Abstract
Metastatic progression, including extravasation and micro-metastatic outgrowth, is the main cause of cancer patient death. Recent studies suggest that cancer cells reprogram their metabolism to support increased proliferation through increased glycolysis and biosynthetic activities, including lipogenesis pathways. However, metabolic changes during metastatic progression, including alterations in regulatory gene expression, remain undefined. We show that transforming growth factor beta 1 (TGFβ1) induced Epithelial-to-Mesenchymal Transition (EMT) is accompanied by coordinately reduced enzyme expression required to convert glucose into fatty acids, and concomitant enhanced respiration. Over-expressed Snail1, a transcription factor mediating TGFβ1-induced EMT, was sufficient to suppress carbohydrate-responsive-element-binding protein (ChREBP, a master lipogenic regulator), and fatty acid synthase (FASN), its effector lipogenic gene. Stable FASN knock-down was sufficient to induce EMT, stimulate migration and extravasation in vitro. FASN silencing enhanced lung metastasis and death in vivo. These data suggest that a metabolic transition that suppresses lipogenesis and favors energy production is an essential component of TGFβ1-induced EMT and metastasis.
Keywords: Transforming growth factor beta 1 - TGFβ1, Epithelial-to-Mesenchymal Transition, Metastasis, Fatty acid synthase, Lipogenesis
Introduction
Metastatic disease is the primary cause of death in many cancers, especially in patients with breast and non-small cell lung (NSCLC) cancers (1, 2). Tumor metastasis is a complex process, starting with primary tumor invasion through endothelial barriers by a process known as EMT, characterized by loss of cell-cell adhesion and increased cell motility (3–5).
Transforming growth factor beta1 (TGFβ1) production within the tumor microenvironment is frequently enhanced in breast and NSCLCs (6). Chronic TGFβ1 exposure stimulates cancer cell EMT, increasing cell motility and metastasis through its downstream signaling pathways (7), including induction of several transcriptional factors, like Snail1 (8, 9).
In general, cancer cells maintain a high level of glycolysis in the presence or absence of oxygen (i.e., the Warburg effect). Elevated glucose metabolism serves a variety of functions, including energy production, provision of macromolecular precursors, and establishment of an NADPH pool to enable cells to resist oxidative stress (10, 11). In addition to higher glycolytic rates, rapidly proliferating cancer cells commonly have substantial increases in de novo fatty acid synthesis for lipogenesis and membrane production (12). Several lipogenic enzymes are required for cancer cell growth, including ATP citrate lyase (ACLY), FASN and acetyl-CoA carboxylase (ACC) (13–17).
Although glucose metabolism has been intensively investigated in rapidly growing cancer cells, metabolic changes that occur during EMT are poorly understood. Here, we demonstrate that transcriptional regulators of lipogenesis, ChREBP and SREBP, are dramatically down-regulated in A549 adenocarcinoma NSCLC cells during TGFβ1-induced EMT, a mechanism driven by increased SNAIL1 expression. Accordingly, cancer cells undergoing EMT have increased respiration, accompanied by elevated oxygen consumption and corresponding increases in ATP content. Importantly, Snail1 plays a key role in regulating this metabolic reprogramming, since cells forced over-expression of Snail1 strongly suppressed ChREBP expression, a key lipogenic transcription factor. In turn, suppressed ChREBP levels reduced expression of FASN, an essential enzyme in de novo fatty acid synthesis, and enhanced EMT. Furthermore, stable FASN silencing by shRNA knockdown was sufficient to enhance EMT, accompanied by prototypic changes in expression of key functional mesenchymal marker genes, vimentin and E-cadherin, with stimulated cell migration in vitro. When examined in vivo, stable FASN knockdown cells showed enhanced metastatic potential and caused increased lethality due to metastatic spreading in NOD/SCID mice. Together, these data suggest that suppressing lipogenesis is an essential metabolic component of EMT, and a requirement for successful establishment of distant metastases by diverting energy needed for cell migration and metastases.
