Abstract
Phosphatidylinositol 3-kinase (PI3K) promotes cancer cell survival, migration, growth, and proliferation by generating phosphatidylinositol 3,4,5-trisphosphate (PIP3) in the inner leaflet of the plasma membrane. PIP3 recruits pleckstrin homology (PH) domain-containing proteins to the membrane to activate oncogenic signaling cascades. Anti-cancer therapeutics targeting the PI3K/AKT/mTOR pathway are in clinical development. In a mass spectrometric screen to identify PIP3-regulated proteins in breast cancer cells, levels of the Rac activator PIP3-dependent Rac exchange factor 1 (P-REX1) increased in response to PI3K inhibition, and decreased upon loss of the PI3K antagonist PTEN. P-REX1 mRNA and protein levels were positively correlated with ER expression, and inversely correlated with PI3K pathway activation in breast tumors as assessed by gene expression and phosphoproteomic analyses. P-REX1 increased activation of Rac1, PI3K/AKT, and MEK/ERK signaling in a PTEN-independent manner, and promoted cell and tumor viability. Loss of P-REX1 or inhibition of Rac suppressed PI3K/AKT and MEK/ERK, and decreased viability. P-REX1 also promoted insulin-like growth factor-1 receptor (IGF-1R) activation, suggesting that P-REX1 provides positive feedback to activators upstream of PI3K. In support of a model where PIP3-driven P-REX1 promotes both PI3K/AKT and MEK/ERK signaling, high levels of P-REX1 mRNA (but not phospho-AKT or a transcriptomic signature of PI3K activation) were predictive of sensitivity to PI3K inhibitors among breast cancer cell lines. P-REX1 expression was highest in ER+ breast tumors compared to many other cancer subtypes, suggesting that neutralizing the P-REX1/Rac axis may provide a novel therapeutic approach to selectively abrogate oncogenic signaling in breast cancer cells.
Keywords: P-REX1, PI3K, MEK, IGF-1R
Introduction
The phosphatidylinositol 3-kinase (PI3K)/AKT/mTOR pathway is the most commonly aberrantly activated pathway in human cancer, promoting cell growth, migration, survival, and proliferation. Genes encoding proteins in this pathway are mutated in >70% of breast cancers, and pathway activation has been linked with resistance to anti-estrogen and anti-HER2 therapies in estrogen receptor α (ER)-positive and HER2-positive breast cancers, respectively [1]. PI3K pathway-targeted therapeutics are in clinical development. Class IA PI3Ks are activated by receptor tyrosine kinases (RTKs) such as members of the epidermal growth factor receptor family (EGFR, HER3) and the insulin receptor family (InsR, IGF-1R), and by Gβγ subunits released from activated G-protein coupled receptors (GPCRs) [2]. PI3K phosphorylates phosphatidylinositol (4,5)-bisphosphate (PIP2) in the inner leaflet of the plasma membrane to yield phosphatidylinositol (3,4,5)-trisphosphate (PIP3) [3]. Lipid phosphatase activity of the tumor suppressor PTEN antagonizes PI3K by dephosphorylating PIP3 [4]. PIP3 recruits AKT and other pleckstrin homology (PH) domain-containing proteins to the membrane to promote their interaction and initiate signaling cascades.
In a proteomic screen to identify PIP3-regulated proteins as effectors and markers of PI3K activation in breast cancer cells, we found that levels of PIP3-dependent Rac exchange factor 1 (P-REX1) were inversely correlated with PI3K activation. This association was confirmed in human breast tumors. P-REX1 was most highly expressed in ER+ breast cancer compared with various other cancer subtypes, and P-REX1 mRNA levels were predictive of sensitivity to PI3K inhibition. We also found that P-REX1 activates IGF-1R/InsR, Rac1, PI3K/AKT, and MEK/ERK signaling, and promotes cell and tumor viability. These findings place P-REX1 both downstream and upstream of PI3K, suggesting that P-REX1 is a key component of a positive feedback loop that drives oncogenic signaling.
Results
PI3K activity regulates P-REX1 expression
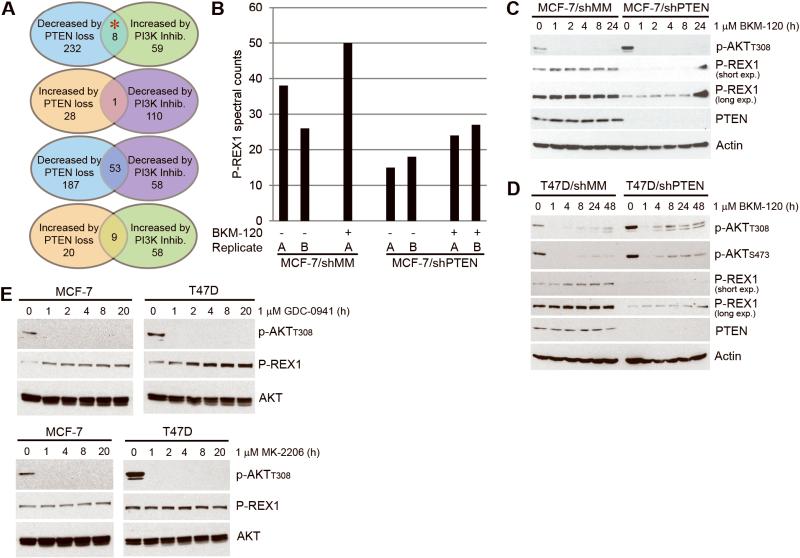
We performed a mass spectrometry (MS)-based proteomic screen to identify proteins modulated by PI3K pathway activity in breast cancer cells. MCF-7 cells stably expressing shRNA targeting PTEN or mismatch control (shMM) were treated ± the pan-PI3K inhibitor BKM-120 [5] for 16 h. Lysates were analyzed by multi-dimensional protein identification technology (MudPIT) coupled with tandem MS/MS. Proteins exhibiting altered abundance in response to PI3K inhibition and/or PTEN knockdown are listed in Table S1. Comparing the lists of proteins altered ≥1.5 fold in abundance, we identified P-REX1 as a protein downregulated by PTEN knockdown (which activates PI3K), and upregulated by PI3K inhibition (Fig. 1A-B). P-REX1 is a guanine exchange factor (GEF) that integrates inputs from GPCRs via Gβγ, and from RTKs and GPCRs via PIP3 to activate the Rho family GTPases Rac1/2/3 [6]. Given that P-REX1 was previously implicated in prostate cancer metastasis [7] and shown to promote oncogenic signaling [8, 9], we further investigated its role in PI3K signaling in breast cancer.
