Abstract
XRCC1 is an essential protein required for the maintenance of genomic stability through its implication in DNA repair. The main function of XRCC1 is associated with its role in the single-strand break (SSB) and base excision repair (BER) pathways that share several enzymatic steps. We show here that the polymorphic XRCC1 variant R194W presents a defect in its interaction with the DNA glycosylase OGG1 after oxidative stress. While proficient for single-strand break repair (SSBR), this variant does not colocalize with OGG1, reflecting a defect in its involvement in BER. Consistent with a role of XRCC1 in the coordination of the BER pathway, induction of oxidative base damage in XRCC1-deficient cells complemented with the R194W variant results in increased genetic instability as revealed by the accumulation of micronuclei. These data identify a specific molecular role for the XRCC1-OGG1 interaction in BER and provide a model for the effects of the R194W variant identified in molecular cancer epidemiology studies.
INTRODUCTION
Cellular DNA is continuously exposed to oxidative stress arising from both endogenous and exogenous sources. As a consequence, lesions such as modified bases, abasic (AP) sites, and single-strand breaks (SSBs) are generated (1). One of the major base lesions induced by oxidative stress is 8-oxoguanine (8-oxoG), which is recognized and excised by a specific DNA glycosylase, OGG1, initiating the base excision repair (BER) pathway (2). The AP site produced by OGG1 DNA glycosylase activity is then cleaved by the AP endonuclease APE1, resulting in a SSB. The subsequent synthesis and ligation steps are carried out by polymerase β (POLβ) and ligase 3 (LIG3), respectively, to restore an intact DNA molecule (3). SSBs can also be directly induced in genomic DNA, and most of the enzymatic steps required for their repair are common to the single-strand break repair (SSBR) and BER pathways. Besides the enzymes mentioned above, other proteins participate in the efficient repair of modified bases and SSBs. Of these proteins, XRCC1, which is essential for embryonic development in mice (4), is a protein with no known enzymatic activity that acts as a scaffolding platform for SSBR and BER activities (5, 6). Cells deficient in XRCC1 exhibit increased frequencies of sister chromatid exchanges and chromosomal rearrangements. XRCC1 function is based in its capacity to interact with multiple enzymes and DNA intermediates in various DNA repair pathways (7, 8), coordinating the rate and sequence of the enzymatic activities and thus avoiding the exposure of toxic DNA intermediates to the cellular milieu (9). The various XRCC1 domains responsible for the interactions with BER or SSBR enzymes have been identified. XRCC1 is composed of three structured domains, interspaced by two flexible/nonstructured linkers (10) (see Fig. 1A). The NTD (N-terminal domain) is responsible for the interaction with POLβ (11, 12), the BRCT1 (BRCA1 carboxyl-terminal protein interaction domain 1) is involved in the interaction with poly(ADP-ribose) polymerase 1 (PARP1) and PARP2 (13), and BRCT2 is required for the interaction with and stabilization of LIG3 (14, 15).
FIG 1.
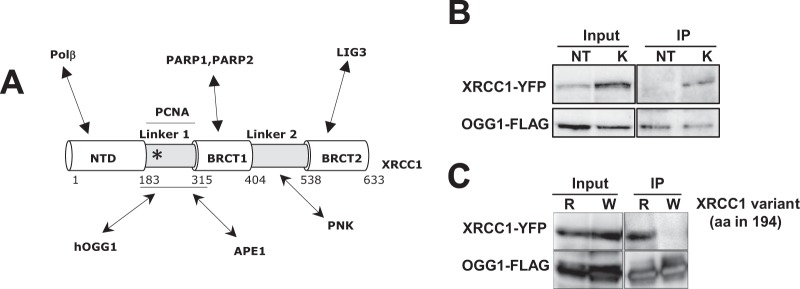
Interaction between OGG1 and XRCC1 is impaired in XRCC1(R194W). (A) Schematic representation of the different domains of XRCC1, the highly structured N-terminal domain (NTD) and BRCT1 and BRCT2 domains, separated by the two linkers. The domains involved in the interaction with different protein partners are indicated. The position of the R194W substitution is indicated by an asterisk. hOGG1, human OGG1; PNK, polynucleotide kinase. (B) HeLa cells cotransfected with plasmids expressing XRCC1-YFP and OGG1-FLAG were treated with KBrO3 (K) or not treated with KBrO3 (NT). Three hours after the treatment, protein extracts were used for immunoprecipitation with the anti-FLAG antibody. Immunoprecipitated (IP) proteins were analyzed by Western blotting with anti-FLAG and anti-GFP antibodies. The left blots show the levels of the proteins in the extract (10% of the input). (C) HeLa cells cotransfected with plasmids expressing XRCC1-YFP (R) or the variant XRCC1(R194W)-YFP (W) and OGG1-FLAG were treated and processed as described above for panel B. aa in 194, amino acid at position 194.
Protein-protein interactions are crucial events for the recruitment of BER factors to the site of repair. After induction of direct SSBs, XRCC1 is rapidly assembled in small nuclear foci through a PARP1-dependent mechanism (16, 17). The XRCC1-L360D mutation results in the perturbation of the BRCT1 domain, thus abolishing the interaction with PARP (13) and consequently, the recruitment of XRCC1 to SSB repair foci (17, 18). Furthermore, disruption of the interaction between POLβ and XRCC1 by the introduction of the V86R substitution in XRCC1, impairs the recruitment of POLβ to the site of the damage (19). Ligation efficiency of BER intermediates is also reduced in cells expressing the XRCC1 mutant V86R, suggesting a defect in the recruitment of later BER factors, such as LIG3 (20).
Taking into account the direct interaction of XRCC1 with several DNA glycosylases and with APE1 (6, 21, 22), it has been proposed that XRCC1 could be recruited during the very first steps of BER, independently of PARP activity (18, 23). Interestingly, PARP activity does not seem to be required for the efficient completion of BER (24). Taken together, these data suggest that a defect in the interaction between XRCC1 and a DNA glycosylase could have an impact on the recruitment of XRCC1 to BER and therefore on the downstream steps of the pathway.
In vitro interaction experiments have shown that both linker 1 and BRCT1 domains of XRCC1 are involved in the interaction with several DNA glycosylases, including OGG1 (6, 21). The facts that the BRCT1 domain mutant (L360D) does not affect the capacity of XRCC1 to interact with OGG1 and is recruited as efficiently as the wild-type XRCC1 to BER sites (18) suggest that linker 1 is sufficient for the interaction between XRCC1 and the DNA glycosylase.
Several naturally occurring genetic polymorphisms have been reported for the XRCC1 gene, and molecular epidemiology studies have linked the presence of XRCC1 variant alleles with altered cancer susceptibility (25, 26). For several of those variants found in the human population, functional analyses have failed to find alterations in their SSBR capacity that could explain the epidemiological observations. One of the most abundant XRCC1 polymorphisms found in the human population is R194W, situated in linker 1. We thus decided to evaluate the impact of this amino acid substitution on XRCC1 interaction with OGG1 and its recruitment to BER after induction of oxidative base damage. We show here that the XRCC1(R194W) variant, while able to repair SSBs, as suggested by previous studies (27), is defective in the interaction with the DNA glycosylase OGG1 and is not correctly recruited to BER of 8-oxoG. Furthermore, we observed an increase in genetic instability in cells expressing the XRCC1(R194W) variant, suggesting an accumulation of BER intermediates. Our findings provide a mechanism for the recruitment of XRCC1 during BER and suggest a model to explain the impact of this XRCC1 polymorphism in cancer development and treatment.
MATERIALS AND METHODS
Plasmid construction.