Results
Changes in metabolic enzyme expression during EMT
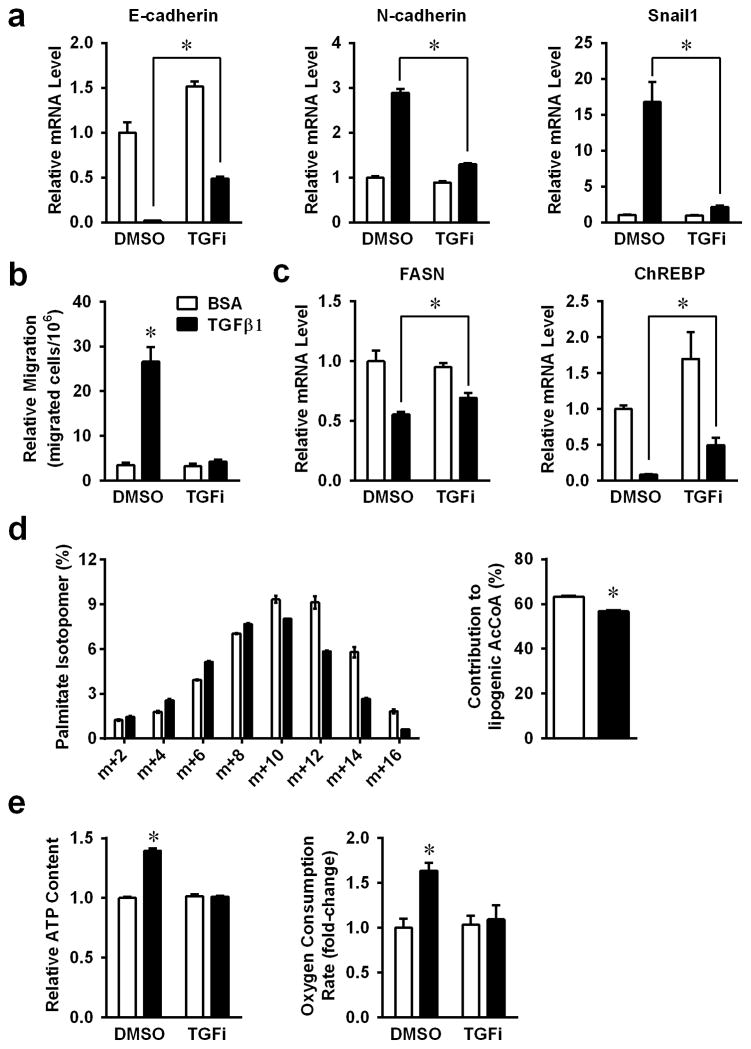
Significant EMT was noted in A459 non-small cell lung carcinoma cells after low dose exposure to TGFβ1, as previously described (18). RT-PCR analysis showed that E-cadherin, N-cadherin and Snail1 were regulated by TGFβ1 treatment, and all changes were blocked by addition of TGFβ1 type I kinase inhibitor (TGF inhibitor) (Figure 1a), strongly suggesting that the observed changes in EMT gene expression were specifically mediated by downstream TGFβ1 signaling. TGFβ1-induced EMT was further confirmed by increased cell motility in transwell assays (Figure 1b).
Figure 1. Changes of de novo lipogenesis during TGFβ1 induced EMT.
A549 cells were treated with 2ng/ml TGFβ1, with or without 400nM TGF inhibitor for 48h. (a) EMT functional proteins E-cadherin, N-cadherin and Snail1, were analyzed by real-time PCR (RT-PCR). (b) Transwell assays of A549 cells were treated with TGFβ1, with or without TGF inhibitor. (c) mRNA expression levels of FASN and ChREBP were analyzed by RT-PCR as in panels A. (d) Labeling of palmitate in A549, after culture in medium containing [U-13C] glucose for 24 hours, and calculated glucose contributed lipogenic acetyl-CoA. (e) Intra-cellular ATP levels and OCRs were measured in TGFβ1 treated cells. (*P<0.05 comparing to BCA control.) (Data are represented as mean ± SEM.)
Most cancer cells have high levels of glycolysis and fatty acid synthesis (10). However, in cancer cells undergoing an EMT, cells become more mobile and may alter their overall metabolism. Indeed, expression level of fatty acid synthase (FASN) was dramatically decreased upon TGFβ1-induced EMT (Figures 1c). Consistently, the fractional contribution of glucose to the fatty acid palmitate was reduced upon TGFβ1 treatment. This reduction was manifested in both the isotopomer distribution of palmitate and in the fraction of the lipogenic acetyl-CoA pool derived from [U-13C]glucose (Figures 1d). Several transcription factors known to control lipogenesis, including PPARα, PPARγ, SREBP1a and SREBP1c, were also down-regulated in A549 NSCLC cells in response to TGFβ1 (Supplementary Figure 1), with ChREBP demonstrating the most dramatic down-regulation (Figure 1c). All TGFβ1-induced metabolic changes were significantly suppressed by TGF inhibitor co-administration. Concomitantly, we noted dramatic increases in intracellular ATP content and oxygen consumption in TGFβ1-treated A549 NSCLC cells (Figures 1e). The down-regulation of lipogenesis was independent of TGFβ1 induced cell cycle arrest (Supplementary Figure 2). TGFβ1 regulated lipogenic gene expression changes were also tested in mouse mammary epithelial cells (NMuMG) (Supplementary Figure 3), in which we previously demonstrated dramatic EMT responses (19).
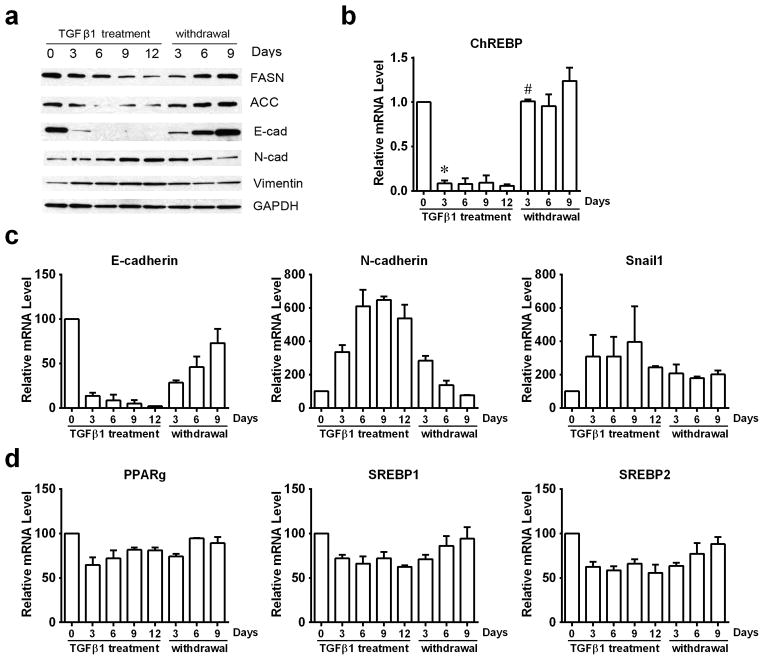
TGFβ1-induced EMT in A549 cells is a reversible process (20), whereby TGFβ1 withdrawal causes a mesenchymal-to-epithelial transition (MET). While decreases in E-cadherin, ACC, and FASN were noted during TGFβ1 treatment with concomitant increases in N-cadherin and Snail1 protein levels, these responses were quickly reversed upon TGFβ1 withdrawal (Figure 2a and 2c). Changes in FASN and ACC protein levels corresponded to dramatic decrease and rebounding of ChREBP mRNA levels after TGFβ1 exposure and withdrawal (Figure 2b), and mRNA levels of other lipogenic transcription factors also showed reversible expression (Figure 2d).
