
Insight from Max Crispin
Allergic reactions arise when people become sensitized to otherwise harmless environmental antigens. In this issue, Shade et al. reveal that the immunoglobulin ε (IgE) antibodies that mediate these reactions have a key vulnerability. They report that the ability of IgE to trigger an allergic reaction through its interaction with mast cells is dependent on a single site of antibody glycosylation. With the in vivo targeting of specific glycoprotein glycans emerging as a viable strategy for modulating endogenous glycoprotein function, these findings are of significant interest.
IgE is found tightly bound to mast cells through the interaction with the high-affinity Fcε receptor (FcεRI). This interaction is sufficiently tight that IgE antibodies are stably immobilized on the immune cells, sensitizing them to specific antigens for extended periods of time. The IgE–FcεRI interaction occurs through the IgE Fc domain. Glycosylation of antibody Fc domains has been known to be important in the interaction of IgG with cellular Fcγ receptors, but there has been conflicting evidence on the role of the equivalent IgE Fc glycans. Shade et al. show that glycosylation of IgE is essential for both FcεRI binding and the triggering of mast cell degranulation. Using a panel of mutants, they demonstrate that IgE effector function is dependent on the glycan at a single glycosylation site in the Cε3 domain at Asn394 and Asn384 in human and mouse IgE, respectively.
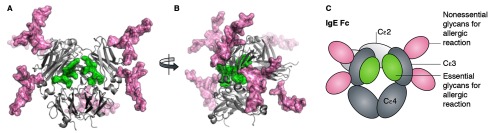
(A and B) Molecular model of the glycosylated IgE Fc domain. The IgE Fc glycans are shown as sticks with surface representation. (C) The structure of panel A is illustrated schematically and the glycans are colored throughout according to their requirement for FcεRI-mediated immune response. Molecular model courtesy of Dr. Mark Wormald.
Typically mammalian glycans are efficiently processed during glycoprotein folding and secretion from the oligomannose-type glycans added cotranslationally to a highly heterogeneous mix of complex-type glycans. The Cε3 glycosylation site identified to be critical for receptor binding has been shown by Shade and others to be trapped in the oligomannose state. Examination of the previously reported crystal structure reveals that these glycans are located in the interstitial space between opposing Cε3 of the homodimeric IgE Fc. The efficient processing to complex glycans is likely to be limited by either the extensive glycan–protein interactions or by steric blocking of the processing enzymes by the wider proteinous environment. Importantly, Shade et al. suggest that the unusual presence of oligomannose glycans at this critical site offers an opportunity to specifically target this structure to shut down the allergic response. They show that a bacterial endoglycosidase from Flavobacterium meningosepticum (EndoF1) that specifically hydrolyses oligomannose-type glycans can disrupt IgE effector functions. Whether bacterial enzymes can ever be used clinically is a point of some debate. Despite their ability to turn off elements of the immune response, it is likely that the immunogenicity of bacterial enzymes will limit any clinical applications to a single dose. The IgG-degrading enzyme from Streptococcus pyogenes, IdeS, is being investigated as a single-dose treatment to enable kidney transplantation for patients who have antibodies that target prospective kidneys and, encouragingly, has recently been shown to be well tolerated in humans in a small-scale safety trial.
These clinical considerations aside, this study demonstrates the importance of IgE glycosylation in allergic responses and provides a route to the selective disruption of this mechanism in allergic diseases.
References
- Shade, K.-T.C., et al. 2015. J. Exp. Med. 10.1084/jem.20142182. [DOI] [Google Scholar]


