressor (2). To date, the only known E3 ligase target of Hace1 is the small Rho-GTPase, Rac1 (9-10). In response to cytotoxic necrotizing factor-1 or hepatocyte growth factor, Hace1 ubiquitylates and targets GTP-bound (activated) Rac1 for proteosomal degradation to block Rac1-dependent bacterial invasion (9) and cell migration (10), respectively. Rac1 is involved in multiple regulatory processes, including reactive oxygen species (ROS) generation by NADPH oxidases, as GTP-bound Rac1 is an essential subunit for activation of Nox1-3-containing NADPH oxidases (11, 12). We recently reported that loss of Hace1 in mice, zebra fish, human Wilms’ tumor tissues, as well as in other human tumor cell lines, leads to increased cellular ROS levels due to high Rac1 activity, resulting in uncontrolled ROS production by Rac1-dependent NADPH oxidases (13). Furthermore, Hace1 indirectly promotes activity of nuclear factor erythroid 2-related factor 2 (NRF2), a master regulator of the antioxidative stress response (14). Hace1 is therefore emerging as a key regulator of oxidative stress.
Altered cellular metabolism is a well-known consequence of malignant transformation (15-18). In addition to glucose, glutamine (Gln) is a major nutrient source for tumor cells in vitro and in vivo (19, 20). Although not an essential amino acid, diverse cancer cell types depend on extracellular Gln for survival, a phenomenon known as Gln addiction (21). Oncogenes such as Myc and K-Ras depend on Gln for transformation and lead to upregulated Gln metabolism (22-24). Recent studies reported that the loss of the tumor suppressor retinoblastoma protein (pRB) is also associated with increased Gln metabolism and renders cells Gln addicted (25, 26). While the Gln amine groups are used in the synthesis of most nonessential amino acids, the carbon skeleton of GLN is used to replenish tricarboxylic acid (TCA) cycle intermediates for bioATP production (27). In addition to supporting the TCA cycle, a significant fraction of Gln-derived carbon leaves the TCA cycle as malate and is converted to pyruvate by NADP+ dependent malic enzyme (ME-1), thus producing NADPH for redox balance (28). Gln-derived glutamate is also directly used for synthesis of the anti-oxidant, glutathione (GSH) (27). Therefore Gln metabolism is crucial for cancer cells to maintain redox balance and to cope with the toxic effects of high ROS. Given that Hace1 deficiency leads to high cellular ROS, we wondered whether Hace1 loss is linked to altered Gln metabolism.
Here we show that Hace1-/- MEFs are highly sensitive to Gln starvation compared to wt control MEFs. Hace1-/- MEFs exhibit increased Gln uptake and metabolism, and are dependent on Gln for soft agar colony formation. Gln deprivation induces cell death in Hace1-/- MEFs by increasing cellular ROS levels. The antioxidant compound N-acetyl cysteine (NAC) or the TCA cycle intermediate oxaloacetate (OAA) efficiently rescues Gln starvation-induced ROS elevation and cell death. Moreover, reduction of superoxide production by inhibition of Rac1-dependent NADPH oxidases in Hace1-/- MEFs reduces superoxide levels and cell death in the absence of Gln. These results indicate that inactivation of the Hace1 tumor suppressor leads to Gln addiction secondary to increased cellular ROS levels.
RESULTS AND DISCUSSION
Hace1 deficient cells are highly sensitive to Gln starvation
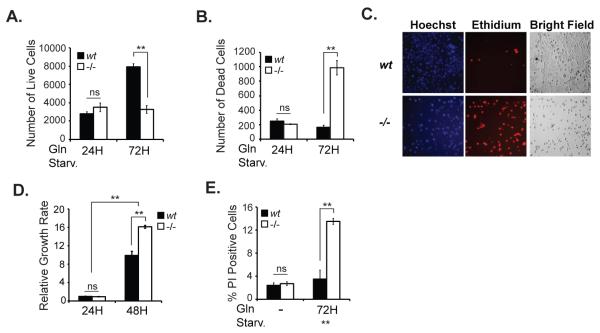
To determine potential differences in glutamine starvation-induced cell death in Hace1-/- versus wt MEFs, we stained cells with Hoechst or ethidium after 24 hours and 72 hours of Gln starvation and quantified the number of live and dead cells, respectively, using an IN Cell Analyzer. While live cell numbers actually increased in Hace1 wt MEFs after 72 hours of Gln starvation (Fig. 1A), there was no increase in dead cell counts for this cell line over the same time period (Fig. 1B), indicating that Hace1 wt MEFs do not depend on Gln for growth or survival. In marked contrast, while live cell counts were unchanged, there was a significant increase in the dead cell count for Hace1-/- MEFs after 72 hours of Gln starvation (Fig. 1A-C). We also compared growth rates of wt and Hace1-/-MEFs in the presence of glutamine. As shown in Fig. 1D, growth of Hace1-/- MEFs is similar (24 hours after seeding) to or greater (48 hours after seeding) than that of wt MEFs. These data indicate that Hace1-/- MEFs are not growth-deficient when Gln is present and the observed effects of Gln starvation on Hace1-/- MEFs is due to the dependency of these cells on Gln. To support these findings, we also quantified cell death by propidium iodide (PI) incorporation into dead cells by flow cytometry. Consistent with the aforementioned results, Gln starvation induced significant cell death only in Hace1-/- MEFs (Fig. 1E). These data establish that loss of Hace1 renders cells dependent on Gln for survival.