Figure 2. TGFβ1 induced reversible EMT responses in A549 cells.
A549 cells were treated with TGFβ1 (2 ng/ml) for the indicated days (marked TGFβ1 treatment). After 12 days, TGFβ1-treated A549 cells were washed with PBS and subsequently cultured in normal growth medium without TGFβ1 for 3, 6, or 9 days. (a) Protein levels of key EMT functional proteins and metabolic enzymes were analyzed by western-blotting. (b) mRNA levels of ChREBP were monitored by RT-PCR. (c) mRNA levels of EMT functional proteins (E-cadherin, N-cadherin, and Snail1) and (d) lipogenic transcriptional regulatory factors (PPAR-gamma, SREBP1, and SREBP2) were monitored by RT-PCR. (*P<0.05 comparing to day0. #P<0.05 comparing to day12 TGFβ1 treatment.) (Data are represented as mean ± SEM.)
Snail1-mediated metabolic regulation during EMT
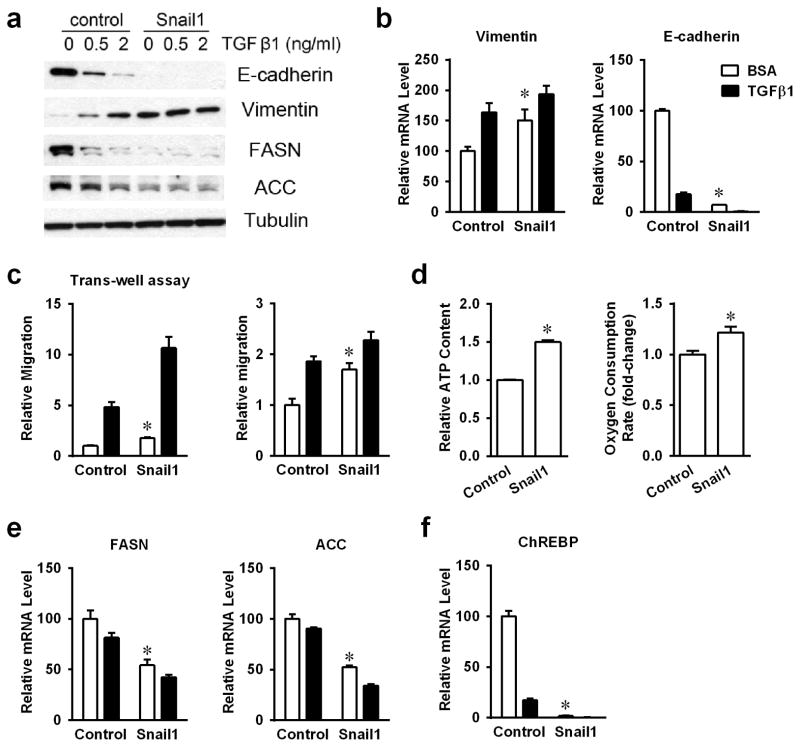
Snail1 is an important transcription factor that mediates the TGFβ1-induced EMT response (8, 9). To explore the role of Snail1 in metabolic regulation stimulated by TGFβ1 exposure, Snail 1 was over-expressed in A549 cells by infection with an adenoviral expression vector (Supplementary Figure 4). As reported previously, Snail1 over-expression resulted in classical features of mesenchymal cells, lower E-cadherin, with concomitantly higher vimentin expression (Figures 3a and 3b). These cells showed significantly enhanced mobility, shown by scratch and transwell assays, with or without TGFβ1 treatments (Figures 3c). Furthermore, Snail1 overexpression exhibited concomitant elevated intracellular ATP levels and oxygen consumption (Figures 3d). Thus, Snail1 over-expression stimulated EMT responses, and associated metabolic shifts in the absence of TGFβ1 exposure.
Figure 3. Snail1 involved in TGFβ1 regulated lipogenesis.
(a) Protein levels of E-cadherin, Vimentin, FASN and ACC were analyzed by western-blotting in stable Snail1 over-expressed A549 cells. (b) The mRNA expression levels of Vimentin and E-cadherin were analyzed by RT-PCR. (c) Transwell assay and scratch assay of control and Snail1 over-expressed A549 cells. (d) Intracellular ATP content and OCRs in Snail1 over-expressed cells with or without TGFβ1 treatment. (e) mRNA levels of lipogenic genes FASN and ACC were analyzed by RT-PCR. (f) mRNA levels of ChREBP were shown by RT-PCR. (*P<0.05 comparing to control A549 cells.) (Data are represented as mean ± SEM.)
Similar to TGFβ1-induced EMT changes, Snail1 over-expressed cells expressed significantly lowered mRNA and protein levels of FASN and ACC (Figures 3a and 3e), which were not significantly affected by TGFβ1 treatments. Furthermore, ChREBP mRNA levels were also dramatically decreased in Snail1 over-expressed cells (Figure 3f). Thus, over-expression of Snail1 closely mimicked metabolic regulation of lipogenic genes in TGFβ1-exposed A549 cells. As both FASN and ACC are known ChREBP target genes (21), the metabolic regulatory role of Snail1 might be mediated by transcriptional control of ChREBP on FASN and ACC.