XRCC1-YFP (YFP stands for yellow fluorescent protein) and OGG1-DsRED monomer plasmids used in this study have been previously described (28, 29). The XRCC1(R194W)-YFP variant was obtained by site-directed mutagenesis of the XRCC1-YFP construct, using the QuikChange II XL site-directed mutagenesis kit (Stratagene). For the plasmid expressing OGG1-FLAG, the OGG1 coding sequence was amplified by PCR using a 3′ primer containing one copy of the FLAG sequence and subsequently cloned into pCDNA(3.1). Plasmid expressing LIG3-RFP (RFP stands for red fluorescent protein) was a kind gift from Heinrich Leonhardt (University of Munich).
Small interfering RNA (siRNA) design and cloning in pEBV siRNA vectors carrying a hygromycin B resistance cassette and establishment of stable knockdown clones were carried out as previously described (30). The RNA interference (RNAi) sequence for XRCC1 (GenBank accession no. NM_006297) spans nucleotides 1832 to 1850. In order to avoid silencing of the exogenously expressed XRCC1-YFP proteins, five silent mutations were introduced by site-directed mutagenesis in the sequence targeted by the short hairpin RNA (shRNA). Thus, the sequence TGGATCTACAGTTGCAATGAG was replaced by TGGATATATAGCTGTAACGAG, in which the modifications are underlined.
Cell lines, culture, and treatments.
CHO cell line EM9 was obtained from E. Sage (Institut Curie, Orsay, France). Cell lines used in this study were cultured in Dulbecco modified Eagle medium (DMEM) (GIBCO-BRL, Invitrogen) containing 10% fetal bovine serum at 37°C with 5% CO2. L132 cells expressing the shRNA against XRCC1 were maintained in DMEM supplemented with 125 μg/ml hygromycin B. For stable cell lines expressing XRCC1-YFP variants, the culture medium was supplemented with 400 μg/ml of G418.
MoFlo and InFlux cell sorters were used for generation of populations of CHO and L132 cells expressing equivalent levels of the XRCC1-YFP-tagged proteins.
Cells were grown on coverslips for microscopy experiments and on petri dishes for biochemical analysis. Transient transfections were done with Lipofectamine 2000 (Life Technologies) according to the manufacturer's instructions. Experiments were performed 24 h after transfection. Cells at about 80% confluence were treated for 30 min at 37°C with 40 mM potassium bromate (KBrO3) (Sigma) diluted in Dulbecco phosphate-buffered saline (DPBS) (Cambrex). The cells were then allowed to recover in DMEM for the times indicated in the figures before fixation or extraction. For the removal of soluble proteins, cells were washed for 5 min on ice with cold CSK buffer [100 mM NaCl, 300 mM glucose, 10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 6.8), 3 mM MgCl2, 0.5% Triton X-100, and protease inhibitors]. The cells were washed twice on ice-cold PBS before fixation in 4% paraformaldehyde (PFA) for 30 min at room temperature. Nuclear DNA was counterstained with 1 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI). The cells on the coverslips were mounted in Dako fluorescence mounting medium.
Determination of cell survival, cell cycle distribution, and quantification of micronucleus formation.
Cell survival was measured 24 h after methyl methanesulfonate (MMS) treatment (diluted in DMEM, 1 h at 37°C) or KBrO3 (diluted in DPBS, 30 min at 37°C). Cell viability was measured with the neutral red uptake assay. Cells were incubated for 1 h at 37°C in medium containing 0.07% (wt/vol) neutral red dissolved in cell culture medium and washed twice with DPBS. Cell lysis was then carried out by adding 1% acetic acid in 50% ethanol solution, and the optical density was measured at 540 nm.
To evaluate cell cycle distribution after KBrO3 treatment, cells were recovered with TrypLE express reagent (Life Technologies) and washed. One million cells were fixed in 2% PFA, and DNA was stained with 0.5 μg/ml Hoechst 33258 for 15 min at room temperature (diluted in DPBS). Cells were washed with DPBS and analyzed on a LSRII flow cytometer (BD Biosciences) for YFP and Hoechst fluorescence. Quantification of the percentage of cells in the G1, S, and G2 phases of the cell cycle was performed with FlowJo_V10 software.
For the quantification of micronucleus formation, cells grown on coverslips were fixed 24 h after KBrO3 treatment and stained with DAPI. Image acquisition was performed with an automated scanner workstation (MetaSystems). A minimum of 1,000 cells was considered for each measurement.
Microscopy and image treatment and analysis.
Image acquisition was performed with a Leica confocal microscope SPE (Wetzlar, Germany), using an ACS APO 40.0×, 1.15-numerical-aperture (NA) oil immersion or ACS APO 63.0×, 1.30-NA oil immersion lens. Plot profiles, cytofluorograms, and image treatment were done with the ImageJ software (W. S. Rasband, ImageJ, U.S. National Institutes of Health, Bethesda, MD, 1997–2014 [http://rsb.info.nih.gov/ij/]). Pearson's and Manders' correlation coefficients, to measure colocalization between green and red signals, were calculated with the ImageJ plug-in JACOP (31). The ROI color-coded plug-in was used for the association of particle-size measurements with a color.
Quantitative reverse transcription-PCR (qRT-PCR) analysis.
Total RNA was prepared from frozen cell pellets using RNeasy Plus kit (Qiagen) and quantified by Nanodrop spectrophotometer. Reverse transcription was performed with Superscript VILO cDNA synthesis kit (Invitrogen). PCRs were performed on a PRISM 7300 PCR system (Applied Biosystems) by using the Absolute quantitative PCR (QPCR) ROX mix (Abgene). Quantification of gene expression was calibrated using a reference standard curve obtained by serial dilutions of a control cDNA. The expression of the constitutive gene rplpo (large ribosomal protein) was used to normalize the expression of XRCC1. XRCC1 was amplified with TaqMan probes purchased from Applied Biosystems (Hs00959834).
Immunoprecipitation and Western blot analysis.
Protein extracts were prepared by sonicating cells with the Bioruptor bath (pulses 30 s on and 30 s off for 10 min at maximum intensity) in NP-40 buffer (0.1% NP-40, 1 mM EDTA, 20 mM Tris-HCl [pH 8], 150 mM NaCl) and centrifugation for 20 min at 13,000 rpm. For immunoprecipitations, 1 mg of protein extract was incubated with anti-FLAG antibody (Sigma), and protein complexes were recovered using Dynabeads goat anti-mouse IgG (Invitrogen Dynal AS) by following the manufacturer's instructions. Protein extracts and immunoprecipitated proteins were separated on a 10% SDS-PAGE gel, blotted, and incubated with anti-FLAG (Sigma), anti-XRCC1 (Neomarkers), antitubulin (Invitrogen), anti-GFP (Roche) or anti-LIG3 (GeneTex) antibodies. Horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Amersham Biosciences, and blots were developed with the Amersham ECL Advance Western blotting detection kit.
Repair kinetics assays.