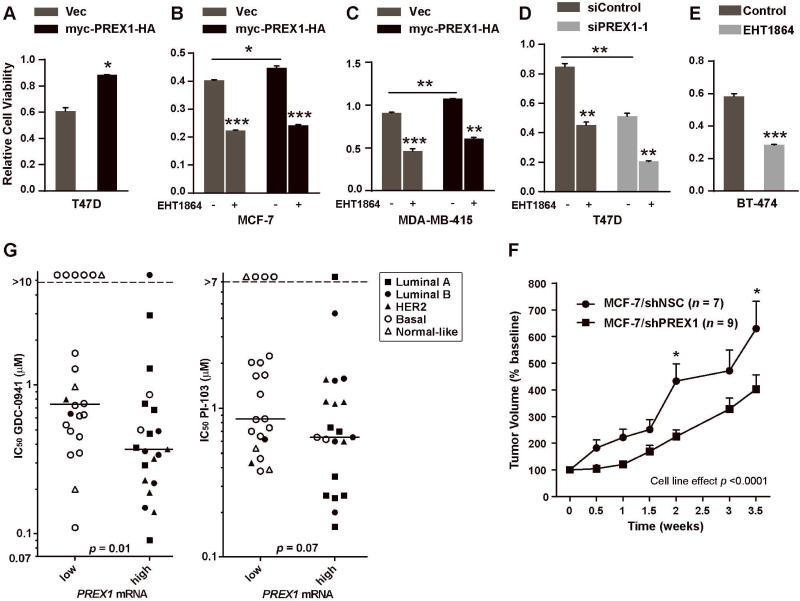
Figure 1. P-REX1 levels decrease upon PTEN loss and increase with PI3K inhibition.
A) Summary of results from mass spectrometry-based proteomic screen in MCF-7 cells (red asterisk indicates P-REX1). B) P-REX1 spectral counts from mass spectrometry shown for each of duplicate runs (run B for MCF-7/shMM + BKM-120 sample failed and was excluded from analysis). C-E) Immunoblots of lysates from cells treated with inhibitors were probed with the indicated antibodies.
We validated MS results by immunoblotting of lysates from breast cancer cells treated with inhibitors of PI3K or AKT. MCF-7/shMM and T47D/shMM cells treated with 1 μM BKM-120 showed reduced p-AKT and increased P-REX1 protein levels over time (Fig. 1C-D). The reason that P-REX1 levels did not increase as markedly in shPTEN cells compared to shMM cells upon PI3K inhibition is unclear but may be due in part to residual PIP3 in shPTEN cells, as evidenced by residual p-AKT in BKM-120-treated T47D/shPTEN cells (Fig. 1D). Since the P-REX1 homologue P-REX2a was found to bind PTEN [10], we considered that PTEN may bind and stabilize P-REX1. However, we detected only very low levels of P-REX1/PTEN interaction in cells (data not shown). Treatment with the pan-PI3K inhibitor GDC-0941 [11] similarly increased P-REX1 levels in parental MCF-7 and T47D cells, while AKT inhibition with MK-2206 did not (Fig. 1E). Thus, P-REX1 levels are modulated downstream of PI3K, but independently of AKT.
We also explored whether PI3K activity modulates P-REX1 gene expression. PI3K inhibition for 4 or 24 h increased P-REX1 mRNA levels in MCF-7 cells (Fig. S1A-B). Mining of prior gene expression microarray data from isogenic MCF-7/shPTEN, T47D/shPTEN, and MDA-MB-361/shPTEN cells compared to respective shMM controls [12] revealed that P-REX1 mRNA levels were significantly downregulated in all shPTEN lines. In addition, PI3K inhibition increased P-REX1 protein levels in cells pretreated with the translation inhibitor cycloheximide (Fig. S1C). These data suggest that P-REX1 levels are modulated by PI3K signaling at the transcriptional and post-translational levels: PIP3-mediated activation of P-REX1 may induce P-REX1 protein turnover as a form of negative feedback, and PI3K inhibition blocks negative feedback to induce transcription of genes encoding proteins that activate and transduce PI3K signaling [e.g., ERBB3, IGF1R [13], TSC1 [14], and PREX1].
Tumor levels of P-REX1 are inversely correlated with PI3K/AKT/mTOR pathway activation, and positively correlated with ER expression in breast tumors
Since PI3K activity modulates P-REX1 levels in breast cancer cells, we explored this relationship in human tumors. Using reverse-phase protein array (RPPA) analysis of lysates from 712 primary breast tumors, we measured levels of P-REX1, phosphoprotein markers of PI3K pathway activation (p-AKTT308, p-AKTS473, p-4E-BP1T37/46, p-4E-BP1S65, p-p70S6KT389, p-mTORS2448, p-TSC2T1462, p-GSK3α/βS21/9), ER, and HER2. P-REX1 expression was inversely correlated with all PI3K pathway markers, positively correlated with ER levels, and modestly negatively correlated with HER2 levels (Fig. S2). Hierarchical clustering of P-REX1 and phosphoprotein data revealed 1) two (phospho)protein clusters, where P-REX1 clustered independently from PI3K pathway markers, and 2) several tumor clusters, where the two most distinct clusters exhibited high P-REX1 with low PI3K activation, and low P-REX1 with high PI3K activation (Fig. 2A).
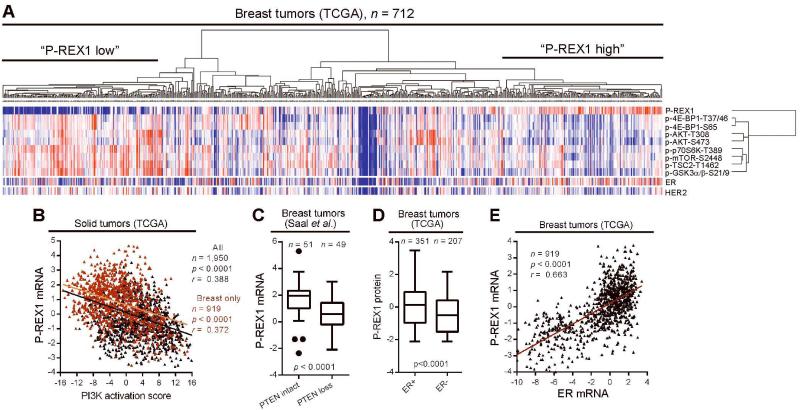
Figure 2. P-REX1 levels are inversely correlated with PI3K activation, and positively correlated with ER expression in human tumors.
A) Hierarchical clustering of signal values of P-REX1 and markers of PI3K pathway activation quantified by RPPA of 712 breast tumors. Levels of ER and HER2 are shown below, but were excluded from clustering. Red = high; blue = low. B) Correlation of P-REX1 mRNA levels and PI3K activation score in all tumor types (black) and breast tumors (red) from TCGA. Pearson correlation coefficients (r) and p-values are indicated. C) Tukey box plot comparing P-REX1 mRNA levels in breast tumors stratified based on expression of an mRNA signature of PTEN-loss vs. -intact (determined in ref. [17]) by Student’s t-test. D) Comparison of P-REX1 protein levels [from tumors in (A)] in ER+ vs. ER− breast tumors as in (C) (clinical ER status determined by IHC). E) Comparison of P-REX1 and ER mRNA levels in breast tumors from TCGA as in (B).
We then evaluated patterns of P-REX1 gene expression in human tumors. P-REX1 mRNA and protein levels were well-correlated (r2=0.6, p<0.0001)) in a set of 667 breast tumors (Fig. S3A). P-REX1 mRNA and protein levels were inversely correlated with a PI3K activation score derived from an expression signature of 1,793 genes (Table S2, derived from ref. [15]; signature does not include P-REX1) across 1,950 solid tumors and 919 breast tumors from The Cancer Genome Atlas (TCGA; Figs. 2B, S3B), and within two other datasets of 420 and 553 breast tumors [16] (Fig. S3C-D). In a fourth dataset of 100 breast tumors [17], P-REX1 mRNA levels were lower in tumors exhibiting a gene expression signature of PTEN loss compared to “PTEN-intact” tumors (Fig. 2C). These findings validate our observations in breast cancer cells (Figs. 1, S1), and confirm that P-REX1 levels are inversely correlated with PI3K activity.