FASN silencing enhances metastatic capacity
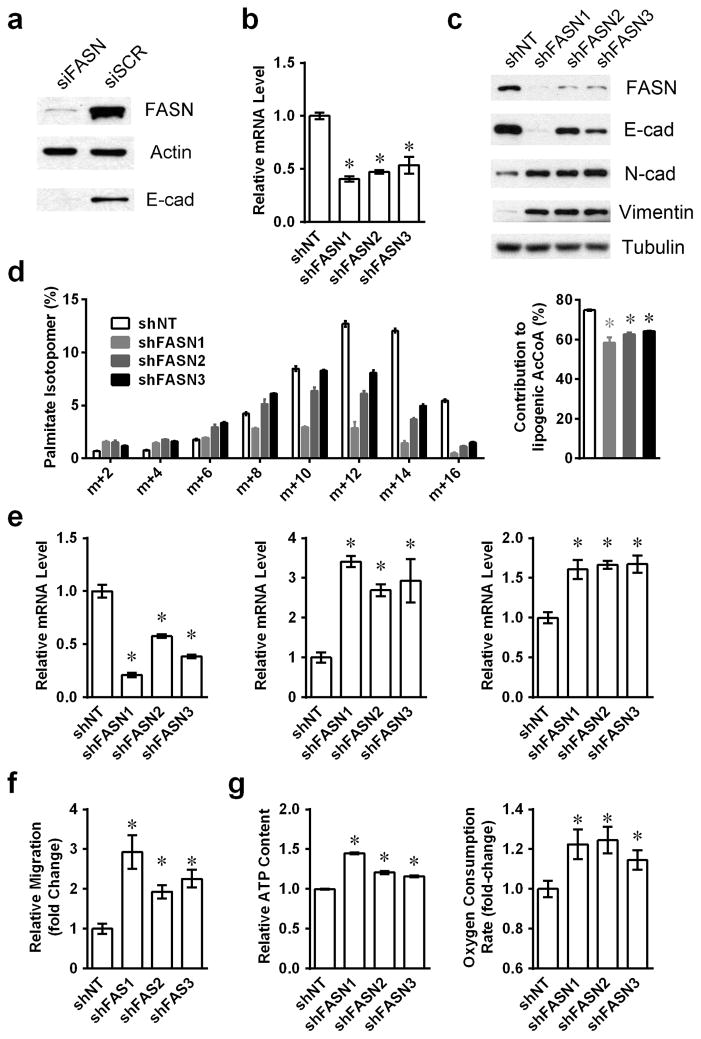
Fatty acid synthesis is required for rapidly proliferating cancer cells, and prior studies have shown that knock-down of FASN expression can impair cancer cell growth and overall survival (22). To examine the roles of diminished FASN expression on cell motility, we transiently knocked down FASN using siRNA transfection (siFASN) and noted significantly lower E-cadherin expression (Figure 4a), which suggested an EMT and possibly increased mobility and metastatic capacity.
Figure 4. Fatty acid synthase knockdown mimic TGFβ1 induced EMT.
(a) Whole A549cell lysates were harvested 48h after siRNA transfection, and protein levels of FASN and E-cadherin were shown by western-blotting. (b) mRNA levels of FASN were monitored by RT-PCR in three FASN knockdown cell populations. (c) Protein levels of FASN and EMT functional proteins were shown by western-blotting in FASN knockdown cell lines. (d) Labeling of palmitate in control or FASN-silenced cell lines cultured in medium containing [U-13C] glucose for 24 hours, and calculated glucose contributed lipogenic acetyl-CoA. (e) mRNA levels of EMT functional proteins E-cadherin, N-cadherin and Vimentin were analyzed by RT-PCR. (f) Cell mobility was measured by transwell migration assays. (g) Intracellular ATP content and ORCs were monitored for stable shFASN knockdown and shNT A549 cells. (*P<0.05 comparing to shNT A549 cells.) (Data are represented as mean ± SEM.)
To further explore the functions of FASN expression on EMT and metastasis, stable FASN knockdown A549 cells were generated by lentiviral-mediated shRNA infection using three separate FASN targeting oligomers. A pooled population of shFASN cells demonstrated enhanced mobility in transwell assays, which was further increased by TGFβ1 treatment (Supplementary Figure 5). Three stable shFASN knockdown cell clones were separately isolated and each showed over fifty-percent decreased FASN mRNA expression levels (Figure 4b), similar to TGFβ1-exposed cells, with significantly suppressed FASN protein levels (Figure 4c). As expected, FASN silencing resulted in a reduced fractional contribution of glucose carbon to fatty acid synthesis (Fig. 4d). Compared to non-target controls, shFASN knockdown clones expressed significantly lower E-cadherin levels, with concomitantly higher N-cadherin and vimentin expression (Figure 4e). All shFASN clones showed dramatically elevated mobility in transwell assays (Figure 4f).