A modified version (32) of the alkaline elution assay originally described by Kohn et al. in 1976 (33) was used to quantify SSBs and oxidative purine lesions sensitive to repair by the Fpg DNA glycosylase. The assay makes use of the fact that the elution rate of chromosomal DNA from a polycarbonate membrane filter (2-μm pore size) depends on the length of the DNA molecules and therefore the number of strand breaks. Briefly, the sum of SSBs and DNA modifications sensitive to the repair glycosylase Fpg was obtained from elution rates in experiments in which the cellular DNA was incubated with Fpg protein (1 μg/ml) immediately after cell lysis, before the elution of the DNA at pH 12.15. Complete incision (at Fpg-sensitive modifications) by the enzyme was shown to be achieved under these conditions. To quantify SSBs, the incubation was carried out without the repair enzyme. The number of Fpg-sensitive modifications was obtained by subtraction of the number of SSBs. The slope of an elution curve obtained with γ-irradiated cells was used for calibration (6 Gy = 1 SSB/106 bp) (33). To obtain repair kinetics, approximately 1.7 Fpg-sensitive sites per 106 bp were induced by incubation with 40 mM bromate for 30 min at 37°C. The numbers of residual (unrepaired) modifications were determined in cells incubated after damage induction for various times in culture medium at 37°C and indicated as percentages of the induced lesions. In some experiments, the number of AP sites was determined as well, using endonuclease IV instead of Fpg.
RESULTS
The XRCC1 variant R194W presents a defect in its interaction and colocalization with OGG1.
The polymorphism R194W is located in the first linker region of XRCC1 (Fig. 1A), a domain of the protein implicated in the interaction with OGG1 (6). To assess the interaction between XRCC1 and the DNA glycosylase, we performed immunoprecipitation experiments on extracts from cells expressing XRCC1-YFP and OGG1 tagged with a FLAG epitope. The assays were done on untreated cells or 3 h after KBrO3 treatment. The oxidant KBrO3 mainly induces the formation of 8-oxoG in genomic DNA. SSBs and AP sites are also generated but in much lower yields and are rapidly repaired. In agreement with the hypothesis that DNA repair complexes are assembled in a dynamic way depending on the DNA lesion to be repaired (35, 36), XRCC1 and OGG1 are found in the same protein complex after treatment with KBrO3, while almost no interaction can be detected between those proteins in untreated cells (18) (Fig. 1B). In order to determine whether the R194W substitution affected the interaction with OGG1, we performed the same experiments in cells expressing either XRCC1 variant and treated with KBrO3. While we confirmed the presence of XRCC1 in the protein complexes precipitated with the anti-FLAG antibody, replacing the arginine with a tryptophan in position 194 of XRCC1 resulted in the loss of the association between XRCC1 and OGG1 (Fig. 1C).
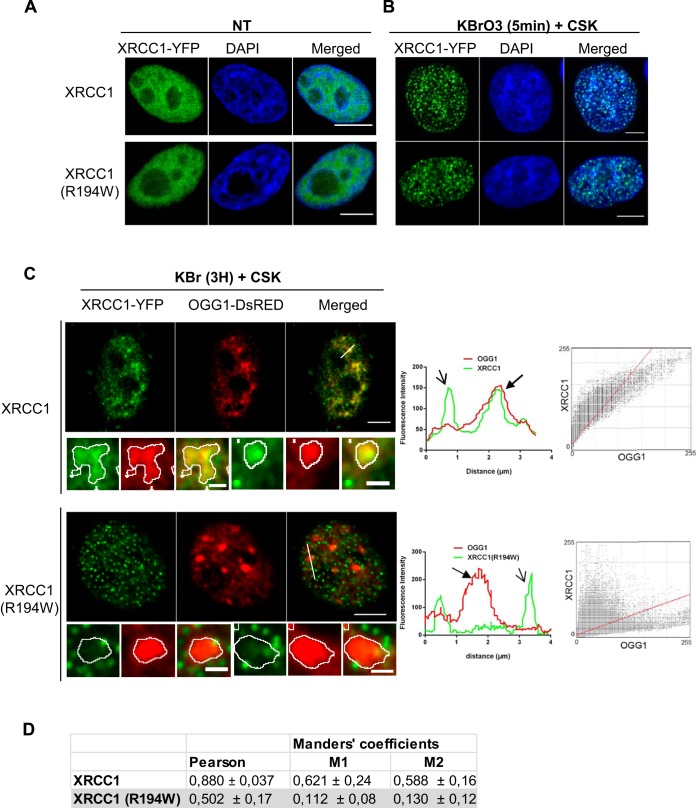
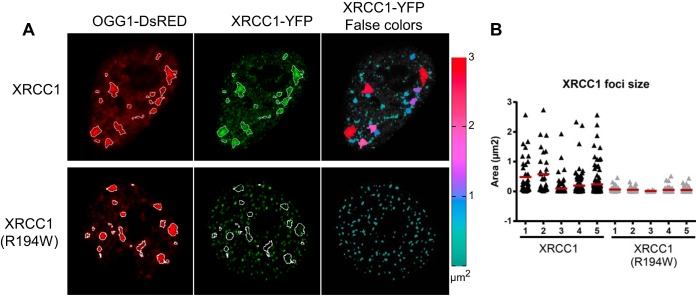
We have previously shown that after induction of oxidative DNA damage, OGG1 is recruited to chromatin, together with other BER proteins such as APE1, XRCC1, and LIG3 (18, 28). The BER-specific recruitment of repair factors can be distinguished from the recruitment to SSBR through several criteria related to their relocalization within the nucleus: (i) the kinetics of formation of detergent-resistant foci or patches; (ii) their morphological parameters, such as size and circularity; and (iii) the requirement of PARP1 activity for their assembly. Indeed, for XRCC1 and LIG3, involved in both SSBR and BER late steps, their distribution presents two types of patterns. They are rapidly—within a few minutes—recruited to small foci, the formation of which depends on PARP activity. BER-specific proteins, such as OGG1 and APE1, are not detected in those structures. Three hours after treatment, however, XRCC1 and LIG3, now together with the DNA glycosylase and APE1, are retained independently of PARP activity in larger patches found in euchromatin regions (18). In order to evaluate the impact of the R194W polymorphism on the recruitment of XRCC1 to chromatin, we compared the subcellular localization of XRCC1-YFP with that of the polymorphic variant 5 min and 3 h after KBrO3 treatment. In mock-treated cells, both proteins showed the same expected pattern of nuclear localization (Fig. 2A), the proteins being soluble in the nucleoplasm as reflected by the fact that no fluorescence could be detected if a detergent extraction was performed before fixation (data not shown). Five minutes after KBrO3 treatment, both XRCC1 variants were assembled in small and bright detergent-resistant foci, characteristic of SSBR (Fig. 2B). As previously shown (18), no recruitment of OGG1 was detected at this early time. At 3 h after the treatment, OGG1-DsRED and XRCC1-YFP colocalized in larger nuclear regions typical of BER sites (18), but we did not observe accumulation of the XRCC1(R194W) variant in those regions (Fig. 2C, outlined in white in the insets and indicated by a thick black arrow in the plot profile). In contrast, both XRCC1 variants were detected in smaller foci devoid of OGG1 (indicated by a thin black arrow in the plot profile and in the insets of Fig. 2C). The cytofluorograms presented in Fig. 2C show a clear dissociation between OGG1-DsRED and XRCC1(R194W)-YFP signals as opposed to the good correlation found for the major form of XRCC1. Pearson's and Manders' coefficients to evaluate colocalization between red (OGG1) and green (XRCC1) signals were calculated. While both parameters indicated very good colocalization between OGG1 and XRCC1, values were significantly reduced in the case of OGG1 and the XRCC1(R194W) variant (Fig. 2D). Further image analysis confirmed the difference in recruitment patterns for the polymorphic variant. In order to facilitate the visualization of both patterns, false colors were applied to the XRCC1-YFP image according to the size of the particles (Fig. 3). As we have previously reported, 3 h after the KBrO3 treatment, XRCC1 displayed a wide range of fluorescent particle sizes that included both large heterogeneously shaped patches overlapping with the OGG1 signal (BER) and small foci where OGG1 was not detected. In contrast, the same analysis of the XRCC1(R194W)-YFP-expressing cells showed that, while OGG1 displayed essentially the same distribution of large patches associated with BER, XRCC1(R194W) was found only in small circular foci corresponding to the SSBR sites (Fig. 3).