Stratifying breast tumors by clinical ER status or molecular subtype, we found that P-REX1 protein and mRNA levels were significantly higher in ER+/luminal tumors compared with ER−/basal tumors, and compared with a diversity of other tumor subtypes (Figs. 2D, S3E-J). P-REX1 and ER mRNA levels were strongly correlated in breast tumors (Fig. 2E). These observations suggest that P-REX1 may an ER+ breast tumor-specific protein.
Genetic lesions in PREX1 are enriched in cancers with mutations in the PI3K pathway
Co-existent mutations in genes encoding proteins that lie in the same signaling cascade (e.g., HER2 and PIK3CA [18]) are thought to provide robustness to promote oncogenic phenotypes and fitness [19]. We analyzed genomic datasets to determine whether PREX1 is genetically altered in human tumors, and whether PREX1 lesions co-exist with other PI3K pathway alterations. PREX1 is amplified or mutated in 3.65% (163/4,462) of cancers, and in 3.65% (25/685) of primary breast tumors (Table S3). PREX1 gene copy number significantly correlates with P-REX1 mRNA and protein levels in breast tumors (Fig. S3K-L), suggesting that PREX1 amplification confers increased expression. We then compared the genetic status of PREX1 and 79 PI3K pathway-related genes across 1,523 solid tumors (482 breast, 212 colorectal, 143 glioblastoma, 179 lung, 207 ovarian, 93 prostate, 207 sarcoma). PREX1 lesions significantly co-occurred with lesions in 51 of 79 PI3K pathway-related genes (Fisher’s exact test p<0.05; Table S4); however, PREX1 lesions were not significantly enriched in tumors with lesions in PIK3CA or PTEN. These results suggest that PREX1 alterations may be enriched in PI3K pathway-driven tumors to enhance PI3K pathway robustness, and/or vice versa.
P-REX1 activates IGF-1R/InsR, PI3K/AKT, and MEK/ERK signaling
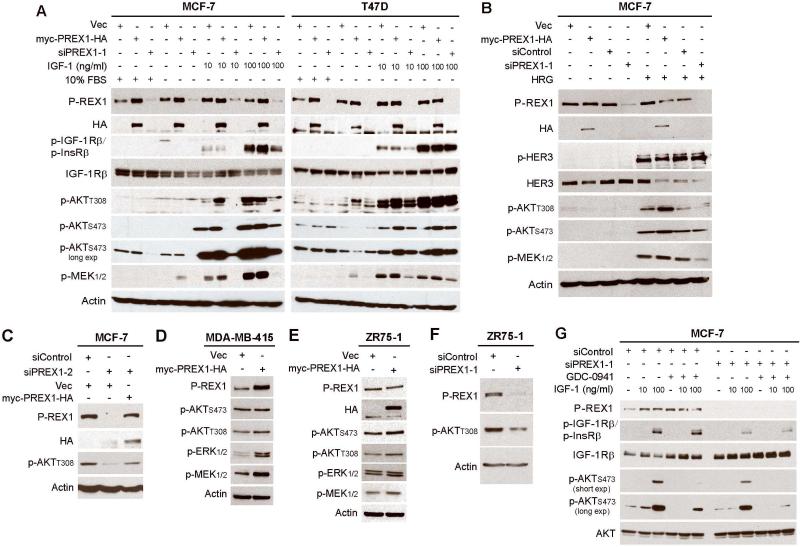
Since PIP3 activates P-REX1 [6] and PI3K inhibition increases P-REX1 levels (Figs. 1, S1), we hypothesized that P-REX1 levels are regulated by negative feedback signaling from PI3K. To determine whether P-REX1 provides feedback to PI3K to form a complete circuit, we overexpressed exogenous myc-PREX1-HA or knocked-down endogenous P-REX1 using RNA interference in MCF-7 and T47D cells. P-REX1 overexpression increased AKT phosphorylation compared to control under IGF-1-stimulated and heregulin (HER3 ligand)-stimulated conditions (Fig. 3A-B). Conversely, P-REX1 knockdown decreased growth factor-induced and steady-state P-AKT in MCF-7 cells (Fig. 3A). These effects were confirmed using a second siRNA against the 3’ UTR of P-REX1, and restoration of P-REX1 expression using the myc-PREX1-HA cDNA construct (not targeted by siPREX1-2) rescued P-AKT levels (Fig. 3C).
Figure 3. P-REX1 activates IGF-1R/InsR, PI3K/AKT, and MEK/ERK signaling in breast cancer cells.
Immunoblot analysis of lysates from cells transfected with vectors encoding myc-PREX1-HA, vector (vec) control, siRNA against P-REX1, or siControl. Panels A-C and E-G describe transiently transfected cells. Panel D describes stably transfected cells. Cells were maintained in 10% FBS (A,C-F), or serum-starved for 24 hours then stimulated ± 10-100 ng/ml IGF-1 for 10 min. (A,G), or 10 ng/ml heregulin (HRG) for 5 min. (B). In panel G, cells were pretreated ± 1 μM GDC-0941 for 2 h, then stimulated ± IGF-1.
Since P-REX2a directly inhibits PTEN lipid phosphatase activity to increase PIP3 levels [10], we also evaluated P-REX1 effects in intrinsically PTEN-deficient breast cancer cells. P-REX1 overexpression in ZR75-1 and MDA-MB-415 cells increased p-AKT, while P-REX1 knockdown reduced p-AKT in ZR75-1 cells (Fig. 3D-F; note- ZR75-1 cells express high levels of endogenous P-REX1, so levels of myc-PREX1-HA overexpression are modest). Thus, the effects of P-REX1 on AKT activation are PTEN-independent. In contrast to reported effects of P-REX2a [10], we did not detect an effect of P-REX1 on PTEN lipid phosphatase activity (data not shown).
Since P-REX1 activates the PI3K/AKT pathway (Fig. 3A-F) that can crosstalk with Rac/Pak/Raf/MEK/ERK pathways [20], and P-REX1 GEF function activates Rac1/2/3 [6, 9] that promote Pak/Raf/MEK/ERK activation [21-24], we tested whether P-REX1 also modulates MEK/ERK signaling in breast cancer. Transfected MCF-7 and T47D cells were serum-starved, then stimulated with IGF-1 or heregulin. P-REX1 overexpression increased and knockdown decreased basal and growth factor-induced p-MEK (Fig. 3A-B). Similar effects on p-MEK and p-ERK were observed under steady-state conditions in MDA-MB-415 and ZR75-1 cells (Fig. 3D-F).
We observed that P-REX1 knockdown in MCF-7 and T47D cells reduced levels of p-IGF-1Rβ/p-InsRβ (Fig. 3A). To determine whether P-REX1 effects on IGF-1R/InsR activation were PI3K-dependent, we knocked-down P-REX1 expression in MCF-7 cells, pre-treated ± GDC-0941 (to inhibit PI3K), then stimulated ± IGF-1. While P-REX1 knockdown reduced p-IGF-1Rβ/p-InsRβ and increased total IGF-1Rβ, PI3K inhibition had no effect on receptor phosphorylation but decreased p-AKT (Fig. 3G). This suggests that P-REX1 promotes IGF-1Rβ/InsRβ activation independently of PI3K/AKT activity.