As observed with TGFβ1-exposed cells, shFASN clones exhibited significant gradient increases in intracellular ATP pools correlating with the extent of FASN level knockdown. Consistently, FASN knockdown clones also had higher oxygen consumption rates (Figure 4g).
shFASN knockdown enhances A549 NSCLC cell lung colonization and lethality in NOD/SCID mice in vivo
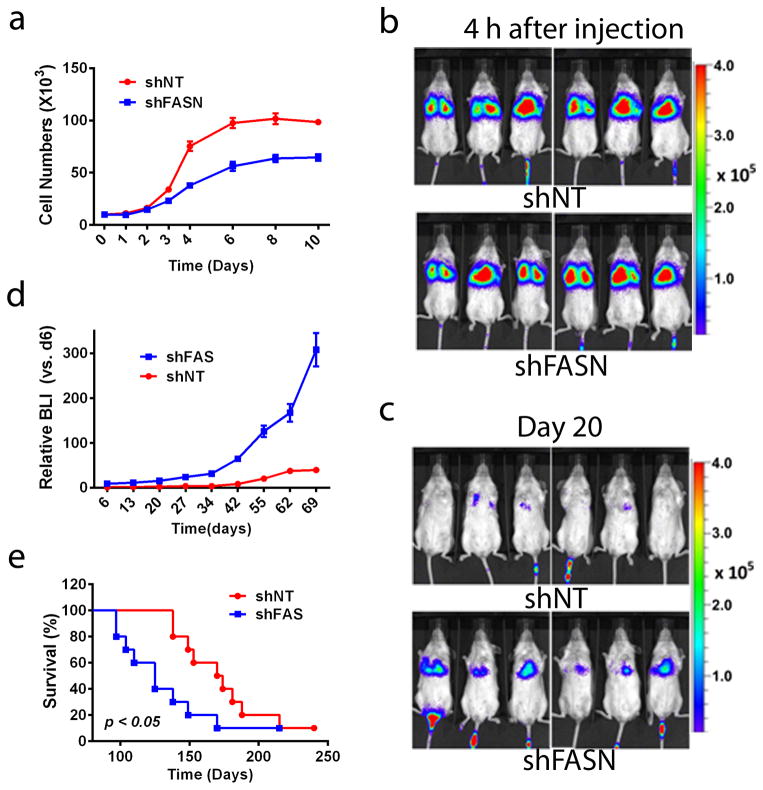
Next, we investigated the role of FASN loss in metastasis and lethality in NOD/SCID mice in vivo. Stable shNT and shFASN1 cells, which has slower growth rates and showed enhanced mobility in vitro (Figure 5a), were injected into the tail veins of female NOD/SCID mice. Initially, the deposition of cells into the lungs within was not different between the two cell injections as measured 4 h post-injection (Figure 5b). While many of the initially deposited NSCLC cells eventually died out in both conditions, later (>20 days) bioluminescent (BLI) images of mice revealed enhanced metastatic tumor colonization derived from luciferase-tagged shFASN cells (Figure 5c), with subsequent increased tumor volumes over time (Figure 5d). Mice injected with shFASN cells also displayed significantly decreased overall survival than animals injected with shNT A549 cells (Figure 5e). Autopsies of animals with initial lung cancers derived from shNT or shFASN only showed lung cancer colonization within the first 200 days. In contrast, mice that eventually died from injected shFASN cells not only demonstrated larger tumor volumes increased involvement of the overall lung, and decreased survival, but also dramatically increased metastatic spread to various other tissues, including adjacent lymph nodes, colon, liver and thymus (Table 1). In contrast, NOD/SCID mice that died from control or shNT A549 cells showed cancer involvement only in their lungs, with no effective spreading to adjacent lymph nodes or other tissues at the time of euthanasia due to large tumor volumes (Supplementary Figure 6).
Figure 5. Stable shFASN knockdown increases metastasis in A549 NSCLC cells.
(a) Growth curve of A549 shFASN knockdown cells in vitro. (b) Stable knockdown shNT (top) or shFASN (bottom) A549 cells (1×106) were injected into the tail veins of anesthetized NOD/SCID female mice, and cell deposition into the lungs were analyzed 4h post-injection. Note that identical viable cell numbers (measured by BLI) deposited into the lungs. (c) Representative images of mice injected with stable shNT or shFASN A549 cells as in ‘A’, but analyzed 20 days later. (d) Relative tumor volumes were measured by BLI intensities at the indicated times. (e) Kaplan-Meier survival for mice bearing stable shNT vs shFASN knockdown A549 lung xenografts. (Data are represented as mean ± SEM.)
Table 1.
shFASN knockdown leads to enhanced metastatic spread weeks after lung colonization.
| #Animals/Condition of 20 total1 | ||
|---|---|---|
| Distal organ affacted | shNT | shFASN |
| a. Gastrointestinal | 0 | 1 |
| b. Liver | 0 | 2 |
| c. Bone marrow | 0 | 0 |
| d. Kidney | 0 | 1 |
| e. Spleen | 0 | 0 |
| f. Heart | 0 | 0 |
| g. Thymus | 0 | 10 |
| h. Brain | 0 | 0 |
| Total affected mice: | 0/20 | 14/202 |
Experiments were performed three times. Two experiments were performed with n=5 per group, and a third experiment was performed with n=10. Data in this table summarizes results of all 20 animals treated similarly with no one experiment having statistically more metastatic spread than the others. The presence of tumor tissue was confirmed by H&E staining.
p≤0.001
Discussion
Although cancer cells have higher glycolytic rates than normal cells, mitochondria of cancer cells are usually fully functional (23, 24). During TGFβ1-induced EMT, increased oxygen consumption strongly suggests that mitochondrial oxidative phosphorylation is stimulated, to generate ATP. Recent study showed that reduced migration is associated with reduced OXPHOS and ATP levels in viable cancer cells (25).
