To the Editor: The Cancer Genome Atlas (TCGA) has been generating multi-modal genomics, epigenomics, and proteomics data for thousands of tumor samples across more than 20 types of cancer. While the access to most level-1 and -2 TCGA data is restricted, the entire level-3 TCGA data as well as some level-1 clinical data (e.g., survival and drug treatments) are publicly available. Included in the public data are genome-wide measurements of different genetic characterizations, such as DNA copy number, DNA methylation, and mRNA expression for the same genes, providing unprecedented opportunities for systematic investigation of cancer mechanisms at multiple molecular and regulatory layers [1-3]. Few tools of integrative data mining for TCGA are present, partly due to lack of tools to acquire and assemble the large scale TCGA data. Specifically, the level-3 TCGA data are stored as hundreds of thousands of sample- and platform-specific files, accessible through HTTP directories on the servers of TCGA Data Coordinating Center (DCC) [4]. Navigating through all of the files manually is impossible. Although Firehose [5] nicely assemble and publish TCGA data, it does not share the program code for data assembly. Currently the community does not have access to open-source data retrieving tools for automatic and flexible data acquisition, hence severely hindering the progress in systemic data integration and reproducible computational analysis using TCGA data. To meet these challenges, we introduce TCGA-Assembler, a software package that automates and streamlines the retrieval, assembly, and processing of public TCGA data. TCGA-Assembler equips users the ability to produce Firehose-type of TCGA data, with open-source and freely available program script. TCGA-Assembler opens a door for the development of data-mining and data-analysis tools that generate fully reproducible results, including data acquisition.
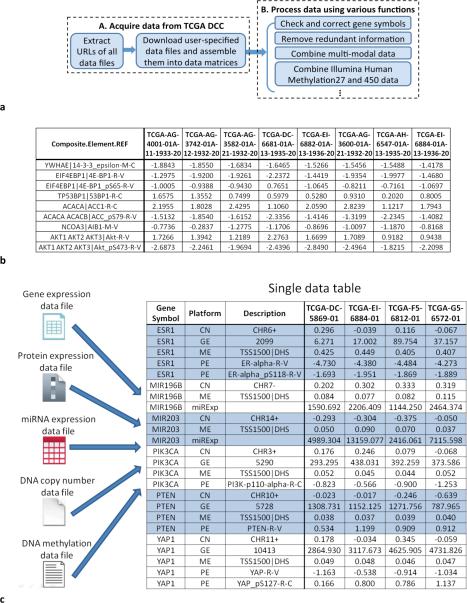
TCGA-Assembler consists of two modules (Fig. 1a), both written in R (http://www.r-project.org). Module A streamlines data downloading and quality check, and module B processes the downloaded data for subsequent analyses (Supplementary Methods). In particular, module A takes advantage of the informative naming mechanism of TCGA data file system (Supplementary Fig. 1) and applies a recursive algorithm to retrieve the URLs of all data files. By string matching on the URLs, module A allows users to download most of TCGA public data (Supplementary Table 1) across genomic features and cancer types. For each genomics feature (such as gene expression from RNA-Seq) a data matrix combining multiple samples (Fig. 1b) is produced, with rows representing genomics units (such as genes) and columns representing samples. Module B provides convenient and important data preprocessing functions, such as mega-data assembly, data cleaning, and quantification of various measurements. For users interested in integrative analysis [6], a mega data matrix (Fig. 1c) is required that matches different types of genomics measurements for the same genes across samples. Module B provides a function “CombineMultiPlatfomData” to fulfill this requirement (Supplementary Methods), which involves intricate data-matching steps to overcome the feature-labeling discrepancies caused by different lab protocols and biotechnologies in the experiments. Other data-processing functions are also provided to facilitate downstream analysis (Supplementary Methods).
Figure 1.
TCGA-Assembler as a tool for acquiring, assembling, and processing public TCGA data. (a) Flowchart of TCGA- Assembler. Module A acquires data from TCGA DCC. Module B processes the obtained data using various functions. (b) Illustration of a data matrix file using protein expressions generated by Reverse Phase Protein Array (RPPA). Each row corresponds to a protein or phosphorylated protein, and each column corresponds to a sample. The first column shows the gene symbol (before “|”) and the name of the protein antibody (after “|”) used in RPPA. (c) Illustration of combining multi- modal data. After combination, a single mega data table is obtained, in which each column (except the first three columns) corresponds to a patient sample and each row corresponds to a genomic/epigenomic feature. All multi-modal data of a gene are adjacent in the table and are indicated by alternating blue/white color. In the second column, GE represents gene expression, PE protein expression, ME DNA methylation, CN copy number, and miRExp miRNA expression. In the third column, the description of GE platform is the Entrez ID of gene; the description of PE platform is the name of the protein antibody used in RPPA assay; “TSS1500|DNS” for the description of ME platform indicates that the values are average methylation measurements of CpG sites that are within 1,500 nucleotide base pairs of transcription start site and are DNAse hypersensitive; the description of CN platform gives the chromosome ID and strand of a gene.
Other big data tools for TCGA are available [5, 7, 8]. In particular, level-3 TCGA data can also be obtained from Firehose [5] at the MIT Broad Institute in the same format as in Fig. 1b, one for each cancer type and genomics platform. Module A of TCGA-Assembler not only provides the same type of data matrices, but also distributes R functions and associated computer program that produce the data matrices. Equipped with the open-source tool, users will be independent and control what and when TCGA data will be acquired locally. More importantly, quantitatively advanced users may integrate our open-source programs with downstream data analysis tools to realize reproducible and automated data analysis for TCGA. Unique to TCGA-Assembler is module B that provides critical functions for data cleaning and processing. For example, the mega data table (Fig. 1c) can be obtained with a single function, behind which substantial efforts have been directed to ensure the validity of process, such as to check and correct gene symbol discrepancies. Lastly, TCGA-Assembler is fully compatible with Firehose in that the data processing functions in Module B can directly process data files downloaded from Firehose. This compatibility is crucial to those who want to take advantage of both software pipelines.
TCGA-Assembler will remain freely available and open-source. In the future, more data processing and analysis functions will be continuously added to TCGA-Assembler based on user feedback and new research needs. The authors request acknowledgment of the use of TCGA-Assembler in published works.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the US National Cancer Institute (R01 CA132897 and R01 CA163481).
Footnotes
AUTHOR CONTRIBUTIONS
Y.Z., P.Q., and Y.J. initialized the original idea. Y.Z. and Y.J. designed and implemented the software, and Y.Z., P.Q., and Y.J. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Supplementary information is available in the online version of the paper (doi)
REFERENCES
- 1.The Cancer Genome Atlas https://tcga-data.nci.nih.gov/tcga/.
- 2.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Open-access HTTP directory on the data server of TCGA Data Coordination Center https://tcga-data.nci.nih.gov/tcgafiles/ftp_auth/distro_ftpusers/anonymous/tumor/
- 5. Firehose website, https://confluence.broadinstitute.org/display/GDAC/Home.
- 6.Xu Y, et al. A Bayesian graphical model for integrative analysis of TCGA data.. IEEE International Workshop on Genomic Signal Processing and Statistics (GENSIPS); 2-4 Dec. 2012; Washington, DC: pp. 135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robbins DE, et al. A self-updating road map of The Cancer Genome Atlas. Bioinformatics. 2013 May 15;29(10):1333–40. doi: 10.1093/bioinformatics/btt141. doi: 10.1093/bioinformatics/btt141. Epub 2013 Apr 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Signaling. 2013 Apr 2;6(269):l1. doi: 10.1126/scisignal.2004088. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.