P-REX1 promotes PI3K/AKT and MEK/ERK signaling via Rac activation
Since P-REX1 activates Rac1/2/3 [6, 9], which can promote Pak/Raf/MEK/ERK activation [21-24], and Rac1 was recently found to activate the p110β isoform of Class IA PI3K [25, 26], we postulated that P-REX1 acts through Rac to drive activation of both PI3K/AKT and MEK/ERK signaling in ER+ breast cancer cells. We first assessed the role of P-REX1 in Rac1 activation in T47D cells with or without RNAi-induced P-REX1 knock-down. In an activated Rac1 pull-down assay, cell lysates treated with GDP (to inactivate Rac1) or GTPγS (non-hydrolyzable GTP to activate Rac1) were used as negative and positive controls, respectively. Immunoblot analysis of pull-down eluates showed that levels of activated Rac1 were lower in GDP-treated than GTPγS-treated lysates, confirming assay specificity for activated (GTP-bound) Rac1 (Fig. S4A). Surprisingly, synthetic Rac1 activation with GTPγS increased P-REX1/Rac1 interaction compared to synthetic Rac1 inactivation with GDP, suggesting that P-REX1 preferentially interacts with activated Rac1. P-REX1 knock-down decreased activated Rac1, which was found in complex with p-ERK. Rac1 has been found in a triple-complex with Pak1 and MEK1/2 [21], so the MEK1/2 substrates ERK1/2 could also be part of this complex. These findings confirm that P-REX1 activates Rac1 in ER+ breast cancer cells, and loss of P-REX1-induced Rac1 activation decreases p-ERK1/2.
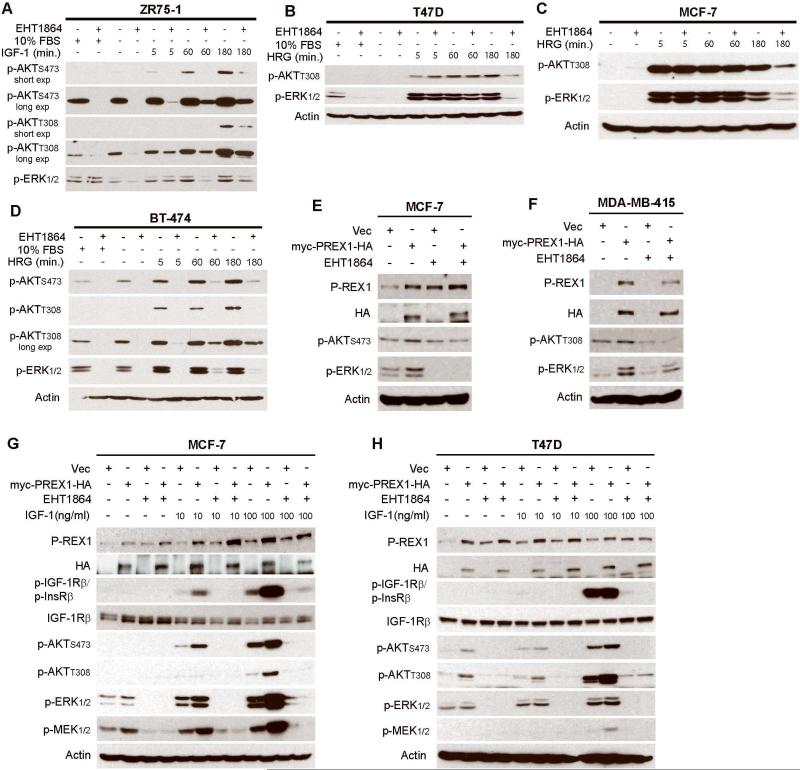
We then tested whether Rac signaling is required for activation of PI3K/AKT and MEK/ERK. Treatment with the Rac1/2/3 small molecule inhibitor EHT1864 [27] reduced steady-state, basal, IGF-1-induced, and heregulin-induced p-AKT and p-ERK1/2 in PTEN-deficient (ZR75-1, MDA-MB-415), PIK3CA-mutant (T47D, BT-474, MCF-7), and HER2+ (BT-474) breast cancer cells, all of which are ER+ (Figs. 4A-D, S4B). These results suggest that Rac GTPases drive activation of the PI3K/AKT and MEK/ERK pathways in ER+ breast cancer cells harboring genetic lesions in the PI3K pathway. Since P-REX1 activates Rac, these findings place P-REX1/Rac upstream of these signaling cascades.
Figure 4. Rac promotes activation of PI3K/AKT and MEK/ERK and is required for P-REX1-induced activation of these pathways in breast cancer.
A-D) Immunoblot analysis of lysates from cells treated as indicated ± serum-starvation for 24 h, then pretreated ± 50 μM EHT1864 × 2 h, and stimulated ± 20 ng/mL HRG or 100 ng/mL IGF-1. E-H) Immunoblot analysis of lysates from cells transiently (E,G,H) or stably (F) transfected with myc-PREX1-HA or vector (vec) control, maintained in 10% FBS (E-F) or serum-starved for 24 hours (G-H), pretreated ± 50 μM EHT1864 × 2 h (E-H), then stimulated ± IGF-1 for 10 min. (G-H).
To confirm the involvement of Rac activation in P-REX1-induced oncogenic signaling, we tested the effects of Rac inhibition with EHT1864 on cells overexpressing myc-PREX1-HA or control. While P-REX1 overexpression increased p-MEK, p-ERK, and p-AKT, such effects were abrogated by treatment with EHT1864 (Fig. 4E-H). Interestingly, Rac inhibition increased levels of endogenous P-REX1 (Fig. 4E), suggesting that Rac signaling provides negative feedback to control P-REX1 levels.
P-REX1 promotes viability of breast cancer cells and tumors
Since P-REX1 increases oncogenic signaling, we tested whether P-REX1 also promotes cell viability. Cells transfected with siRNA against P-REX1, vectors encoding myc-PREX1-HA, or controls were assayed using XTT assay, which measures mitochondrial enzymatic activity. PREX1 overexpression in T47D, MCF-7, and MDA-MB-415 cells increased viable cell number, while PREX1 knockdown decreased viability in T47D cells (Fig. 5A-D). Similarly, inhibition of Rac with EHT1864 decreased viability in all lines tested (Fig. 5A-E). In agreement with prior findings [8], knockdown of P-REX1 also decreased MCF-7 xenograft growth in immunodeficient mice (Figs. 5F, S5).
Figure 5. P-REX1 increases viability of breast cancer cells.
A-E) Cells transiently (A-B) or stably (C) transfected with vectors encoding myc-PREX1-HA or vector (vec) control, transiently transfected with siRNA (D), or untransfected (E), were evaluated by XTT assay. In (B-E), cells were treated ± 50 μM EHT1864 for 3 days prior to assay. Mean of 3-4 replicates ± SEM is shown. *p<0.05, **p<0.01 by Student’s t-test vs. control. F) Mice were subcutaneously implanted with MCF-7/shPREX1 or MCF-7/shNSC cells. Once tumor volumes reached ~150 mm3, tumors were measured twice weekly. Data are presented as % baseline volume (mean + SEM). Data were analyzed by two-way ANOVA followed by Bonferonni post-hoc comparison between cell lines at each time point (*p<0.05; **p<0.01).G) Forty-three breast cancer cell lines were dichotomized based on P-REX1 mRNA levels, and IC50 values were compared by Mann-Whitney U-test. Legend indicates molecular subtypes. Horizontal bars indicate median values.