Increased de novo fatty acid synthesis is a general metabolic feature of cancer cells (26). Prior studies suggested that enzymes in this pathway might be potential targets for cancer therapy, as genetic knockdown or chemical inhibition of these enzymes decreased cancer cell proliferation in vitro and subcutaneous xenograft tumor growth in vivo (13–17). In response to TGFβ1, we discovered that ACC and FASN were coordinately decreased in A549 NSCLC cells, through a Snail1-mediated transcriptional regulatory network. Our data strongly suggested that Snail controls metabolic reprogramming of cancer cells in response to TGFβ1 by regulating several key metabolic transcriptional factors, including SREBP and ChREBP (21, 27). Here, we discovered that ChREBP was dramatically down-regulated by TGFβ1 in A549 cells, and forced overexpression of Snail1 mimicked this response. From promoter analyses, we identified three potential Snail1 binding sites (CACCTG at −130, −682, and −1295 bp from the transcription start site) in the human ChREBP promoter that were identical to those in the human E-cadherin promoter, suggesting that Snail1 could directly bind to the ChREBP promoter and negatively regulate its expression, which will be further investigated by ChIP assay.
TGFβ1 exposure stimulates cell migration, a process requiring a re-balancing of energy from fatty acid synthesis to EMT promotion. Since Snail1 forced overexpression simulated the same responses, this essential transcription factor appears important for both the morphological as well as the metabolic reprogramming that occurs during EMT. Indeed, methods to target Snail would be a potentially important means to inhibiting cancer progression at both the levels of metabolic and morphologic re-programming.
Our observation that decreased fatty acid synthesis regulates and ultimately stimulates cell migration and metastases raises considerable caution in targeting FASN or lipogenesis to prevent cancer. FASN knockdown A549 cells grew slower than control shNT A549 cells, consistent with prior results (22), where the authors concluded that targeting these pathways could be an effective anticancer strategy. However, our results indicate that cells with decreased FASN also showed enhanced metastatic potential, similar to TGFβ1-exposed cells. Thus, while targeting FASN or lipogenesis in general, may suppress growth of primary tumors, a major consequence is enhanced metastasis in NOD/SCID mice injected with shFASN A549 cells to mimic extravasation and cancer progression. One possibility is that loss of FASN expression increases the availability of substrates to supply oxidative phosphorylation. The resulting shift from anabolism to energy production may help support migration and ultimately metastasis (28).
These results strongly suggest TGFβ1, elevated in many cancer microenvironments (29) represents a new paradigm in cancer metabolic reprogramming, central for EMT-derived cell migration and metastases. TGFβ1 exposure shifts metabolism from fatty acids synthesis to enhanced oxidative phosphorylation, which generates sufficient ATP needed for cell migration and metastasis. The data suggest that targeting FASN or other lipogenic enzymes in cancer, while potentially eliciting temporary growth inhibition, might have the untoward consequence of increasing long-term risk of metastasis.
Materials and methods
Reagents
Recombinant Human TGFβ1 was purchased from R&D systems (Minneapolis, MN). The TGFβ1 type I kinase inhibitor (TGF inhibitor, cat. # HTS-466284) was purchased from Calbiochem (Hessen, Darmstadt).
Cell lines
A549 NSCLC cells were purchased from the American Type Culture Collection (ATCC). Normal murine epithelial mammary gland (NMuMG) cells were kindly provided by Dr. Gray Pearson (UT Southwestern) and grown and used for EMT analyses as described(19). All cells were MAP tested and were free of mycoplasma contamination.
Stable Snail1 over-expressing A549 cells were generated using retroviral-mediated pBABE-Snail1 expression vectors kindly provided by Dr. Sendurai A. Mani (University of Texas, M.D. Anderson Cancer Center, Houston, TX). A pooled cell population was used for experiments after puromycin selection. Stable A549 non-targeted (shNT) and FASN knockdown (shFASN) cells were generated by lentiviral-mediated expression of three separate targeting shRNAs for the FASN gene, and stable clones isolated. The shFASN1 clone was selected for the in vivo study.
Palmitate de novo synthesis assay using [U-13C]glucose
In brief, cells were cultured in medium containing [U-13C]glucose for 24 hours. The cells were harvested with 0.4ml 0.1% Triton-X 100, followed by methanol:chloroform extraction. The chloroform phase was dried down with nitrogen gas resuspended with a 2ml methanol:toluene (4:1 V/V) mixture containing 0.01% butylated hydroxytoluene and 2μl acetyl chloride, and heated to 100°C for 1 hour to generate methyl esters of palmitate and other fatty acids. After cooling down the sample, 5 ml of 6% K2CO3 was added. The samples were centrifuged to separate the phases, and the upper toluene phase was transferred to a GC/MS vial. Metabolites were analyzed using an Agilent 6970 gas chromatograph networked to an Agilent 5973 mass selective detector. Retention times and mass fragmentation signatures of methylpalmitate were validated using a pure standard. The mass isotopomer distribution analysis measured the fraction of each metabolite pool that contained every possible number of 13C atoms; that is, a metabolite could contain 0, 1, 2, … n 13C atoms, where n = the number of carbons in the metabolite. For each metabolite, an informative fragment ion containing all carbons in the parent molecule was analyzed using MSDChem software (Agilent), integrating the abundance of all mass isotopomers from m+0 to m+n, where m = the mass of the fragment ion without any 13C. For palmitate, m=270, n=16. The abundance of each mass isotopomer was then corrected mathematically to account for natural abundance isotopes and finally converted into a percentage of the total pool. Glucose contributed lipogenic acetyl-CoA was calculated from the enrichment data by regression mathematical model.