Figure 1. Hace1-/- MEFs show increased sensitivity to Gln starvation.
Approximately 4000 cells were seeded in triplicate in 2x 96-well plates and the following day the culture medium was exchanged with Gln-free medium (DMEM, lacking Gln, pyruvate, and phenol red (Gibco)), supplemented with 10% dialyzed FBS (Invitrogen) and 1% streptomycin/penicillin (Invitrogen). After 24 hours or 72 hours of Gln starvation, live cells were stained with 150 μM Hoechst 33342 (Molecular Probes) (A), and dead cells were stained with 10 μM ethidium homodimer-1 (Molecular Probes) (B) for 1 h at 37 °C. Wells were imaged on an IN Cell Analyzer (GE Healthcare) and analyzed by IN Cell Developer software (GE Healthcare). Cells with overlapping staining were also considered non-viable. C. Representative images of the stained wt and Hace1-/- MEFs after 72 hours of Gln starvation from A and B. D. Growth rates of wt and Hace1-/- MEFs in the presence of 2 mM Gln were measured 24 hours and 48 hours after seeding cells (40.000 cells/well - 96-well plates) using Cyquant Cell Proliferation Assay kit (Life Technologies) according to kit instructions. E. Cells were seeded in 6-well plates in triplicates and the next day the culture medium was exchanged to Gln-free medium with/without 2 mM Gln. After 72 hours, both detached and attached cells were pooled, centrifuged, resuspended in cold PBS containing 1μg/mL propidium iodide (PI) to stain dead cells, and analyzed immediately using FACSCalibur flow cytometer (BD Biosciences) in FL-3 channel. Error bars represent S.D. of three independent experiments. A two-tailed Student’s t-test was performed to determine the significance. *p<0.05, **p<0.001.
Hace1-/- MEFs take up and metabolize more Gln compared to wt MEFs
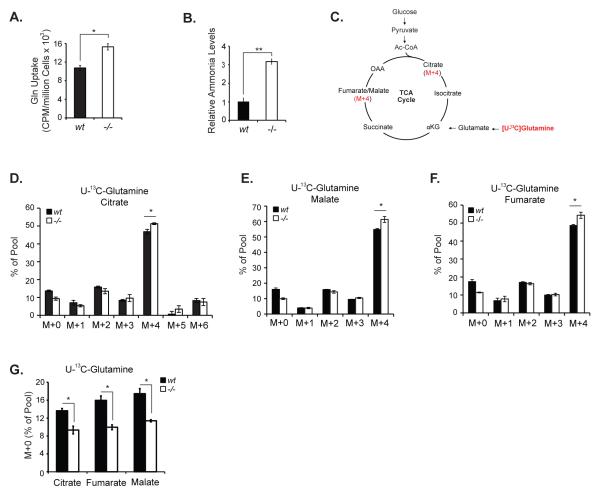
Since Hace1-/- MEFs depend on Gln for survival, we predicted that these cells might have increased Gln uptake and metabolism compared to their wt counterparts. Using radioactively labeled glutamine (U-14C Gln), we measured Gln uptake and found that Hace1-/- MEFs take up significantly more Gln than wt MEFs (Fig. 2A). We confirmed the effect of Hace1 expression on cellular Gln uptake in HEK293 cells stably expressing control or Hace1 specific shRNAs. HEK293 cells express high levels of Hace1 as compared to various human cancer cell lines and therefore they are considered as a good model system to study effects of Hace1 on cellular processes (13). HEK293 cells with stable Hace1 shRNA-mediated knockdown exhibited increased Gln uptake as compared with cells stably expressing control shRNAs (suppl. Fig. 1). Conversion of Gln to glutamate is the first step of Gln metabolism and is associated with ammonia production. Consistent with increased Gln uptake, Hace1-/- MEFs generate significantly higher levels of ammonia in vitro (Fig. 2B). Many transformed cell lines use Gln as the major anaplerotic precursor for synthesis of the TCA cycle intermediates (18, 28, 29). Since Hace1-/- MEFs uptake more Gln, we next investigated whether Gln is metabolized differently in Hace1-/- versus wt MEFs. We therefore cultured cells in media containing [U-13C]-Gln and analyzed the incorporation of Gln carbons into the TCA cycle intermediates citrate, malate, and fumarate using mass spectrometry. Processing of [U-13C]-Gln in the TCA cycle generates M+4 isotopomers (Fig. 2C), whereas non-Gln sources (e.g. glucose) produce M+0 isotopomers (un-labeled) of these intermediates. We found that both cell lines efficiently incorporated Gln carbons into TCA cycle intermediates (Fig. 2D-F), suggesting that Hace1 loss does not cause a major change in the pathways by which these cells utilize Gln for anaplerosis. However, Hace1-/- MEFs displayed significantly higher levels of M+4 isotopomers of citrate, malate, and fumarate, indicating increased Gln metabolism. In contrast, levels of unlabeled intermediates (M+0 isotopomers) were higher in Hace1 wt MEFs (Fig. 2G), suggesting a modest enhancement in the fraction of TCA cycle intermediate pools labeled by Gln in Hace1-/- MEFs over this time course. This data implies that loss of Hace1 results in a slight shift towards Gln as a more favored anaplerotic precursor.