FIG 2.
Colocalization between OGG1 and XRCC1 is impaired in the variant XRCC1(R194W). (A) HeLa cells were transfected with plasmids carrying genes coding for XRCC1 or the variant XRCC1(R194W) fused to the YFP (green). Twenty-four hours after transfection, cells were fixed, and DNA was stained with DAPI (blue). The cells were not treated with KBrO3 (NT). Bars, 5 μm. (B) HeLa cells expressing XRCC1-YFP were treated with KBrO3 for 30 min and allowed to recover in DMEM for 5 min. The cells were extracted with CSK buffer prior to fixation in order to remove soluble proteins. DNA was stained with DAPI (blue). Bars, 5 μm. (C) HeLa cells cotransfected with plasmids expressing OGG1-DsRED and the XRCC1-YFP or XRCC1(R194W)-YFP variant and were treated with KBrO3. Three hours after treatment, soluble proteins were extracted with CSK buffer prior to fixation. Bars, 5 μm and 2 μm (insets). The positions of the line scans used for the plot profiles are indicated in the merged images. Correlations between green and red fluorescence signals are presented in two-dimensional (2D) cytofluorograms. (D) Pearson's and Manders' correlation coefficients between XRCC1 (green) and OGG1 (red) signals were calculated as indicators of colocalization and presented as the means ± standard errors of the means (SEMs) for 10 cells.
FIG 3.
Characterization of foci for the XRCC1 and XRCC1(R194W) variants. (A) HeLa cells were cotransfected with XRCC1-YFP (green) variants and OGG1-DsRED (red) and treated with KBrO3. Three hours after treatment, the cells were washed with CSK buffer to remove soluble proteins, fixed, and observed by using a confocal microscope. The OGG1 patches are outlined with a white line in both the OGG1 and XRCC1 images. Only the major variant of XRCC1 was detected in those regions. False colors were applied to the XRCC1-YFP image according to the size of the XRCC1 particles using the ice LUT (lookup table) ramp depicted at the right of the images. (B) Sizes of XRCC1 foci in 5 cells for each XRCC1 variant.
Cells expressing the XRCC1(R194W) variant are not affected in their direct single-strand sealing and base excision rates.
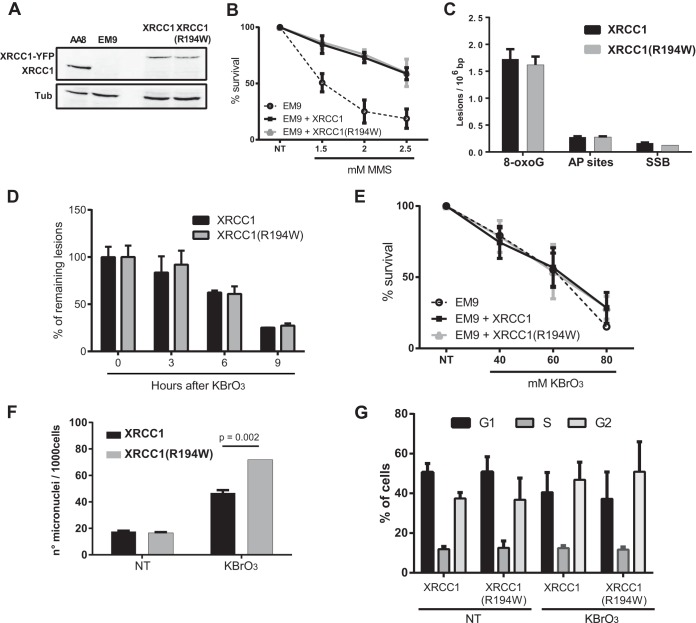
EM9 cells, derived from the AA8 CHO cell line, lack XRCC1, are hypersensitive to alkylating agents and ionizing radiation, and display a marked genetic instability reflected in high levels of chromosomal rearrangements, sister chromatid exchanges, and micronuclei (34, 37). All these phenotypes can be efficiently complemented by the human XRCC1 protein (34, 37). Because the defect in repair of SSBs potentially explains most phenotypes described for XRCC1-deficient cells, little is known on the specific role of XRCC1 in BER. The fact that the XRCC1(R194W) variant is recruited to SSBR but not to BER provides a powerful tool to analyze the impact of the absence of XRCC1 specifically on BER. We thus complemented EM9 CHO cells with either human XRCC1 variant. Plasmids carrying genes coding for the fusion proteins XRCC1-YFP, XRCC1(R194W)-YFP, or YFP alone were transfected into EM9 cells, and cell populations expressing similar levels of the YFP-tagged proteins were selected by cell sorting and used in further experiments (Fig. 4A). Comet assays were performed on the EM9 populations to measure the repair kinetics of DNA strand breaks induced by ionizing radiation. In agreement with previous reports (27, 38), cells expressing XRCC1(R194W) did not show a defect in their capacity to repair strand breaks (data not shown). Moreover, both variants were equally efficient in reversing the sensitivity of EM9 cells to the alkylating agent MMS (Fig. 4B), confirming the proficiency of the XRCC1(R194W)-YFP for SSBR.
FIG 4.
Cellular responses of EM9 cells complemented with either XRCC1 or the XRCC1(R194W) variant. (A) Protein extracts of AA8 (wild-type) cells and EM9 cells (XRCC1 deficient) complemented with YFP alone or with either of the two XRCC1 variants fused to the YFP were analyzed by Western blotting using anti-XRCC1 and antitubulin antibodies. (B) Cell survival after exposure to increasing concentrations of MMS. (C) Levels of induced 8-oxoG/Fpg-sensitive sites, AP sites, and SSBs measured by alkaline elution just after the KBrO3 treatment (0-h time point). Values are means plus SEMs (error bars) from three independent experiments. (D) Cell survival after exposure to increasing concentrations of KBrO3. (E) Fpg-sensitive sites were measured by alkaline elution at different times after KBrO3 treatment. The percentage of induced lesions remaining is shown on the y axis. Values are means ± SEMs from three independent experiments. (F) Micronuclei were counted 24 h after treatment with KBrO3. Values are means plus SEMs from three independent experiments, and 1,000 cells were counted in each experiment. (G) Cell cycle was evaluated in nontreated cells and 24 h after KBrO3 treatment for both complemented cells. The percentages of cells in G1, S, and G2 in a representative experiment are indicated.
In order to evaluate the impact of the XRCC1(R194W) variant on the cell responses to oxidative stress, cells were exposed to KBrO3. As expected, the major lesion induced by KBrO3 was 8-oxoG, while minor amounts of AP sites and SSBs could be detected. For all three types of lesions, the levels of induced damage were very similar in the two cell lines, independently of the expressed XRCC1 variant (Fig. 4C). In order to assess the capacity of the cells expressing the different XRCC1 variants to excise 8-oxoG, we determined repair kinetics by using the alkaline elution technique. Cells were treated with KBrO3 and directly collected (0-h time point) or allowed to repair in DMEM for different times. The rates of removal of 8-oxoG were indistinguishable in EM9 cells complemented with either human XRCC1 variant, with about 50% of the lesions repaired 6 h after treatment (Fig. 4D). AP sites and SSBs directly induced by the KBrO3 treatment were efficiently repaired in both cell lines and were undetectable 3 h after the treatment (data not shown).
These results confirmed that the R194W substitution does not affect the removal of 8-oxoG by OGG1 or the repair efficiency of directly formed SSBs.