Oxygen consumption rate (OCR) analyses
OCRs were measured using the XF24 Analyzer (Seahorse Bioscience) as described(30). OCRs were normalized by cell number and data presented as relative X-fold changes vs control cells.
Intracellular ATP measurements
Intracellular ATP levels were measured using Cell Titer-Glo® Luminescent Cell Viability Assays (Promega). ATP concentrations were normalized by cell number and data expressed as relative X-fold changes vs. untreated control cells.
Transwell migration assays
Log-phase Snail1 overexpressing, shFASN knockdown, or appropriate control (vector alone or shNT) A549 cells (2 × 104) were pretreated with TGFβ1 for 24 h, with or without TGF inhibitor (400 nM, 1h) and seeded into matrigel pre-coated trans-well chambers in 0.1% FBS DMEM. Cells migrating through membranes and attaching to the bottom of transwell plates were counted after 48 h using BLI.
Wound healing assay in vitro
Stable vector control and Snail1 overexpression A549 cells were treated with TGFβ1, with or without TGF inhibitor (added 1 h prior to TGFβ1) and assessed for wound healing as described(19). Relative X-fold changes compared to untreated or vector alone control cells were measured.
Cell cycle analyses
A549 cells were treated with or without TGFβ1 for 48 h. Trypsinized cells were stained with PI, and analyzed for cell cycle distribution by flow cytometry as described (31).
Ionizing Radiation
A549 cells were exposed to 2.5, 5.0 or 7.5 Gy of ionizing radiation (IR), cultured for 24 h under normal growth conditions, and then harvested for Western blot analyses of cell cycle markers and metabolic enzymes.
Metastatic analyses
Female NOD/SCID mice (10 animals/group, 6–8 weeks old, 18–20 grams) were tail-vein injected with log-phase luciferase-labeled shFASN or shNT A549 cells (1 × 106) and imaged 4h (to assess initial cells lodged into lungs) and at various times (days) post-injection. BLI measurements, animal weights, and survival were measured over time. Mice were sacrificed when weights decreased 10% due to tumor formation. Sacrificed mice were assessed for metastatic spread to lymph nodes, and other normal tissues by BLI signals ex vivo, and confirmed for tumor tissue using H & E staining as described (32).
Immunohistochemistry (IHC)
Normal (n=2/group) tissues were processed, parafin embedded, sectioned (3 um) and stained with H&E to confirm tumor versus normal tissue as described (19).
Immunoblot analyses
Protein extraction and immunoblot analyses were performed as described(33). Primary antibodies used in this study were FASN, ACC, E-cadherin, Vimentin, Snail1 (Cell Signaling Technology, Beverley, CA), N-cadherin, p53, p21 (Santa Cruz Biotechnology, Dallas, TX), a-Tubulin, Actin (Sigma-Aldrich, St. Louis, MO), and GAPDH (Calbiochem).
Statistical analyses
All experiments were performed at least two times in duplicate, with many experiments performed three times in triplicate. Data in vitro were analyzed by two-tailed student’s T tests. Tumor volumes, monitored by BLI, and survival (Kaplan-Meier) curves were statistically analyzed by ANOVA analyses. Data are represented as mean ± SEM.
Supplementary Material
Acknowledgments
We are grateful to Dr. Xiuquan Luo for their aid in the BLI measurements and animal injections. This work was supported by DOE/NASA grant DE-FG-022179-18-21 and by NIH R01 CA139217 to D.A.B, by NIH RO1 CA157996 and CPRIT RP130272 to R.J.D. We are grateful to the imaging core of the Simmons Cancer Center Support Grant (5P30 CA142543-03).
References
- 1.Semenza GL. Molecular mechanisms mediating metastasis of hypoxic breast cancer cells. Trends in molecular medicine. 2012 Sep;18(9):534–43. doi: 10.1016/j.molmed.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pallis AG, Syrigos K. Targeted (and chemotherapeutic) agents as maintenance treatment in patients with metastatic non-small-cell lung cancer: current status and future challenges. Cancer treatment reviews. 2012 Nov;38(7):861–7. doi: 10.1016/j.ctrv.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005 Aug 29;24(37):5764–74. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 4.Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. The Journal of clinical investigation. 2009 Jun;119(6):1438–49. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang L, Moses HL. Transforming growth factor beta: tumor suppressor or promoter? Are host immune cells the answer? Cancer research. 2008 Nov 15;68(22):9107–11. doi: 10.1158/0008-5472.CAN-08-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim WS, Park C, Jung YS, Kim HS, Han J, Park CH, et al. Reduced transforming growth factor-beta type II receptor (TGF-beta RII) expression in adenocarcinoma of the lung. Anticancer research. 1999 Jan-Feb;19(1A):301–6. [PubMed] [Google Scholar]