Figure 2. Hace1-/- MEFs take up and catabolize more Gln as compared with the wt MEFs.
A. Hace1-/- MEFs uptake more Gln than wt cells. Hace1-/- and wt MEFs were seeded in triplicate in 6-well plates and Gln uptake was determined as in (32) with modifications. Cells were incubated in Gln-free medium for 20 min. and 0.2 μCi/mL [U-14C]Gln was added. After a 15 minute incubation at room temperature, cells were washed 3x with PBS, lyzed with 250 μL of 0.2% SDS in 0.2 N NaOH, incubated for 30 minutes at room temperature, transferred into eppendorf tubes and incubated in a heating block at 60 °C for another 20 minutes. A 25 μL aliquot of 1 N HCl was added into each tube to neutralize NaOH, and 200 μL of the lysate was transferred into scintillation vials containing 6 mL scintillation liquid (Scintisafe Econo, Fisher Scientific), and the total radioactivity was determined using a β-scintillation counter (Perkin Elmer). Radioactivity was normalized to protein concentration. B. Hace1-/- MEFs secrete increased ammonia. Hace1 wt and KO MEFs were seeded in triplicate in 6-well plates. Two days after, the culture media from the wells were collected and ammonia levels were analyzed using an ammonia assay kit (BioVision) according to the kit instructions. C. Schematic of Gln metabolism in the TCA cycle. D-F. Analysis of the TCA cycle intermediates citrate, malate, and fumarate derived from 13C-Gln. 13C-labeling experiments were performed essentially as described (38). Cells were cultured in regular media to 80-90% confluence in 10 cm dishes, and after rinsing with ice-cold PBS, overlaid with medium containing 10 mM glucose and 4 mM [U-13C]-glutamine. After 8 hours incubation, labeled cells were rinsed with ice-cold PBS, lysed in cold 50% methanol, and subjected to three freeze-thaw cycles. The lysates were centrifuged to remove precipitated protein, then evaporated and derivatized by trimethylsilylation (Tri-Sil HTP reagent, Thermo). Three μL of the derivatized material were injected into an Agilent 6970 gas chromatograph equipped with a fused silica capillary GC column (30 m length, 0.25 mm diameter) and networked to an Agilent 5973 Mass Selective Detector. Abundance of the following ions was monitored: m/z 245-249 for fumarate; m/z 335-339 for malate; and m/z 465-471 for citrate. The measured distribution of mass isotopomers was corrected for natural abundance of 13C (39). G. Comparison of unlabeled TCA cycle intermediate pools in Hace1-/-and wt MEFs. A two-tailed Student’s t-test was performed to determine statistical significance. *p<0.05, **p<0.001.