Increased genetic instability in cells complemented with the XRCC1(R194W) variant after the induction of 8-oxoG.
To analyze the consequences of the defective interaction of XRCC1(R194W) with OGG1, we first compared the variants with respect to the cytotoxicity of KBrO3. No difference on cell survival could be detected after exposure of the cells to increasing amounts of KBrO3. Importantly, however, even the complete absence of XRCC1 did not sensitize the cells to the oxidative stress induced by this treatment (Fig. 4E). This indicates that the main cause of cell death under these conditions is not due to DNA lesions.
We next quantified the micronuclei generated in the two cell lines after treatment with KBrO3. Micronucleus formation is a very sensitive indicator of genotoxicity, in particular of the generation of strand breaks and collapsed replication forks (39). Consistent with a deficiency in the latter steps of BER, EM9 cells complemented with XRCC1(R194W) showed a higher number of micronuclei than cells complemented with the major form of XRCC1 (Fig. 4F). In order to discard the possibility that the difference in the number of cells containing a micronucleus was due to an alteration in the cell cycle progression, we determined the percentage of cells in different cell cycle phases. Neither the XRCC1 polymorphism nor the treatment affected the cell cycle distribution (Fig. 4G). Taken together, these results are consistent with the hypothesis that the higher levels of micronuclei in the EM9 cells expressing XRCC1(R194W) are the result of a defect in DNA repair.
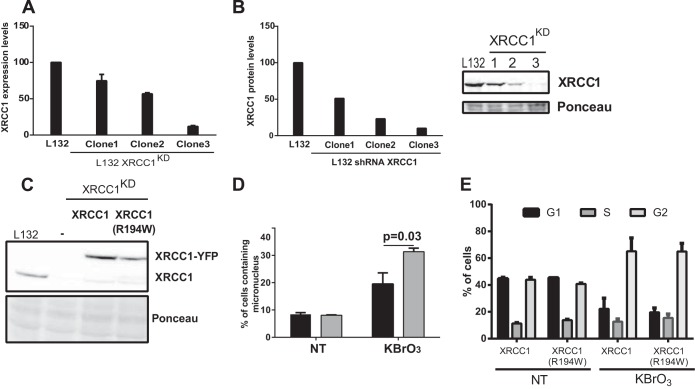
To confirm the above results in human cells, we established a human cell line in which the XRCC1 was downregulated by expression of an shRNA. Previous studies using this strategy have shown that deficiency of XRCC1 in human cells has very similar consequences to the ones described in mouse or CHO cellular models (40, 41). We therefore stably transfected L132 cells with a plasmid expressing an shRNA against XRCC1, and different clones were isolated. Clone 3, with less than 10% of XRCC1 expression as determined both by RT-PCR (Fig. 5A) and by Western blot analysis (Fig. 5B) was selected for further experiments and named L132-XRCC1KD (KD stands for knockdown). The increased sensitivity of L132-XRCC1KD cells to the alkylating agent MMS confirmed the functional efficiency of the endogenous XRCC1 knockdown (data not shown). Plasmids carrying genes coding for the fusion proteins XRCC1-YFP and XRCC1(R194W)-YFP but carrying silent mutations in the sequence targeted by the shRNA were then transfected into L132-XRCC1KD cells. Cell populations expressing similar levels of the YFP-tagged proteins were selected by cell sorting, and expression levels of the XRCC1-YFP variants were checked by Western blotting (Fig. 5C). We then analyzed the induction of micronuclei in both cell lines 24 h after KBrO3 treatment. In agreement with our observations in the EM9 cellular model, high levels of micronuclei were detected in human cells complemented with the XRCC1 variant R194W after induction of 8-oxoG compared to those expressing the major variant (Fig. 5D). While the treatment induced an accumulation of L132 cells in G2, the cell cycle distribution was again essentially identical for both XRCC1 genotypes. These results confirm the importance of XRCC1 recruitment to BER for the maintenance of genetic stability in response to oxidative DNA damage.
FIG 5.
Cellular responses of L132 cells complemented with either XRCC1 or the XRCC1(R194W) variant after exposure to oxidative stress. (A) L132 cells were transfected with a plasmid expressing a shRNA against XRCC1, and different clones were selected. Levels of XRCC1 mRNA were measured by qRT-PCR. (B) XRCC1 protein levels in different clones (clones 1 to 3) of L132 cells expressing an shRNA against XRCC1. (C) Western blots showing expression levels of endogenous XRCC1 and exogenous XRCC1-YFP in L132 cells. Clone 3 was selected for the following experiments. (D) Micronuclei were counted 24 h after treatment with KBrO3. Data are represented as mean plus SEM from three independent experiments, and 1,000 cells were counted. (E) Cell cycle was evaluated in nontreated (NT) cells and 24 h after KBrO3 treatment for both complemented cells. The percentages of cells in G1, S, and G2 in a representative experiment are indicated.
Recruitment of LIG3 to BER is impaired in cells expressing the XRCC1(R194W) variant.
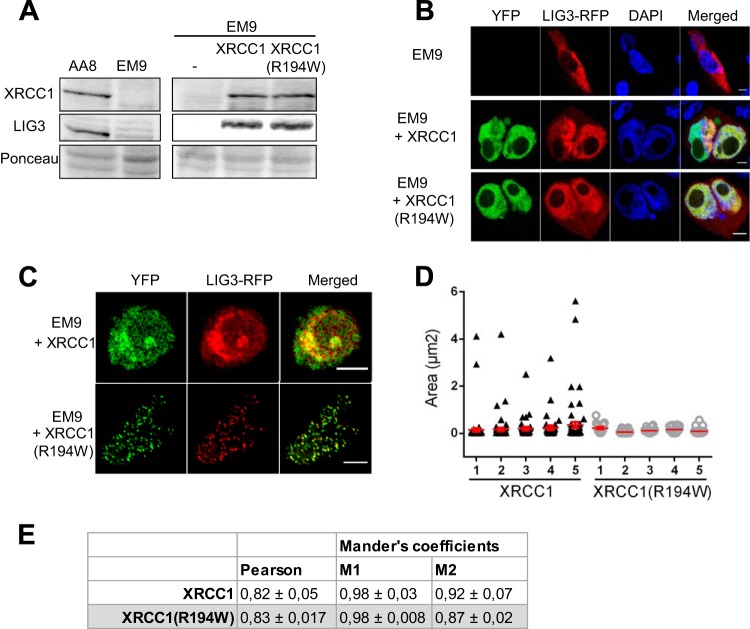
DNA ligase IIIα (LIG3) plays a central role in the repair of SSBs, either formed directly or generated as intermediates in the processing of damaged bases. LIG3 stability in cells requires its interaction with XRCC1 (42). As shown in Fig. 6A and B, while LIG3 was almost undetectable in EM9 cells, both XRCC1 variants allowed the recovery of the levels of LIG3 found in the XRCC1-proficient CHO cells (AA8). This is consistent with in vitro interaction experiments that showed that the R194W substitution does not affect XRCC1 interactions with PARP1, POLβ, and LIG3, involving the BRCT1, NTD, and BRCT2 domains of XRCC1, respectively (27). Furthermore, it has been shown that the BRCT2 domain of XRCC1 is required for the recruitment of LIG3 to the sites of repair in order to perform the last step of BER (43). In order to analyze whether there was a defect in the recruitment of LIG3 to BER in cells expressing the XRCC1(R194W) variant, we monitored the localization of LIG3-RFP fluorescent protein in EM9 cells expressing either of the XRCC1 variants. The cells were treated with KBrO3 and fixed 3 h after the treatment. LIG3-RFP colocalized with both XRCC1 variants in small and circular foci corresponding to the SSBR sites. In cells complemented with the major form of XRCC1, larger patches where XRCC1 and LIG3 colocalize were also observed, but they were not observed in cells complemented with the XRCC1(R194W) variant (Fig. 6C and D). Furthermore, correlation coefficients between XRCC1 and LIG3 were very high (Fig. 6E), confirming the colocalization of the two proteins, independently of the nuclear substructures where they were found. This is in agreement with the idea that LIG3 is present in the cells in a complex with XRCC1, and thus, recruitment of LIG3 depends on that of XRCC1. Furthermore, they are consistent with a model in which the increased genomic stability observed in the cells expressing the R194W variant of XRCC1 is due to an impaired recruitment of LIG3 to BER processes initiated by OGG1, resulting in a defect in the ligation of SSBs generated as intermediates.