- 7.Elliott RL, Blobe GC. Role of transforming growth factor Beta in human cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005 Mar 20;23(9):2078–93. doi: 10.1200/JCO.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 8.Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nature cell biology. 2000 Feb;2(2):76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 9.Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nature cell biology. 2000 Feb;2(2):84–9. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 10.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009 May 22;324(5930):1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell metabolism. 2008 Jan;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Shao W, Espenshade PJ. Expanding roles for SREBP in metabolism. Cell metabolism. 2012 Oct 3;16(4):414–9. doi: 10.1016/j.cmet.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanai J, Doro N, Sasaki AT, Kobayashi S, Cantley LC, Seth P, et al. Inhibition of lung cancer growth: ATP citrate lyase knockdown and statin treatment leads to dual blockade of mitogen-activated protein kinase (MAPK) and phosphatidylinositol-3-kinase (PI3K)/AKT pathways. Journal of cellular physiology. 2012 Apr;227(4):1709–20. doi: 10.1002/jcp.22895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer cell. 2005 Oct;8(4):311–21. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005 Sep 15;24(41):6314–22. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- 16.Brusselmans K, De Schrijver E, Verhoeven G, Swinnen JV. RNA interference-mediated silencing of the acetyl-CoA-carboxylase-alpha gene induces growth inhibition and apoptosis of prostate cancer cells. Cancer research. 2005 Aug 1;65(15):6719–25. doi: 10.1158/0008-5472.CAN-05-0571. [DOI] [PubMed] [Google Scholar]
- 17.Lupu R, Menendez JA. Pharmacological inhibitors of Fatty Acid Synthase (FASN)--catalyzed endogenous fatty acid biogenesis: a new family of anti-cancer agents? Current pharmaceutical biotechnology. 2006 Dec;7(6):483–93. doi: 10.2174/138920106779116928. [DOI] [PubMed] [Google Scholar]
- 18.Kim JH, Jang YS, Eom KS, Hwang YI, Kang HR, Jang SH, et al. Transforming growth factor beta1 induces epithelial-to-mesenchymal transition of A549 cells. Journal of Korean medical science. 2007 Oct;22(5):898–904. doi: 10.3346/jkms.2007.22.5.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Araki S, Eitel JA, Batuello CN, Bijangi-Vishehsaraei K, Xie XJ, Danielpour D, et al. TGF-beta1-induced expression of human Mdm2 correlates with late-stage metastatic breast cancer. The Journal of clinical investigation. 2010 Jan;120(1):290–302. doi: 10.1172/JCI39194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X, Sun H, Qi J, Wang L, He S, Liu J, et al. Sequential introduction of reprogramming factors reveals a time-sensitive requirement for individual factors and a sequential EMT-MET mechanism for optimal reprogramming. Nature cell biology. 2013 May 26; doi: 10.1038/ncb2765. [DOI] [PubMed] [Google Scholar]
- 21.Uyeda K, Yamashita H, Kawaguchi T. Carbohydrate responsive element-binding protein (ChREBP): a key regulator of glucose metabolism and fat storage. Biochemical pharmacology. 2002 Jun 15;63(12):2075–80. doi: 10.1016/s0006-2952(02)01012-2. [DOI] [PubMed] [Google Scholar]
- 22.De Schrijver E, Brusselmans K, Heyns W, Verhoeven G, Swinnen JV. RNA interference-mediated silencing of the fatty acid synthase gene attenuates growth and induces morphological changes and apoptosis of LNCaP prostate cancer cells. Cancer research. 2003 Jul 1;63(13):3799–804. [PubMed] [Google Scholar]
- 23.Ferreira LM, Hebrant A, Dumont JE. Metabolic reprogramming of the tumor. Oncogene. 2012 Sep 6;31(36):3999–4011. doi: 10.1038/onc.2011.576. [DOI] [PubMed] [Google Scholar]
- 24.Metallo CM, Vander Heiden MG. Understanding metabolic regulation and its influence on cell physiology. Molecular cell. 2013 Feb 7;49(3):388–98. doi: 10.1016/j.molcel.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou H, Zhang B, Zheng J, Yu M, Zhou T, Zhao K, et al. The inhibition of migration and invasion of cancer cells by graphene via the impairment of mitochondrial respiration. Biomaterials. 2014 Feb;35(5):1597–607. doi: 10.1016/j.biomaterials.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 26.Yuan HX, Xiong Y, Guan KL. Nutrient sensing, metabolism, and cell growth control. Molecular cell. 2013 Feb 7;49(3):379–87. doi: 10.1016/j.molcel.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004 Nov;86(11):839–48. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 28.Thompson CB. Metabolic enzymes as oncogenes or tumor suppressors. The New England journal of medicine. 2009 Feb 19;360(8):813–5. doi: 10.1056/NEJMe0810213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikushima H, Miyazono K. TGFbeta signalling: a complex web in cancer progression. Nature reviews Cancer. 2010 Jun;10(6):415–24. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- 30.Huang X, Dong Y, Bey EA, Kilgore JA, Bair JS, Li LS, et al. An NQO1 substrate with potent antitumor activity that selectively kills by PARP1-induced programmed necrosis. Cancer research. 2012 Jun 15;72(12):3038–47. doi: 10.1158/0008-5472.CAN-11-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goetz EM, Shankar B, Zou Y, Morales JC, Luo X, Araki S, et al. ATM-dependent IGF-1 induction regulates secretory clusterin expression after DNA damage and in genetic instability. Oncogene. 2011 Sep 1;30(35):3745–54. doi: 10.1038/onc.2011.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klokov D, Leskov K, Araki S, Zou Y, Goetz EM, Luo X, et al. Low dose IR-induced IGF-1-sCLU expression: a p53-repressed expression cascade that interferes with TGFbeta1 signaling to confer a pro-survival bystander effect. Oncogene. 2013 Jan 24;32(4):479–90. doi: 10.1038/onc.2012.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurosu H, Choi M, Ogawa Y, Dickson AS, Goetz R, Eliseenkova AV, et al. Tissue-specific expression of betaKlotho and fibroblast growth factor (FGF) receptor isoforms determines metabolic activity of FGF19 and FGF21. The Journal of biological chemistry. 2007 Sep 14;282(37):26687–95. doi: 10.1074/jbc.M704165200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.