Gln starvation induces cell death in Hace1 deficient cells by augmenting ROS levels
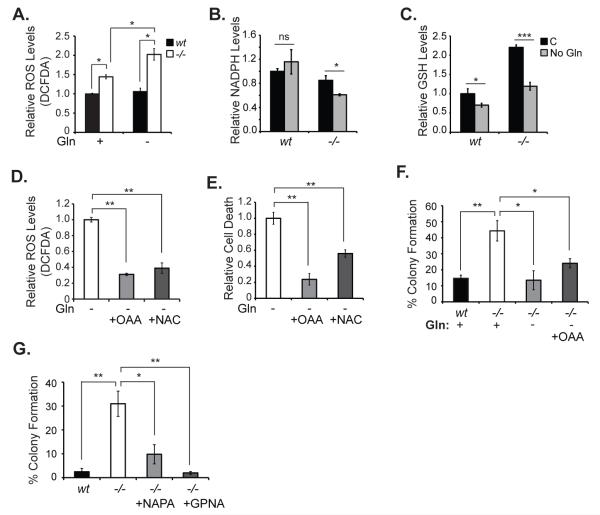
We previously demonstrated that Hace1 loss leads to increased ROS levels both in vitro and in vivo (13). Since Gln is a precursor for the synthesis of the antioxidant GSH, we investigated the possibility that Gln is required for survival of Hace1-/- MEFs by mitigating the toxic effects of increased ROS production. We compared ROS levels in Hace1-/- and wt MEFs with or without Gln starvation using the general ROS indicator CM-H2DCFDA followed by flow cytometry. We found that Hace1-/- MEFs have significantly higher ROS compared to wt MEFs, and that Gln starvation enhances ROS levels in Hace1-/- but not in wt MEFs (Fig. 3A). Furthermore, Gln starvation caused a significant reduction in both NADPH and reduced GSH levels in Hace1-/-MEFs (Figs. 3B and 3C). Of note, GSH levels were also slightly decreased in wt MEFs upon Gln starvation, but to a much lesser extend compared with the marked decrease observed in Hace1-/-MEFs (Fig. 3C), suggesting that a severe depletion of reduced GSH pools is required for ROS elevation in these cells. These results provide strong evidence that Hace1-/- MEFs depend on Gln for ROS homeostatis. Accordingly, we predicted that glutamine starvation-induced cell death may be due to elevated ROS levels in Hace1-/- MEFs and that a ROS scavenger, or a TCA cycle intermediate that can be converted to glutamate and eventually to GSH, could rescue cell death in the absence of Gln. Indeed, addition of the anaplerotic precursor oxaloacetate (OAA), which can be utilized by cells to derive glutamate and GSH, or the antioxidant N-acetyl cysteine (NAC), to the culture medium efficiently reduced Gln starvation-induced ROS elevation and cell death in Hace1-/- MEFs (Fig. 3D, 3E). This strongly suggests that Gln starvation induces cell death in Hace1 deficient cells by augmenting cellular ROS levels.
Figure 3. Gln starvation induces cell death in Hace1-/- MEFs by augmenting ROS.
A. Cells were seeded in 6-well plates in triplicate and the next day the culture medium was exchanged with Gln-free medium with or without 2 mM Gln. After 48 hours, ROS levels were measured using the general ROS indicator CM-H2DCFDA (5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester) (Molecular Probes). Cells were incubated with 5 μM DCFDA for 40 minutes. Medium was removed and the cells were washed with PBS. After trypsinization, cells were centrifuged and resuspended in PBS containing 1μg/mL PI, and analyzed immediately by flow cytometry using the FL-1 channel for DCFDA and the FL-3 channel for PI fluorescence. B. Cells were seed in 6 cm dishes in triplicates and the next day the culture medium was exchanged with Gln-free medium with or without 2 mM Gln. After 38 hours, NADPH levels were measured using NADPH-Glo assay kit (Promega). Data was normalized to protein concentration. C. Cells were seeded and treated as in B and reduced GSH levels were measured using GSH assay kit (Biovision). Data was normalized to protein concentration. D. Cells were seeded as in A and the culture medium was replaced the next day by Gln-free medium. 5 mM OAA or NAC was added as indicated and ROS was measured after 48 hours as described in A. E. Cells were seeded and treated as in B, and after 72 hours Gln starvation-induced cell death was measured as described in Fig. 1E. Error bars represent S.D. of three independent experiments. F. Soft agar colony formation assays. Approximately 8000 cells/well were mixed with 1 mL of 0.4% agar in DMEM without Gln, supplemented with Gln or OAA as indicated, and layered on 6-well plates covered with 0.8% agar in DMEM in triplicate. The wells were supplemented with 2-4 drops of the corresponding medium every 2 days. After 2 weeks, wells were imaged and colony numbers were counted using ImageJ. G. Soft agar colony formation assays were performed as described in F, except that DMEM contained Gln and 10 mM NaPA or 100 μM GPNA was included as indicated. Error bars represent standard deviations (SD) of three independent experiments. A two-tailed Student’s t-test was performed to determine statistical significance. *p<0.05, **p<0.001.