FIG 6.
Recruitment of LIG3 to BER is impaired in cells expressing the XRCC1 (R194W) variant. (A) Protein extracts were prepared from XRCC1-deficient CHO (EM9) and parental (AA8) cell lines, as well as from the EM9 cells complemented with the different XRCC1-YFP variants. Protein levels for XRCC1 and LIG3 were determined by Western blotting. (B) LIG3-RFP (red) was transfected in EM9 cells or in cells complemented with XRCC1-YFP or XRCC1(R194W)-YFP (green). The cells were fixed 24 h after transfection, and DNA was stained with DAPI (blue). The cells were analyzed by confocal microscopy. (C) EM9 cells were cotransfected with XRCC1-YFP (green) variants and LIG3-RFP (red). The cells were treated with KBrO3, and 3 h after treatment, soluble proteins were removed, and the cells were fixed and observed with a confocal microscope. (D) Sizes of detergent-resistant LIG3 foci. The sizes of foci in five representative cells for each genotype are displayed. (E) Pearson's and Manders' M1 and M2 correlation coefficients between XRCC1 (green) and LIG3 (red) signals were calculated as indicators of colocalization. Results are presented as means ± SEMs for at least 10 cells. Bars, 5 μm.
DISCUSSION
After induction of DNA damage, DNA repair proteins are dynamically recruited to sites of repair inside the nucleus. Depending on the type of lesion induced, and therefore the DNA repair pathway analyzed, the kinetics of recruitment of the participating proteins to the sites of repair can vary from a few seconds to a few hours (19, 36, 44, 45). XRCC1 has been implicated in both SSBR and BER pathways. The protein is recruited within minutes to a SSB (16, 17, 46), and the formation of XRCC1 foci under those circumstances correlates with the repair kinetics for SSBs. Impairment of the assembly of XRCC1 foci is associated with an inefficient recruitment of the downstream activities of SSBR (19). The use of an oxidative treatment inducing mainly 8-oxoG lesions has allowed us to characterize the recruitment of XRCC1 to the repair of oxidative base damage by the BER pathway. Our previous observations have shown that assembly of BER complexes takes place in euchromatin regions independently of PARP activity and with slower kinetics than the assembly to SSBR foci (18). The kinetics of relocalization of XRCC1 to BER centers is in good correlation with the slow repair kinetics of oxidized bases. This is likely due to the time required by the glycosylase to initiate the repair pathway, as the removal of the base by the glycosylase is considered the rate-limiting step in BER (47). Both in vitro and in vivo experiments (6, 21, 22, 48) have suggested that it is the initiating enzyme, APE1 for AP sites and the DNA glycosylases for modified bases, that after recognition of the lesion, recruits XRCC1 through protein-protein interactions to coordinate the BER process. The later steps of the pathway would then be, as in the case of SSBR, very rapid to avoid the accumulation of toxic DNA intermediates (5).
The identification of a defect of the XRCC1(R194W) variant in its capacity to interact with the DNA glycosylase OGG1 and in its recruitment to the BER pathway after the induction of 8-oxoG allowed us to explore for the first time the specific role of XRCC1 in BER. Indeed, we observe a defect in the recruitment of the XRCC1(R194W) variant to the subnuclear regions where OGG1 relocalizes after induction of 8-oxoG by KBrO3 treatment (Fig. 2 and 3). Those particular nuclear subdomains where OGG1 accumulates most likely represent regions where BER of 8-oxoG takes place (18, 28). The data presented here demonstrate that the interaction of XRCC1 with the DNA glycosylase is necessary for the recruitment of the scaffolding protein to the BER pathway (6, 22). Since the 8-oxoG excision rates (Fig. 4D) were very similar in cells complemented with XRCC1 or XRCC1(R194W), we propose that the OGG1-XRCC1 interaction is mainly required to ensure the recruitment and coordination of downstream steps of repair and avoid the accumulation of toxic DNA intermediates (6, 9, 22). The detection limits of the techniques available did not allow us to see potential differences in the levels of 8-oxoG repair intermediates, such as AP sites or SSBs. However, we show here that cells expressing XRCC1(R194W) fail to recruit LIG3 to the sites of BER and accumulate higher numbers of micronuclei after exposure to oxidative stress, a likely consequence of the persistence of repair intermediates. Efficient sealing of SSBs in chromosomal DNA during BER is indeed crucial for maintaining genome integrity (49, 50). It has been previously observed that a defect in XRCC1 affects a postincision step, the single-strand break rejoining, probably due to a defect in LIG3 (51). Deletions or mutations of the BRCT2 of XRCC1 abolishing the interaction with LIG3 result in a defect in SSB ligation/repair, suggesting that XRCC1 is important not only for the stabilization of the ligase (42, 52) but also for its recruitment to sites of strand breaks (43). The results presented here show that in the case of 8-oxoG repair, impairment of the OGG1-XRCC1 interaction, as achieved by the R194W substitution, can also lead to a defective LIG3 recruitment and the consequent increase in genetic instability after induction of base damage. The discrepancy between the increased genomic instability observed in cells expressing XRCC1(R194W) and the lack of effect of the XRCC1 variant on the cytotoxicity of KBrO3 is likely due to the relatively small number of unprocessed DNA breaks that are needed to induce micronuclei. This is supported by the observation that even the complete absence of XRCC1 does not lead to an increased sensitivity to the treatment, suggesting that the cytotoxicity induced by this treatment is largely the consequence of damage to targets other than DNA.
It has been previously shown that the interaction of XRCC1 with PARP1 through the BRCT1 domain of XRCC1 is essential for its recruitment to SSBs (17, 18). The finding that replacement of single amino acids in the XRCC1 sequence specifically impairs recruitment of XRCC1 to sites of SSBR (BRCT1 mutants such as L360D) or to sites of BER (R194W) clearly indicates that specific domains of XRCC1 are involved in its recruitment to the different DNA repair pathways. This is consistent with the idea that the role of XRCC1 in DNA repair is defined by protein-protein interactions. It is worth noticing that although the linker 1 region of XRCC1 presents a high variability between species, the presence of the arginine at position 194 is extremely conserved throughout evolution. This region is rich in positively charged residues, and the substitution of a positively charged arginine by a hydrophobic tryptophan may affect protein-protein interactions and the formation of complexes effective in DNA repair.