P-REX1 contains amino-terminal tandem DH/PH domains characteristic of GEFs (Fig. S6A). The DH domain is required for binding Gβγ and GEF activity. The PH domain is needed for PIP3 binding and intracellular membrane localization [28]. Prior evidence indicates that PH domain binding to PIP3 may increase GTPase (i.e., Rac) binding activity of the DH domain in GEFs, although this remains controversial and may differ between GEFs (reviewed in ref. [29]). Adding to this understanding, we observed that deletion of the DH domain (ΔDH) increases P-REX1 binding to PIP3, and facilitates interaction with other phosphatidylinositol species (Figs. S6B-C), suggesting that the DH domain regulates interactions of the PH domain with lipid membranes. Adjacent to the DH/PH domains are two Dishevelled, Egl-10, and Pleckstrin (DEP) domains reported to be required for interaction with mTOR [30]; however, we did not detect an interaction of P-REX1 with mTOR in immunoprecipitation experiments (data not shown). Overexpression in T47D cells revealed that full-length P-REX1 increased viability, while the ΔDH and ΔDH/PH mutants were less effective (Fig. S6D). These findings suggest that P-REX1 GEF activity and membrane localization are critical for P-REX1-mediated promotion of cell viability.
Since P-REX1 requires PIP3 for activation [6], P-REX1 promotes PI3K/AKT and MEK/ERK signaling (Fig. 3), and combined inhibition of PI3K and MEK has strong anti-tumor effects (reviewed in ref. [20]), we hypothesized that P-REX1-expressing cells would be sensitized to pharmacologic inhibition of PI3K. Breast cancer cell lines (n=43) were dichotomized based on P-REX1 mRNA levels (extracted from the Cancer Cell Line Encyclopedia [31]), and IC50 values in response to GDC-0941 and the PI3K/mTOR dual inhibitor PI-103 (IC50 values extracted from [32]) were compared between P-REX1-high vs. -low cell lines. P-REX1-high cells were significantly more sensitive to GDC-0941 (p=0.01), and showed a trend towards sensitivity to PI-103 (p=0.07) compared to P-REX1-low cells (Fig. 5G). In agreement with P-REX1 levels being highest in ER+ breast cancers (Figs. 2D-E, S3), P-REX1-high cells classified mainly as luminal or HER2-enriched by molecular subtyping, while P-REX1-low cells were mostly basal or normal-like. In a multivariate model including molecular subtype, P-REX1 status (high/low) was not independently predictive of sensitivity to PI3K inhibition, suggesting that P-REX1 expression is tightly linked with molecular subtype. Transcriptomic PI3K activation score and P-AKTT308 levels were not predictive of sensitivity to PI3K inhibition (Fig. S7).
Discussion
Herein, we globally identified proteins quantitatively modulated by PI3K pathway inhibition and activation in breast cancer cells. We found that P-REX1 levels are inversely correlated with PI3K activation and positively correlated with ER expression in human breast tumors. P-REX1 activates Rac GTPases to promote activation of IGF-1R/InsR, PI3K/AKT, and MEK/ERK to increase breast cancer cell viability in a PI3K/PIP3-dependent, PTEN-independent manner. Therefore, P-REX1 lies both downstream and upstream of PI3K, and creates a positive feedback loop to drive oncogenic signaling (Fig. 6). In support of a model where P-REX1/Rac drives both PI3K/AKT and MEK/ERK signaling, breast cancer cells expressing high levels of P-REX1 are more sensitive to PI3K inhibition than P-REX1-low cells.
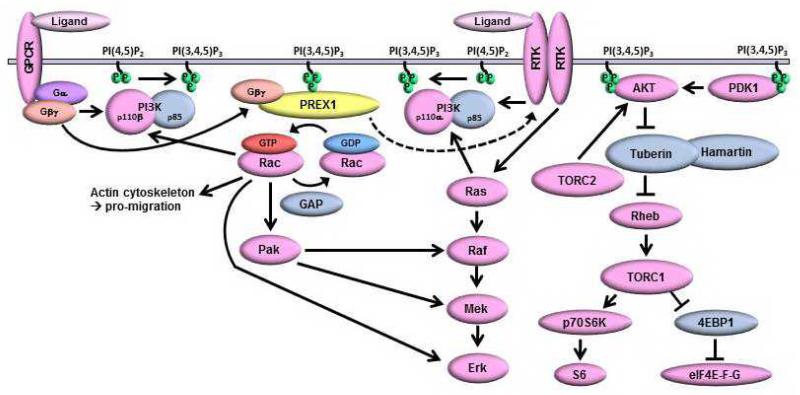
Figure 6. Model of PI3K-dependent P-REX1 positive feedback to activate Rac, IGF-1R/InsR, PI3K/AKT, and MEK/ERK signaling.
Growth factor receptor tyrosine kinases (RTKs) and G-protein coupled receptors (GPCRs) activate PI3K, which produces PIP3. P-REX1 is recruited to the membrane via PH domain binding to PIP3. Activation of GPCRs induces dissociation of Gβγ subunits, which bind the P-REX1 DH domain to promote its GEF activity and Rac activation. Rac activates PI3K/p110β to increase PIP3 production, stimulates Raf/MEK signaling via Pak, interacts with p-ERK, and promotes actin cytoskeleton remodeling. P-REX1 also promotes PI3K-independent activation of IGF-1R/InsR, which can lead to further activation of PI3K and MEK signaling.
We and others found that P-REX1 knockdown reduces levels of activated Rac in breast cancer cells (Fig. S4A and refs. [9, 33]). In addition, we demonstrated that P-REX1 activates IGF-1R/InsR, PI3K/AKT, and MEK/ERK signaling in breast cancer cells in a Rac activation-dependent manner (Figs. 3-5, S4). Recent studies have implicated Ras and Rho family GTPases in the regulation of PI3K activity. Fritsch et al. showed that activated Rac1 binds and activates PI3K/p110β to increase PIP3 production [25, 26]. Therefore, P-REX1 may activate PI3K/p110β via Rac in breast cancer cells. P-REX1 also promotes activation of IGF-1R/InsR in a PI3K/PIP3-independent, Rac-dependent manner (Figs. 3G, 4G-H), which in turn can activate PI3K/p110α [34]. We did not consistently detect P-REX1/IGF-1Rβ or P-REX1/IRS-1 interaction in immunoprecipitation trials (data not shown), and the mechanism underlying P-REX1-mediated receptor activation requires further study.