The pentose phosphate and ME-1 pathways are considered as the major NADPH generating mechanisms in cells (30, 31). As mentioned earlier, ME-1 contributes to cellular NADPH pools by converting Gln-derived malate to pyruvate in an NADP+ dependent manner, releasing NADPH as a side product (28). To address whether inhibition of ME-1 expression in wt MEFs can induce a similar effect to that of Gln starvation observed in Hace1-/- MEFs, we knocked down ME-1 in wt MEFs using specific siRNAs and measured cell death in the presence or absence of Gln. ME-1 knock down increased the number of dead cells slightly but significantly in the wt MEFs under both glutamine replete and glutamine deplete conditions (suppl. Figure 2A). We confirmed successful knockdown of ME-1 by qRT-PCR analysis (suppl. Fig. 2B). A marked increase in cell death is not expected upon knockdown of ME-1 because cells likely compensate for the NADPH reduction caused by ME-1 loss by upregulating NADPH production from the pentose phosphate pathway. This may be the reason why knock down of ME-1 in wt MEFs is not as effective as Gln starvation in inducing cell death as seen with Hace1-/- MEFs.
Transformed cells feed Gln into the TCA cycle to maintain sufficient pools of biosynthetic precursors to support oncogenic processes (28). We therefore tested if Gln withdrawal could block the ability of Hace1-/- MEFs to form colonies in soft agar, a well-established read-out of transformation activity. As reported (13), Hace1-/- MEFs formed strikingly higher number of colonies than wt MEFs in medium containing regular Gln levels, but this was completely inhibited in the absence of Gln (Fig. 3F). Addition of the anaplerotic molecule OAA in soft agar medium did not restore colony formation efficiency of Hace1-/-MEFs in the absence of Gln (Fig. 3F) despite its ability to rescue ROS elevation and cell death under these conditions. This is expected because in addition to its role as an anaplerotic precursor for biosynthesis of TCA cycle intermediates and GSH, Gln is also directly utilized for hexosamine and nucleotide biosynthesis, which are essential for proliferation (32, 33). Thus, Hace1-/- MEFs are reliant on Gln for transformation activity.
We next evaluated chemical inhibitors of cellular Gln uptake to assess the potential use of pharmacologic depletion of Gln to inhibit growth of Hace1 deficient tumors. Two such inhibitors are sodium phenylacetate (NaPA) and gamma-L-glutamyl-p-nitroanilide hydrochloride (GPNA). NaPA is known to block tumor growth in vivo by depleting Gln in the circulation (34), and can block growth of cancer cells in vitro (35), although the mechanism for the latter is not clear. GPNA is an inhibitor of the Gln transporter ASCT2 and therefore reduces cellular Gln uptake (36). NaPA and GPNA both very effectively blocked soft agar colony formation of Hace1-/-MEFs (Fig. 3G). Together, these results suggest that Hace1-/- MEFs take up and metabolize more Gln and are dependent on Gln for ROS homeostasis, survival, and cellular transformation, providing preliminary evidence that Gln depletion may be an effective strategy to block growth of Hace1 deficient tumors.
Increased ROS generation by NADPH oxidases contributes to Gln starvation-induced cell death in Hace1-/- MEFs
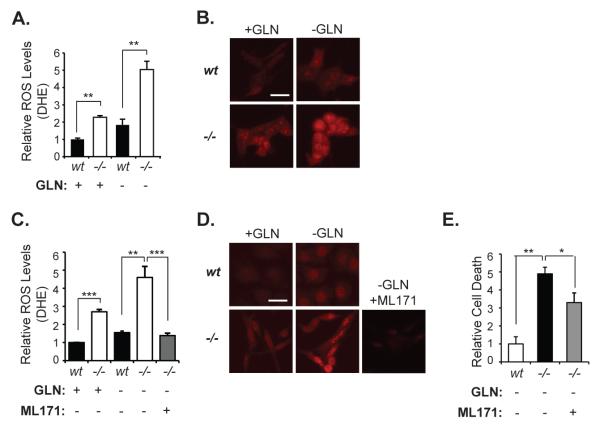
To confirm that increased superoxide generation by NADPH oxidase complexes is the source of ROS augmentation and cell death in Hace1-/- MEFs after Gln withdrawal, we first analyzed superoxide levels in Hace1-/- and wt MEFs with or without Gln starvation, using the superoxide specific dye dihydroethidum (DHE). As previously observed (13), Hace1-/- MEFs had significantly higher superoxide levels compared to wt MEFs (Figs. 4A and 4B). Gln starvation only modestly increased superoxide levels in Hace1 wt MEFs, whereas it caused a dramatic increase in Hace1-/- MEFs (Fig. 4A, 4B). Blocking NADPH oxidase activity using the NOX1-containing NADPH oxidase inhibitor ML171 (37) completely abrogated Gln starvation-induced superoxide production (Figs. 4C and 4D), and significantly reduced Gln starvation-induced cell death in Hace1-/- MEFs (Fig. 4E). It should be noted that the rescue effect of ML171 treatment on cell viability in the absence of Gln was not as striking as its ability to abrogate superoxide levels in Hace1-/- MEFs, indicating that other sources of ROS may also contribute to Gln starvation-induced cell death. These results strongly argue that Hace1 deficiency leads to increased superoxide generation by NADPH oxidases, and suggest that Hace1 deficient cells may depend on Gln to cope with increased cellular ROS levels to avoid ROS-induced cell death.