Epidemiological studies have suggested an association of certain human XRCC1 polymorphisms with genetic instability and cancer susceptibility. However, the association between the XRCC1(R194W) variant and either an increased or reduced cancer risk is largely dependent on the population considered and the kind of cancer analyzed (25, 53, 54). The absence of a SSBR defect for the XRCC1(R194W) variant (38) (Fig. 4B) together with its proficient protein-protein interactions with its SSBR protein partners (27) (Fig. 6) and its normal recruitment kinetics to the site of SSBs (27) (Fig. 2B) rule out a straightforward explanation linking the epidemiological data to a defect in SSBR. Different kinds of ROS can give rise to an extremely diverse spectrum of genotoxic damage such as SSBs, base modifications, sugar damage, or abasic sites that in combination with the polymorphisms of DNA repair proteins would dictate the overall response of the cell. This could explain the inconclusive results obtained in epidemiological studies. It is interesting to note that the persistent oxidative stress found in tumors may result in an up to a 10-fold increase in the levels of 8-oxoG in the tumor compared to adjacent healthy tissues (55–57). Our results unveiled a specific defect of XRCC1(R194W) in BER, thus providing a plausible explanation for the impact of this polymorphism on genetic stability and cancer development.
ACKNOWLEDGMENTS
We thank the members of our laboratories for critical reading, discussions, and suggestions. We also thank the IRCM microscopy and cytometry facilities, Françoise Hoffschir for help with micronucleus determination, and Heinrich Leonhardt (University of Munich) for kindly sending us the LIG3-RFP plasmid.
This work was funded by INSERM and grants from the Association pour la Recherche sur le Cancer (PJA 20131200165 to J.P.R.), the CEA Radiobiology program (to J.P.R. and A.C.), and the Deutsche Forschungsgemeinschaft (EP11/8-2 to B.E.). Exchanges between the laboratories of J.P.R. and B.E. were supported by the PROCOPE program (28345TK).
REFERENCES
- 1.Cadet J, Delatour T, Douki T, Gasparutto D, Pouget JP, Ravanat JL, Sauvaigo S. 1999. Hydroxyl radicals and DNA base damage. Mutat Res 424:9–21. doi: 10.1016/S0027-5107(99)00004-4. [DOI] [PubMed] [Google Scholar]
- 2.Radicella JP, Dherin C, Desmaze C, Fox MS, Boiteux S. 1997. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes DE, Lindahl T. 2004. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet 38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 4.Tebbs RS, Flannery ML, Meneses JJ, Hartmann A, Tucker JD, Thompson LH, Cleaver JE, Pedersen RA. 1999. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev Biol 208:513–529. doi: 10.1006/dbio.1999.9232. [DOI] [PubMed] [Google Scholar]
- 5.Caldecott KW. 2003. XRCC1 and DNA strand break repair. DNA Repair (Amst) 2:955–969. doi: 10.1016/S1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 6.Marsin S, Vidal AE, Sossou M, Menissier-de Murcia J, Le Page F, Boiteux S, de Murcia G, Radicella JP. 2003. Role of XRCC1 in the coordination and stimulation of oxidative DNA damage repair initiated by the DNA glycosylase hOGG1. J Biol Chem 278:44068–44074. doi: 10.1074/jbc.M306160200. [DOI] [PubMed] [Google Scholar]
- 7.Caldecott KW. 2003. Protein-protein interactions during mammalian DNA single-strand break repair. Biochem Soc Trans 31:247–251. [DOI] [PubMed] [Google Scholar]
- 8.Nazarkina ZK, Khodyreva SN, Marsin S, Lavrik OI, Radicella JP. 2007. XRCC1 interactions with base excision repair DNA intermediates. DNA Repair (Amst) 6:254–264. doi: 10.1016/j.dnarep.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Wilson SH, Kunkel TA. 2000. Passing the baton in base excision repair. Nat Struct Biol 7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 10.Marintchev A, Robertson A, Dimitriadis EK, Prasad R, Wilson SH, Mullen GP. 2000. Domain specific interaction in the XRCC1-DNA polymerase beta complex. Nucleic Acids Res 28:2049–2059. doi: 10.1093/nar/28.10.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caldecott KW, Aoufouchi S, Johnson P, Shall S. 1996. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res 24:4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kubota Y, Nash RA, Klungland A, Schar P, Barnes DE, Lindahl T. 1996. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J 15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 13.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. 1998. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol 18:3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nash RA, Caldecott KW, Barnes DE, Lindahl T. 1997. XRCC1 protein interacts with one of two distinct forms of DNA ligase III. Biochemistry 36:5207–5211. doi: 10.1021/bi962281m. [DOI] [PubMed] [Google Scholar]
- 15.Taylor RM, Wickstead B, Cronin S, Caldecott KW. 1998. Role of a BRCT domain in the interaction of DNA ligase III-alpha with the DNA repair protein XRCC1. Curr Biol 8:877–880. doi: 10.1016/S0960-9822(07)00350-8. [DOI] [PubMed] [Google Scholar]
- 16.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. 2003. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res 31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okano S, Lan L, Caldecott KW, Mori T, Yasui A. 2003. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol Cell Biol 23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campalans A, Kortulewski T, Amouroux R, Menoni H, Vermeulen W, Radicella JP. 2013. Distinct spatiotemporal patterns and PARP dependence of XRCC1 recruitment to single-strand break and base excision repair. Nucleic Acids Res 41:3115–3129. doi: 10.1093/nar/gkt025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A. 2004. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc Natl Acad Sci U S A 101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dianova II, Sleeth KM, Allinson SL, Parsons JL, Breslin C, Caldecott KW, Dianov GL. 2004. XRCC1-DNA polymerase beta interaction is required for efficient base excision repair. Nucleic Acids Res 32:2550–2555. doi: 10.1093/nar/gkh567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campalans A, Marsin S, Nakabeppu Y, O'Connor RT, Boiteux S, Radicella JP. 2005. XRCC1 interactions with multiple DNA glycosylases: a model for its recruitment to base excision repair. DNA Repair (Amst) 4:826–835. doi: 10.1016/j.dnarep.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 22.Vidal AE, Boiteux S, Hickson ID, Radicella JP. 2001. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. EMBO J 20:6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanssen-Bauer A, Solvang-Garten K, Sundheim O, Pena-Diaz J, Andersen S, Slupphaug G, Krokan HE, Wilson DM III, Akbari M, Otterlei M. 2011. XRCC1 coordinates disparate responses and multiprotein repair complexes depending on the nature and context of the DNA damage. Environ Mol Mutagen 52:623–635. doi: 10.1002/em.20663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strom CE, Johansson F, Uhlen M, Szigyarto CA, Erixon K, Helleday T. 2011. Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic Acids Res 39:3166–3175. doi: 10.1093/nar/gkq1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goode EL, Ulrich CM, Potter JD. 2002. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev 11:1513–1530. [PubMed] [Google Scholar]
- 26.Ladiges W, Wiley J, MacAuley A. 2003. Polymorphisms in the DNA repair gene XRCC1 and age-related disease. Mech Ageing Dev 124:27–32. doi: 10.1016/S0047-6374(02)00166-5. [DOI] [PubMed] [Google Scholar]