Cross-talk between the PI3K/AKT and Pak/Raf/MEK/ERK pathways exists at several levels [20], and we demonstrated that P-REX1 modulates both pathways in ER+ breast cancer cells. P-REX1 activates MEK/ERK signaling via activation of Rac (Figs. 4, S4). The Rac effector Pak1 can serve as an intermediary to activate Raf/MEK/ERK signaling (Fig. 6 & refs. [21-24]). In support of our findings, a recent study showed that P-REX1 knock-down reduces Rac1/Pak/Raf/MEK/ERK signaling [33]. Furthermore, Rac/Pak1-mediated activation of ERK and AKT is required for HER2-induced transformation of breast cancer cells [35]. We thus infer that P-REX1 GEF activity promotes Rac/Pak-induced activation of MEK/ERK and PI3K/AKT in ER+ breast cancer cells. Additionally, we detected binding of activated Rac1 to p-ERK1/2, and synthetic Rac1 activation with GTPγS increased P-REX1/Rac1 interaction (Fig. S4A). A Rac1/Pak1/MEK1/2 complex has been described [21]; we speculate that this complex may also bind ERK1/2. Whether P-REX1 interacts with Rac1 when complexed with Pak1/MEK1/2/ERK1/2 is unclear. Given that GEFs typically interact with GDP-bound GTPases, GTPase-activating proteins (GAPs) interact with GTP-bound GTPases, and other GEF/GAP/Rho GTPase complexes have been described [36], it is conceivable that P-REX1 interacts with a complex containing activated Rac1 and a GAP.
In support of a role for P-REX1 driving both the PI3K/AKT and MEK/ERK pathways, we observed that high P-REX1 mRNA levels, but not PI3K activation score (by gene expression) or P-AKTT308 level, are predictive of increased sensitivity to PI3K inhibitors (Figs. 5G, S7). Drug combinations targeting PI3K and MEK elicit dramatic anti-tumor effects in preclinical studies, but such combinations are accompanied by considerable toxicity in patients [20]. Thus, P-REX1 may be a marker of cells in which PI3K inhibitors alone will block both PI3K and MEK. Indeed, PI3K inhibition decreases P-ERK levels which, in turn, induces upregulation of the pro-apoptotic protein Bim in P-REX1-expressing cells [33]. Interestingly, the two luminal/ER+ breast cancer cell lines most resistant to GDC-0941 (HCC-1500 and MDA-MB-134VI; Fig. 5G) harbor PAK1 amplification, and Pak1 knock-down or treatment with the Pak inhibitor PF-3758309 was shown to induce apoptosis and decrease MEK/ERK phosphorylation in these cells [37]. PAK1 is amplified in 8.4%, 13.8%, and 3.7% of luminal, HER2-enriched, and basal breast cancers, respectively (extracted from ref. [38] using cBio [39]). Thus, P-REX1 expression and PAK1 amplification may serve as respective positive and negative biomarkers of sensitivity to PI3K inhibitors in breast cancer.
The P-REX1 homolog P-REX2a binds PTEN and inhibits its lipid phosphatase activity [10]. While we detected a low level of P-REX1/PTEN binding in situ, in vitro and in situ analyses did not reveal an effect of P-REX1 on PTEN phosphatase activity (data not shown). Furthermore, we observed that P-REX1 activates PI3K/AKT and MEK/ERK signaling in PTEN-null cells (MDA-MB-415 and ZR75-1; Figs. 3D-F, 4F). Thus, P-REX1 and P-REX2a may promote PI3K pathway activation via different mechanisms.
In summary, we report that P-REX1 mediates a PI3K/PIP3-dependent positive feedback loop to promote activation of Rac, IGF-1R/InsR, PI3K/AKT, and MEK/ERK signaling in ER+ breast cancer. Combined targeting of two key signaling nodes in cancer, PI3K and MEK, is being investigated in several ongoing clinical studies [20]. Thus, targeting the P-REX1/Rac axis may inhibit both PI3K and MEK signaling to elicit anti-cancer effects. Our mechanistic findings on P-REX1 are from ER+ breast cancer models harboring genetic lesions in the PI3K pathway; P-REX1 effects in ERBB2/PIK3CA/PTEN-wild-type ER+ models, and in ER− models remain to be determined. Furthermore, P-REX1 mRNA expression is predictive of sensitivity to PI3K inhibitors in breast cancer cells, offering P-REX1 as a biomarker to identify tumors likely to respond to these agents. P-REX1 is most highly expressed in ER+ breast cancers, suggesting that P-REX1 is an oncoprotein mainly in this subtype of cancer. Since P-REX1 knock-out mice are generally healthy with mild neutrophilia [40], targeting of P-REX1 may provide a high therapeutic index in patients.
Material and Methods
Cell culture
Cell lines (ATCC) were grown in DMEM (Life Technologies) with 10% FBS (Hyclone). MCF-7 and T47D cells stably expressing shRNA targeting PTEN or mismatch control (shMM) were described previously [41]. Generation of other stable cell lines is described in Supplementary Methods. In transient transfection experiments, cells were transfected with pcDNA plasmid encoding myc-PREX1-HA (from Atanasio Pandiella, CSIC-Universidad de Salamanca) or FLAG-HA control (Addgene) plus Lipofectamine 2000. Cells were transfected with siRNA targeting P-REX1 or AllStars Negative Control (Qiagen, Dharmacon) plus Lipofectamine RNAiMAX (Life Technologies). Cells were treated with IGF-1 (PeproTech), heregulin (HRG), BKM-120, GDC-0941, MK-2206 (Selleckchem), or EHT1864 (Tocris Bioscience) as indicated in figures.
Mass spectrometry (MS)
Lysates from MCF-7/shPTEN and MCF-7/shMM cells treated with 1 μM BKM-120 or DMSO (control) for 16 h were analyzed by MudPIT coupled with tandem MS/MS [42, 43]. Proteins were identified using the Sequest algorithm [44] and assembled into groups using IDPicker [45] (details in Supplementary Methods).
Immunoblotting
Cells were lysed in RIPA buffer containing protease and phosphatase inhibitors. Lysates were sonicated for 15 sec., centrifuged at 18k × g for 10 min. at 4°C, and protein in supernatants was quantified using BCA assay (Pierce). Reduced/denatured lysates were separated by SDS-PAGE, and proteins were transferred to nitrocellulose. Even protein loading across lanes was confirmed with Ponceau S staining. Blots were probed with antibodies against P-REX1 (Abcam), HA, HER3 (Santa Cruz), Rac1 (Cytoskeleton), p-AKTS473, p-AKTT308, AKT, p-S6S240/4, p-ERK1/2T202/Y204, p-MEK1/2S217/221, p-IGF-1RβY1135/6/p-InsRβY1150/1, IGF-1R, IRS-1, p-HER3Y1289, or Actin (Cell Signaling). HRP-labeled secondary antibodies (GE Healthcare) and ECL substrate (Pierce) were used for signal detection.
Cell viability assay
XTT cell viability assay (Biotium) was performed per manufacturer’s instructions. Cells were transfected with siRNA or plasmid, then reseeded 48 h later at 1-1.5 × 104/well in 96-well plates. Assay was performed 48-72 h later by adding 25 μL activated XTT solution, and reading absorbance at 450 nm (reflective of live cells) and 655 nm (background) after 2-6 hours. Background-subtracted values (Abs450 - Abs655) of 3-4 replicate wells were compared by Student’s t-test. Similarly, cells were seeded, then treated the next day ± 50 μM EHT1864, and XTT assay was performed 48 h later.
Breast cancer xenografts
Animal studies were approved by the Dartmouth College IACUC. Female NOD-scid IL2Rγ−/− (NSG; NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice (6-7 wks old; obtained from the Norris Cotton Cancer Center Transgenic & Genetic Construct Shared Resource) were implanted subcutaneously with a 60-day-release 17β-estradiol pellet (0.72 mg; Innovative Research of America) and 5 × 106 cells (MCF-7/shPREX1 or /shNSC; described in Supplementary Methods) suspended in 50% matrigel (BD Biosciences). Tumor diameters were measured twice per week using calipers (volume = length2 × width/2), and volumes were analyzed as percent baseline by two-way ANOVA followed by Bonferroni post-hoc comparison.