Figure 4. Increased superoxide production by NADPH oxidase contributes to GLN starvation-induced cell death in Hace1-/- MEFs.
A. Gln starvation augments superoxide levels in Hace1-/- MEFs. Cells were cultured with or without Gln for 36 hours and analyzed for superoxide content by the DHE assay as described previously (13). B. Representative fluorescent images of DHE stained cells from A 40X magnification was used. Scale bar represents 10 μm. C. DHE staining of the Hace1-/- and wt MEFs, with/without Gln starvation. Cells were Gln- starved for 36 hours, and 3 μM ML171 was added into culture medium of Hace1-/- MEFs as indicated. D. Representative fluorescent images of DHE-stained cells from C. 40X magnification was used. Scale bar represents 10 μm. E. Hace1-/- MEFs were cultured with or without GLN for 72 hours and cell death was measured using IN Cell analyzer (as described for Figure 1B). ML171 at 3 μM was added as indicated. Error bars represent standard deviations (SD) of three independent experiments (PI assays) or standard errors of the mean (SEM) of at least 120 cells from three different experiments (DHE assays). A two-tailed Student’s t-test was performed to determine statistical significance. *p<0.05, **p<0.001.
In summary, our data indicate that Hace1 deficient cells become addicted to Gln to adapt to the increased oxidative stress characteristic of these cells (13). In the absence of Gln, ROS levels are further enhanced, resulting in potentially toxic levels of oxidative stress and cell death. These results provide preliminary evidence that blocking Gln uptake or metabolism may represent a tractable therapeutic strategy for treatment of cancers in which Hace1 is inactivated.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by funds from the Canadian Institute of Health Research (MOP-123416) and British Columbia Cancer Foundation through generous donations from Team Finn and Ride to Conquer Cancer (to PHS), and from the National Institutes of Health (R01 CA157996) and Damon-Runyon Cancer Research Foundation (to RJD). NC was funded by the Canadian Institutes of Health Research, Frederick Banting and Charles Best Canada Graduate Scholarship Doctoral Award.
Footnotes
CONFLICTS OF INTEREST The authors have no conflicts of interest to declare.
REFERENCES
- 1.Anglesio MS, Evdokimova V, Melnyk N, Zhang L, Fernandez CV, Grundy PE, et al. Differential expression of a novel ankyrin containing E3 ubiquitin-protein ligase, Hace1, in sporadic Wilms’ tumor versus normal kidney. Human molecular genetics. 2004 Sep 15;13(18):2061–74. doi: 10.1093/hmg/ddh215. PubMed PMID: 15254018. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L, Anglesio MS, O’Sullivan M, Zhang F, Yang G, Sarao R, et al. The E3 ligase HACE1 is a critical chromosome 6q21 tumor suppressor involved in multiple cancers. Nature medicine. 2007 Sep;13(9):1060–9. doi: 10.1038/nm1621. PubMed PMID: 17694067. [DOI] [PubMed] [Google Scholar]
- 3.Hibi K, Sakata M, Sakuraba K, Shirahata A, Goto T, Mizukami H, et al. Aberrant methylation of the HACE1 gene is frequently detected in advanced colorectal cancer. Anticancer research. 2008 May-Jun;28(3A):1581–4. PubMed PMID: 18630515. [PubMed] [Google Scholar]
- 4.Thelander EF, Ichimura K, Corcoran M, Barbany G, Nordgren A, Heyman M, et al. Characterization of 6q deletions in mature B cell lymphomas and childhood acute lymphoblastic leukemia. Leukemia & lymphoma. 2008 Mar;49(3):477–87. doi: 10.1080/10428190701817282. PubMed PMID: 18297524. [DOI] [PubMed] [Google Scholar]
- 5.Sakata M, Kitamura YH, Sakuraba K, Goto T, Mizukami H, Saito M, et al. Methylation of HACE1 in gastric carcinoma. Anticancer research. 2009 Jun;29(6):2231–3. PubMed PMID: 19528486. [PubMed] [Google Scholar]
- 6.Huang Y, de Reynies A, de Leval L, Ghazi B, Martin-Garcia N, Travert M, et al. Gene expression profiling identifies emerging oncogenic pathways operating in extranodal NK/T-cell lymphoma, nasal type. Blood. 2010 Feb 11;115(6):1226–37. doi: 10.1182/blood-2009-05-221275. PubMed PMID: 19965620. Pubmed Central PMCID: 2826234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slade I, Stephens P, Douglas J, Barker K, Stebbings L, Abbaszadeh F, et al. Constitutional translocation breakpoint mapping by genome-wide paired-end sequencing identifies HACE1 as a putative Wilms tumour susceptibility gene. Journal of medical genetics. 2010 May;47(5):342–7. doi: 10.1136/jmg.2009.072983. PubMed PMID: 19948536. [DOI] [PubMed] [Google Scholar]