- 27.Berquist BR, Singh DK, Fan J, Kim D, Gillenwater E, Kulkarni A, Bohr VA, Ackerman EJ, Tomkinson AE, Wilson DM III. 2010. Functional capacity of XRCC1 protein variants identified in DNA repair-deficient Chinese hamster ovary cell lines and the human population. Nucleic Acids Res 38:5023–5035. doi: 10.1093/nar/gkq193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amouroux R, Campalans A, Epe B, Radicella JP. 2010. Oxidative stress triggers the preferential assembly of base excision repair complexes on open chromatin regions. Nucleic Acids Res 38:2878–2890. doi: 10.1093/nar/gkp1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campalans A, Amouroux R, Bravard A, Epe B, Radicella JP. 2007. UVA irradiation induces relocalisation of the DNA repair protein hOGG1 to nuclear speckles. J Cell Sci 120:23–32. doi: 10.1242/jcs.03312. [DOI] [PubMed] [Google Scholar]
- 30.Biard DS, Despras E, Sarasin A, Angulo JF. 2005. Development of new EBV-based vectors for stable expression of small interfering RNA to mimick human syndromes: application to NER gene silencing. Mol Cancer Res 3:519–529. doi: 10.1158/1541-7786.MCR-05-0044. [DOI] [PubMed] [Google Scholar]
- 31.Bolte S, Cordelieres FP. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 32.Pflaum M, Will O, Epe B. 1997. Determination of steady-state levels of oxidative DNA base modifications in mammalian cells by means of repair endonucleases. Carcinogenesis 18:2225–2231. doi: 10.1093/carcin/18.11.2225. [DOI] [PubMed] [Google Scholar]
- 33.Kohn KW, Erickson LC, Ewig RA, Friedman CA. 1976. Fractionation of DNA from mammalian cells by alkaline elution. Biochemistry 15:4629–4637. doi: 10.1021/bi00666a013. [DOI] [PubMed] [Google Scholar]
- 34.Sossou M, Flohr-Beckhaus C, Schulz I, Daboussi F, Epe B, Radicella JP. 2005. APE1 overexpression in XRCC1-deficient cells complements the defective repair of oxidative single strand breaks but increases genomic instability. Nucleic Acids Res 33:298–306. doi: 10.1093/nar/gki173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kowalczykowski SC. 2000. Some assembly required. Nat Struct Biol 7:1087–1089. doi: 10.1038/81923. [DOI] [PubMed] [Google Scholar]
- 36.Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. 2001. Sequential assembly of the nucleotide excision repair factors in vivo. Mol Cell 8:213–224. doi: 10.1016/S1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 37.Thompson LH, West MG. 2000. XRCC1 keeps DNA from getting stranded. Mutat Res 459:1–18. doi: 10.1016/S0921-8777(99)00058-0. [DOI] [PubMed] [Google Scholar]
- 38.Takanami T, Nakamura J, Kubota Y, Horiuchi S. 2005. The Arg280His polymorphism in X-ray repair cross-complementing gene 1 impairs DNA repair ability. Mutat Res 582:135–145. doi: 10.1016/j.mrgentox.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 39.Iarmarcovai G, Bonassi S, Botta A, Baan RA, Orsiere T. 2008. Genetic polymorphisms and micronucleus formation: a review of the literature. Mutat Res 658:215–233. doi: 10.1016/j.mrrev.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 40.Brem R, Hall J. 2005. XRCC1 is required for DNA single-strand break repair in human cells. Nucleic Acids Res 33:2512–2520. doi: 10.1093/nar/gki543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan J, Wilson PF, Wong HK, Urbin SS, Thompson LH, Wilson DM III. 2007. XRCC1 down-regulation in human cells leads to DNA-damaging agent hypersensitivity, elevated sister chromatid exchange, and reduced survival of BRCA2 mutant cells. Environ Mol Mutagen 48:491–500. doi: 10.1002/em.20312. [DOI] [PubMed] [Google Scholar]
- 42.Caldecott KW, Tucker JD, Stanker LH, Thompson LH. 1995. Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res 23:4836–4843. doi: 10.1093/nar/23.23.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor RM, Moore DJ, Whitehouse J, Johnson P, Caldecott KW. 2000. A cell cycle-specific requirement for the XRCC1 BRCT II domain during mammalian DNA strand break repair. Mol Cell Biol 20:735–740. doi: 10.1128/MCB.20.2.735-740.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim JS, Krasieva TB, Kurumizaka H, Chen DJ, Taylor AM, Yokomori K. 2005. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J Cell Biol 170:341–347. doi: 10.1083/jcb.200411083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagy Z, Soutoglou E. 2009. DNA repair: easy to visualize, difficult to elucidate. Trends Cell Biol 19:617–629. doi: 10.1016/j.tcb.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 46.Mortusewicz O, Leonhardt H. 2007. XRCC1 and PCNA are loading platforms with distinct kinetic properties and different capacities to respond to multiple DNA lesions. BMC Mol Biol 8:81. doi: 10.1186/1471-2199-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cappelli E, Degan P, Frosina G. 2000. Comparative repair of the endogenous lesions 8-oxo-7,8-dihydroguanine (8-oxoG), uracil and abasic site by mammalian cell extracts: 8-oxoG is poorly repaired by human cell extracts. Carcinogenesis 21:1135–1141. doi: 10.1093/carcin/21.6.1135. [DOI] [PubMed] [Google Scholar]
- 48.Chou WC, Wang HC, Wong FH, Ding SL, Wu PE, Shieh SY, Shen CY. 2008. Chk2-dependent phosphorylation of XRCC1 in the DNA damage response promotes base excision repair. EMBO J 27:3140–3150. doi: 10.1038/emboj.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ensminger M, Iloff L, Ebel C, Nikolova T, Kaina B, Löbrich M. 2014. DNA breaks and chromosomal aberrations arise when replication meets base excision repair. J Cell Biol 206:29–43. doi: 10.1083/jcb.201312078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okano S, Kanno S, Nakajima S, Yasui A. 2000. Cellular responses and repair of single-strand breaks introduced by UV damage endonuclease in mammalian cells. J Biol Chem 275:32635–32641. doi: 10.1074/jbc.M004085200. [DOI] [PubMed] [Google Scholar]
- 51.Cappelli E, Taylor R, Cevasco M, Abbondandolo A, Caldecott K, Frosina G. 1997. Involvement of XRCC1 and DNA ligase III gene products in DNA base excision repair. J Biol Chem 272:23970–23975. doi: 10.1074/jbc.272.38.23970. [DOI] [PubMed] [Google Scholar]
- 52.Caldecott KW, McKeown CK, Tucker JD, Ljungquist S, Thompson LH. 1994. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol Cell Biol 14:68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng YZ, Liu YL, He XF, Wei W, Shen XL, Xie DL. 2014. Association between the XRCC1 Arg194Trp polymorphism and risk of cancer: evidence from 201 case-control studies. Tumour Biol 35:10677–10697. doi: 10.1007/s13277-014-2326-x. [DOI] [PubMed] [Google Scholar]
- 54.Zhou X, Gu L, Zeng Y, Wei L, Ying M, Wang N, Su C, Wang Y, Liu C. 2014. The XRCC1 Arg194Trp and Arg280His polymorphisms in head and neck cancer susceptibility: a meta-analysis. Tumour Biol 35:10665–10676. doi: 10.1007/s13277-014-2247-8. [DOI] [PubMed] [Google Scholar]
- 55.Jaruga P, Zastawny TH, Skokowski J, Dizdaroglu M, Olinski R. 1994. Oxidative DNA base damage and antioxidant enzyme activities in human lung cancer. FEBS Lett 341:59–64. doi: 10.1016/0014-5793(94)80240-8. [DOI] [PubMed] [Google Scholar]
- 56.Olinski R, Zastawny T, Budzbon J, Skokowski J, Zegarski W, Dizdaroglu M. 1992. DNA base modifications in chromatin of human cancerous tissues. FEBS Lett 309:193–198. doi: 10.1016/0014-5793(92)81093-2. [DOI] [PubMed] [Google Scholar]
- 57.Toyokuni S, Okamoto K, Yodoi J, Hiai H. 1995. Persistent oxidative stress in cancer. FEBS Lett 358:1–3. doi: 10.1016/0014-5793(94)01368-B. [DOI] [PubMed] [Google Scholar]