RPPA analysis
Tumors analyzed by RPPA were part of TCGA project. P-REX1 was quantified specifically for this study. Other (phospho)proteins were quantified as part of TCGA [46]. All tumor proteomic data has been deposited at The Cancer Proteome Atlas (TCPA; http://app1.bioinformatics.mdanderson.org/tcpa/_design/basic/index.html). Tumor lysates were analyzed as described in Supplementary Methods.
Comparing P-REX1 gene expression with sensitivity to PI3K inhibition
RMA-normalized P-REX1 mRNA expression values from cancer cell lines were obtained from the Cancer Cell Line Encyclopedia [31]. Molecular subtypes of 43 breast cancer cell lines were determined by PAM50 subtyping using R software with package ‘genefu,’ median-centered data, and the 306 intrinsic subtype genes and centroids identified in ref. [47]. Cell lines were dichotomized based on high vs. low P-REX1 mRNA levels, and IC50 values in response to treatment with GDC-0941 and PI-103 (obtained from ref. [32]) were compared between groups by Mann-Whitney U-test.
Supplementary Material
Acknowledgements
We thank Chad Creighton for advice on gene expression analysis, and the Norris Cotton Cancer Center Transgenic & Genetic Construct Shared Resource and Genomics & Molecular Biology Shared Resource for assistance. Financial support was provided by NIH: K99/R00CA142899 (TWM), Dartmouth College Norris Cotton Cancer Center Support Grant P30CA023108, Vanderbilt-Ingram Cancer Center Breast Cancer Specialized Program of Research Excellence Grant P50CA98131, and Vanderbilt-Ingram Cancer Center Support Grant P30CA68485; the American Cancer Society IRG-58-009-50 (TWM) and 121329-RSG-11-187-01-TBG (AMG); and The Commonwealth Foundation for Cancer Research (AMG).
Footnotes
Conflict of interest: All authors have reported no financial conflicts of interest relevant to this study.
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
References
- [1].Miller TW, Rexer BN, Garrett JT, Arteaga CL. Mutations in the phosphatidylinositol 3-kinase pathway: role in tumor progression and therapeutic implications in breast cancer. Breast Cancer Res. 2011;13(6):224. doi: 10.1186/bcr3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9(8):550–62. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- [3].Whitman M, Downes CP, Keeler M, Keller T, Cantley L. Type I phosphatidylinositol kinase makes a novel inositol phospholipid, phosphatidylinositol-3-phosphate. Nature. 1988;332(6165):644–6. doi: 10.1038/332644a0. [DOI] [PubMed] [Google Scholar]
- [4].Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. Journal of Biological Chemistry. 1998;273(22):13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- [5].Maira SM, Pecchi S, Huang A, Burger M, Knapp M, Sterker D, et al. Identification and characterization of NVP-BKM120, an orally available pan-class I PI3-kinase inhibitor. Mol Cancer Ther. 2012;11(2):317–28. doi: 10.1158/1535-7163.MCT-11-0474. [DOI] [PubMed] [Google Scholar]
- [6].Welch HC, Coadwell WJ, Ellson CD, Ferguson GJ, Andrews SR, Erdjument-Bromage H, et al. P-Rex1, a PtdIns(3,4,5)P3- and Gbetagamma-regulated guanine-nucleotide exchange factor for Rac. Cell. 2002;108(6):809–21. doi: 10.1016/s0092-8674(02)00663-3. [DOI] [PubMed] [Google Scholar]
- [7].Qin J, Xie Y, Wang B, Hoshino M, Wolff DW, Zhao J, et al. Upregulation of PIP3-dependent Rac exchanger 1 (P-Rex1) promotes prostate cancer metastasis. Oncogene. 2009;28(16):1853–63. doi: 10.1038/onc.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Montero JC, Seoane S, Ocana A, Pandiella A. P-Rex1 participates in Neuregulin-ErbB signal transduction and its expression correlates with patient outcome in breast cancer. Oncogene. 2011;30(9):1059–71. doi: 10.1038/onc.2010.489. [DOI] [PubMed] [Google Scholar]
- [9].Sosa MS, Lopez-Haber C, Yang C, Wang H, Lemmon MA, Busillo JM, et al. Identification of the Rac-GEF P-Rex1 as an essential mediator of ErbB signaling in breast cancer. Mol Cell. 2010;40(6):877–92. doi: 10.1016/j.molcel.2010.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Fine B, Hodakoski C, Koujak S, Su T, Saal LH, Maurer M, et al. Activation of the PI3K pathway in cancer through inhibition of PTEN by exchange factor P-REX2a. Science. 2009;325(5945):1261–5. doi: 10.1126/science.1173569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Folkes AJ, Ahmadi K, Alderton WK, Alix S, Baker SJ, Box G, et al. The identification of 2-(1H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer. Journal of Medicinal Chemistry. 2008;51(18):5522–5532. doi: 10.1021/jm800295d. [DOI] [PubMed] [Google Scholar]
- [12].Miller TW, Perez-Torres M, Narasanna A, Guix M, Stal O, Perez-Tenorio G, et al. Loss of Phosphatase and Tensin Homologue Deleted on Chromosome 10 Engages ErbB3 and Insulin-Like Growth Factor-I Receptor Signaling to Promote Antiestrogen Resistance in Breast Cancer. Cancer Research. 2009;69(10):4192–4201. doi: 10.1158/0008-5472.CAN-09-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chakrabarty A, Sanchez V, Kuba MG, Rinehart C, Arteaga CL. Feedback upregulation of HER3 (ErbB3) expression and activity attenuates antitumor effect of PI3K inhibitors. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1018001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Khatri S, Yepiskoposyan H, Gallo CA, Tandon P, Plas DR. FOXO3a regulates glycolysis via transcriptional control of tumor suppressor TSC1. J Biol Chem. 2010;285(21):15960–5. doi: 10.1074/jbc.M110.121871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Creighton CJ, Fu X, Hennessy BT, Casa AJ, Zhang Y, Gonzalez-Angulo AM, et al. Proteomic and transcriptomic profiling reveals a link between the PI3K pathway and lower estrogen-receptor (ER) levels and activity in ER+ breast cancer. Breast Cancer Res. 2010;12(3):R40. doi: 10.1186/bcr2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Saal LH, Johansson P, Holm K, Gruvberger-Saal SK, She QB, Maurer M, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci U S A. 2007;104(18):7564–9. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hanker AB, Pfefferle AD, Balko JM, Kuba MG, Young CD, Sanchez V, et al. Mutant PIK3CA accelerates HER2-driven transgenic mammary tumors and induces resistance to combinations of anti-HER2 therapies. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(35):14372–14377. doi: 10.1073/pnas.1303204110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27(41):5497–510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Saini KS, Loi S, de Azambuja E, Metzger-Filho O, Saini ML, Ignatiadis M, et al. Targeting the PI3K/AKT/mTOR and Raf/MEK/ERK pathways in the treatment of breast cancer. Cancer Treat Rev. 2013;39(8):935–46. doi: 10.1016/j.ctrv.2013.03.009. [DOI] [PubMed] [Google Scholar]