- 8.Diskin SJ, Capasso M, Schnepp RW, Cole KA, Attiyeh EF, Hou C, et al. Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nature genetics. 2012 Oct;44(10):1126–30. doi: 10.1038/ng.2387. PubMed PMID: 22941191. Pubmed Central PMCID: 3459292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torrino S, Visvikis O, Doye A, Boyer L, Stefani C, Munro P, et al. The E3 ubiquitinligase HACE1 catalyzes the ubiquitylation of active Rac1. Developmental cell. 2011 Nov 15;21(5):959–65. doi: 10.1016/j.devcel.2011.08.015. PubMed PMID: 22036506. [DOI] [PubMed] [Google Scholar]
- 10.Castillo-Lluva S, Tan CT, Daugaard M, Sorensen PH, Malliri A. The tumour suppressor HACE1 controls cell migration by regulating Rac1 degradation. Oncogene. 2013 Mar 28;32(13):1735–42. doi: 10.1038/onc.2012.189. PubMed PMID: 22614015. [DOI] [PubMed] [Google Scholar]
- 11.Cheng G, Diebold BA, Hughes Y, Lambeth JD. Nox1-dependent reactive oxygen generation is regulated by Rac1. The Journal of biological chemistry. 2006 Jun 30;281(26):17718–26. doi: 10.1074/jbc.M512751200. PubMed PMID: 16636067. [DOI] [PubMed] [Google Scholar]
- 12.Ueyama T, Geiszt M, Leto TL. Involvement of Rac1 in activation of multicomponent Nox1- and Nox3-based NADPH oxidases. Molecular and cellular biology. 2006 Mar;26(6):2160–74. doi: 10.1128/MCB.26.6.2160-2174.2006. PubMed PMID: 16507994. Pubmed Central PMCID: 1430270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daugaard M, Nitsch R, Razaghi B, McDonald L, Jarrar A, Torrino S, et al. Hace1 controls ROS generation of vertebrate Rac1-dependent NADPH oxidase complexes. Nature communications. 2013 Jul 17;4:2180. doi: 10.1038/ncomms3180. PubMed PMID: 23864022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rotblat B, Southwell AL, Ehrnhoefer DE, Skotte NH, Metzler M, Franciosi S, et al. HACE1 reduces oxidative stress and mutant Huntingtin toxicity by promoting the NRF2 response. Proceedings of the National Academy of Sciences of the United States of America. 2014 Feb 10; doi: 10.1073/pnas.1314421111. PubMed PMID: 24516159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer discovery. 2012 Oct;2(10):881–98. doi: 10.1158/2159-8290.CD-12-0345. PubMed PMID: 23009760. Pubmed Central PMCID: 3491070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeBerardinis RJ. Is cancer a disease of abnormal cellular metabolism? New angles on an old idea. Genetics in medicine : official journal of the American College of Medical Genetics. 2008 Nov;10(11):767–77. doi: 10.1097/GIM.0b013e31818b0d9b. PubMed PMID: 18941420. Pubmed Central PMCID: 2782690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008 Feb;18(1):54–61. doi: 10.1016/j.gde.2008.02.003. PubMed PMID: 18387799. Pubmed Central PMCID: 2476215. Epub 2008/04/05. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008 Jan;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. PubMed PMID: 18177721. Epub 2008/01/08. eng. [DOI] [PubMed] [Google Scholar]
- 19.Klimberg VS, McClellan JL, Claude H, Organ Honorary Lectureship. Glutamine, cancer, and its therapy. American journal of surgery. 1996 Nov;172(5):418–24. doi: 10.1016/s0002-9610(96)00217-6. PubMed PMID: 8942537. [DOI] [PubMed] [Google Scholar]
- 20.Rajagopalan KN, DeBerardinis RJ. Role of glutamine in cancer: therapeutic and imaging implications. J Nucl Med. 2011 Jul;52(7):1005–8. doi: 10.2967/jnumed.110.084244. PubMed PMID: 21680688. Epub 2011/06/18. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010 Aug;35(8):427–33. doi: 10.1016/j.tibs.2010.05.003. PubMed PMID: 20570523. Pubmed Central PMCID: 2917518. Epub 2010/06/24. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proceedings of the National Academy of Sciences of the United States of America. 2008 Dec 2;105(48):18782–7. doi: 10.1073/pnas.0810199105. PubMed PMID: 19033189. Pubmed Central PMCID: 2596212. Epub 2008/11/27. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009 Apr 9;458(7239):762–5. doi: 10.1038/nature07823. PubMed PMID: 19219026. Pubmed Central PMCID: 2729443. Epub 2009/02/17. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proceedings of the National Academy of Sciences of the United States of America. 2010 May 11;107(19):8788–93. doi: 10.1073/pnas.1003428107. PubMed PMID: 20421486. Pubmed Central PMCID: 2889315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reynolds MR, Lane AN, Robertson B, Kemp S, Liu Y, Hill BG, et al. Control of glutamine metabolism by the tumor suppressor Rb. Oncogene. 2013 Jan 28; doi: 10.1038/onc.2012.635. PubMed PMID: 23353822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicolay BN, Gameiro PA, Tschop K, Korenjak M, Heilmann AM, Asara JM, et al. Loss of RBF1 changes glutamine catabolism. Genes & development. 2013 Jan 15;27(2):182–96. doi: 10.1101/gad.206227.112. PubMed PMID: 23322302. Pubmed Central PMCID: 3566311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shanware NP, Mullen AR, DeBerardinis RJ, Abraham RT. Glutamine: pleiotropic roles in tumor growth and stress resistance. J Mol Med (Berl) 2011 Mar;89(3):229–36. doi: 10.1007/s00109-011-0731-9. PubMed PMID: 21301794. Epub 2011/02/09. eng. [DOI] [PubMed] [Google Scholar]
- 28.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2007 Dec 4;104(49):19345–50. doi: 10.1073/pnas.0709747104. PubMed PMID: 18032601. Pubmed Central PMCID: 2148292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010 Jan 21;29(3):313–24. doi: 10.1038/onc.2009.358. PubMed PMID: 19881548. Pubmed Central PMCID: 2809806. Epub 2009/11/03. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014 Jun 12;510(7504):298–302. doi: 10.1038/nature13236. PubMed PMID: 24805240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013 Apr 4;496(7443):101–5. doi: 10.1038/nature12040. PubMed PMID: 23535601. Pubmed Central PMCID: 3656466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wellen KE, Lu C, Mancuso A, Lemons JM, Ryczko M, Dennis JW, et al. The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev. 2010 Dec 15;24(24):2784–99. doi: 10.1101/gad.1985910. PubMed PMID: 21106670. Pubmed Central PMCID: 3003197. Epub 2010/11/26. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tong X, Zhao F, Thompson CB. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr Opin Genet Dev. 2009 Feb;19(1):32–7. doi: 10.1016/j.gde.2009.01.002. PubMed PMID: 19201187. Pubmed Central PMCID: 2707261. Epub 2009/02/10. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moldave K, Meister A. Synthesis of phenylacetylglutamine by human tissue. The Journal of biological chemistry. 1957 Nov;229(1):463–76. PubMed PMID: 13491597. [PubMed] [Google Scholar]
- 35.Li XN, Parikh S, Shu Q, Jung HL, Chow CW, Perlaky L, et al. Phenylbutyrate and phenylacetate induce differentiation and inhibit proliferation of human medulloblastoma cells. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004 Feb 1;10(3):1150–9. doi: 10.1158/1078-0432.ccr-0747-3. PubMed PMID: 14871995. [DOI] [PubMed] [Google Scholar]
- 36.Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009 Feb 6;136(3):521–34. doi: 10.1016/j.cell.2008.11.044. PubMed PMID: 19203585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gianni D, Taulet N, Zhang H, DerMardirossian C, Kister J, Martinez L, et al. A novel and specific NADPH oxidase-1 (Nox1) small-molecule inhibitor blocks the formation of functional invadopodia in human colon cancer cells. ACS chemical biology. 2010 Oct 15;5(10):981–93. doi: 10.1021/cb100219n. PubMed PMID: 20715845. Pubmed Central PMCID: 2955773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng T, Sudderth J, Yang C, Mullen AR, Jin ES, Mates JM, et al. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proceedings of the National Academy of Sciences of the United States of America. 2011 May 24;108(21):8674–9. doi: 10.1073/pnas.1016627108. PubMed PMID: 21555572. Pubmed Central PMCID: 3102381. Epub 2011/05/11. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandez CA, Des Rosiers C, Previs SF, David F, Brunengraber H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J Mass Spectrom. 1996 Mar;31(3):255–62. doi: 10.1002/(SICI)1096-9888(199603)31:3<255::AID-JMS290>3.0.CO;2-3. PubMed PMID: 8799277. Epub 1996/03/01. eng. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.