- [21].Wang ZP, Fu M, Wang LF, Liu JJ, Li YH, Brakebusch C, et al. p21-Activated Kinase 1 (PAK1) Can Promote ERK Activation in a Kinase-independent Manner. Journal of Biological Chemistry. 2013;288(27):20093–20099. doi: 10.1074/jbc.M112.426023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].King AJ, Sun HY, Diaz B, Barnard D, Miao WY, Bagrodia S, et al. The protein kinase Pak3 positively regulates Raf-1 activity through phosphorylation of serine 338. Nature. 1998;396(6707):180–183. doi: 10.1038/24184. [DOI] [PubMed] [Google Scholar]
- [23].Slack-Davis JK, Eblen ST, Zecevic M, Boerner SA, Tarcsafalvi A, Diaz HB, et al. PAK1 phosphorylation of MEK1 regulates fibronectin-stimulated MAPK activation. J Cell Biol. 2003;162(2):281–91. doi: 10.1083/jcb.200212141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Manser E, Leung T, Salihuddin H, Zhao ZS, Lim L. A Brain Serine Threonine Protein-Kinase Activated by Cdc42 and Rac1. Nature. 1994;367(6458):40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- [25].Fritsch R, de Krijger I, Fritsch K, George R, Reason B, Kumar MS, et al. RAS and RHO families of GTPases directly regulate distinct phosphoinositide 3-kinase isoforms. Cell. 2013;153(5):1050–63. doi: 10.1016/j.cell.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yang HW, Shin MG, Lee S, Kim JR, Park WS, Cho KH, et al. Cooperative Activation of PI3K by Ras and Rho Family Small GTPases. Molecular Cell. 2012;47(2):281–290. doi: 10.1016/j.molcel.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Onesto C, Shutes A, Picard V, Schweighoffer F, Der CJ. Characterization of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. Small Gtpases in Disease, Pt B. 2008;439:111. doi: 10.1016/S0076-6879(07)00409-0. [DOI] [PubMed] [Google Scholar]
- [28].Hill K, Krugmann S, Andrews SR, Coadwell WJ, Finan P, Welch HC, et al. Regulation of P-Rex1 by phosphatidylinositol (3,4,5)-trisphosphate and Gbetagamma subunits. J Biol Chem. 2005;280(6):4166–73. doi: 10.1074/jbc.M411262200. [DOI] [PubMed] [Google Scholar]
- [29].Hoffman GR, Cerione RA. Signaling to the Rho GTPases: networking with the DH domain. FEBS Lett. 2002;513(1):85–91. doi: 10.1016/s0014-5793(01)03310-5. [DOI] [PubMed] [Google Scholar]
- [30].Hernandez-Negrete I, Carretero-Ortega J, Rosenfeldt H, Hernandez-Garcia R, Calderon-Salinas JV, Reyes-Cruz G, et al. P-Rex1 links mammalian target of rapamycin signaling to Rac activation and cell migration. J Biol Chem. 2007;282(32):23708–15. doi: 10.1074/jbc.M703771200. [DOI] [PubMed] [Google Scholar]
- [31].Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].O'Brien C, Wallin JJ, Sampath D, GuhaThakurta D, Savage H, Punnoose EA, et al. Predictive biomarkers of sensitivity to the phosphatidylinositol 3' kinase inhibitor GDC-0941 in breast cancer preclinical models. Clin Cancer Res. 2010;16(14):3670–83. doi: 10.1158/1078-0432.CCR-09-2828. [DOI] [PubMed] [Google Scholar]
- [33].Ebi H, Costa C, Faber AC, Nishtala M, Kotani H, Juric D, et al. PI3K regulates MEK/ERK signaling in breast cancer via the Rac-GEF, P-Rex1. Proc Natl Acad Sci U S A. 2013;110(52):21124–9. doi: 10.1073/pnas.1314124110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Guillermet-Guibert J, Bjorklof K, Salpekar A, Gonella C, Ramadani F, Bilancio A, et al. The p110 beta isoform of phosphoinositide 3-kinase signals downstream of G protein-coupled receptors and is functionally redundant with p110 gamma. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(24):8292–8297. doi: 10.1073/pnas.0707761105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Arias-Romero LE, Villamar-Cruz O, Pacheco A, Kosoff R, Huang M, Muthuswamy SK, et al. A Rac-Pak signaling pathway is essential for ErbB2-mediated transformation of human breast epithelial cancer cells. Oncogene. 2010;29(43):5839–49. doi: 10.1038/onc.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Premont RT, Perry SJ, Schmalzigaug R, Roseman JT, Xing Y, Claing A. The GIT/PIX complex: an oligomeric assembly of GIT family ARF GTPase-activating proteins and PIX family Rac1/Cdc42 guanine nucleotide exchange factors. Cell Signal. 2004;16(9):1001–11. doi: 10.1016/j.cellsig.2004.02.002. [DOI] [PubMed] [Google Scholar]
- [37].Ong CC, Jubb AM, Haverty PM, Zhou W, Tran V, Truong T, et al. Targeting p21-activated kinase 1 (PAK1) to induce apoptosis of tumor cells. Proc Natl Acad Sci U S A. 2011;108(17):7177–82. doi: 10.1073/pnas.1103350108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Welch HC, Condliffe AM, Milne LJ, Ferguson GJ, Hill K, Webb LM, et al. P-Rex1 regulates neutrophil function. Curr Biol. 2005;15(20):1867–73. doi: 10.1016/j.cub.2005.09.050. [DOI] [PubMed] [Google Scholar]
- [41].Miller TW, Perez-Torres M, Narasanna A, Guix M, Stal O, Perez-Tenorio G, et al. Loss of Phosphatase and Tensin homologue deleted on chromosome 10 engages ErbB3 and insulin-like growth factor-I receptor signaling to promote antiestrogen resistance in breast cancer. Cancer Res. 2009;69(10):4192–201. doi: 10.1158/0008-5472.CAN-09-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].MacCoss MJ, McDonald WH, Saraf A, Sadygov R, Clark JM, Tasto JJ, et al. Shotgun identification of protein modifications from protein complexes and lens tissue. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(12):7900–7905. doi: 10.1073/pnas.122231399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Martinez MN, Emfinger CH, Overton M, Hill S, Ramaswamy TS, Cappel DA, et al. Obesity and altered glucose metabolism impact HDL composition in CETP transgenic mice: a role for ovarian hormones. J Lipid Res. 2012;53(3):379–89. doi: 10.1194/jlr.M019752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Eng JK, McCormack AL, Yates JR., III An Approach to Correlate Tandem Mass Spectral Data of Peptides with Amino Acid Sequences in a Protein Database. J Am Soc Mass Spectrom. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- [45].Ma ZQ, Dasari S, Chambers MC, Litton MD, Sobecki SM, Zimmerman LJ, et al. IDPicker 2.0: Improved protein assembly with high discrimination peptide identification filtering. J Proteome Res. 2009;8(8):3872–81. doi: 10.1021/pr900360j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Li J, Lu Y, Akbani R, Ju Z, Roebuck PL, Liu W, et al. TCPA: a resource for cancer functional proteomics data. Nat Methods. 2013;10(11):1046–7. doi: 10.1038/nmeth.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hu ZY, Fan C, Oh DS, Marron JS, He XP, Qaqish BF, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7 doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.